Background: Bile acid synthesis plays an important role in nutrient absorption and maintaining metabolic homeostasis under normal physiology.
Results: Glucose induces cholesterol 7α-hydroxylase activity and postprandial bile acid synthesis via insulin signaling and epigenetic mechanisms.
Conclusion: Glucose and insulin are major postprandial factors that stimulate bile acid synthesis to maintain hepatic metabolic homeostasis.
Significance: Nutrient regulation of bile acid synthesis is impaired in diabetes and obesity.
Keywords: Bile Acid, Bile Acid Metabolism, Lipid Metabolism, Metabolic Diseases, Metabolic Regulation, Nuclear Receptors, CYP7A1, Epigenetic Regulation, FGF15, FXR
Abstract
Bile acids facilitate postprandial absorption of nutrients. Bile acids also activate the farnesoid X receptor (FXR) and the G protein-coupled receptor TGR5 and play a major role in regulating lipid, glucose, and energy metabolism. Transgenic expression of cholesterol 7α-hydroxylase (CYP7A1) prevented high fat diet-induced diabetes and obesity in mice. In this study, we investigated the nutrient effects on bile acid synthesis. Refeeding of a chow diet to fasted mice increased CYP7A1 expression, bile acid pool size, and serum bile acids in wild type and humanized CYP7A1-transgenic mice. Chromatin immunoprecipitation assays showed that glucose increased histone acetylation and decreased histone methylation on the CYP7A1 gene promoter. Refeeding also induced CYP7A1 in fxr-deficient mice, indicating that FXR signaling did not play a role in postprandial regulation of bile acid synthesis. In streptozocin-induced type I diabetic mice and genetically obese type II diabetic ob/ob mice, hyperglycemia increased histone acetylation status on the CYP7A1 gene promoter, leading to elevated basal Cyp7a1 expression and an enlarged bile acid pool with altered bile acid composition. However, refeeding did not further increase CYP7A1 expression in diabetic mice. In summary, this study demonstrates that glucose and insulin are major postprandial factors that induce CYP7A1 gene expression and bile acid synthesis. Glucose induces CYP7A1 gene expression mainly by epigenetic mechanisms. In diabetic mice, CYP7A1 chromatin is hyperacetylated, and fasting to refeeding response is impaired and may exacerbate metabolic disorders in diabetes.
Introduction
Bile acids are the end products of cholesterol catabolism in the liver (1). In addition to the classic function of bile acids in facilitating intestine absorption and transport of nutrients, drugs, and steroids, bile acids also play important roles in regulating the lipids, drugs, glucose, and energy metabolism (2). Recently, we reported that mice overexpressing cholesterol 7α-hydroxylase (CYP7A), which encodes the rate-limiting enzyme in the classic bile acid biosynthetic pathway, have increased bile acid pool and are resistant to high fat diet-induced insulin resistance and obesity (3). This is consistent with the current concept that bile acid signaling is important in maintaining metabolic homeostasis. Impaired bile acid homeostasis could lead to adverse metabolic consequences, such as cholestasis, liver injury, diabetes, and obesity (2, 3).
Bile acid synthesis is mainly controlled by the transcriptional regulation of the CYP7A1 gene. It is well established that the CYP7A1 gene is repressed by bile acids returning to the liver via enterohepatic circulation. This bile acid feedback mechanism allows the liver to efficiently increase or decrease bile acid synthesis in response to changes in bile acid levels and thus to maintain a constant bile acid pool. It is thought that CYP7A1 gene transcription is inhibited by a mechanism involving the bile acid-activated farnesoid X receptor (FXR)2 in hepatocytes and enterocytes. It was first discovered that activation of FXR induced small heterodimer partner (SHP) in the liver to inhibit CYP7A1 gene transcription (4). More recently, it has been proposed that FXR induces mouse intestine fibroblast growth factor 15 (FGF15) or a human ortholog, FGF19, which may act as an endocrine hormone to repress CYP7A1 gene transcription in hepatocytes (5). Adenovirus-mediated transduction of FGF19 activates hepatocyte membrane FGF receptor 4, which activates the c-Jun N-terminal kinase (JNK) and extracellular signal-regulated kinase 1/2 (ERK1/2) of the mitogen-activated protein kinase (MAPK) pathway to inhibit CYP7A1 gene transcription in mice (5, 6). The physiological relevance of these mechanisms in the regulation of bile acid homeostasis remained to be established.
CYP7A1 gene expression and hepatic bile acid synthesis rate exhibit a marked circadian rhythm. In rodents, bile acid synthesis and CYP7A1 expression peaked in the dark period and decreased in the light period (7–9). In humans, CYP7A1 activity, as reflected by serum bile acid synthesis marker 7α-hydroxy-4-cholesten-3-one levels, shows two peaks that coincide with meal intake (at 1 p.m. and 9 p.m.), reduces at night, and returns to basal levels in the morning (10, 11). Because liver metabolism is highly active during the postprandial period and humans undergo fasting-to-feeding cycles several times a day, these observations indicate a potential link between induction of bile acid synthesis and regulation of postprandial nutrient metabolism. Hence, it is likely that nutrient intake governs bile acid synthesis, whereas bile acids control postprandial nutrient absorption and metabolism. Unfortunately, clear evidence supporting such nutrient regulation of liver bile acid metabolism is still lacking.
It has been reported that bile acid pool and fecal bile acids are elevated in diabetic patients with uncontrolled hyperglycemia and are decreased upon insulin treatment (12). CYP7A1 activity is increased in streptozocin (STZ)-treated diabetic rats, suggesting that insulin represses CYP7A1, whereas a lack of insulin induces CYP7A1 (13). Furthermore, it has been reported that peroxisome proliferator activated receptor γ coactivator-1α (PGC-1α) activates cyp7a1 gene transcription in mice after overnight fasting (14). In contrast to these studies in mice, we reported that insulin and glucose induced, whereas glucagon repressed, the CYP7A1 gene in primary human hepatocytes (15–17). It was thought that species differences in regulation of human and rodent CYP7A1 gene transcription might explain the observed discrepancy (1).
In this study, we created a humanized CYP7A1-transgenic (tg) mouse model to study nutrient regulation of bile acid synthesis. We demonstrated that glucose and insulin are key physiological regulators of CYP7A1 gene expression during the postprandial state. Glucose induction of bile acid synthesis may be an important mechanism by which the liver regulates postprandial glucose and lipid homeostasis. In both type I and type II diabetic mice, hyperglycemia increased basal levels of Cyp7a1 gene expression, but these mice were not responsive to fasting and refeeding.
EXPERIMENTAL PROCEDURES
Animal
Humanized CYP7A1-tg mice were generated as follows. A ∼167-kb bacteria artificial chromosome (clone 114M5) containing the entire human CYP7A1 gene was purchased from Invitrogen. This clone was purified and used to generate human CYP7A1-transgenic mice using standard microinjection at the mouse facility at the University of California, Irvine. A founder mouse on a mixed C57BL/6J and BALB/c (CB6) strain background was obtained and was cross-bred with heterozygote cyp7a1 knock-out (Cyp7a1+/−) mice on congenic C57BL/6J background (bred by Dr. Sandra K. Erickson, University of California, San Francisco) to generate humanized CYP-7A1-tg founders harboring one copy of the human CYP7A1 transgene on a mouse cyp7a1 knock-out background. Humanized CYP7A1-tg mice used in the study were generated from subsequent cross-breeding using F1 littermates. Male wild type C57BL/6J mice and ob/ob mice were purchased from the Jackson Laboratory (Bar Harbor, ME). The fxr−/− mice on C57BL/6J background were a gift from Dr. Yanqiao Zhang (Northeast Ohio Medical University). To induce type I diabetes, male C57BL/6 mice were intraperitoneally injected with 7.5 mg/kg streptozocin once daily for 5 consecutive days. Control mice were injected with vehicle only (sodium citrate buffer, pH 4.5). One week after the last injection, mice with hyperglycemia (non-fasting blood glucose >400 mg/dl) were used for further study. All mice were maintained on a standard chow diet and water ad libitum and housed in a room with a 12-h light (6 a.m. to 6 p.m.) and 12-h dark (6 p.m. to 6 a.m.) cycle. For the fasting and refeeding study, mice were fasted from 9 a.m. and refed with a standard laboratory chow diet at midnight. For restricted feeding, regular chow was given at 12 a.m. (Zeitgeber time 18 (ZT18), lights on at ZT0 and off at ZT12), for 3 h, and mice were moved to clean cages without food at ZT21. Intestine cholesterol absorption was measured by a dual isotope plasma ratio method as described previously (18). Animal protocols were approved by the Institutional Animal Care and Use Committee.
CYP7A1 Enzyme Activity Assay
Mouse liver microsomes were isolated for the analysis of CYP7A1 enzyme activity with a high performance liquid chromatography-based method as described previously (19).
Bile Acid Analysis
Bile acids in liver, intestine (with content), gallbladder, and feces were extracted with 95% ethanol once overnight, 80% ethanol once for 2 h, and methanol/chloroform (2:1) once for 2 h at 50 °C. Serum samples were used directly. Total bile acids were determined with a bile acid assay kit (Genzyme Diagnostic, Framingham, MA). The bile acid pool was determined as the total amount of bile acids in liver, intestine, and gallbladder. Gallbladder and fecal bile acid compositions were analyzed using liquid chromatography (LC)/mass spectrometry (MS/MS) as described previously (20).
RNA Isolation and Quantitative Real-time PCR
Total RNA was isolated with TRI reagent (Sigma). All primers/probe sets for real-time PCR were ordered from TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA). Amplification of ubiquitin C was used as an internal control. Relative mRNA expression was quantified using the comparative CT (Ct) method and expressed as 2−ΔΔCt.
Immunoblots
Antibodies used in immunoblots were purchased from Cell Signaling Biotechnology, Inc. (Danvers, MA), except for a monoclonal antibody to mouse cyp7a1 (Cosmo Bio USA Inc., Carlsbad, CA). Total tissue lysates were prepared in radioimmune precipitation assay buffer (Cell Signaling Biotec, Inc.) and used in SDS-PAGE for immunoblotting. Mouse liver microsomes were used for assay of CYP7A1 protein.
Epigenetic Analysis
Histone acetylation and methylation status of CYP7A1 gene promoter were analyzed by chromatin immunoprecipitation (ChIP) assay. Nuclei were isolated from mouse livers, and ChIP assays were performed with a kit (Millipore, Billerica, MA) as described previously (21). Antibodies against acetyl-histone 3, acetyl-histone 4, and dimethylated or trimethylated histone 3 lysine 9 (H3K9) (Cell Signaling Technology, Inc.) were used to immunoprecipitate chromatin. Real-time PCR was used to quantify ChIP assay results. TaqMan primers/probe sets detecting the human CYP7A1 proximal promoter containing the bile acid response elements were described previously (22). The following SYBR primers were used to detect mouse cyp7a1 promoter regions: −3k region, −2205GGTTAGAGACCTGGATTGCTTAGC (forward) and −2093ATAGCACATCGTCTTCTCAAATGG (reverse); −2k region, −1483GAGGGTCGCTTGGCTTTAAA (forward) and −1400TCTGAGGTAAGGAGAAAGGAAAACAT (reverse); −1k region, −219ACCTTCGGCTTATCGACTATTGC (forward) and −163TATCTGGCCTTGAACTAAGTCCATCT (reverse); +1k region, +518GTGAGAGCACAGAGCCTGAGTTT (forward) and +590TTCCCAGTCACACAGTTGGTTAAC (reverse). Non-immune IgG was used as a negative control for subtracting background.
Measurement of Serum Glucose and Insulin
Serum glucose was measured with a One-touch Ultra glucometer (LifeScan, Mountain View, CA). Serum insulin was determined by a mouse insulin ELISA kit (Crystal Chem Inc., Downers Grove, IL).
Recombinant Adenovirus
Adenovirus expressing a dominant negative form of AKT (Ad-DN-AKT) was purchased from Vector Biolabs (Philadelphia, PA). Adeno-FoxO1 expressing a constitutively active nuclear form of FoxO1 (Ad-FoxO1) was a generous gift from Dr. D. Accili (Columbia University, New York). Adenovirus was purified from HEK293A cells by CsCl centrifugation. Viral titers were determined by an Adeno-X rapid titer kit (Stratagene). Adenovirus was administered at 2 × 109 pfu/mouse via tail vein. Experiments were carried out 7 days postinjection.
Statistical Analysis
Results are expressed as mean ± S.E. except where indicated. Statistical analysis was performed by Student's t test. p < 0.05 is considered statistically significant.
RESULTS
Feeding-induced cyp7a1 Gene Expression in Wild Type C57BL6J Mice
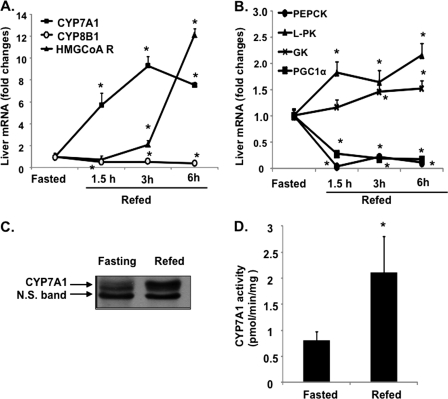
For studying nutrient regulation of cyp7a1 gene expression, mice were fasted for 15 h and then refed regular chow for 1.5, 3, and 6 h. Food was given to mice at midnight because mice are actively feeding during the dark period, and thus, such conditions may resemble human daytime activities. Fig. 1A shows that CYP7A1 mRNA expression levels were markedly and rapidly induced upon refeeding. Also 3-hydroxy-3-methyl-glutaryl Co-A reductase in the de novo cholesterol synthesis pathway was induced as a compensatory response to decreased cholesterol in hepatocytes. In contrast, the mRNA levels of cholesterol 12α-hydroxylase (CYP8B1) involved in cholic acid synthesis decreased ∼50% (Fig. 1A). As controls, mRNA levels of glycolytic genes, glucokinase, and liver pyruvate kinase increased after refeeding (Fig. 1B), whereas the gluconeogenic and fasting-induced gene phosphoenolpyruvate carboxykinase and fasting-induced PGC-1α were rapidly reduced by refeeding as expected (Fig. 1B). Immunoblot analysis showed that CYP7A1 protein levels (Fig. 1C) and enzyme activity (Fig. 1D) in liver microsomes were induced ∼3-fold by refeeding.
FIGURE 1.
Feeding induces CYP7A1 in wild type C57BL6J mice. Male C57BL6J mice (12 weeks old) were fasted for 15 h from 9 a.m. to 12 a.m. and were either fasted or refed a regular chow diet for the time indicated. A and B, liver mRNA expression levels were measured by real-time PCR. C, CYP7A1 protein levels were determined by immunoblot of isolated liver microsomes from fasted and refed (3 h) mice. D, microsomal CYP7A1 enzyme activity was determined using an HPLC-based method as described under “Experimental Procedures.” Results are expressed as mean ± S.E. (error bars) (n = 4). *, statistical significance, p < 0.05 versus fasted mice. N.S., not significant.
Glucose Induces cyp7a1 Gene Expression via Insulin Signaling
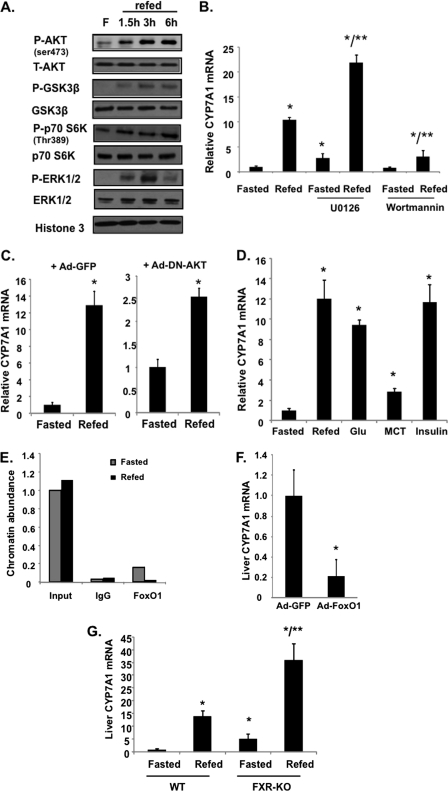
Because our previous studies have shown that treatment of primary human hepatocytes with insulin or glucose induced CYP7A1 mRNA (15, 17), we therefore tested the potential roles of glucose and/or insulin signaling in mediating feeding induction of the cyp7a1 gene in mice. Fig. 2A shows that refeeding of fasted mice resulted in rapid phosphorylation of AKT (protein kinase B) and insulin target genes glycogen synthase kinase 3β (GSK3β) and p70S6 kinase (p70S6K). Phosphorylation of ERK1/2 of the MAPK pathway was rapidly increased at 1.5 h, peaking at 3 h, and then decreased to low levels at 6 h after refeeding (Fig. 2A). ERK is a downstream target of insulin, bile acids, and FGF15/19 signaling (6). When mice were pretreated with an ERK inhibitor, U0126, the basal CYP7A1 mRNA expression was increased, but refeeding did not affect the -fold induction of CYP7A1 mRNA by refeeding (Fig. 2B). In contrast, when mice were pretreated with wortmannin (phosphatidylinositol 3-kinase inhibitor) to block AKT activity, the refeeding induction of CYP7A1 mRNA was significantly blunted (Fig. 2B). Similarly, adenovirus-mediated transduction of a liver-specific dominant negative form of AKT also attenuated the feeding induction of CYP7A1 mRNA (Fig. 2C). These results support a role of insulin signaling in the postprandial induction of cyp7a1 gene expression. To study if glucose or insulin directly induces CYP7A1 in mice, we performed oral gavage of a single dose of glucose or intraperitoneal injection of insulin to fasted mice. Both insulin and glucose administration rapidly induced CYP7A1 mRNA expression to levels comparable with that induced by a chow diet. Interestingly, oral gavage of medium chain triglycerides had much less effect (Fig. 2D). These data further confirm that glucose activation of insulin/AKT signaling plays a major role in induction of CYP7A1 by feeding.
FIGURE 2.
Insulin signaling mediates feeding induction of CYP7A1 in wild type C57BL6J mice. A, feeding effects on total and phosphorylated AKT, GSK3β p70 S6K, ERK1/2, and AMP-activated protein kinase in mouse liver were determined by immunoblot; samples were pooled from four mice. B, mice were fasted from 9 a.m. and given a single dose of the ERK inhibitor U0126 (5 mg/kg) or the PI3K inhibitor, wortmannin (0.8 mg/kg) by intravenous injection at 11 p.m. One h later, mice were either fasted or refed chow for an additional 3 h. Liver mRNA expression was determined by real-time PCR. C, mice were injected (intravenously) with adenovirus (2−9 pfu/mouse) expressing GFP (left) or a dominant negative form of AKT (Ad-DN-AKT) (right). Seven days later, mice were fasted for 15 h or refed chow for an additional 3 h, and liver CYP7A1 mRNA was measured. D, mice fasted for 15 h were refed chow or given a single dose of glucose (8 g/kg, oral gavage), medium chain triglycerides (MCT) (4 mg/kg, oral gavage), or insulin (0.8 unit/kg, intraperitoneal injection). Liver CYP7A1 mRNA levels were determined 3 h later. E, ChIP assay of FoxO1 binding to the cyp7a1 gene promoter in mouse livers. Liver nuclei were isolated from fasted and refed (3 h) mice. Pooled samples from four mice were used. ChIP primers detecting the proximal cyp7a1 promoter region (−1k) were used. Real-time PCR was done in triplicate, and mean values were plotted. F, effect of adenovirus-mediated transduction of constitutively active nuclear form of FoxO1 on CYP7A1 mRNA expression levels in mouse liver. Mice were injected (intravenously) with adenovirus (2−9 pfu/mouse) expressing either GFP as control or expressing a constitutively active nuclear form of FoxO1 (Ad-FoxO1). Seven days later, liver CYP7A1 mRNA was measured in overnight fasted mice. G, liver CYP7A1 mRNA was determined in fasted and refed (3 h) wild type and FXR knock-out (KO) mice. Results are expressed as mean ± S.E. (error bars) (n = 4). *, statistical significance versus fasted mice; **, statistical significance versus fasted + U0126 or fasted FXR knock-out mice (p < 0.05).
Insulin signaling is known to inactivate the forkhead box transcription factor O1 (FoxO1) by phosphorylation and nuclear exclusion (23). ChIP assays show that refeeding markedly reduced FoxO1 occupancy on the CYP7A1 gene promoter (Fig. 2E), and adenovirus-mediated transduction of a constitutively active nuclear form of FoxO1 strongly reduced Cyp7a1 mRNA levels in mouse liver (Fig. 2F). These results are consistent with our previous reports that FoxO1 inhibits CYP7A1 by blocking HNF4α interaction with PGC-1α (3, 15), suggesting that insulin inhibits FoxO1 binding to the CYP7A1 gene promoter and results in induction of CYP7A1 gene expression.
A recent study reports that FXR induces FGF15, which is a postprandial activator that stimulates hepatic protein and glycogen synthesis independent of insulin in mice (24). We thus studied the role of FXR in postprandial regulation of CYP7A1 using fxr−/− mice. Fig. 2G shows that refeeding strongly induced CYP7A1 mRNA expression in fxr−/− mice as in wild type mice, suggesting that postprandial FXR signaling had little effect on induction of CYP7A1 by refeeding.
Glucose Induction of Cyp7a1 Gene Expression by Epigenetic Mechanisms
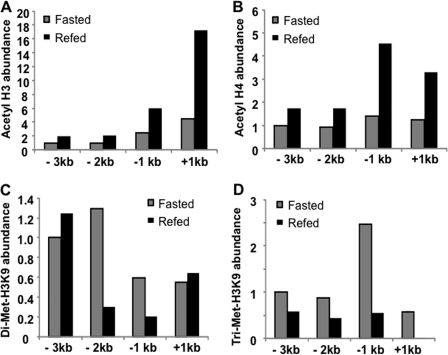
Recent studies suggest that glucose, as an indicator of nutrient availability, directly controls many cellular, physiological, and disease pathways via activation of gene expression. One such mechanism of glucose action is acetylation of the histone code (25). We have reported previously that high glucose induced CYP7A1 via epigenetic modification of CYP7A1 chromatin in primary human hepatocytes (17). ChIP assays show that refeeding resulted in increasing histone 3 (Fig. 3A) and histone 4 acetylation (markers of gene activation) (Fig. 3B) and decreasing histone 3 lysine 9 (H3K9) di- and trimethylation (markers of gene silencing) (Fig. 3, C and D, respectively) in the mouse cyp7a1 gene promoter, with more dramatic histone modification occurring within the −1 to +1 kb region of the cyp7a1 gene. These results provided in vivo evidence that glucose induces CYP7A1 expression via epigenetic mechanisms.
FIGURE 3.
Feeding caused histone hyperacetylation and histone hypomethylation in the cyp7a1 gene chromatin in wild type C57BL6J mice. Mice were fasted or refed chow for 3 h. ChIP assays were used to determine the histone 3 (H3) acetylation (A), histone 4 (H4) acetylation (B), H3K9 dimethylation (C), and H3K9 trimethylation (D). Pooled samples from four mice were used for real-time PCR to detect acetylated histones 3 and 4, dimethylated H3K9, and trimethylated H3K9 in CYP7A1 chromatin. Assays were done in triplicate, and mean values were plotted.
Nutrients Control Circadian Variations of cyp7a1 Gene Expression and Bile Acid Homeostasis
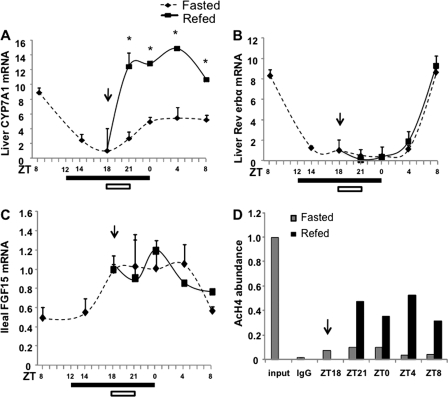
It is well documented that CYP7A1 activity and bile acid synthesis exhibit distinct circadian rhythms (7), which are regulated by liver clock genes, such as Rev-erbα (8, 26). We thus studied the effects of restricted feeding on CYP7A1 mRNA expression in mice under a 12-h dark-light cycle (lights on at ZT0 and lights off at ZT12). Mice were fasted for 12 h. One group of fasted mice was fed at ZT18 (6 h into the dark period) for 3 h, whereas another group of mice were continuously fasted. CYP7A1 mRNA levels in mouse liver were measured over a 24-h period. Without food intake, CYP7A1 mRNA levels decreased during the dark cycle and slightly increased prior to the beginning of the light cycle, suggesting fasting dampening circadian rhythm (Fig. 4A, broken line). Restricted feeding markedly induced CYP7A1 mRNA expression (Fig. 4, solid line). The CYP7A1 mRNA expression pattern in fasted mice correlated with that of Rev-erbα (Fig. 4B, dotted line), which positively regulates cyp7a1 mRNA expression in mice (8, 26). It is noted that Rev-erbα mRNA levels were not affected by feeding (Fig. 4B). FXR is known to induce FGF15 in mice when fed an FXR agonist or cholic acid, and FGF15/FGFR4 signaling inhibited cyp7a1 gene transcription in mouse hepatocytes (5). In fasted mice, ileal FGF15 mRNA levels increased in the dark period and decreased in the light period (Fig. 4C, broken line), which was inversely correlated with CYP7A1 mRNA as expected. However, refeeding did not alter FGF15 mRNA expression levels. As a result, the CYP7A1 mRNA expression pattern was similar to that of ileal FGF15 in refed mice. Together, these data suggest that fasting alters circadian variations of CYP7A1 mRNA expression and that feeding is the major cause of increasing Cyp7a1 mRNA levels in the dark. Thus, fasting-refeeding may entrain (synchronize) the circadian rhythm of bile acid synthesis. Furthermore, restricted feeding rapidly increased histone 4 acetylation in the cyp7a1 gene promoter (Fig. 4D), suggesting that hyperacetylation of CYP7A1 chromatin induces CYP7A1 gene expression. The lack of a refeeding effect on intestinal FGF15 mRNA expression further supports the conclusion that dietary glucose plays a major role in postprandial induction of CYP7A1, and FXR signaling does not have any effect on CYP7A1 gene expression during restricted feeding.
FIGURE 4.
Nutrients control the circadian variations of cyp7a1 gene expression in wild type C57BL6J mice. Mice were fasted at 9 a.m. (ZT3). For restricted feeding, regular chow was given at 12 a.m. (ZT18) for 3 h, and mice were moved to a clean cage without food. Liver CYP7A1 mRNA (A), liver Rev-erbα mRNA (B), and ileum FGF16 mRNA (C) expression levels were measured at the times indicated. A filled bar below each panel indicates the dark period. An arrow and open bar indicate the start of the restricted feeding period (3 h). Results are expressed as mean ± S.E. (error bars) (n = 4). *, statistical significance versus fasted mice at the same time point (p < 0.05). D, ChIP assays were used to determine histone 4-acetylation (AcH4) in the cyp7a1 gene promoter in mouse liver. Pooled samples from four mice were used. Real-time PCR was used to detect acetylated H4 in CYP7A1 chromatin. Assays were done in triplicate, and mean values were plotted.
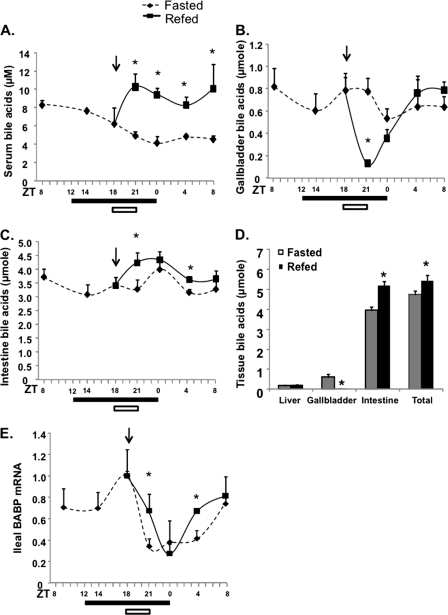
We next determined the effects of restricted feeding on bile acid homeostasis. Mice under fasting conditions showed a gradual decrease of serum bile acids (Fig. 5A), and restricted feeding for 3 h rapidly increased serum bile acid levels. As expected, restricted feeding rapidly reduced bile acids in the gallbladder, which was refilled gradually (Fig. 5B). Interestingly, intestine bile acid contents remained constant during fasting, and restricted feeding significantly increased intestine bile acid content (Fig. 5C). Fig. 5D shows that the intestine retained ∼80% of the total bile acid pool, which was increased by ∼18% after restricted feeding. It appears that the modest increase in intestine bile acid levels did not induce the intestine FXR target gene, ileum bile acid-binding protein (Fig. 5E). These results suggest that the bile acid pool size remains relatively constant during the fasting and refeeding cycle. Increasing de novo bile acid synthesis during the postprandial period may contribute to increasing intestine and serum bile acid levels.
FIGURE 5.
Effects of restricted feeding on tissue and serum bile acid homeostasis. Serum bile acids (A), gallbladder bile acids (B), and intestine bile acids (C) were measured in fasted and food-restricted mice at different time points, as indicated. D, tissue bile acids were quantified in fasted and refed (3 h) mice. E, ileum bile acid-binding protein mRNA was determined by real-time PCR. A filled bar below each panel indicates the dark period. An arrow and open bar indicate the start of restricted feeding and period (3 h). Results are expressed as mean ± S.E. (error bars) (n = 4). *, statistical significance versus fasted mice at the same time point (p < 0.05).
Diabetes Is Associated with Increased Bile Acid Pool and Altered Bile Acid Composition
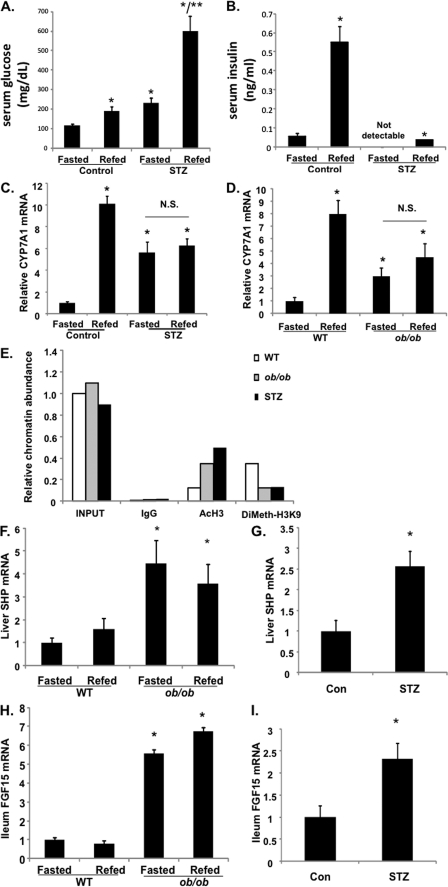
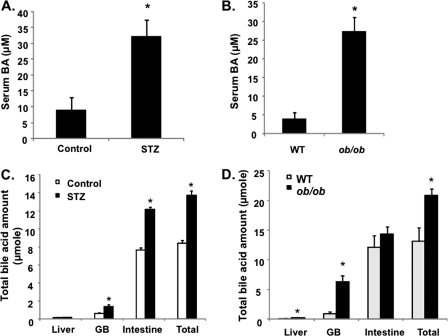
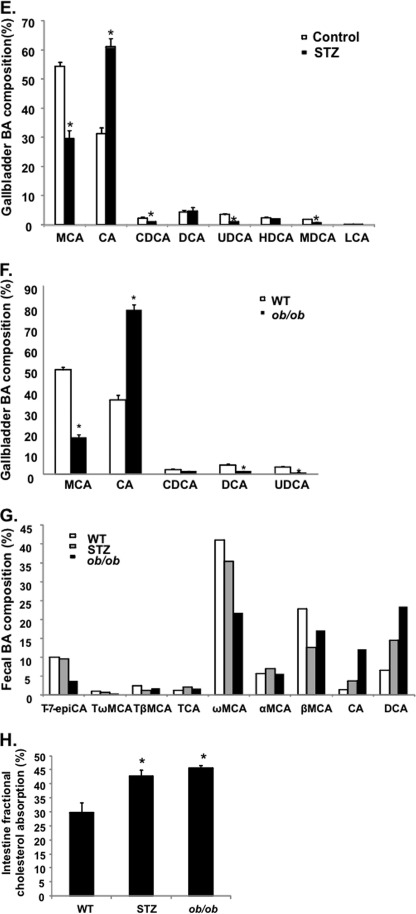
We then studied the refeeding effect on CYP7A1 gene expression and bile acid homeostasis in diabetic mice. STZ-induced type I diabetic mice are hyperglycemic and insulin-insufficient; thus, they were used as a model to study the glucose effect independent of insulin. Genetically obese ob/ob mice are insulin-resistant and hyperglycemic; thus, they were used as a type II diabetic model. As shown in Fig. 6, A and B, STZ treatment caused hyperglycemia and abolished insulin secretion in mice. Both the STZ-treated mice (Fig. 6C) and ob/ob mice (Fig. 6D) expressed markedly higher basal levels of CYP7A1 mRNA, and refeeding did not further induce CYP7A1 mRNA in these mice. Furthermore, the ChIP assay shows that both STZ-treated and ob/ob mice had increased histone 3-acetylation and reduced H3K9 dimethylation (Fig. 6E), suggesting that hyperglycemia induced CYP7A1 expression in diabetic mice by epigenetic mechanisms. It is interesting to note that both type I and type II diabetic mice had significantly higher hepatic SHP (Fig. 6, F and G) and ileal FGF15 (Fig. 6, H and I) expression than those of wild type mice. These data indicate that in diabetic conditions, the positive effect of glucose may dominate over the negative effect of FGF15 and SHP in controlling cyp7a1 gene transcription. Both STZ-treated and ob/ob mice had 3–5-fold higher serum bile acids (Fig. 7, A and B) and a ∼30% larger bile acid pool (Fig. 7, C and D). Interestingly, analysis of gallbladder bile acid composition revealed a trend of increase of taurocholic acid and decrease of tauromuricholic acids, thus resulting in a higher bile acid hydrophobicity index (calculated according to Heuman (27)), from −0.41 in wild type to −0.2 in both STZ mice (Fig. 7E) and ob/ob mice (Fig. 7F). Analysis of fecal bile acid composition showed that diabetic mice excreted more cholic acid and deoxycholic acid and less muricholic acids than wild type mice (Fig. 7G). Cholic acid has a lower critical micelle concentration (50 μm); thus, it is more efficient in facilitating intestinal cholesterol absorption (28). This may explain the significantly higher intestinal fractional cholesterol absorption of diabetic mice compared with wild type mice (Fig. 7H) and may contribute to hypercholesterolemia in diabetic mice.
FIGURE 6.
Diabetes is associated with elevated CYP7A1 mRNA expression and histone hyperacetylation in cyp7a1 gene chromatin. STZ-treated mice, ob/ob mice, and their respective controls were fasted for 15 h and refed for 3 h. A, serum glucose in STZ mice. B, serum insulin in STZ mice. C, liver CYP7A1 mRNA in STZ mice. D, liver CYP7A1 mRNA expression levels in ob/ob mice. E, ChIP assays of histone 3 acetylation and H3K9 methylation in CYP7A1 gene promoter of ob/ob and STZ mice. F, liver SHP mRNA in ob/ob mice. G, liver SHP mRNA in STZ mice. H, ileum FGF15 mRNA in ob/ob mice. I, intestine FGF15 mRNA in STZ mice. Liver and ileum mRNA expression was determined by real-time PCR. Results are expressed as mean ± S.E. (error bars) (n = 4). *, statistical significance, p < 0.05 versus fasted control mice. **, statistical significance, p < 0.05 versus fasted STZ mice. N.S., not significant. ChIP assays were used to determine H3 acetylation and H3K9 dimethylation in fasted mouse livers using pooled samples (n = 4), and real-time PCR was used to detect acetylated histone 3 (AcH3) and dimethylated H3K9 (DiMeth-H3K9) in CYP7A1 chromatin. Assays were done in triplicate, and mean values were plotted.
FIGURE 7.
Diabetic mice had an enlarged bile acid pool, altered bile acid composition, and increased intestine cholesterol absorption. A, serum bile acid concentration in STZ mice; B, serum bile acid concentration in ob/ob mice; C, total bile acid contents in liver, gallbladder (GB), intestine, and total bile acid pool in STZ mice. D, total bile acid contents in liver, gallbladder (GB), intestine, and total bile acid pool (liver + GB + intestine) in ob/ob mice. E, gallbladder bile acid composition in STZ mice. F, gallbladder bile acid composition in ob/ob mice. G, fecal bile acid composition in STZ and ob/ob mice. Pooled fecal bile acid extracts from 3–4 mice were used. H, intestinal fractional cholesterol absorption in STZ and ob/ob mice. Bile acid composition and intestine fractional cholesterol absorption were determined as described under “Experimental Procedures.” Results are expressed as mean ± S.E. (error bars) (n = 3–4). *, statistical significance versus control mice (p < 0.05). Fecal samples from four mice were pooled, and bile acids were extracted for composition analysis. T, tauro-conjugated bile acids; CA, cholic acid; MCA, α- and β-muricholic acids; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; UDCA, ursodeoxycholic acid; HDCA, hyodeoxycholic acid; LCA, lithocholic acid.
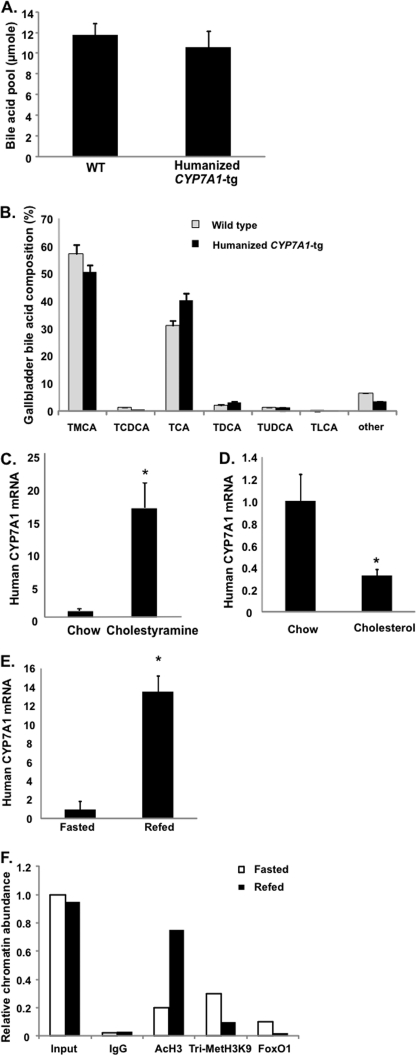
Refeeding Stimulates CYP7A1 Expression in Humanized CYP7A1-tg Mice but Not in Diabetic Mice
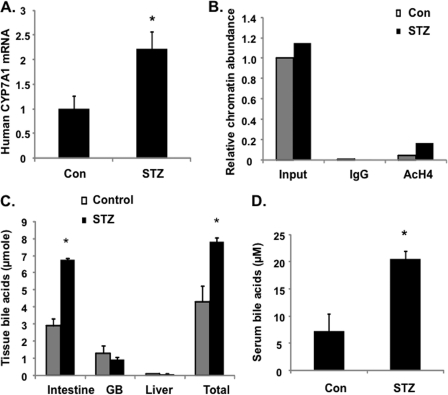
it is now well documented that transcription of the human and mouse CYP7A1 genes is differentially regulated by diet, fasting, insulin/glucagon, and diurnal rhythm (1). There are substantial differences in CYP7A1 gene promoter sequences among different species. However, it is not clear whether or not species differences in CYP7A1 gene regulation are due to the genomic effects (DNA sequences) or epigenetic mechanisms. Current study of human CYP7A1 gene regulation is limited to human hepatocyte models. To study nutrient regulation of human CYP7A1 gene expression in an in vivo system, we created the humanized CYP7A1-tg mice that harbor one copy of the human CYP7A1 gene on a mouse cyp7a1 knock-out background. The humanized CYP7A1-tg mice had a bile acid pool size (Fig. 8A) comparable with that of wild type mice. The gallbladder bile acid composition in humanized CYP7A1-tg mice was somewhat altered, showing a small increase in cholic acid and decrease in α- and β-muricholic acids compared with those of wild type mice (Fig. 8B). Feeding humanized CYP7A1-tg mice the bile acid binding resin cholestyramine markedly induced human CYP7A1 mRNA (Fig. 8C). Unlike mouse cyp7a1, which was induced by cholesterol feeding via activation of LXRα (29), feeding humanized CYP7A1-tg mice with a 2% cholesterol-enriched diet did not induce but instead repressed CYP7A1 mRNA expression (Fig. 8D). This is consistent with the lack of an LXRα binding site on the human CYP7A1 gene promoter (30). Together, these results indicate that the human CYP7A1 transgene in mice is functional in producing an active enzyme. Refeeding markedly induced CYP7A1 mRNA in humanized CYP7A1-tg mice (Fig. 8E), increased histone 3-acetylation, and decreased histone H3K9 trimethylation (Fig. 8F) and decreased FoxO1 occupancy on the human CYP7A1 gene promoter (Fig. 8F). Furthermore, STZ-treated diabetic humanized CYP7A1-tg mice had significantly higher basal CYP7A1 mRNA (Fig. 9A), histone 4-acetylation (Fig. 9B), bile acid pool size (Fig. 9C), and serum bile acids (Fig. 9D) compared with control mice. In summary, these data provided for the first time in vivo evidence that the human CYP7A1 gene was induced by glucose via epigenetic mechanisms to increase bile acid synthesis in response to feeding. However, in diabetic mice, hyperglycemia increased basal CYP7A1 expression by epigenetic mechanisms, but fasting-refeeding did not further increase CYP7A1 expression.
FIGURE 8.
Feeding induces CYP7A1 in humanized CYP7A1-tg mice. Total bile acid pool (A) and gallbladder bile acid composition (B) were determined in humanized CYP7A1-tg mice and wild type controls. C and D, human CYP7A1 mRNA levels in humanized CYP7A1-tg mice. Mice were fed chow or chow containing 5% cholestyramine or 2% cholesterol for 1 week. Human CYP7A1 mRNA was determined by real-time PCR. E, refeeding effect on human CYP7A1 mRNA expression in humanized CYP7A1-tg mice. Mice were fasted or refed for 3 h. Liver CYP7A1 mRNA was determined by real-time PCR. Results are expressed as mean ± S.E. (error bars), n = 3–4. *, statistical significance (p < 0.05). F, ChIP assay of the abundance of acetylated histone 3 (AcH3), trimethylated H3K9, and FoxO1 in the human CYP7A1 gene promoter of humanized CYP7A1-tg mice. Pooled liver samples from 3–4 mice were used in ChIP assays. Real-time PCR was done in triplicate, and mean values were plotted. Abbreviations for bile acids are as in Fig. 7.
FIGURE 9.
Hyperglycemia in STZ-treated humanized CYP7A1-tg mice caused increased human CYP7A1 gene expression and enlarged bile acid pool. STZ-treated humanized CYP7A1-tg mice and their respective controls were used. A, human CYP7A1 mRNA level in STZ-treated diabetic humanized CYP7A1-tg mice. B, ChIP assay of histone 4-acetylation (AcH4) in the CYP7A1 gene promoter. Pooled liver samples from three mice were used in ChIP assays. Real-time PCR was done in triplicate, and mean values were plotted. C, tissue bile acid contents in intestine, gallbladder, liver, and total bile acid pool of STZ-treated and control humanized CYP7A1-tg mice. D, serum bile acid concentrations in STZ-treated and control humanized CYP7A1-tg mice. Results are expressed as mean ± S.E. (error bars) (n = 3–4). *, statistical significance versus control (p < 0.05).
DISCUSSION
In this study, we demonstrated for the first time that in both wild type and humanized CYP7A1-tg mice, glucose and insulin rapidly induced CYP7A1 gene expression and bile acid synthesis, leading to an enlarged bile acid pool size and elevated circulating bile acids. Postprandial stimulation of bile acid synthesis may be viewed as a mechanism to control postprandial glucose and lipid homeostasis by enhancing bile acid signaling. The identification of glucose as a physiological regulator of CYP7A1 directly links activation of bile acid synthesis to nutrient availability. Indeed, liver synthesis of protein, cholesterol, fatty acids, and glycogen are all controlled by glucose and insulin signaling via mechanisms such as stimulating insulin secretion, generating glycolytic intermediates, and altering epigenetic histone codes (25, 31). These processes are stimulated during postprandial periods to control glucose, lipid, and energy metabolism and storage. In this study, we showed that glucose might directly activate CYP7A1 by hyperacetylation of histone on the CYP7A1 gene promoter.
A key role of bile acid-activated FXR signaling in mediating bile acid regulation of hepatic bile acid synthesis has been demonstrated by numerous studies using pharmacological doses of bile acids, FXR agonists, or FGF19 or genetic knock-out of the fxr gene in mice (4, 5). The direct evidence that the FXR-dependent pathways play a role in regulating CYP7A1 gene transcription under normal physiological conditions is still lacking. Our results from this study show little fluctuation of bile acid pool and intestine FGF15 in the fasting to refeeding transition. The modest transient increase of intestine bile acids after food intake may not be sufficient to activate intestinal FXR signaling, as evidenced by the lack of induction of FGF15 and ileum bile acid-binding protein. Recent studies reported that transfusion of human FGF19 activates hepatic MAPK/ERK to stimulate glycogen synthesis and repress glucose production in mouse liver, suggesting that FGF15 may act as an insulin-independent regulator of postprandial glucose homeostasis (24, 32). This current study shows that restricted feeding induced CYP7A1 and bile acid synthesis but not FGF15 mRNA, suggesting that the stimulatory effects exerted by glucose and insulin may dominate over the inhibitory effects of FXR/SHP and FXR/FGF15 signaling in the postprandial state to increase circulating bile acids. It appears that circulating bile acids may activate TGR5 to promote secretion of GLP-1 (glucagon-like peptide-1) in intestine (33) and energy expenditure in brown fats to increase insulin sensitivity and reduce weight gain (34).
A key finding of this study is that the glucose/insulin axis controls hepatic bile acid synthesis upon refeeding of fasted mice. In both mouse models of type I and type II diabetes, bile acid pool sizes are increased, but the fasting-to-feeding regulation of the cyp7a1 gene was lost. In ob/ob mice, both the insulin/FoxO1 pathway and glucose-stimulated epigenetic mechanisms contribute to increased basal cyp7a1 expression, whereas in STZ mice, the insulin/FoxO1 pathway does not exist, and the glucose-stimulated epigenetic mechanism may play a major role in increasing basal cyp7a1 expression. Thus, our current study is consistent with the increasingly appreciated “glycemic memory” effect, whereby long lasting epigenetic changes of genes caused by exposure to high blood glucose underlie many chronic diabetic complications (25, 35, 36). This study demonstrates that bile acid levels are sensitive to serum glucose, and enlarged bile acid pool and fasting serum bile acid levels in diabetes could closely reflect nutritional status and energy imbalance. The implication of bile acids in obesity and diabetes is further supported by a recent clinical study demonstrating that serum bile acids were higher in patients with prior gastric bypass than in overweight and severely obese patients without gastric bypass, and serum bile acids were positively correlated with serum GLP-1 (37). It is likely that in gastric bypass patients, bile acid synthesis may be increased due to reduced bile acid feedback, resulting in improved glucose tolerance in obese patients.
It seems paradoxical that both ob/ob and STZ mice have an increased basal expression of Cyp7a1 and bile acid pool and are insulin-resistant or insulin-insufficient and diabetic, but Cyp7a1-transgenic mice also have increased bile acid pool size but are resistant to high fat diet-induced insulin resistance and obesity (3). This is probably because in Cyp7a1-transgenic mice, increased hydrophobic bile acid pool and signaling stimulates the FXR/SHP pathway to inhibit lipogenesis and gluconeogenesis and the TGR5 pathway to stimulate intestinal GLP-1 secretion and brown adipocyte energy metabolism (34, 38). In diabetic mice, increased bile acid pool size and serum bile acids are the consequences of hyperglycemia, not the cause of diabetes.
Results from the current study are consistent with a recent finding that lowering circulating bile acids worsened diet-induced obesity and diabetes, whereas increasing bile acid pool size improved glucose homeostasis (39). Interestingly, a recent metabolomics study identified bile acids as the most elevated metabolites in human sera after an oral glucose challenge in patients with normal glucose tolerance, but this response was blunted in patients with impaired glucose tolerance (40). Serum bile acid levels have become biomarkers for diagnosis of liver diseases, diabetes, and obesity.
In summary, the current study of CYP7A1 gene regulation under fasting and refeeding cycles and diabetic conditions provides several lines of evidence supporting dietary glucose and insulin as key postprandial regulators that control hepatic bile acid synthesis. Such a regulatory mechanism ensures that bile acids function as postprandial metabolic regulators of glucose and lipid homeostasis. This coordinated regulation of postprandial glucose and bile acid metabolism may play an important role in regulating postprandial lipid and glucose homeostasis. Impaired response of bile acid synthesis to fasting and refeeding may contribute to pathogenesis of diabetes and obesity.
This work was supported, in whole or in part, by National Institutes of Health Grants R01DK-44442 and R37DK-58379 (to J. Y. L. C.), DK-081461 and ES-009649 (to C. D. K.), and DK-072187 (to S. K. E.). This work was also supported by a Veterans Affairs merit award (to S. K. E.).
- FXR
- farnesoid X receptor
- H3K9
- histone 3 lysine 9
- SHP
- small heterodimer partner
- STZ
- streptozocin
- ZT
- Zeitgeber time
- tg
- transgenic
- Ad
- adenovirus.
REFERENCES
- 1. Chiang J. Y. (2009) Bile acids. Regulation of synthesis. J. Lipid Res. 50, 1955–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lefebvre P., Cariou B., Lien F., Kuipers F., Staels B. (2009) Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 89, 147–191 [DOI] [PubMed] [Google Scholar]
- 3. Li T., Owsley E., Matozel M., Hsu P., Novak C. M., Chiang J. Y. (2010) Transgenic expression of cholesterol 7α-hydroxylase in the liver prevents high-fat diet-induced obesity and insulin resistance in mice. Hepatology 52, 678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., Maloney P. R., Willson T. M., Kliewer S. A. (2000) A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6, 517–526 [DOI] [PubMed] [Google Scholar]
- 5. Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., Gerard R. D., Repa J. J., Mangelsdorf D. J., Kliewer S. A. (2005) Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2, 217–225 [DOI] [PubMed] [Google Scholar]
- 6. Song K. H., Li T., Owsley E., Strom S., Chiang J. Y. (2009) Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7α-hydroxylase gene expression. Hepatology 49, 297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chiang J. Y., Miller W. F., Lin G. M. (1990) Regulation of cholesterol 7α-hydroxylase in the liver. Purification of cholesterol 7α-hydroxylase and the immunochemical evidence for the induction of cholesterol 7α-hydroxylase by cholestyramine and circadian rhythm. J. Biol. Chem. 265, 3889–3897 [PubMed] [Google Scholar]
- 8. Noshiro M., Usui E., Kawamoto T., Kubo H., Fujimoto K., Furukawa M., Honma S., Makishima M., Honma K., Kato Y. (2007) Multiple mechanisms regulate circadian expression of the gene for cholesterol 7α-hydroxylase (Cyp7a), a key enzyme in hepatic bile acid biosynthesis. J. Biol. Rhythms 22, 299–311 [DOI] [PubMed] [Google Scholar]
- 9. Zhang Y. K., Guo G. L., Klaassen C. D. (2011) Diurnal variations of mouse plasma and hepatic bile acid concentrations as well as expression of biosynthetic enzymes and transporters. PLoS One 6, e16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gälman C., Angelin B., Rudling M. (2005) Bile acid synthesis in humans has a rapid diurnal variation that is asynchronous with cholesterol synthesis. Gastroenterology 129, 1445–1453 [DOI] [PubMed] [Google Scholar]
- 11. Lundåsen T., Gälman C., Angelin B., Rudling M. (2006) Circulating intestinal fibroblast growth factor 19 has a pronounced diurnal variation and modulates hepatic bile acid synthesis in man. J. Intern. Med. 260, 530–536 [DOI] [PubMed] [Google Scholar]
- 12. Bennion L. J., Grundy S. M. (1977) Effects of diabetes mellitus on cholesterol metabolism in man. N. Engl. J. Med. 296, 1365–1371 [DOI] [PubMed] [Google Scholar]
- 13. Subbiah M. T., Yunker R. L. (1984) Cholesterol 7α-hydroxylase of rat liver. An insulin-sensitive enzyme. Biochem. Biophys. Res. Commun. 124, 896–902 [DOI] [PubMed] [Google Scholar]
- 14. Shin D. J., Campos J. A., Gil G., Osborne T. F. (2003) PGC-1α activates CYP7A1 and bile acid biosynthesis. J. Biol. Chem. 278, 50047–50052 [DOI] [PubMed] [Google Scholar]
- 15. Li T., Kong X., Owsley E., Ellis E., Strom S., Chiang J. Y. (2006) Insulin regulation of cholesterol 7α-hydroxylase expression in human hepatocytes. Roles of forkhead box O1 and sterol regulatory element-binding protein 1c. J. Biol. Chem. 281, 28745–28754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Song K. H., Chiang J. Y. (2006) Glucagon and cAMP inhibit cholesterol 7α-hydroxylase (CYP7A1) gene expression in human hepatocytes. Discordant regulation of bile acid synthesis and gluconeogenesis. Hepatology 43, 117–125 [DOI] [PubMed] [Google Scholar]
- 17. Li T., Chanda D., Zhang Y., Choi H. S., Chiang J. Y. (2010) Glucose stimulates cholesterol 7α-hydroxylase gene transcription in human hepatocytes. J. Lipid Res. 51, 832–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li T., Matozel M., Boehme S., Kong B., Nilsson L. M., Guo G., Ellis E., Chiang J. Y. (2011) Overexpression of cholesterol 7α-hydroxylase promotes hepatic bile acid synthesis and secretion and maintains cholesterol homeostasis. Hepatology 53, 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chiang J. Y. (1991) Reversed-phase high-performance liquid chromatography assay of cholesterol 7α-hydroxylase. Methods Enzymol. 206, 483–491 [DOI] [PubMed] [Google Scholar]
- 20. Zhang Y., Klaassen C. D. (2010) Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. J. Lipid Res. 51, 3230–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li T., Chiang J. Y. (2005) Mechanism of rifampicin and pregnane X receptor inhibition of human cholesterol 7α-hydroxylase gene transcription. Am. J. Physiol. Gastrointest. Liver Physiol. 288, G74–G84 [DOI] [PubMed] [Google Scholar]
- 22. Li T., Chiang J. Y. (2007) A novel role of transforming growth factor β1 in transcriptional repression of human cholesterol 7α-hydroxylase gene. Gastroenterology 133, 1660–1669 [DOI] [PubMed] [Google Scholar]
- 23. Nakae J., Kitamura T., Silver D. L., Accili D. (2001) The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J. Clin. Invest. 108, 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kir S., Beddow S. A., Samuel V. T., Miller P., Previs S. F., Suino-Powell K., Xu H. E., Shulman G. I., Kliewer S. A., Mangelsdorf D. J. (2011) FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science 331, 1621–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wellen K. E., Hatzivassiliou G., Sachdeva U. M., Bui T. V., Cross J. R., Thompson C. B. (2009) ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Duez H., van der Veen J. N., Duhem C., Pourcet B., Touvier T., Fontaine C., Derudas B., Baugé E., Havinga R., Bloks V. W., Wolters H., van der Sluijs F. H., Vennström B., Kuipers F., Staels B. (2008) Regulation of bile acid synthesis by the nuclear receptor Rev-erbα. Gastroenterology 135, 689–698 [DOI] [PubMed] [Google Scholar]
- 27. Heuman D. M. (1989) Quantitative estimation of the hydrophilic-hydrophobic balance of mixed bile salt solutions. J. Lipid Res. 30, 719–730 [PubMed] [Google Scholar]
- 28. Murphy C., Parini P., Wang J., Björkhem I., Eggertsen G., Gåfvels M. (2005) Cholic acid as key regulator of cholesterol synthesis, intestinal absorption, and hepatic storage in mice. Biochim. Biophys. Acta 1735, 167–175 [DOI] [PubMed] [Google Scholar]
- 29. Lehmann J. M., Kliewer S. A., Moore L. B., Smith-Oliver T. A., Oliver B. B., Su J. L., Sundseth S. S., Winegar D. A., Blanchard D. E., Spencer T. A., Willson T. M. (1997) Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J. Biol. chem. 272, 3137–3140 [DOI] [PubMed] [Google Scholar]
- 30. Chiang J. Y., Kimmel R., Stroup D. (2001) Regulation of cholesterol 7α-hydroxylase gene (CYP7A1) transcription by the liver orphan receptor (LXRα). Gene 262, 257–265 [DOI] [PubMed] [Google Scholar]
- 31. Friis R. M., Wu B. P., Reinke S. N., Hockman D. J., Sykes B. D., Schultz M. C. (2009) A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Res. 37, 3969–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Potthoff M. J., Boney-Montoya J., Choi M., He T., Sunny N. E., Satapati S., Suino-Powell K., Xu H. E., Gerard R. D., Finck B. N., Burgess S. C., Mangelsdorf D. J., Kliewer S. A. (2011) FGF15/19 regulates hepatic glucose metabolism by inhibiting the CREB-PGC-1α pathway. Cell Metab. 13, 729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thomas C., Gioiello A., Noriega L., Strehle A., Oury J., Rizzo G., Macchiarulo A., Yamamoto H., Mataki C., Pruzanski M., Pellicciari R., Auwerx J., Schoonjans K. (2009) TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 10, 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Watanabe M., Houten S. M., Mataki C., Christoffolete M. A., Kim B. W., Sato H., Messaddeq N., Harney J. W., Ezaki O., Kodama T., Schoonjans K., Bianco A. C., Auwerx J. (2006) Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 439, 484–489 [DOI] [PubMed] [Google Scholar]
- 35. Pinney S. E., Simmons R. A. (2010) Epigenetic mechanisms in the development of type 2 diabetes. Trends Endocrinol. Metab. 21, 223–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pirola L., Balcerczyk A., Okabe J., El-Osta A. (2010) Epigenetic phenomena linked to diabetic complications. Nat. Rev. Endocrinol. 6, 665–675 [DOI] [PubMed] [Google Scholar]
- 37. Patti M. E., Houten S. M., Bianco A. C., Bernier R., Larsen P. R., Holst J. J., Badman M. K., Maratos-Flier E., Mun E. C., Pihlajamaki J., Auwerx J., Goldfine A. B. (2009) Serum bile acids are higher in humans with prior gastric bypass. Potential contribution to improved glucose and lipid metabolism. Obesity 17, 1671–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Watanabe M., Houten S. M., Wang L., Moschetta A., Mangelsdorf D. J., Heyman R. A., Moore D. D., Auwerx J. (2004) Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J. Clin. Invest. 113, 1408–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Watanabe M., Horai Y., Houten S. M., Morimoto K., Sugizaki T., Arita E., Mataki C., Sato H., Tanigawara Y., Schoonjans K., Itoh H., Auwerx J. (2011) Lowering bile acid pool size with a synthetic farnesoid X receptor (FXR) agonist induces obesity and diabetes through reduced energy expenditure. J. Biol. Chem. 286, 26913–26920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shaham O., Wei R., Wang T. J., Ricciardi C., Lewis G. D., Vasan R. S., Carr S. A., Thadhani R., Gerszten R. E., Mootha V. K. (2008) Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol. Syst. Biol. 4, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]