Background: The Ag85 proteins are fibronectin-binding proteins of mycobacteria.
Results: The binding site is mapped in the C terminus of fibronectin, Fn14.
Conclusion: Mycobacterial Ag85 proteins interact with the C terminus of fibronectin.
Significance: This was the first evidence that Ag85 proteins bind to the C terminus of fibronectin (Fn14).
Keywords: Fibronectin, Microbial Pathogenesis, Microbiology, Mycobacterium, Protein Domains, Ag85 Complex, Fn-binding Protein, Mycobacterium avium subsp. paratuberculosis, Protein-Protein Interaction
Abstract
The members of the antigen 85 protein family (Ag85), consisting of members Ag85A, Ag85B, and Ag85C, are the predominantly secreted proteins of mycobacteria and possess the ability to specifically interact with fibronectin (Fn). Because Fn-binding proteins are likely to be important virulence factors of Mycobacterium spp., Ag85 may contribute to the adherence, invasion, and dissemination of organisms in host tissue. In this study, we reported the Fn binding affinity of Ag85A, Ag85B, and Ag85C from Mycobacterium avium subsp. paratuberculosis (MAP) (KD values were determined from 33.6 to 68.4 nm) and mapped the Ag85-binding motifs of Fn. Fn14, a type III module located on the heparin-binding domain II (Hep-2) of Fn, was discovered to interact with Ag85 from MAP. The peptide inhibition assay subsequently demonstrated that a peptide consisting of residues 17–26 from Fn14 (17SLLVSWQPPR26, termed P17–26) could interfere with Ag85B binding to Fn (73.3% reduction). In addition, single alanine substitutions along the sequence of P17–26 revealed that the key residues involved in Ag85-Fn binding likely contribute through hydrophobic and charge interactions. Moreover, binding of Ag85 on Fn siRNA-transfected Caco2 cells was dramatically reduced (44.6%), implying the physiological significance of the Ag85-Fn interaction between mycobacteria and host cells during infection. Our results indicate that Ag85 binds to Fn at a novel motif and plays a critical role in mycobacteria adherence to host cells by initiating infection. Ag85 might serve as an important colonization factor potentially contributing to mycobacterial virulence.
Introduction
Johne's disease occurs worldwide as a chronic intestinal disease of domestic and wild ruminants that is caused by Mycobacterium avium subsp. paratuberculosis (MAP)2 (1). The disease is characterized clinically by intermittent to persistent diarrhea, progressive weight loss, and eventually death (2). Early studies, based on cultures of ileocecal lymph nodes collected at slaughter, indicate that the prevalence of MAP in culled dairy cows is 2.9% (3). In addition to the agricultural economic impact, MAP has been suspected as a causative agent in Crohn disease in humans (4).
Microbial infections are initiated by molecular interactions between the pathogen and receptor molecules on host cells, resulting in microbial adhesion and, sometimes, subsequent internalization (5). Furthermore, when bacteria adhere to surfaces, they are substantially more resistant to host antimicrobial defenses (6). The extracellular matrix (ECM) of the cell consists of a complex mixture of macromolecules, including collagens, fibronectin, fibrinogen, vitronectin, laminin, and heparin sulfate (6), all of which function as ligands for bacterial adhesion. Fibronectin (Fn), a 220-kDa ECM protein that forms a dimer by disulfide linkage, is composed of three different modules subdivided into several different domains, including an N-terminal domain (NTD), a gelatin-binding domain (GBD), a cell-binding domain (CBD), a 40-kDa domain containing heparin-binding domain II (Hep-2, including Fn12, Fn13, and Fn14) and Fn15, and a fibrin-binding domain II (7, 8). Fn plays a pivotal role in bacteria-host interactions by interacting with microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) (9), a group of microbial surface proteins that interact with ECMs of host cells and initiate infection. MSCRAMMs that can bind to the NTD, the GBD (10–12), or the Hep-2 have been identified (13, 14).
Antigen 85 proteins (Ag85), consisting of members Ag85A, Ag85B, and Ag85C, sharing high sequence and structural homology are secreted and retained in the cell wall of mycobacteria (15). Ag85 expression is essential for intracellular survival of Mycobacterium tuberculosis within macrophage-like cell line models and is therefore likely to be a virulence factor (16). In addition to promoting excellent immunogenicity (17–20), Ag85 possesses mycolyltransferase activity and catalyzes the synthesis of the most abundant glycolipid of the mycobacterial cell wall, trehalose 6,6-dimycolate (21). Ag85 interacts with Fn (22) at a specific Fn-binding motif (23). The Fn-binding motif of Ag85B of Mycobacterium kansasii has been reported previously to contain 11 residues, 98FEWYYQSGLSV108 (23). This motif has high homology to Ag85A and Ag85C (Fig. 1A) (24, 25, 36). In the context of the homologous Ag85 structures from M. tuberculosis (Fig. 1B), the Fn-binding motif includes a partial β-sheet and α-helix connected by a turn (24, 25). The helical region of the motif, 98FEWYYQ103, was demonstrated to be the most important for Fn binding (23) and also contains residues present on the surface of Ag85. The key negatively charged residue Glu99, the polar residue Gln103, and the hydrophobic residues Phe98, Trp100, Tyr101, and Tyr102 suggested that the corresponding binding site on Fn is likely to be composed of electrostatic and hydrophobic components.
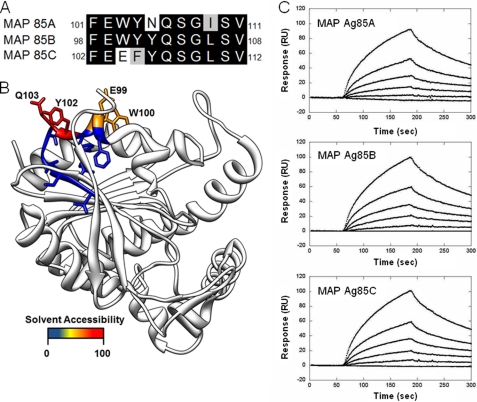
FIGURE 1.
Characterization of MAP Ag85-Fn interaction. A, comparison of the Fn-binding motifs of MAP Ag85, subtypes Ag85A, Ag85B, and Ag85C. B, structure depicts the solvent accessible residues (98–108) on the Fn-interacting region of Ag85B. The homologous structure of M. tuberculosis Ag85B (Protein Data Bank code 1f0n) (25) was used to illustrate the region previously found to interact with Fn as indicated by Naito et al. (23). Residues were colored by the solvent accessible surface area, and the most accessible residues are labeled. The four labeled residues are conserved between Ag85B from M. tuberculosis and MAP. C, SPR analysis of MAP Ag85 interacting with Fn. Fn (50 μg) was immobilized on the surface of a CM5 chip. The concentration of recombinant MAP Ag85 ranged from 0 to 90 nm (from bottom to top). The measured KD values were obtained from the average of duplicate experiments, which were 68.4 ± 4.6, 36.7 ± 5.4, and 33.6 ± 4.2 nm for Ag85A, Ag85B, and Ag85C, respectively.
In this study, the binding of Ag85 to Fn was characterized and localized to the Fn14 fragment of Fn. Furthermore, the Ag85-binding residues of Fn were determined to be SLLVSWQPPR. Finally, the binding of Ag85 to Fn was significantly reduced on Fn siRNA-transfected Caco-2 cells, indicating that Ag85 proteins are important Fn-binding antigens participating in mycobacterial invasion.
MATERIALS AND METHODS
Bacterial Strains and Cell Culture
Mycobacterium avium subsp. paratuberculosis (MAP) strain K-10 was propagated in Middlebrook 7H9 (broth) and 7H10 (agar plate) (BD Biosciences). For medium preparation, 7H9 (4.7 mg) or 7H10 (19 mg) was supplemented with 10% oleic acid/albumin/dextrose/catalase (BD Biosciences), 0.05% Tween 80 (Sigma), 2.5 μg of mycobactin J (Allied Monitor, Fayetteville, MO), and glycerol (0.125 and 0.5%, for 7H9 broth and 7H10 agar plate, respectively) in tissue culture flasks. Caco-2 cells were cultured in Dulbecco's minimum essential medium (DMEM) containing 10% fetal bovine serum (Invitrogen) and were grown at 37 °C in a humidified atmosphere with 5% CO2 (26). Escherichia coli strains were cultured in Luria-Bertani broth (LB) with appropriate antibiotics (Table 1).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristic | Sources |
|---|---|---|
| M. avium subsp. paratuberculosis | ||
| K-10 | Wild type MAP strain | |
| E. coli strains | ||
| DH5α | FΦ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK mK+) supE44 thi−1 gyrA96 relA1λ | Invitrogen |
| BL21 | F−, ompT, hsdSB (rB−, mB−), dcm, gal, λ(DE3) | Promega |
| Plasmids | ||
| pGEX4T-2 | Ampr, GST tag protein expression vector | GE Healthcare |
| pET-16b | Ampr, His tag protein expression vector | Merck |
| pGEX4T-2-Fn12 | pGEX4T2 carrying Fn12 | |
| pGEX4T-2-Fn13 | pGEX4T2 carrying Fn13 | |
| pGEX4T-2-Fn14 | pGEX4T2 carrying Fn14 | |
| pGEX4T-2-Fn15 | pGEX4T2 carrying Fn15 | |
| pET-16b-85A | pET-16b carrying MAP Ag85A | |
| pET-16b-85B | pET-16b carrying MAP Ag85B | |
| pET-16b-85C | pET-16b carrying MAP Ag85C | |
Reagents and Antibodies
Fibronectin (extracted from human plasma), the N-terminal domain of Fn (NTD), the gelatin-binding domain of Fn (GBD), EDTA, sodium chloride, sodium phosphate monobasic, sodium phosphate dibasic, Tris, magnesium chloride, manganese chloride, zinc chloride, copper chloride, and calcium chloride were purchased from Sigma. The Fn cell-binding domain (CBD), the Fn 40-kDa domain, and mouse anti-fibronectin were ordered from Millipore (Billerica, MA). Mouse anti-α-actin antibody, mouse anti-histidine tag, mouse anti-GST antibody, and HRP-conjugated goat anti-mouse antibody were purchased from Invitrogen. HRP-conjugated goat anti-mouse IgG antibody and 3,3′,5,5′-tetramethylbenzidine (TMB) peroxidase substrate were purchased from Kirkegaard & Perry Laboratories (Gaithersburg, MD). Polyclonal M. tuberculosis Ag85 antibody was purchased from Mycobacterial Research Laboratories, Department of Microbiology, Immunology, Pathology, Colorado State University. Synthesized peptides (Table 2) were ordered from Genemed Synthesis (San Antonio, TX).
TABLE 2.
Synthetic peptides used in this study
| Peptide | Sequence |
|---|---|
| P1–16 | AIDAPSNLRFLATTPN |
| P14–26 | TPNSLLVSWQPPR |
| P25–38 | PRARITGYIIKYEK |
| P37–52 | EKPGSPPREVVPRPRP |
| P51–66 | RPGVTEATITGLEPGT |
| P63–78 | EPGTEYTIYVIALKNN |
| P77–90 | NNQKSEPLIGRKKT |
| P17–26 | SLLVSWQPPR |
| P18–26 | LLVSWQPPR |
| P17–25 | SLLVSWQPP |
| PS1a | ALLVSWQPPR |
| PS2a | SALVSWQPPR |
| PS3a | SLAVSWQPPR |
| PS4a | SLLASWQPPR |
| PS5a | SLLVAWQPPR |
| PS6a | SLLVSAQPPR |
| PS7a | SLLVSWAPPR |
| PS8a | SLLVSWQAPR |
| PS9a | SLLVSWQPAR |
| PS10a | SLLVSWQPPA |
a Substituted residues are underlined.
Plasmid Construction and Protein Purification
His-tagged MAP85A, Ag85B, and Ag85C were constructed by using the pET-16b vector (Merck). GST-tagged Fn12, Fn13, Fn14, and Fn15 were generated using the pGEX-4T-2 vector (GE Healthcare). The oligonucleotides used for PCR were shown in Table 3. The recombinant protein plasmids were transformed into E. coli DH5α competent cells and then were streaked on a Luria-Bertani (LB) agar plate containing 100 μg/ml ampicillin. Colonies were selected from the agar plate and grown in 5 ml of LB culture containing 100 μg/ml ampicillin at 37 °C overnight. The correct construct was subsequently transformed into E. coli BL21 for protein expression. The 10-ml overnight culture of a single transformant was used to inoculate 1 liter of fresh LB medium containing 100 μg/ml ampicillin. The cells were grown to A600 = 0.6 and induced with 1 mm isopropyl β-thiogalactopyranoside. After a 16- h post-induction growth at 20 °C, the cells were harvested by centrifugation at 6000 rpm for 15 min. The protein purification was conducted at 4 °C. The cell paste obtained from 1 liter of cell culture was suspended in 50 ml of phosphate-buffered saline (PBS). Cells were disrupted with a French press instrument (AIM-AMINCO Spectronic Instruments) applied at 12,000 p.s.i. The lysis solution was centrifuged (10,000 rpm for 30 min), and the pellet was discarded. The cell-free extract was loaded onto a 10-ml nickel-nitrilotriacetic acid or GST column that was equilibrated with PBS. The protein purification procedures were the same as described previously (27, 28).
TABLE 3.
Oligonucleotides used in this study (restriction enzyme sites were underlined)
| Primer | Sequence (5′ → 3′) |
|---|---|
| MAP Ag85A NdeI-fp | GGAATTCCATATGCGCCCCGGTCtgccg |
| MAP Ag85A BamHI-rp | CGGGATCCTTAGGTGCCCTGGCCG |
| MAP Ag85B NdeI-fp | GGAATTCCATATGCGTCCGGGCCTGCCC |
| MAP Ag85B XhoI-rp | CCGCTCGAGTtATCCGCCGCCGCC |
| MAP Ag85C NdeI-fp | GGAATTCCATATGAAGCCGGGGCTTCCGG |
| MAP Ag85C BamHI-rp | CGGGATCCTtAGGTGGCGGGCTG |
| Fn12 EcoRI-fp | CGGAATTCTTATTCTTGAGCCAACCAATCTG |
| Fn12 XhoI-rp | CCGCTCGAGTTACTCCAGAGTCGTGAC |
| Fn13 EcoRI-fp | CGGAATTCTTAATGTCAGCCCTCCAAGAAG |
| Fn13 XhoI-rp | CCGCTCGAGTTACGTGGAGGCATC |
| Fn14 EcoRI-fp | CGGAATTCTTGCCATTGATGCCCCATCC |
| Fn14 XhoI-rp | CCGCTCGAGTTATGTCTTTTTCCTCCC |
| Fn15 EcoRI-fp | CGGAATTCCCGGGGCTCAATCC |
| Fn15 XhoI-rp | CCGCTCGAGTTACTTGTGCCTCCTC |
Detection of Ag85 Proteins from MAP K-10 Culture
Culture supernatant of MAP K-10 (4 weeks) was subjected to ammonium sulfate precipitation (80% saturation) and then dialyzed by PBS to remove the salt (29). Extracted Ag85 proteins were confirmed by Western blotting using Ag85 antibody.
Protein Binding Assays by ELISA
To measure the binding affinity of MAP Ag85 (Ag85A, Ag85B, and Ag85C) to Fn, human plasma Fn (0.5 μg) was coated on microtiter plate wells using 0.1 m NaHCO3 (pH 9.3) coating buffer at 4 °C for overnight and blocked by PBS plus 3% BSA at 37 °C for 1 h. Subsequently, various concentrations (0.1, 0.2, 0.4, 0.8, and 1.6 μm) of His-tagged MAP Ag85 were added to each well and incubated at 37 °C for 1 h. After three washes with PBST (PBS plus 0.05% Tween 20), bound Ag85 was detected by mouse anti-histidine (1:1000) and HRP-conjugated goat anti-mouse IgG (1:1000), serving as primary and secondary antibodies, respectively (27, 28). To investigate the MAP Ag85B-binding domains of Fn, 0.5 μg of full-length Fn, four Fn proteolytic domains (including NTD, GBD, CBD, and 40-kDa domain) or BSA (negative control) was coated on microtiter plate wells. His-tagged MAP Ag85B (0.1 μm) was added to each well and detected as described previously. To map the MAP Ag85B-binding fragments of the 40-kDa domain, MAP Ag85B (0.5 μg) was coated on microtiter plate wells using the same strategy described above. GST-tagged Fn12, Fn13, Fn14, or Fn15 (0.5 μg) was then added to each well and incubated at 37 °C for 1 h. Mouse anti-GST (1:1000) and HRP-conjugated goat anti-mouse IgG (1:1000) were used as primary and secondary antibodies, respectively. After washing the plates three times with PBST, 100 μl of TMB substrate (Kirkegaard & Perry Laboratories) was added to each well, allowed to react for 5 min, and quenched with the addition of 100 μl of 0.5% hydrofluoric acid. Microtiter plates were read at 450 nm using an ELISA plate reader (Bioteck EL-312). Each value represents the mean ± S.E. of three trials in triplicate samples.
Surface Plasmon Resonance (SPR)
The interactions of Fn to MAP Ag85 and Fn14 or P17–26 to MAP Ag85B were analyzed by an SPR technique using a Biacore 2000 instrument (GE Healthcare). To determine the Fn binding activity of Ag85 complex proteins, Fn (50 μg) was immobilized on a CM5 chip (GE Healthcare). Then 10 μl of MAP Ag85A, Ag85B, or Ag85C (in the concentration of 0, 2.8, 11.2, 22.5, 22.5, 45, and 90 nm) was injected into the flow cell at 10 μl/min at 25 °C. To measure the binding affinities of Fn14 and P17–26 to Ag85B, MAP Ag85B was immobilized on a CM5 chip. A control flow cell was injected with PBS buffer without Fn14 or P17–26. The 10 μl of Fn14 (in the concentration of 0, 62.5, 250, 500, 1000, and 2000 nm) or P17–26 (0, 0.625, 2.5, 5, 10, and 20 μm) was injected into the flow cell at 10 μl/min at 25 °C. The chip surface was regenerated by removal of analyte with a regeneration buffer (10 mm glycine-HCl at pH 2.0). All sensorgram data were subtracted from the negative control flow cell. To obtain the kinetic parameters of the interaction, the data of the sensograms were fitted by BIAevaluation software version 3.0 using the one-step biomolecular association reaction model (1:1 Langmuir model), which resulted in optimum mathematical fits with the lowest χ values.
GST Pulldown Assay
The GST pulldown assay was performed as described previously (30). Purified GST-fused Fn fragments (Fn12–15) or GST (negative control) was loaded onto 0.5 ml of glutathione-Sepharose beads (Amersham Biosciences) at 4 °C overnight. The beads were then washed three times with PBS. A 20-μg aliquot of His-tagged MAP Ag85B was incubated with GST-fused proteins immobilized on glutathione-Sepharose beads at 4 °C for 3 h. After incubation, the beads were separated by centrifugation, washed three times with PBS, and boiled in Laemmli sample loading buffer consisting of 50 mm Tris-HCl (pH 6.8), 100 mm dithiothreitol, 2% SDS, 0.25 mm PMSF, and 0.1% bromphenol blue in 20% glycerol. The eluted proteins were subjected to 12% SDS-PAGE and electroblotted onto polyvinylidene difluoride membranes. The membranes were incubated in 5% skim milk in PBST overnight and then incubated with mouse anti-histidine antibody (1:1000). The immunocomplexes were detected by HRP-conjugated goat anti-mouse IgG antibody (1:5000).
Peptide Inhibition Assay
Fn (0.5 μg) was coated on microtiter plate wells and blocked as described previously. His-tagged MAP Ag85B (0.1 μm) was preincubated with a 50 μm concentration of synthetic peptides in 100 μl of PBS buffer at 4 °C for 1 h. The MAP Ag85B/peptide mixture was then added directly to Fn-coated microtiter wells. Binding was detected by ELISA as described above.
Small Interfering RNA (siRNA) Inhibition of MAP Ag85B Binding
Synthetic siRNA duplexes directed against human Fn (AM11012) and negative siRNA duplex (AM4611) were purchased from Applied Biosystems (Carlsbad, CA). RNA duplexes were introduced into Caco-2 cells by the method of lipofection (31, 32), and 8 × 105 cells were transfected with 0.4 μg of negative siRNA or Fn-siRNA. Adhesion assays were performed 72 h after lipofection. The knockdown efficiency of endogenous Fn expression was determined as described previously (33) with slight modification. The protein content of Caco-2 cells (106) was analyzed using Western immunoblotting. The α-actin derived from Caco-2 cells was measured as a loading control using a mouse anti-actin antibody (1:5000). The band intensity was measured by densitometry using the ImageJ software (National Institutes of Health, Bethesda) (34). The MAP Ag85B binding assay was performed 72 h after lipofection. To determine the binding of MAP Ag85B to Fn, 50 μm MAP Ag85B (100 μl) was added to Caco-2 cells (106) transfected with Fn or negative siRNA. To determine the binding of MAP Ag85B and the expression of Fn on Caco-2 cells, mouse anti-His (1:250) and mouse anti-Fn (1:250) antibodies served as the primary antibodies. All of the experiments were performed in triplicate.
Bacterial and Protein Attachment Assays
To investigate the adhesion of MAP strain K-10 or recombinant MAP Ag85B on Caco-2 cells, the attachment assay was slightly modified from the method reported previously (33). Basically, Caco-2 cells (106) were cultured on microtiter plate wells at 37 °C overnight. Ice-cold PBS buffer was used to wash the cells prior to the assays to prevent MAP K-10 invasion. For bacterial attachment assay, a total of 1.4 × 109 MAP K-10 was suspended in ice-cold medium and then incubated with coated Caco-2 cells at 4 °C for 1 h. The wells incubated without Caco-2 cells served as negative controls. Unattached bacteria were removed by five washes with PBS. PBS containing 1% (v/v) Triton X-100 was applied to resuspend adherent bacteria, and the 104-fold dilutions of adherent bacterial suspension were spread on 7H10 agar plates to determine the number of cell-associated bacteria per well. For the protein attachment assay, filtered (0.45 μm) His-tagged MAP Ag85B (0.1 μm) was added to the wells coated with Caco-2 cells (106) and incubated at 37 °C for 1 h. Wells were washed three times with PBS. Bound MAP Ag85B was detected as mentioned above.
Protein Depiction
Images of protein structures were generated using UCSF Chimera (35). The surface-accessible surface areas for residues were also computed using UCSF Chimera.
RESULTS
Binding Affinity of MAP Ag85 to Fn
Because of the slight variation among the Fn-binding motifs of Ag85 (Fig. 1, A and B), differences in the KD values were measured as 68.4 ± 4.6 nm (MAP Ag85A), 36.7 ± 5.4 nm (MAP Ag85B), and 33.6 ± 4.2 nm (MAP Ag85C), respectively (Fig. 1C). The kon, koff, and KD values are summarized in Table 4. Unlike the interaction of some bacterial adhesins and ECM components (32, 34, 37), divalent metal ions (Ca2+, Cu2+, Co2+, Fe2+, Mg2+, Mn2+, Ni2+, and Zn2+) were not required for the Ag85-Fn interaction (data not shown).
TABLE 4.
The measured kon, koff, and KD values of M. avium subsp. paratuberculosis Ag85 bound to Fn
Values represent the mean ± S.E.
| Fn-binding motifa | kon | koff | KD | |
|---|---|---|---|---|
| m−1s−1 | s−1 | nm | ||
| MAP Ag85A | FEWYNQSGISV | 9.2 × 104 ± 1.1 | 6.3 × 10−3 ± 0.8 | 68.4 ± 4.6 |
| MAP Ag85B | FEWYYQSGLSV | 1.2 × 105 ± 0.2 | 4.4 × 10−3 ± 0.7 | 36.7 ± 5.4 |
| MAP Ag85C | FEEFYQSGLSV | 1.4 × 104 ± 0.3 | 4.7 × 10−3 ± 0.7 | 33.6 ± 4.2 |
a Data were reported by Naito et al. (23).
MAP Ag85B Binds to 40-kDa Domain of Fn
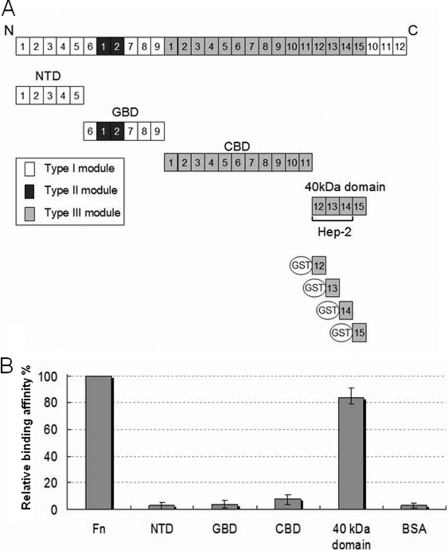
To identify the MAP Ag85B-binding fragments on Fn, NTD, GBD, CBD, and the 40-kDa domains of Fn (Fig. 2A) were examined by using ELISA. As shown on Fig. 2B, the 40-kDa domain was the only domain to strongly interact with MAP Ag85B. No significant interaction between MAP Ag85B and the NTD, GBD, or CBD was observed.
FIGURE 2.
MAP Ag85B interacted with the 40-kDa domain of Fn. A, schematic representation of Fn and truncated Fn constructs used in this study. B, relative binding affinities of His-tagged MAP Ag85B (0.1 μm) to immobilized Fn fragments (containing the NTD, the GBD, the CBD, and the 40-kDa domain) or BSA (0.1 μg). Bound MAP Ag85B was detected by ELISA.
Fn14 Possesses MAP Ag85B-binding Activity
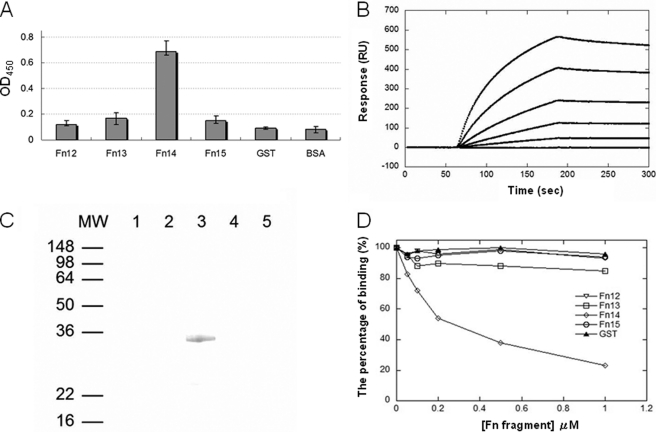
The 40-kDa domain of Fn was subsequently divided into four fragments (Fn12, Fn13, Fn14, and Fn15), and each was fused with GST tags (Fig. 3A). As presented in Fig. 3A, Fn14 significantly bound to immobilized MAP Ag85B, whereas Fn12, Fn13, or Fn15 could not bind. The binding potency (KD) of Fn14 to MAP Ag85B measured by SPR was 48.1 ± 4.7 nm (Fig. 3B). GST pulldown assay further demonstrated that Fn14 was the solitary fragment binding to MAP Ag85B (Fig. 3C). Moreover, Fn14-pretreated MAP Ag85B apparently diminished its affinity to interact with Fn, indicating that MAP Ag85B bound to Fn by targeting to Fn14 (Fig. 3D).
FIGURE 3.
Fn14 bound to MAP Ag85B. A, binding of fragments derived from the 40-kDa domain (including Fn12, Fn13, Fn14, and Fn15) to immobilized MAP Ag85B. GST-tagged Fn12, Fn13, Fn14, or Fn15 (0.1 μm) was added to microtiter plate wells coated with 0.5 μg of MAP Ag85B. Bound proteins were detected by ELISA. B, KD determination of Fn14 to MAP Ag85B by SPR analysis. The measured kon, koff, and KD values were 7.7 × 103 ± 1.8 m−1 s−1, 3.7 × 10−4 ± 0.6 s−1, and 48.1 ± 4.7 nm, respectively. Values represent the mean ± S.E. C, His-tagged MAP Ag85B was applied to the GST beads preimmobilized by recombinant Fn12 (lane 1), Fn13 (lane 2), Fn14 (lane 3), Fn15 (lane 4), or GST (lane 5; negative control). The pulldown complex was analyzed by immunoblot analysis using anti-His antibody. D, Fn14 interfered with the binding of MAP Ag85B to Fn. MAP Ag85B was preincubated with various concentrations (0.05, 0.1, 0.2, 0.5, and 1 μm) of Fn12, Fn13, Fn14, Fn15, or GST (negative control) prior to interaction with immobilized Fn. Bound proteins were detected by ELISA.
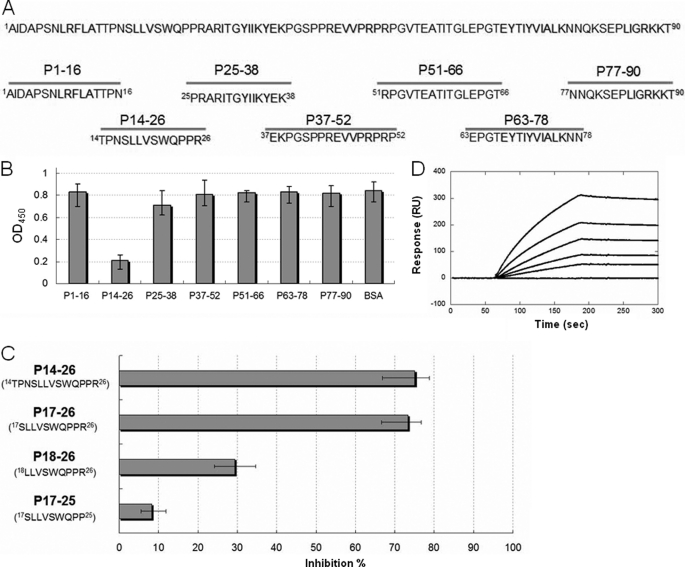
Mapping the Ag85-binding Motif on Fn14
Seven peptides synthesized based on the amino acid sequence of Fn14 were subjected to the peptide inhibition assay to identify the Ag85-binding motif (Fig. 4A). As shown on Fig. 4B, Fn14 residues 14–26, termed P14–26 (14TPNSLLVSWQPPR26), harbored the highest inhibition potency among those peptides. Fn14 residues 17–26, termed P17–26 (17SLLVSWQPPR26), were derived from P14–26 with a three-amino acid truncation at the N terminus to discard the non-β-sheet structure part (38) and had the same inhibition ability as P14–26 (Fig. 4C). Peptides with an additional one-amino acid deletion from either the N terminus (18LLVSWQPPR26) or the C terminus (17SLLVSWQPP25) lost most of the ability to interfere with MAP Ag85B-Fn binding (Fig. 4C). The measured KD value of P17–26 with MAP Ag85B was 414.2 ± 13.6 nm (Fig. 4D). As a result, the motif on Fn14 required for high affinity MAP Ag85B binding was SLLVSWQPPR.
FIGURE 4.
Peptide inhibition assay using synthetic peptides. A, schematic diagram showing the seven peptides (lower panel) synthesized based on the sequence of Fn14 (upper panel). B, peptide inhibition assay measured by ELISA. Peptide 14–26 (P14–26) possessed the highest inhibition potency among all peptides. BSA served as a negative control. C, optimal peptide length determination. Peptide deletion of P14–26 with three residues at the N terminus (P17–26) harbored near-full inhibition activity. Peptide derivatives of P17–26 with one residue deleted at the N terminus (P18–26) and the C terminus (P17–25) showed the reduced inhibition abilities of 61 and 89%, respectively (compared with P14–26). D, KD determination of P17–28 to MAP Ag85B by SPR analysis. The measured kon, koff, and KD values were 33.8 ± 2.6 m−1 s−1, 1.4 ± 0.5 × 10−5 s−1, and 414.2 ± 13.6 nm, respectively. Values represent the mean ± S.E.
Single Residue Substitution of P17–26
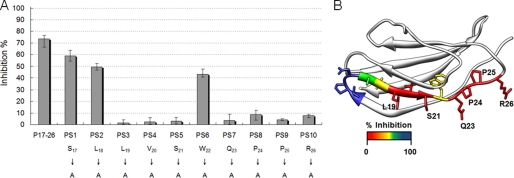
Ten peptides (PS1–10) (Table 2) were derived from P17–26 by single alanine scanning substitution and were applied to determine the key binding residues. Analogous peptides were subjected to the peptide inhibition assay (Fig. 5A). Compared with P17–26, PS3 (L19A), PS4 (V20A), PS5 (S21A), PS7 (Q23A), PS8 (P24A), PS9 (P25A), and PS10 (R26A) showed dramatically reduced inhibition potencies (>50% reduction). However, PS1 (S17A), PS2 (L18A), and PS8 (W22A) showed only minor inhibition of Ag85-Fn binding (20–25% reduction). The data are mapped onto the structure of Fn14 (Fig. 5B) (38). The 10 amino acids corresponding to P17–26 are a part of the second β-sheet and extend into a less structured loop containing adjacent proline residues.
FIGURE 5.
Identification of the key residues of Fn14 involved in MAP Ag85B binding. A, 10 synthetic peptides were derived from P14–26 by single alanine scanning substitution. His-tagged MAP Ag85B (0.1 μm) was preincubated with a 50 μm concentration of synthetic peptides and then subjected to microtiter plate wells coated with Fn (0.5 μg). B, percent inhibition was mapped onto the structure of Fn14 (Protein Data Bank code 1fnh) (38) with red colored residues showing the largest effect of alanine substitution.
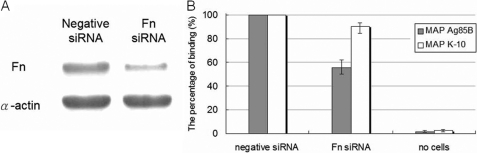
Transfection of Fn siRNA Inhibits MAP Ag85B Binding on Mammalian Cells
To further elucidate the role of Fn as a cell surface receptor for the putative binding partner, Ag85, the binding of MAP Ag85B to Caco-2 cells was examined. Fn is normally expressed on the surface of Caco-2 cells (32). Fn expression levels can be reduced by 62.7% with transfected Fn siRNA duplex as compared with cells transfected with negative siRNA duplex (Fig. 6A). The binding of recombinant MAP Ag85B was 44.6% less for the Fn siRNA-transfected Caco-2 cells with greatly reduced Fn expression (Fig. 6B, solid bar). However, only a 9.7% decreased binding in adhesion of MAP K-10, the wild-type MAP strain expressing Ag85 proteins (supplemental Fig. 1), to Fn siRNA-transfected Caco-2 cells was detected (Fig. 6B, open bars).
FIGURE 6.
Binding of MAP Ag85B and MAP K-10 to Fn siRNA-transfected Caco-2 cells. A, detection of the expression of Fn and α-actin in Caco-2 cells after 72 h of transfection with Fn or negative siRNA. Fn and α-actin were detected by Western blotting using Fn and α-actin antibodies. The relative endogenous Fn expression level of Fn siRNA-transfected Caco-2 cells was 37.3% (estimated by ImageJ). B, binding of MAP Ag85B was dramatically reduced in siRNA-transfected Caco-2 cells (44.6% reduced). His-tagged MAP Ag85B (0.1 μm) was added to the wells coated with Fn or negative siRNA-transfected Caco-2 cells (106) and incubated at 37 °C for 1 h. Bound MAP Ag85B was detected by anti-His antibody (solid bar). A total of 1.4 × 109 MAP K-10 cells was incubated with immobilized Caco-2 cells (106) transfected with negative siRNA or Fn siRNA on microtiter plate wells. The wells incubated without Caco-2 cells served as the negative control. Binding of MAP K-10 to Fn siRNA-transfected Caco-2 cells was slightly abolished (9.7% reduced) (open bar). Each datum is the mean ± S.D. of quadruplicate wells.
DISCUSSION
Adhesion is a pivotal first step that allows pathogenic bacteria, including mycobacteria, to infect host cells. MSCRAMMs, a group of virulence factors located in the outer surface of bacteria, mediate adhesion of a wide variety of pathogenic bacteria, including Staphylococcus, Streptococcus, Enterococcus, Borrelia, Leptospira, and others to ECM components such as Fn, fibrinogen, collagen, elastin, laminin, and peptidoglycan (6, 9, 27, 31, 39–41). Ag85 proteins, including Ag85A, Ag85B, and Ag85C, have been reported to interact with Fn (15, 42). In addition to Ag85, a number of mycobacteria proteins were also identified to bind to Fn such as MPT51 (also called Ag85D or FbpD) (43) and ModD (44, 45). Binding to Fn, likely a significant factor in the virulence of mycobacteria, represents a potential first step in the attachment and entry of mycobacteria into host cells.
The Fn-binding motif, a stretch of 11 residues (FEWYYQSGLSV), of Ag85B from M. kansasii has been reported previously (23). In the three-dimensional structure of Ag85, the residues are located on the surface opposite the substrate and cell wall interacting region providing access for Fn binding while attached to mycobacteria (46). Binding differences between Fn-binding motifs from MAP Ag85A, Ag85B, and Ag85C could be explained by amino acid sequence variations. A hydrophobic residue (Tyr) of MAP Ag85B and Ag85C is a hydrophilic one (Asn) in the corresponding position of MAP Ag85A, resulting in a reduced affinity to Fn (∼2-fold decrease). A similar Fn binding affinity was observed for MAP Ag85B and Ag85C, even though Glu and Phe are substituted for Trp and Tyr, respectively, in Ag85C. The Ag85 structure (24, 25) as well as a previous alanine substitution study (23) suggest that the FEWYYQ helix is the most likely region for direct Fn interaction (supplemental Fig. 2A). In this sequence, the 2nd position Glu, the 3rd position Trp, the 5th position Tyr, and the 6th position Gln are surface-accessible (Fig. 1B). Although the 2nd position Glu and the 6th position Gln are conserved among MAP Ag85 proteins, differences for 3rd and 5th position hydrophobic residues suggest that the 3rd position Trp is less likely to be involved in direct Fn binding than the 5th position Tyr. The residues involved provide a basis for both hydrophobic and charged interactions.
Here, we demonstrated that MAP Ag85B binds to the 40-kDa domain of Fn (Fig. 2B), and we further showed that Fn14 was the only fragment to independently interact with MAP Ag85B (KD = 48.1 ± 4.7 nm), with the similar binding affinity to that of Fn (KD = 36.7 ± 5.4 nm) (Fig. 3, A–D). Previous reports locating the Ag85-binding sites on Fn have differed as the GBD was suggested by Peake et al. (47) and the CBD and the Hep-2 were suggested by Naito et al. (48). The discrepancy might be because the recombinant Mycobacterium bovis Ag85B was prepared under denaturing conditions for the studies conducted by Peake et al. (47), which might have caused the protein to lose its native folded structure. In the study by Naito et al. (48), the Ag85-binding fragments of Fn were identified by a proteolytic strategy (thermolysin digestion); however, following the proteolytic digestion, the real Ag85-binding fragment of Fn (SLLVSWQPPR) was possibly missed because thermolysin preferably cleaves substrates with bulky hydrophobic residues (Leu, Ile, Val, and Phe) at the P1′ position (49). Moreover, the Ag85-binding site was mapped to Hep-2, a huge 30-kDa domain composed by Fn12, Fn13, and Fn14, using recombinant protein that locates a region but does not establish the specific binding site. Meanwhile, we demonstrated that the addition of heparin (100 μm) did not interfere with the binding between the 40-kDa domain (Fn12–15) and MAP Ag85B (data not shown), indicating that the Ag85-binding site on Fn is distinct from the heparin-binding sequence on Fn13. In this study, we divided the 40-kDa domain of Fn into four individual fragments (Fn12, Fn13, Fn14, and Fn15) and then tested each to further establish the specifics of binding. Fn14 was proven to be the MAP Ag85B-interacting fragment.
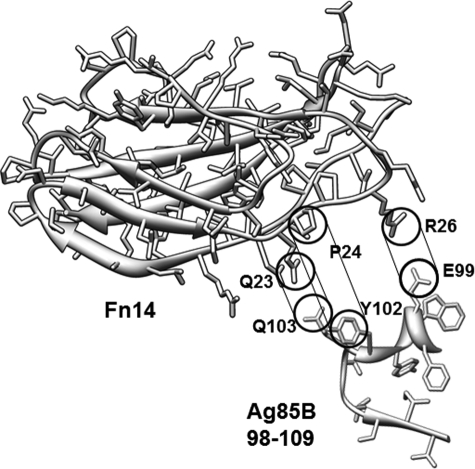
The Ag85-binding motif (17SLLVSWQPPR26) of Fn14 that we have identified is a typical β-strand in the type III Fn core structure (38). The hydrophobic residues (Leu19, Val20, Pro24, and Pro25), polar residues (Ser21 and Gln23), and positively charged residue (Arg26) in the Fn motif were essential for MAP Ag85B binding (Fig. 5A). Mapped onto the Fn14 structure, the Ag85-binding motif residues extend along the length of the domain (Fig. 5B). The most exposed region for a direct interaction with Ag85 is the stretch from Gln23 to Arg26 in Fn14. Positioning the structure of Fn14 near its binding site on Ag85, potential Fn14-Ag85 interactions exist to explain the experimental results (Fig. 7). Matching the negatively and positively charged residues on Ag85 and Fn14, respectively, a hydrophobic and hydrogen-bonding interaction would line up and match the Fn-interacting residues on Ag85. Because all Ag85 crystal structures are very similar overlaying structures for the Fn-binding region, the Fn-interacting helix should maintain its conformation upon interacting with Fn14 (supplemental Fig. 2, B and C). Although the potential Ag85-binding site is near the link between Fn14 and Fn13, the position of Fn13 in the Fn12–Fn14 crystal structure would not sterically inhibit Ag85 binding to Fn14. This docking site fails to explain the effect of mutations at positions Leu19 through Ser21, but because these residues are sandwiched between two other β-sheets, they are less exposed for Ag85 interactions. Also, the N-terminal end of the Ag85-binding region (17SLLVSW22) is more hydrophobic and therefore more likely to be involved in nonspecific binding interactions. In addition, two similar sequences were found in Fn2 (SFVVSWVSAS) and Fn10 (SLLISWDAPA) located within the CBD (data not shown), which might explain that CBD contributed a weak (∼7.8% relative binding affinity) but not major affinity to MAP Ag85B binding, as well as the results from Naito et al. (23) (Fig. 2B). In agreement with a previous report (23), our positioning of the protein-protein interacting surface suggests that both a charge-charge (Glu-Arg) and hydrophobic interactions are required for Ag85-Fn interaction. Interestingly, the determined KD value of P17–26 (414.2 ± 13.6 nm) was significantly larger than that of Fn (36.7 ± 4.2 nm) or Fn14 (48.1 ± 4.7 nm). The three-dimensional structure and folding could be important for Fn binding to Ag85, because P17–26 is likely to be a structurally flexible peptide.
FIGURE 7.
Potential interacting sites between Fn14 and Ag85B. The Fn14-binding residues from Ag85B are shown along Fn14 and suggest a possible orientation of the interacting surfaces. Both electrostatic and hydrophobic interactions (indicated) are thought to drive the protein association.
Binding to components of the ECM, including Fn, facilitates the adhesion of pathogens to host cells (50). Fn also serves as a mediator to induce endocytosis and initiate the entry of bacteria upon binding to bacterial surface proteins (51, 52). We tested whether reducing the expression of Fn on Caco-2 cells would impair the ability of Ag85 or Ag85-expressing MAP K-10 to bind to the Caco-2 cells. The reduced binding (44.6%) of MAP Ag85B to Fn siRNA-treated Caco-2 cells suggests an important adhesive role for the Ag85 protein family. In Fn siRNA-transfected Caco-2 cells, Fn expression remains at 37.3% of endogenous levels allowing MAP Ag85B to still bind but at a reduced level. The corresponding loss of Fn expression and MAP Ag85B binding suggests that Fn is the main cell surface receptor for MAP Ag85B. However, only a 9.7% reduction was observed in MAP K-10 binding to Fn siRNA-transfected Caco-2 cells. These results indicate that the Ag85-Fn interaction accounts for only a portion of the ability of a MAP cell to adhere to a host cell. Other surface antigens such as FbpD, ModD, or unidentified proteins of MAP K-10 probably participate in additional ECM interactions (44, 45, 53–55).
In conclusion, we have characterized the binding affinities of Ag85 to Fn and demonstrated that MAP Ag85B interacts with Fn by targeting to a novel motif (SLLVSWQPPR). Our finding also clarified that the interaction mechanism between Ag85 proteins and Fn relies on residual charge and hydrophobic interactions. Because Ag85 proteins are important colonization factors potentially contributing to mycobacterial virulence and play not only a role in Fn-mediated host attachment but possess the essential acyltransferase activity contributing to the biosynthesis of mycobacterial cell wall, the Ag85 protein family is an important target for further antimycobacterial studies.
Supplementary Material
This work was supported by Agriculture and Food Research Initiative, United States Department of Agriculture Cooperative State Research, Education, and Extension Service Grant 2008-55620-18710 (Johne's Disease Integrated Program), the National Research Initiative, United States Department of Agriculture Cooperative State Research, Education, and Extension Service Grant 2008-35204-04626, the Agriculture and Food Research Initiative, United States Department of Agriculture Cooperative State Research, Education, and Extension Service Grant 2009-65119-05993, the Biotechnology Research and Development Corporation (BRDC), and the Animal Formula Fund Grant NY-478455 and NY-478437.

This article contains supplemental Figs. 1 and 2.
- MAP
- Mycobacterium avium subsp. paratuberculosis
- Hep-2
- heparin-binding domain II
- ECM
- extracellular matrix
- NTD
- N-terminal domain
- GBD
- gelatin-binding domain
- CBD
- cell binding domain
- SPR
- surface plasmon resonance
- Fn
- fibronectin
- MSCRAMM
- microbial surface components recognizing adhesive matrix molecule.
REFERENCES
- 1. Johne H. A., Frothingham L. (1895) Ein Eigentheumlicher Fall von Tuberculose beim Rind. Dtsch. Ztschr. Tier-med. 21, 438–454 [Google Scholar]
- 2. Cocito C., Gilot P., Coene M., de Kesel M., Poupart P., Vannuffel P. (1994) Paratuberculosis. Clin. Microbiol. Rev. 7, 328–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. National Animal Health Monitoring System (1997) Johne's Disease of United States Dairy Operations, #N245.1097, United States Department of Agriculture-APHIS-VC, CEAH, Fort Collins, CO [Google Scholar]
- 4. Hermon-Taylor J. (2009) Mycobacterium avium subspecies paratuberculosis, Crohn's disease, and the Doomsday scenario. Gut Pathog. 1, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Soto G. E., Hultgren S. J. (1999) Bacterial adhesins. Common themes and variations in architecture and assembly. J. Bacteriol. 181, 1059–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patti J. M., Allen B. L., McGavin M. J., Höök M. (1994) MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48, 585–617 [DOI] [PubMed] [Google Scholar]
- 7. Potts J. R., Campbell I. D. (1994) Fibronectin structure and assembly. Curr. Opin. Cell Biol. 6, 648–655 [DOI] [PubMed] [Google Scholar]
- 8. Vakonakis I., Campbell I. D. (2007) Extracellular matrix. From atomic resolution to ultrastructure. Curr. Opin. Cell Biol. 19, 578–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schwarz-Linek U., Höök M., Potts J. R. (2004) The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol. Microbiol. 52, 631–641 [DOI] [PubMed] [Google Scholar]
- 10. Joh D., Speziale P., Gurusiddappa S., Manor J., Höök M. (1998) Multiple specificities of the staphylococcal and streptococcal fibronectin-binding microbial surface components recognizing adhesive matrix molecules. Eur. J. Biochem. 258, 897–905 [DOI] [PubMed] [Google Scholar]
- 11. Joh D., Wann E. R., Kreikemeyer B., Speziale P., Höök M. (1999) Role of fibronectin-binding MSCRAMMs in bacterial adherence and entry into mammalian cells. Matrix Biol. 18, 211–223 [DOI] [PubMed] [Google Scholar]
- 12. Probert W. S., Kim J. H., Höök M., Johnson B. J. (2001) Mapping the ligand-binding region of Borrelia burgdorferi fibronectin-binding protein BBK32. Infect. Immun. 69, 4129–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dabo S. M., Confer A. W., Anderson B. E., Gupta S. (2006) Bartonella henselae Pap31, an extracellular matrix adhesin, binds the fibronectin repeat III13 module. Infect. Immun. 74, 2513–2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kingsley R. A., Keestra A. M., de Zoete M. R., Bäumler A. J. (2004) The ShdA adhesin binds to the cationic cradle of the fibronectin 13FnIII repeat module. Evidence for molecular mimicry of heparin binding. Mol. Microbiol. 52, 345–355 [DOI] [PubMed] [Google Scholar]
- 15. Wiker H. G., Harboe M. (1992) The antigen 85 complex. A major secretion product of Mycobacterium tuberculosis. Microbiol. Rev. 56, 648–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Armitige L. Y., Jagannath C., Wanger A. R., Norris S. J. (2000) Disruption of the genes encoding antigen 85A and antigen 85B of Mycobacterium tuberculosis H37Rv. Effect on growth in culture and in macrophages. Infect. Immun. 68, 767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baldwin S. L., D'Souza C. D., Orme I. M., Liu M. A., Huygen K., Denis O., Tang A., Zhu L., Montgomery D., Ulmer J. B. (1999) Immunogenicity and protective efficacy of DNA vaccines encoding secreted and non-secreted forms of Mycobacterium tuberculosis Ag85A. Tuber. Lung Dis. 79, 251–259 [DOI] [PubMed] [Google Scholar]
- 18. Huygen K., Content J., Denis O., Montgomery D. L., Yawman A. M., Deck R. R., DeWitt C. M., Orme I. M., Baldwin S., D'Souza C., Drowart A., Lozes E., Vandenbussche P., Van Vooren J. P., Liu M. A., Ulmer J. B. (1996) Immunogenicity and protective efficacy of a tuberculosis DNA vaccine. Nat. Med. 2, 893–898 [DOI] [PubMed] [Google Scholar]
- 19. Langermans J. A., Doherty T. M., Vervenne R. A., van der Laan T., Lyashchenko K., Greenwald R., Agger E. M., Aagaard C., Weiler H., van Soolingen D., Dalemans W., Thomas A. W., Andersen P. (2005) Protection of macaques against Mycobacterium tuberculosis infection by a subunit vaccine based on a fusion protein of antigen 85B and ESAT-6. Vaccine 23, 2740–2750 [DOI] [PubMed] [Google Scholar]
- 20. Kamath A. T., Feng C. G., Macdonald M., Briscoe H., Britton W. J. (1999) Differential protective efficacy of DNA vaccines expressing secreted proteins of Mycobacterium tuberculosis. Infect. Immun. 67, 1702–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Belisle J. T., Vissa V. D., Sievert T., Takayama K., Brennan P. J., Besra G. S. (1997) Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276, 1420–1422 [DOI] [PubMed] [Google Scholar]
- 22. Denis O., Lozes E., Huygen K. (1997) Induction of cytotoxic T-cell responses against culture filtrate antigens in Mycobacterium bovis bacillus Calmette-Guérin-infected mice. Infect. Immun. 65, 676–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Naito M., Ohara N., Matsumoto S., Yamada T. (1998) The novel fibronectin-binding motif and key residues of mycobacteria. J. Biol. Chem. 273, 2905–2909 [DOI] [PubMed] [Google Scholar]
- 24. Ronning D. R., Klabunde T., Besra G. S., Vissa V. D., Belisle J. T., Sacchettini J. C. (2000) Crystal structure of the secreted form of antigen 85C reveals potential targets for mycobacterial drugs and vaccines. Nat. Struct. Biol. 7, 141–146 [DOI] [PubMed] [Google Scholar]
- 25. Anderson D. H., Harth G., Horwitz M. A., Eisenberg D. (2001) An interfacial mechanism and a class of inhibitors inferred from two crystal structures of the Mycobacterium tuberculosis 30-kDa major secretory protein (Antigen 85B), a mycolyl transferase. J. Mol. Biol. 307, 671–681 [DOI] [PubMed] [Google Scholar]
- 26. Janvilisri T., Scaria J., Chang Y. F. (2010) Transcriptional profiling of Clostridium difficile and Caco-2 cells during infection. J. Infect. Dis. 202, 282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin Y. P., Greenwood A., Nicholson L. K., Sharma Y., McDonough S. P., Chang Y. F. (2009) Fibronectin binds to and induces conformational change in a disordered region of leptospiral immunoglobulin-like protein B. J. Biol. Chem. 284, 23547–23557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin Y. P., Lee D. W., McDonough S. P., Nicholson L. K., Sharma Y., Chang Y. F. (2009) Repeated domains of leptospira immunoglobulin-like proteins interact with elastin and tropoelastin. J. Biol. Chem. 284, 19380–19391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nagai S., Wiker H. G., Harboe M., Kinomoto M. (1991) Isolation and partial characterization of major protein antigens in the culture fluid of Mycobacterium tuberculosis. Infect. Immun. 59, 372–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu Q., Yan B., Li S., Duan C. (2004) Fibronectin binds insulin-like growth factor-binding protein 5 and abolishes Its ligand-dependent action on cell migration. J. Biol. Chem. 279, 4269–4277 [DOI] [PubMed] [Google Scholar]
- 31. Lin Y. P., Chang Y. F. (2008) The C-terminal variable domain of LigB from Leptospira mediates binding to fibronectin. J. Vet. Sci. 9, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin Y. P., Kuo C. J., Koleci X., McDonough S. P., Chang Y. F. (2011) Manganese binds to Clostridium difficile Fbp68 and is essential for fibronectin binding. J. Biol. Chem. 286, 3957–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chessa D., Winter M. G., Nuccio S. P., Tükel C., Bäumler A. J. (2008) RosE represses Std fimbrial expression in Salmonella enterica serotype typhimurium. Mol. Microbiol. 68, 573–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin Y. P., Raman R., Sharma Y., Chang Y. F. (2008) Calcium binds to leptospiral immunoglobulin-like protein, LigB, and modulates fibronectin binding. J. Biol. Chem. 283, 25140–25149 [DOI] [PubMed] [Google Scholar]
- 35. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 36. Sanki A. K., Boucau J., Umesiri F. E., Ronning D. R., Sucheck S. J. (2009) Design, synthesis, and biological evaluation of sugar-derived esters, α-ketoesters and α-ketoamides as inhibitors for Mycobacterium tuberculosis antigen 85C. Mol. Biosyst. 5, 945–956 [DOI] [PubMed] [Google Scholar]
- 37. Tung J. Y., Yang C. W., Chou S. W., Lin C. C., Sun Y. J. (2010) Calcium binds to LipL32, a lipoprotein from pathogenic Leptospira, and modulates fibronectin binding. J. Biol. Chem. 285, 3245–3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sharma A., Askari J. A., Humphries M. J., Jones E. Y., Stuart D. I. (1999) Crystal structure of a heparin- and integrin-binding segment of human fibronectin. EMBO J. 18, 1468–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Santos R., Franza T., Laporte M. L., Sauvage C., Touati D., Expert D. (2001) Essential role of superoxide dismutase on the pathogenicity of Erwinia chrysanthemi strain 3937. Mol. Plant-Microbe Interact. 14, 758–767 [DOI] [PubMed] [Google Scholar]
- 40. Schwarz-Linek U., Pilka E. S., Pickford A. R., Kim J. H., Höök M., Campbell I. D., Potts J. R. (2004) High affinity streptococcal binding to human fibronectin requires specific recognition of sequential F1 modules. J. Biol. Chem. 279, 39017–39025 [DOI] [PubMed] [Google Scholar]
- 41. Vazquez V., Liang X., Horndahl J. K., Ganesh V. K., Smeds E., Foster T. J., Hook M. (2011) Fibrinogen is a ligand for the Staphylococcus aureus microbial surface components recognizing adhesive matrix molecules (MSCRAMM) bone sialoprotein-binding protein (Bbp). J. Biol. Chem. 286, 29797–29805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Janicki B. W., Chaparas S. D., Daniel T. M., Kubica G. P., Wright G. L., Yee G. S. (1971) A reference system for antigens of Mycobacterium tuberculosis. Am. Rev. Respir. Dis. 104, 602–604 [DOI] [PubMed] [Google Scholar]
- 43. Ohara N., Kitaura H., Hotokezaka H., Nishiyama T., Wada N., Matsumoto S., Matsuo T., Naito M., Yamada T. (1995) Characterization of the gene encoding the MPB51, one of the major secreted protein antigens of Mycobacterium bovis BCG, and identification of the secreted protein closely related to the fibronectin binding 85 complex. Scand. J. Immunol. 41, 433–442 [DOI] [PubMed] [Google Scholar]
- 44. Schorey J. S., Holsti M. A., Ratliff T. L., Allen P. M., Brown E. J. (1996) Characterization of the fibronectin-attachment protein of Mycobacterium avium reveals a fibronectin-binding motif conserved among mycobacteria. Mol. Microbiol. 21, 321–329 [DOI] [PubMed] [Google Scholar]
- 45. Schorey J. S., Li Q., McCourt D. W., Bong-Mastek M., Clark-Curtiss J. E., Ratliff T. L., Brown E. J. (1995) A Mycobacterium leprae gene encoding a fibronectin-binding protein is used for efficient invasion of epithelial cells and Schwann cells. Infect. Immun. 63, 2652–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ronning D. R., Vissa V., Besra G. S., Belisle J. T., Sacchettini J. C. (2004) Mycobacterium tuberculosis antigen 85A and 85C structures confirm binding orientation and conserved substrate specificity. J. Biol. Chem. 279, 36771–36777 [DOI] [PubMed] [Google Scholar]
- 47. Peake P., Gooley A., Britton W. J. (1993) Mechanism of interaction of the 85B secreted protein of Mycobacterium bovis with fibronectin. Infect. Immun. 61, 4828–4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Naito M., Fukuda T., Sekiguchi K., Yamada T. (2000) The domains of human fibronectin mediating the binding of α antigen, the most immunopotent antigen of mycobacteria that induces protective immunity against mycobacterial infection. Biochem. J. 347, 725–731 [PMC free article] [PubMed] [Google Scholar]
- 49. Ambler R. P., Meadway R. J. (1968) The use of thermolysin in amino acid sequence determination. Biochem. J. 108, 893–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Holmes A. R., McNab R., Millsap K. W., Rohde M., Hammerschmidt S., Mawdsley J. L., Jenkinson H. F. (2001) The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence. Mol. Microbiol. 41, 1395–1408 [DOI] [PubMed] [Google Scholar]
- 51. Peacock S. J., Foster T. J., Cameron B. J., Berendt A. R. (1999) Bacterial fibronectin-binding proteins and endothelial cell surface fibronectin mediate adherence of Staphylococcus aureus to resting human endothelial cells. Microbiology 145, 3477–3486 [DOI] [PubMed] [Google Scholar]
- 52. Mempel M., Schnopp C., Hojka M., Fesq H., Weidinger S., Schaller M., Korting H. C., Ring J., Abeck D. (2002) Invasion of human keratinocytes by Staphylococcus aureus and intracellular bacterial persistence represent haemolysin-independent virulence mechanisms that are followed by features of necrotic and apoptotic keratinocyte cell death. Br. J. Dermatol. 146, 943–951 [DOI] [PubMed] [Google Scholar]
- 53. Ratliff T. L., McCarthy R., Telle W. B., Brown E. J. (1993) Purification of a mycobacterial adhesin for fibronectin. Infect. Immun. 61, 1889–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhao W., Schorey J. S., Groger R., Allen P. M., Brown E. J., Ratliff T. L. (1999) Characterization of the fibronectin binding motif for a unique mycobacterial fibronectin attachment protein, FAP. J. Biol. Chem. 274, 4521–4526 [DOI] [PubMed] [Google Scholar]
- 55. Secott T. E., Lin T. L., Wu C. C. (2001) Fibronectin attachment protein homologue mediates fibronectin binding by Mycobacterium avium subsp. paratuberculosis. Infect. Immun. 69, 2075–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.