Background: The highly regulated hypoxia-inducible factor 1α (HIF-1α) is a key player in the cellular response to hypoxia.
Results: The ubiquitin-specific protease 19 (USP19) rescues HIF-1α from degradation in a non-catalytic manner.
Conclusion: USP19 is required for cells to mount an appropriate response to hypoxia.
Significance: Learning about HIF-1α regulation is essential for understanding the physiological and pathophysiological conditions of the hypoxic response.
Keywords: Deubiquitination, Deubiquitylation, Hypoxia, Hypoxia-inducible Factor (HIF), Ubiquitin
Abstract
A proper cellular adaptation to low oxygen levels is essential for processes such as development, growth, metabolism, and angiogenesis. The response to decrease in oxygen supply, referred to as hypoxia, is also involved in numerous human diseases including cancer, inflammatory conditions, and vascular disease. The hypoxia-inducible factor 1-α (HIF-1α), a key player in the hypoxic response, is kept under stringent regulation. At normoxia, the levels are kept low as a consequence of the efficient degradation by the ubiquitin-proteasome system, and in response to hypoxia, the degradation is blocked and the accumulating HIF-1α promotes a transcriptional response essential for proper adaptation and survival. Here we show that the ubiquitin-specific protease-19 (USP19) interacts with components of the hypoxia pathway including HIF-1α and rescues it from degradation independent of its catalytic activity. In the absence of USP19, cells fail to mount an appropriate response to hypoxia, indicating an important role for this enzyme in normal or pathological conditions.
Introduction
Cells have evolved sophisticated mechanisms to sense and adapt to the natural fluctuations of oxygen levels. This adaptation is crucial for normal physiology such as adaptation to high altitude or proper embryogenesis but is also involved in numerous pathophysiological conditions such as inflammation, cardiovascular diseases, stroke, and cancer (1, 2). Limitation in oxygen triggers a chain of events that leads to the activation of hypoxia-inducible factors (HIF).3 HIFs are transcription factors formed by a dimer consisting of an unstable α-subunit and a stable β-subunit, also referred to as aryl hydrocarbon receptor nuclear translocator (ARNT). Human HIF-α has three isoforms, HIF-1α, HIF-2α, and HIF-3α, of which the first two are closely related and have been extensively studied, whereas HIF-3α is subject to extensive splicing, and the role of its different forms remain largely unknown (3, 4). HIF-1α plays a role in the acute hypoxic response and is known to promote the expression of more than 60 genes associated with processes such as erythropoiesis, angiogenesis, cell growth, differentiation, survival, or apoptosis (5). HIF-1α is kept under tight regulation, and in normoxia, it is one of the most short-lived proteins known (6). The steady-state levels are kept low due to its degradation by the ubiquitin-proteasome system. The detailed mechanisms by which HIF-1α stability and activity are regulated are under intense investigation and may withhold yet unidentified players and therapeutic targets (7).
Protein modifications by ubiquitin regulate numerous cellular processes by affecting protein stability or function (8). Covalent linkage of ubiquitin to target proteins is directed by the orchestrated activity of a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a substrate-specific ubiquitin ligase (E3) that mediates the transfer of ubiquitin to the target. Like most posttranslational modifications, ubiquitination is also reversible. This process is performed by the family of ∼100 deubiquitylating enzymes (DUBs), which are cysteine or metallo-proteases emerging as important regulators in numerous molecular signaling pathways (9). DUBs are categorized into five subclasses based on homology between their catalytic domains: ubiquitin-specific protease (USPs), ubiquitin C-terminal hydrolases, Otubain proteases, Machado-Joseph disease proteases, and JAB1/MPN/Mov34 metallo-proteases (9).
The functional outcome of ubiquitylation depends on the type of ubiquitin chain formed. For HIF-1α, it typically triggers degradation by the proteasome (10). The instability of HIF-1α at normoxia is mainly due to the activity of prolyl hydroxylases (PHDs) that use molecular oxygen as a co-substrate for catalysis to hydroxylate HIF-1α (11). This increases the affinity for the von Hippel-Lindau (VHL) ubiquitin ligase, which promotes HIF-1α ubiquitylation and subsequent degradation (10, 12). Three PHDs have been identified, and their abundance varies greatly between cell types. Although the role of PHD1 is still unclear, PHD2 has been reported as the major regulator of HIF-1α hydroxylation during normoxia, and PHD3 has been appointed a function in mild or prolonged hypoxic conditions (13, 14). The PHDs are also regulated by their interaction with the family of Siah ubiquitin ligases (for Seven in absentia homologue). Although Siah2 controls PHD1 and PHD3 ubiquitination during mild hypoxic conditions, the role for Siah1 is less clear (15, 16). Upon oxygen deprivation, HIF-1α rapidly accumulates and dimerizes with ARNT to form an active transcription factor complex in the nucleus. The activity of the HIF-1α/ARNT heterodimer can be further regulated by the oxygen-dependent factor inhibiting HIF (FIH), an asparaginyl hydroxylase, acting on nuclear HIF-1α inhibiting the recruitment of transcriptional co-activators such as cAMP-response element-binding protein (CREB)-binding protein and p300 (17, 18).
Key players in essential pathways are often subject to ubiquitin regulation mediated by one or several ubiquitin ligases or DUBs. The prime example is probably the tumor suppressor p53, which is subject for regulation by more than 10 ubiquitin ligases and three DUBs (19). The VHL-interacting deubiquitylating enzyme (VDU)-2 is to our knowledge so far the only reported DUB in the hypoxia pathway (20), rendering it possible that there are unidentified DUBs in this pathway still to be discovered.
Here we show that USP19 interacts with HIF-1α and describe a non-catalytic role for this enzyme in stabilizing HIF-1α after cellular exposure to hypoxia. The presence of USP19 is required to mount a proper hypoxic response, and we therefore suggest that USP19 is a previously unknown regulator of HIF-1α stability.
EXPERIMENTAL PROCEDURES
Plasmids and Yeast Two-hybrid Screen
For the yeast two-hybrid bait construct, 1–1485 nucleotides of USP19 were cloned in-frame with the GAL4 DNA binding domain (DBD) and Myc tag in the yeast expression vector pGBKT7 (Clontech), generating pGBKT7-GAL4(DBD)-USP19(1–495aa)-Myc. The yeast two-hybrid screen was performed using the MatchmakerTM pretransformed HeLa library (Clontech) according to manufacturer's protocol. The short hairpin RNA (shRNA)-expressing plasmids pRETRO-SUPER-USP19A and -D were generated by cloning the target sequences GAGACAGGGTCTCGATATG and GATCAATGACTTGGTGGAG of USP19 mRNA in the pRETRO-SUPER vector (21). Plasmids overexpressing Myc-USP19, Myc-USP19(C506S), USP19ΔTM-Myc, and FLAG-tagged HIF-1α, HIF-2α, HIF-3α (splice form: HIF-3α1), PHD1, PHD2, PHD3, and VHL have been previously described (4, 22, 23).
Tissue Culture, Transfections, and Immunostainings
The human cervical cancer cell line HeLa, the human embryonic kidney cell line HEK293T, the human melanoma cell line M2, and the human osteosarcoma cell line U2OS were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (v/v), 2 mm glutamine, and 100 units/ml penicillin and 100 μg/ml streptomycin (Sigma). All exposures to hypoxia were performed with 1% O2 for indicated time. Transfections were performed using jetPEI (Polyplus Transfection) according to manufacturer's protocol or by the calcium phosphate method. To achieve an efficient knockdown of USP19 in HeLa cells transfected with the pRETRO-SUPER plasmids, the cells were treated with 0.5 μg/ml puromycin to kill untransfected cells. For immunostainings, the cells were grown and transfected on glass coverslips and fixed in 4% formaldehyde in phosphate-buffered saline (PBS) (w/v) and stained with the appropriate antibodies diluted in 50 mm Tris, pH 7.5, 0.9% NaCl (w/v), 0.1% gelatin (w/v), and 0.5% Triton X-100 (v/v).
Immunoprecipitations
Co-immunoprecipitations were performed in a buffer containing 50 mm Tris (pH 7.4), 150 mm NaCl, 0.05 mm EDTA, and 1% IGEPAL CA-630 (Sigma) (v/v) supplemented with protease inhibitor mixture (Roche Applied Science). Immunoprecipitations with USP19 antibodies were performed over-night followed by binding to GammaBind Sepharose (GE Healthcare, Uppsala, Sweden) and subsequent washings in lysis buffer. Immunoprecipitations of FLAG-tagged proteins was performed with FLAG(M2) affinity gel (Sigma) according to the manufacturer's protocol.
Western Blot and Antibodies
Proteins were fractionated in precast polyacrylamide Bis-Tris 4–12% gradient gels (Invitrogen). After transfer to polyvinylidene fluoride membranes (Millipore, Billerica, MA), the filters were blocked in PBS containing 5% fat-free milk (w/v) and 0.1% Tween 20 (v/v) for 1 h. The membranes were incubated with primary antibodies for 1 h at room temperature or overnight at 4 °C followed by washing steps and 1 h of incubation with the appropriate horseradish peroxidase-conjugated secondary antibodies. The results were revealed by enhanced chemiluminescence (ECL; GE Healthcare).
The antibodies used were: FLAG(M2) (Sigma); β-actin(AC-15), Myc(9E10), or Myc(A14) (Santa Cruz Biotechnology, Santa Cruz, CA); USP19(A301-586A) or USP19(A301-587A) (Bethyl Laboratories, Montgomery, TX); or HIF-1α (BD Biosciences). Secondary antibodies for immunostainings were: donkey anti-rabbit Alexa Fluor 488 and donkey anti-mouse Alexa Fluor 555 (Invitrogen). Secondary antibodies for Western blot were: donkey anti-rabbit and sheep anti-mouse (Zymed Laboratories Inc., San Francisco, CA).
Quantitative PCR (qPCR)
Total RNA was obtained using an RNeasy mini kit (Qiagen, Valencia, CA), and 1 μg of RNA was used for reverse transcription using M-MuLV reverse transcriptase (New England Biolabs) according to the manufacturer's protocol. qPCR was performed with Power SYBR Green PCR master mix (Applied Biosystems) using the 7500 real-time PCR system (Applied Biosystems). The primer sequences used are: β-actin, 5′-CCTGGCACCCAGCACAAT-3′ and 5′-GGGCCGGACTCGTCATACT-3′; USP19, 5′-CGGCACAAGATGAGGGA-3′ and 5′-GGCACCGGCAGATAAAGAAA-3′; glucose transporter 1 (GLUT1), 5′-CAGCAGCAAGAAGCTGAC-3′ and 5′-GGGCATTGATGACTCCAG-3′; and vascular endothelial growth factor α (VEGFα), 5′-ATTATGCGGATCAAACCTCAC-3′ and 5′TCTTTCTTTGGTCTGCATTCAC-3. The expression of β-actin was used as internal control.
RESULTS
USP19 Interacts with HIF-1α
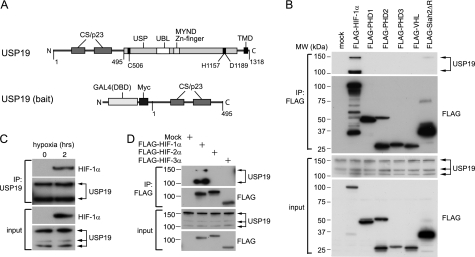
We have previously shown that USP19 is a DUB with a C-terminal transmembrane domain (TMD) anchoring it to the endoplasmic reticulum (ER) (22). At the ER, USP19 can affect the degradation of ER-associated degradation (ERAD) substrates and appears up-regulated in response to ER stress (22). USP19 is also up-regulated during catabolic stress causing skeletal muscle atrophy (24) and affects cell cycle progression (25). This motivated us to further investigate USP19 function and identify interacting partners by performing a yeast two-hybrid screen. The bait was restricted to the N-terminal part of USP19 (1–495 amino acids) harboring a bipartite CS domain named after CHORD-containing proteins (for cysteine- and histidine-rich domain) and SGT1 (for suppressor of G-two allele of SKP1). This region shares high homology to the p23 protein and is therefore occasionally also referred to as a p23 domain (26) (Fig. 1A). CS domains are frequently found in co-chaperones of Hsp90; however, the impact of its presence in USP19 is still unclear.
FIGURE 1.
USP19 interacts with HIF-1α. A, schematic representation of the full-length and the 1–495-amino acid long USP19, which was used as bait in a yeast two-hybrid screen. CS domain, p23 protein domain (p23), USP domain, ubiquitin-like domain (UBL), myeloid translocation protein 8, Nervy protein, Deaf-1 zinc finger (MYND Zn-finger), transmembrane domain (TMD), and the positions of the amino acids Cys, His, and Asp in the catalytic triad are indicated (DNA binding domain (DBD)). B, immunoprecipitations (IP) using FLAG(M2) affinity gel from HEK293T cells transfected with FLAG-tagged components of the hypoxia pathway as indicated. The co-immunoprecipitated endogenous USP19 was detected with the anti-USP19(A301-587A) antibody (Bethyl Laboratories) as indicated. Note that USP19 appears in multiple forms, indicated by arrows, likely representing splice variants or processed forms. MW, molecular weight markers. C, co-immunoprecipitation of endogenous proteins from HeLa cells in normoxia and hypoxia using the anti-USP19(A301-587A) antibody as indicated. D, co-immunoprecipitation using anti-FLAG(M2) affinity gel showing selective interaction between Myc-USP19 and FLAG-HIF-1α but not FLAG-HIF-2α and FLAG-HIF-3α.
In the yeast two-hybrid screen, Siah1 and Siah2, the vertebrate homologs of the Drosophila “seven in absentia” gene (Sina) (27), appeared as interacting partners of USP19. Apart from their role in hypoxia, they have been ascribed functions in diverse cellular processes including cell proliferation, apoptosis, and tumor suppression (16, 28, 29). Among these vast functions of Siah, we chose to investigate any potential involvement of USP19 in the hypoxia pathway.
To test whether USP19 could interact with additional components within the hypoxia pathway, we performed co-immunoprecipitation experiments from HEK293T cells transiently overexpressing HIF-1α, the hydroxylases PHD1, PHD2, and PHD3, and the ubiquitin ligase VHL. The hypoxia components were FLAG-tagged, and FLAG-Siah2ΔRING was included as a positive control. A deletion mutant lacking the RING domain was used to avoid the inherent instability brought to these ligases by the RING. Immunoprecipitations using FLAG affinity gel confirmed the interaction between USP19 and Siah2 as expected, although it appeared with low efficiency. More striking was a solid interaction between USP19 and FLAG-HIF-1α (Fig. 1B). As illustrated with arrows, the endogenous USP19 appears as multiple bands in Western blot; most dominant are bands around 100, 130, and 150 kDa (Fig. 1B). The 100- and 150-kDa forms repeatedly co-immunoprecipitated with FLAG-HIF-1α, although with a clear preference for the 100-kDa variant.
The interaction was validated by co-immunoprecipitation of endogenous proteins using an anti-USP19 antibody. To accumulate detectable HIF-1α levels, HeLa cells were exposed to hypoxia for 2 h prior to the immunoprecipitation (Fig. 1C). This result was reproduced with two different antibodies against USP19 (supplemental Fig. S1). The interaction was specific to HIF-1α and neither to a long splice form of HIF-3α containing all the major domains (bHLH, PASa, PASb, ODD/NTAD (oxygen-dependent degradation/N-terminal transactivation domains), and a leucine zipper) nor to HIF-2α (Fig. 1D). HIF-1α and HIF-2α are highly homologous and bind to similar HIF-response elements, but HIF-2α is suggested to play a more prominent role in the chronic adaptation to hypoxia (30), which may imply a function for USP19 in the acute hypoxic response.
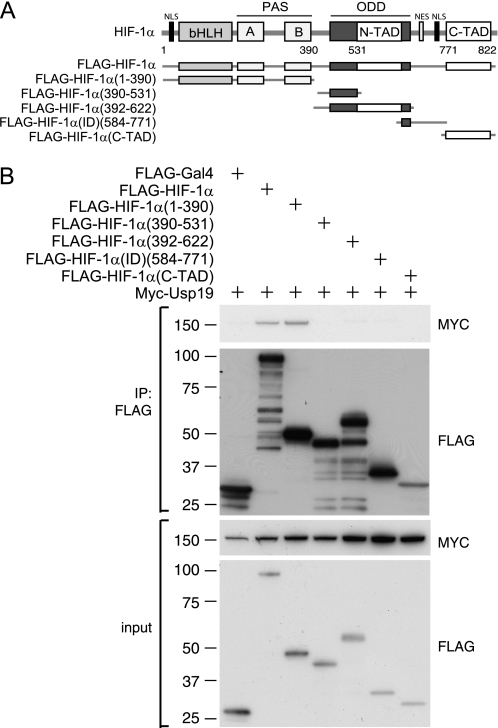
Mapping Interaction Domains of HIF-1α
HIF-1α belongs to the family of basic-loop-helix (bHLH)- and PER-ARNT-SIM (PAS) domain-containing transcription factors. The bHLH domain near the N terminus is required for the binding to HIF-response element sequences present in HIF target genes, and the PAS domains mediate dimerization to ARNT. The transactivation domains (TADs) and the ODD domain, which is the target of hydroxylation-dependent ubiquitylation, reside in the C-terminal region (Fig. 2A). To pinpoint which region of HIF-1α is responsible for the interaction with USP19, co-immunoprecipitations with different FLAG-HIF-1α fragments were performed. Interaction was detected between USP19 and the N-terminal part of HIF-1α containing the PAS and bHLH domains (Fig. 2B).
FIGURE 2.
Mapping HIF-1α interaction domain. A, schematic illustration of truncated, FLAG-tagged, HIF-1α constructs. bHLH, PAS domain, ODD, N/C-terminal transactivation domain (N-TAD and C-TAD), nuclear localization signal (NLS), and nuclear export signal (NES) are indicated. B, co-immunoprecipitations (IP) using a FLAG(M2) affinity gel from lysates of HEK293T cells co-transfected with the truncated forms of HIF-1α or FLAG-GAL4 as control, together with Myc-USP19.
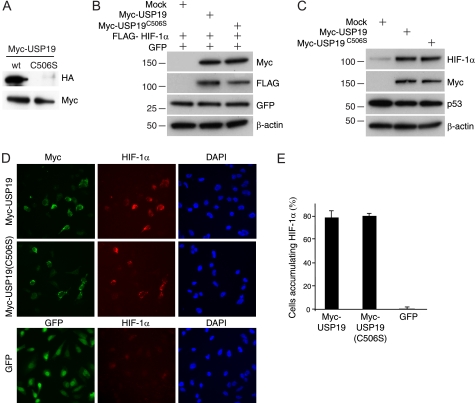
USP19 Stabilizes HIF-1α Independent of DUB Activity
Previous studies on USP19 have shown that it can rescue proteins such as Kip1 ubiquitylation-promoting complex (KPC1) and ERAD substrates from proteasomal degradation (22, 25). For this reason, we tested whether USP19 activity was also able to rescue HIF-1α from degradation. First we performed labeling experiments using the active site-directed DUB probe, HA-ubiquitin-VME (vinyl methylester), which gives a measure of the DUB activity (31), and certified that the catalytic mutant Myc-USP19(C506S) was indeed inactive (Fig. 3A). The interaction between USP19 and HIF-1α remained, or occurred even more efficiently, with the catalytic mutant USP19 (supplemental Fig. S2). Next we tested whether overexpression of Myc-USP19 and the inactive Myc-USP19(C506S) influenced the levels of co-transfected FLAG-HIF-1α. Interestingly, both the wild-type and the inactive USP19 stabilized FLAG-HIF-1α to a similar extent (Fig. 3B). This non-catalytic rescue of HIF-1α was confirmed by also looking at endogenous HIF-1α levels (Fig. 3C). To test whether the stabilizing effect was limited to HIF-1α and not to other short-lived proteins in general, the same samples were probed against p53, which did not appear specifically regulated under these conditions (Fig. 3C). Immunostainings of HeLa cells transiently transfected with Myc-USP19 and Myc-USP19(C506S) confirmed the rescue of endogenous HIF-1α (Fig. 3D). Scoring revealed that ∼80% of the USP19-overexpressing cells accumulated endogenous HIF-1α in normoxia (Fig. 3E). This effect of USP19 was reproduced in additional cell lines, suggesting that it is not cell type-specific, but a rather general effect (Fig. 4). The HIF-1α accumulating in response to USP19 overexpression, however, appeared transcriptionally inactive under these conditions, suggesting that additional events priming its activity were absent.
FIGURE 3.
USP19 stabilizes HIF-1α independent of catalytic activity. A, active site labeling with the HA-ubiquitin-VME probe in lysates from cells expressing Myc-USP19 and the mutant Myc-USP19(C506S). The upper blot illustrates the enzymatically active Myc-USP19 covalently linked to the probe as detected using an anti-HA antibody. The lower blot illustrates the expression of both Myc-USP19 and Myc-USP19(C506S) using anti-Myc(9E10) antibody. B, Western blot to test the effect of overexpressed USP19 on co-transfected FLAG-HIF-1α steady-state levels as indicated. GFP was included as a co-transfection control. C, Western blot analysis of U2OS cells overexpressing Myc-USP19 and Myc-USP19(C506S), probed as indicated. D, micrographs of cells transfected with Myc-USP19, Myc-USP19(C506S), or GFP as control. Immunostainings are shown with anti-HIF-1α (red) and anti-Myc(A14) (green) and nuclear counterstaining using DAPI as indicated. E, quantification of results in D where 100 USP19-positive cells were scored for positive HIF-1α co-staining. Values show mean ± S.D. of triplicates.
FIGURE 4.
USP19 stabilizes HIF-1α independent of ER localization. A, graphs illustrating HeLa cells, the melanoma cell line M2, and HEK293T cells overexpressing Myc-USP19, Myc-USP19(C506S), and Myc-USP19ΔTM, which lacks ER localization. The number of cells accumulating HIF-1α in USP19-overexpressing cells was scored by counting cells positive for HIF-1α immunostaining. B, Western blot of HeLa cells transiently overexpressing Myc-USP19, Myc-USP19(C506S), and Myc-USP19ΔTM in normoxia. The blots were probed against Myc, HIF-1α, and β-actin as indicated.
USP19 Rescues HIF-1α from Degradation Independent of ER Localization
Our previous study spatially and functionally placed USP19 to the ER (22). This raises the question whether ER localization is of significance for stabilizing HIF-1α. We therefore tested whether deletion of the C-terminal anchor of USP19, which is required for ER localization and the ability to stabilize ERAD substrates, influenced the stabilizing effect on HIF-1α in normoxia. However, USP19 with deletion of the TM domain behaved similar to full-length USP19, suggesting that ER localization is not required for the rescue of HIF-1α from degradation (Fig. 4, A and B).
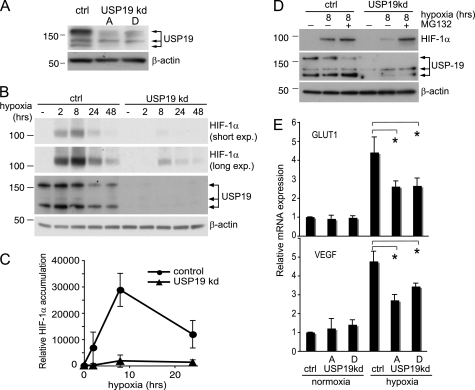
Loss of USP19 Impairs Cellular Response to Hypoxia
Our findings that USP19 can stabilize and interact with HIF-1α and not HIF-2α suggest that it may be involved in the regulation of the acute cellular response to hypoxia. To address this possibility, we analyzed the hypoxic response in HeLa cells with suppressed USP19 expression. The knockdown was performed by transfection of plasmids expressing shRNAs targeting two different regions within the USP19 mRNA (Fig. 5A). Three days after transfection of the shRNA-expressing plasmid pRETRO-SUPER and pRETRO-SUPER-USP19A, the cells were exposed to hypoxia for the indicated time. Interestingly, cells with low USP19 expression dramatically failed to accumulate HIF-1α after exposure to hypoxia (Fig. 5, B and C). Because the main regulation of HIF-1α steady-state levels is mediated by proteasomal degradation rather than at transcriptional level (32), our data suggest that in the absence of USP19, HIF-1α is continuously degraded by the proteasome, disregarding the hypoxic conditions. To test this, USP19 was knocked down in HeLa cells and exposed to hypoxia or exposed to hypoxia in combination with treatment with the proteasome inhibitor MG132 (10 μm). Indeed HIF-1α accumulated in response to the treatment with MG132 in USP19 knockdown cells, illustrating that USP19 is important for the natural rescue of HIF-1α from proteasomal degradation in response to hypoxia (Fig. 5D).
FIGURE 5.
Loss of USP19 impairs hypoxic response. A, Western blot experiment assessing the efficiency of two different shRNA-expressing vectors, pRETRO-SUPER-USP19A (indicated by A) and pRETRO-SUPER-USP19D (indicated by D), in suppressing USP19 protein expression. ctrl, control; kd, knockdown. B, 3 days after transfection, HeLa cells transfected with the empty control plasmid pRETRO-SUPER or pRETRO-SUPER-USP19A were exposed to hypoxia or kept in normoxia as indicated. Western blots were probed anti-HIF-1α (short and long exposure (exp.)) to investigate the effect by USP19 knockdown on HIF-1α accumulation. β-Actin was included as control. C, quantification by densitometry from short exposure Western blots of three independent experiments performed as in Fig. 4B. Values represent relative induction of HIF-1α during hypoxia as compared with normoxia, mean ± S.D. D, same experimental setup as in Fig. 4B, but cells exposed to hypoxia for 8 h were treated in parallel with the proteasome inhibitor MG132 (10 μm). The results illustrate a continuous proteasomal degradation of HIF-1α during hypoxia in cells with suppressed USP19 expression, E, knockdown of USP19 impairs the HIF-1α transcriptional response during hypoxia. Relative mRNA expression was assessed by qPCR of the HIF-1α target genes GLUT1 and VEGF in cells with or without USP19 knockdown ± hypoxia. Values represent expression levels relative to β-actin mRNA ± S.D. from four independent experiments. *, p values < 0.05 for indicated comparisons.
To test whether the lack of HIF-1α accumulation in response to hypoxia was of functional significance, we performed qPCR in USP19 knockdown cells assaying the expression of the well established HIF-1α target genes: the GLUT1 and the VEGF. Because loss of USP19 delays progression of the cell cycle (25) and appears slightly toxic to cells, we performed the qPCR with limited levels of USP19 knockdown. A ∼50% reduction of USP19 mRNA was used and was expected to provide a good representation of a physiological condition (supplemental Fig. S3). Indeed the HIF-1α transcriptional response was significantly reduced in USP19 knockdown cells during hypoxia (Fig. 5E). Taken together our data strongly support a role for USP19 in stabilizing HIF-1α and promoting a proper transcriptional response during hypoxia.
DISCUSSION
During the last years, substantial progress has been made to delineate the molecular mechanisms that resolve reduced oxygen levels into an adjusting cellular response (33). Here we show that USP19 and HIF-1α interact with each other and that USP19 regulates HIF-1α stability in a non-catalytic manner. In the absence of USP19, cells fail to mount an appropriate response to hypoxia.
USP19 is a DUB reported to protect certain proteasomal substrates from degradation by virtue of its enzymatic activity (22, 25). However, our findings suggest that USP19 exerts a non-catalytic mode of regulation because overexpression of the inactive USP19 stabilized HIF-1α to the same extent as the wild type. Similar observations for USP19 were made studying its effect on the turnover of particular ERAD substrates (22) and the cellular inhibitors of apoptosis (c-IAPs), c-IAP-1 and c-IAP2 (34). Non-catalytic functions of DUBs are not unprecedented and have recently emerged as an important means for these enzymes to increase their functions (35, 36). Hence it seems as if USP19 belongs to these DUBs that have developed this non-canonical way of regulation, possibly by their ability to recognize ubiquitin or by mere protein interactions or competitive bindings with additional partners.
HIF-1α and HIF-2α are highly homologous and bind similar HIF-response element motifs; however, they exert different functions. This is best illustrated by their dissimilar embryonic deletion phenotypes, their role in tumor angiogenesis or in adaptation to chronic hypoxia (37–40). Although HIF-1α and HIF-2α share some common interacting partners, we found that USP19 interacts specifically with HIF-1α. It is likely that the differences between HIF-1α and HIF-2α in part are mediated through their selective protein interactions, and possibly USP19 by this means contributes to their functional differences.
HIF-1α interacts with USP19 via its N-terminal region harboring the PAS and bHLH domains. Although this region is typically involved in DNA binding and dimerization to ARNT, other interactions taking place here include binding to the molecular chaperone Hsp90 and RACK1 associated with O2-independent regulation of HIF-1α (41) or to the minichromosome maintenance protein 7 (MCM7) involved in the O2-dependent regulation (42). The region within USP19 important for interacting or stabilizing HIF-1α remains to be determined and may hold interesting mechanistic insight. p23/CS domains have been proposed to play a role in Hsp90 binding (26) or suggested to function as a general binding module recruiting heat shock proteins to multiprotein complexes (43). This raises the question whether USP19 is part of such a stabilizing complex and whether it in addition could be relevant for O2-independent regulation of HIF-1α. MYND (myeloid translocation protein 8, Nervy protein, Deaf-1) domains are believed to mediate protein-protein interactions, and interestingly USP19 shares this domain with numerous other proteins, including PHD2 (44, 45). Any significance of this for the role of USP19 in hypoxia remains to be discovered.
We have previously identified the TMD of USP19 to be indispensable for USP19 stabilization of ERAD substrates (22). However, here we found that the TMD was perfectly dispensable for the stabilization of HIF-1α (Fig. 4). In agreement with this is our observation that the preferred form of USP19 (the 100-kDa form) interacting with HIF-1α (Fig. 1B) is likely lacking the TMD because this is only present in three of the 12 documented splice forms, all expected to migrate around 150 kDa (Ensembl Genome Browser). Thus, ER localization is not a likely criterion for USP19 regulation of HIF-1α and may represent a multifunctionality of this protein directed by its subcellular localization. However, it should be noted that the regulation of HIF-1α is complex and that conditions of severe hypoxia (<0.01% O2) induce ER stress. This is due to defects in the protein-folding capacity of the ER emerging at these O2 levels (46). Thus, possibly USP19 is induced after such ER stress stimulation and plays an integrated role in the hypoxic and ER stress response under conditions of extreme hypoxia.
By summarizing USP19 functions, a general impression of stress involvement arises. Here we show that USP19 is involved in hypoxic stress, and we previously showed involvement in ER stress (22). Other studies have shown an up-regulation of USP19 in rat skeletal muscle in response to different stress including streptozotocin-induced diabetes, dexamethasone treatment, and cancer (24). In addition, considering the ability of USP19 to rescue c-IAPs from degradation after apoptotic stimulation (34), it is tempting to speculate that the general biological function of USP19 is of a cytoprotective nature intended to adapt cells to different types of stress.
In conclusion, we have found that USP19 is important for regulating HIF-1α and that loss of USP19 expression dramatically impaired the cellular response to hypoxia. Hence we suggest that the role of USP19 in the hypoxia pathway may have important implications for normal physiology or pathophysiology.
Supplementary Material
Acknowledgments
We thank Ze'ev Ronai (Burnham Institute for Medical Research) for kindly providing the Siah2 plasmid, René Bernards (The Netherlands Cancer Institute) for the pRetroSuper plasmid, and Johanna Myllyharju (University of Oulu, Finland) for the FLAG-HIF-3α plasmid.
This work was supported by grants from the Swedish Research council (to K. L., T. P., and M. A.), Karolinska Institutet (to K. L., B. Z., and M. A.), Magn Bergvalls stiftelse (to K. L.), Åke Wibergs Stiftelse (to K. L.), The Swedish Cancer Society (to T. P.), and ERACOL (to K. V.).

This article contains supplemental Figs. S1–S3.
- HIF
- hypoxia-inducible factor
- ARNT
- aryl hydrocarbon receptor nuclear translocator
- bHLH
- basic helix-loop-helix
- CS
- CHORD and SGT1 domain
- DUB
- deubiquitylating enzyme
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- GLUT1
- glucose transporter 1
- c-IAP
- cellular inhibitor of apoptosis
- ODD
- oxygen-dependent domain
- PAS
- PER-ARNT-SIM domain
- PHD
- prolyl hydroxylase
- qPCR
- quantitative PCR
- TAD
- transactivation domain
- TMD
- transmembrane domain
- USP19
- ubiquitin-specific protease 19
- VHL
- von Hippel-Lindau
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Cramer T., Yamanishi Y., Clausen B. E., Förster I., Pawlinski R., Mackman N., Haase V. H., Jaenisch R., Corr M., Nizet V., Firestein G. S., Gerber H. P., Ferrara N., Johnson R. S. (2003) HIF-1α is essential for myeloid cell-mediated inflammation. Cell 112, 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bertout J. A., Patel S. A., Simon M. C. (2008) The impact of O2 availability on human cancer. Nat. Rev. Cancer 8, 967–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gu Y. Z., Moran S. M., Hogenesch J. B., Wartman L., Bradfield C. A. (1998) Molecular characterization and chromosomal localization of a third α-class hypoxia-inducible factor subunit, HIF3α. Gene Expr. 7, 205–213 [PMC free article] [PubMed] [Google Scholar]
- 4. Heikkilä M., Pasanen A., Kivirikko K. I., Myllyharju J. (2011) Roles of the human hypoxia-inducible factor (HIF)-3α variants in the hypoxia response. Cell Mol. Life Sci. 68, 3885–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ratcliffe P. J., O'Rourke J. F., Maxwell P. H., Pugh C. W. (1998) Oxygen sensing, hypoxia-inducible factor 1, and the regulation of mammalian gene expression. J. Exp. Biol. 201, 1153–1162 [DOI] [PubMed] [Google Scholar]
- 6. Huang L. E., Gu J., Schau M., Bunn H. F. (1998) Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 7987–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilson W. R., Hay M. P. (2011) Targeting hypoxia in cancer therapy. Nat. Rev. Cancer 11, 393–410 [DOI] [PubMed] [Google Scholar]
- 8. Glickman M. H., Ciechanover A. (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 9. Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., Bernards R. (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 [DOI] [PubMed] [Google Scholar]
- 10. Tanimoto K., Makino Y., Pereira T., Poellinger L. (2000) Mechanism of regulation of the hypoxia-inducible factor 1 α by the von Hippel-Lindau tumor suppressor protein. EMBO J. 19, 4298–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P. H., Pugh C. W., Schofield C. J., Ratcliffe P. J. (2001) Caenorhabditis elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- 12. Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., Kriegsheim Av, Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 13. Appelhoff R. J., Tian Y. M., Raval R. R., Turley H., Harris A. L., Pugh C. W., Ratcliffe P. J., Gleadle J. M. (2004) Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 279, 38458–38465 [DOI] [PubMed] [Google Scholar]
- 14. Berra E., Benizri E., Ginouvès A., Volmat V., Roux D., Pouysségur J. (2003) HIF prolyl hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 22, 4082–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakayama K., Qi J., Ronai Z. (2009) The ubiquitin ligase Siah2 and the hypoxia response. Mol. Cancer Res. 7, 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nakayama K., Frew I. J., Hagensen M., Skals M., Habelhah H., Bhoumik A., Kadoya T., Erdjument-Bromage H., Tempst P., Frappell P. B., Bowtell D. D., Ronai Z. (2004) Siah2 regulates stability of prolyl hydroxylases, controls HIF1α abundance, and modulates physiological responses to hypoxia. Cell 117, 941–952 [DOI] [PubMed] [Google Scholar]
- 17. Carrero P., Okamoto K., Coumailleau P., O'Brien S., Tanaka H., Poellinger L. (2000) Redox-regulated recruitment of the transcriptional co-activators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1α. Mol. Cell. Biol. 20, 402–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., Bruick R. K. (2002) FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hock A., Vousden K. H. (2010) Regulation of the p53 pathway by ubiquitin and related proteins. Int. J. Biochem. Cell Biol. 42, 1618–1621 [DOI] [PubMed] [Google Scholar]
- 20. Li Z., Wang D., Messing E. M., Wu G. (2005) VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1α. EMBO Rep. 6, 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brummelkamp T. R., Bernards R., Agami R. (2002) Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2, 243–247 [DOI] [PubMed] [Google Scholar]
- 22. Hassink G. C., Zhao B., Sompallae R., Altun M., Gastaldello S., Zinin N. V., Masucci M. G., Lindsten K. (2009) The ER-resident ubiquitin-specific protease 19 participates in the UPR and rescues ERAD substrates. EMBO Rep. 10, 755–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. André H., Pereira T. S. (2008) Identification of an alternative mechanism of degradation of the hypoxia-inducible factor 1α. J. Biol. Chem. 283, 29375–29384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Combaret L., Adegoke O. A., Bedard N., Baracos V., Attaix D., Wing S. S. (2005) USP19 is a ubiquitin-specific protease regulated in rat skeletal muscle during catabolic states. Am. J. Physiol. Endocrinol. Metab. 288, E693–700 [DOI] [PubMed] [Google Scholar]
- 25. Lu Y., Adegoke O. A., Nepveu A., Nakayama K. I., Bedard N., Cheng D., Peng J., Wing S. S. (2009) USP19 deubiquitinating enzyme supports cell proliferation by stabilizing KPC1, a ubiquitin ligase for p27Kip1. Mol. Cell. Biol. 29, 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Garcia-Ranea J. A., Mirey G., Camonis J., Valencia A. (2002) p23 and HSP20/α-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett. 529, 162–167 [DOI] [PubMed] [Google Scholar]
- 27. Carthew R. W., Rubin G. M. (1990) seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell 63, 561–577 [DOI] [PubMed] [Google Scholar]
- 28. Matsuzawa S., Takayama S., Froesch B. A., Zapata J. M., Reed J. C. (1998) p53-inducible human homologue of Drosophila seven in absentia (Siah) inhibits cell growth: suppression by BAG-1. EMBO J. 17, 2736–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roperch J. P., Lethrone F., Prieur S., Piouffre L., Israeli D., Tuynder M., Nemani M., Pasturaud P., Gendron M. C., Dausset J., Oren M., Amson R. B., Telerman A. (1999) SIAH-1 promotes apoptosis and tumor suppression through a network involving the regulation of protein folding, unfolding, and trafficking: identification of common effectors with p53 and p21Waf1. Proc. Natl. Acad. Sci. U.S.A. 96, 8070–8073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Holmquist-Mengelbier L., Fredlund E., Löfstedt T., Noguera R., Navarro S., Nilsson H., Pietras A., Vallon-Christersson J., Borg A., Gradin K., Poellinger L., Påhlman S. (2006) Recruitment of HIF-1α and HIF-2α to common target genes is differentially regulated in neuroblastoma: HIF-2α promotes an aggressive phenotype. Cancer Cell 10, 413–423 [DOI] [PubMed] [Google Scholar]
- 31. Borodovsky A., Ovaa H., Kolli N., Gan-Erdene T., Wilkinson K. D., Ploegh H. L., Kessler B. M. (2002) Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 9, 1149–1159 [DOI] [PubMed] [Google Scholar]
- 32. Laughner E., Taghavi P., Chiles K., Mahon P. C., Semenza G. L. (2001) HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1α (HIF-1α) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol. Cell. Biol. 21, 3995–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Semenza G. L. (2011) Oxygen sensing, homeostasis, and disease. N. Engl. J. Med. 365, 537–547 [DOI] [PubMed] [Google Scholar]
- 34. Mei Y., Hahn A. A., Hu S., Yang X. (2011) The USP19 deubiquitinase regulates the stability of c-IAP1 and c-IAP2. J. Biol. Chem. 286, 35380–35387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hanna J., Hathaway N. A., Tone Y., Crosas B., Elsasser S., Kirkpatrick D. S., Leggett D. S., Gygi S. P., King R. W., Finley D. (2006) Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127, 99–111 [DOI] [PubMed] [Google Scholar]
- 36. Peth A., Besche H. C., Goldberg A. L. (2009) Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol. Cell 36, 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Loboda A., Jozkowicz A., Dulak J. (2010) HIF-1 and HIF-2 transcription factors: similar but not identical. Mol. Cells 29, 435–442 [DOI] [PubMed] [Google Scholar]
- 38. Wiesener M. S., Jürgensen J. S., Rosenberger C., Scholze C. K., Hörstrup J. H., Warnecke C., Mandriota S., Bechmann I., Frei U. A., Pugh C. W., Ratcliffe P. J., Bachmann S., Maxwell P. H., Eckardt K. U. (2003) Widespread hypoxia-inducible expression of HIF-2α in distinct cell populations of different organs. FASEB J. 17, 271–273 [DOI] [PubMed] [Google Scholar]
- 39. Hu C. J., Wang L. Y., Chodosh L. A., Keith B., Simon M. C. (2003) Differential roles of hypoxia-inducible factor 1α (HIF-1α) and HIF-2α in hypoxic gene regulation. Mol. Cell. Biol. 23, 9361–9374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Patel S. A., Simon M. C. (2008) Biology of hypoxia-inducible factor-2α in development and disease. Cell Death Differ. 15, 628–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu Y. V., Baek J. H., Zhang H., Diez R., Cole R. N., Semenza G. L. (2007) RACK1 competes with HSP90 for binding to HIF-1α and is required for O2-independent and HSP90 inhibitor-induced degradation of HIF-1α. Mol. Cell 25, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hubbi M. E., Luo W., Baek J. H., Semenza G. L. (2011) MCM proteins are negative regulators of hypoxia-inducible factor 1. Mol Cell 42, 700–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee Y. T., Jacob J., Michowski W., Nowotny M., Kuznicki J., Chazin W. J. (2004) Human Sgt1 binds HSP90 through the CHORD-Sgt1 domain and not the tetratricopeptide repeat domain. J. Biol. Chem. 279, 16511–16517 [DOI] [PubMed] [Google Scholar]
- 44. Barth S., Edlich F., Berchner-Pfannschmidt U., Gneuss S., Jahreis G., Hasgall P. A., Fandrey J., Wenger R. H., Camenisch G. (2009) Hypoxia-inducible factor prolyl-4-hydroxylase PHD2 protein abundance depends on integral membrane anchoring of FKBP38. J. Biol. Chem. 284, 23046–23058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barth S., Nesper J., Hasgall P. A., Wirthner R., Nytko K. J., Edlich F., Katschinski D. M., Stiehl D. P., Wenger R. H., Camenisch G. (2007) The peptidyl prolyl cis/trans-isomerase FKBP38 determines hypoxia-inducible transcription factor prolyl-4-hydroxylase PHD2 protein stability. Mol. Cell. Biol. 27, 3758–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tu B. P., Weissman J. S. (2004) Oxidative protein folding in eukaryotes: mechanisms and consequences. J. Cell Biol. 164, 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.