Background: The isoform importin α5 recognizes a subset of cargos such as phosphorylated STAT1 and the influenza virus polymerase subunit PB2.
Results: Nucleoporin Nup50 stabilizes the closed conformation of armadillo repeat 10 in importin α5.
Conclusion: We suggest that Nup50 primary function is to prevent cargo rebinding in preparation for recycling.
Significance: Nup50 modulates the import reaction by directly altering the three-dimensional structure of import α5.
Keywords: Influenza Virus, Nuclear Pore, Nuclear Transport, Signaling, X-ray Crystallography, Armadillo Repeat, PB2, Importin Alpha5, Nup50
Abstract
The human genome encodes six isoforms of importin α that show greater than 60% sequence similarity and remarkable substrate specificity. The isoform importin α5 can bind phosphorylated cargos such as STAT1 and Epstein-Barr Virus Nuclear Antigen 1, as well as the influenza virus polymerase subunit PB2. In this work, we have studied the interaction of the nucleoporin Nup50 with importin α5. We show that the first 47 residues of Nup50 bind to the C terminus of importin α5 like a “clip,” stabilizing the closed conformation of ARM 10. In vitro, Nup50 binds with high affinity either to empty importin α5 or to a preassembled complex of importin α5 bound to the C-terminal domain of the import cargo PB2, resulting in a trimeric complex. By contrast, PB2 can only bind with high affinity to importin α5 in the absence of Nup50. This suggests that Nup50 primary function may not be to actively displace the import cargo from importin α5 but rather to prevent cargo rebinding in preparation for recycling. This is the first evidence for a nucleoporin modulating the import reaction by directly altering the three-dimensional structure of an import adaptor.
Introduction
Trafficking of most cellular proteins between the nucleus and the cytoplasm is mediated by soluble transport factors known as karyopherins that shuttle constantly through the nuclear pore complex (NPC)2 (1). The human genome encodes at least 20 karyopherins related to importin β (also named β-karyopherins), which are classified as importins and exportins depending on their involvement in nuclear import or export (2), respectively. Cargos flagged for nuclear import usually expose a highly basic patch of residues known as the nuclear localization signal (NLS), which is exemplified by the SV40 large T-antigen NLS (126PKKKRKV132) (3). In the classical import pathway, the NLS is recognized and bound by the adaptor importin α, which heterodimerizes with the receptor importin β via an N-terminal importin β binding (IBB) domain (4). The import complex translocates through the NPC rapidly in a process that involves importin β binding to phenylalanine/glycine-containing nucleoporins lining the NPC (5). Binding of the small GTPase RanGTP to importin β dissociates it from phenylalanine/glycine-containing nucleoporins and breaks up the importin α/β heterodimer. Importin β complexed to RanGTP is then recycled back to the cytoplasm, where GTP is hydrolyzed to GDP. Consequently, Ran plays a pivotal role both in energizing and conferring directionality to nuclear transport (1).
β-Karyopherins are extraordinarily flexible molecules composed of 19–21 tandem HEAT repeats (6–8). Each HEAT repeat is comprised of two α-helices connected by a turn. Similarly, the adaptor importin α presents a repetitive structure of 10 consecutive armadillo (ARM) motifs (9). Each ARM consists of three α-helices (named H1-H2-H3) that stack together to form an extended NLS-binding surface (the “ARM core”), flexibly connected to the N-terminal IBB domain (4). The concave surface of the core of importin α contains two NLS-binding sites: a major site between ARM repeats 2–4 and a minor site between repeats 7 and 8 (see Fig. 1A). In all crystal structures of importin α1 bound to classical monopartite NLSs determined to date, the NLS is bound at the major NLS-binding site, and a partially occupied peptide is also seen at the minor binding site (10, 11). In contrast, the bipartite NLS of nucleoplasmin spans the entire NLS-binding groove, occupying both major and minor NLS-binding sites (11, 12). Other nonclassical NLSs, such as those found in the mitotic regulator TPX2 (13) or phospholipid scramblase 4 (14), bind largely or exclusively to the minor NLS-binding site on importin α1. The IBB domain of importin α can also bind the NLS-binding groove to function as an internal autoinhibitory signal (4). This ensures that only fully assembled import complexes (consisting of NLS-importin α/β) are imported and aids in their disassembly once in the nucleus (4, 9).
FIGURE 1.
Structure of the ΔIBB-importin α5 bound to Nup50 (residues 1–47). A, schematic diagram of importin α5 domain structure that consists of an N-terminal IBB domain, an ARM core, and a C-terminal tail. The major and minor NLS-binding sites as well as the Nup50-binding region and ARM 10 are indicated. B, ribbon representation of the human ΔIBB-importin α5 (in cyan) bound to residues 1–47 of Nup50 (in red) shown in sticks. The nucleoporin wraps around the C terminus of importin α making contacts with ARMs 4–10. C, magnified view of a region of the Nup50 refined model (residues 1–7) superimposed to a final 2Fo − Fc electron density map (contoured at 1.1 σ above noise).
The human genome encodes six isoforms of importin α that fall into three phylogenetically distinct groups: α1s, α2s, and α3s (9). All importin α isoforms contain 10 stacked ARMs and share greater than 60% sequence similarity (9). At the functional level, different importin α isoforms exhibit different substrate specificity while maintaining the ability to bind and import classical NLS substrates (15). A well characterized isoform is importin α5, which is involved in the nuclear import of the phosphorylated transcription factor STAT1 (16), the Epstein-Barr virus nuclear antigen 1 (17), and influenza virus RNA polymerase subunit PB2 (18, 19). A crystal structure of importin α5 in complex with the C-terminal core of influenza PB2 (residues 678–759; herein referred to as PB2) (18) revealed that PB2 occupies the entire binding groove of importin α5, including the major and minor NLS recognition sites. In this structure, the C-terminal helix of importin α5 ARM 10 is unpacked and mediates domain swap dimer formation in the crystal (18). This conformation of ARM 10 reflects the unusual flexibility of this isoform, which represents an important determinant both for cargo specificity and high affinity binding (20).
Nup50 (also known as Npap60 or in yeast Nup2p) is a mobile (21) nucleoporin localized at steady state primarily in the nucleoplasmic fibrils of the NPC (22, 23). It functions as a cofactor for the importin α1-β nuclear import complex and was hypothesized to play a role in cargo disassembly (24–26). The N terminus of Nup50 (residues 1–109) binds the C terminus of importin α1 at two distinct sites, corresponding to the minor NLS-binding site and ARM 10 (27). Despite the high level of specify for importin α1, disagreement exists in the literature as to the functional role of this interaction. Lindsay et al. (28) proposed that Nup50 functions as a tri-stable switch that stimulates nuclear protein import through the classical import pathway. This function was shown to be dependent on Nup50 simultaneous binding to importin α1 and an NLS cargo during nuclear import (28). In contrast, Matsuura and Stewart (27) found that binding of Nup50 and an NLS (both monopartite and bipartite) is mutually exclusive, and thus the main role of Nup50 may be related to displacing import complexes and recycling importin α to the cytoplasm. This idea is corroborated by the evidence that some of the sites bound by Nup50 in importin α1 are also critical for the assembly of CAS (27, 29), the export receptor that, in concert with RanGTP, exports importin α back to the cytoplasm. However, the putative role of Nup50 in dissociating the import complex was recently challenged by single-molecule observations in permeabilized cells that revealed Nup50 is insufficient to dissociate an importin α-NLS cargo complex on the time scale of nuclear transport (30). Finally, Nup50 was shown to be expressed in two splicing isoforms that play opposite roles in regulating the import reaction (31). Although the longer isoform (residues 1–469) promotes release of NLS cargo from importin α, the shorter isoform (residues 29–469), found only in humans, accelerates nuclear import of classical NLS cargos. In the present study, we have determined the structure of the isoform importin α5 in complex with the Nup50 and characterized how this nucleoporin modulates importin α5 binding to the representative import cargo PB2.
EXPERIMENTAL PROCEDURES
Molecular Cloning and Recombinant Proteins
Cloning of ΔIBB-importin α5 (residues 66–538) and PB2 (residues 678–759) were previously described (18). A FLAG-tagged version of ΔIBB-importin α5 was constructed by introducing the FLAG epitope (DYKDDDDK) upstream of residue 66 of importin α5 using long PCR (construct FLAG-ΔIBB-importin α5). His-MBP-PB2 was constructed by inserting the fragment of the PB2 gene encoding residues 678–759 into an engineered pET28a vector that also contains the MBP gene downstream of the His6 tag. A synthetic gene encoding residues 1–109 of human Nup50 was inserted between the BamHI and NotI sites of a pGEX-6p-1 vector (GE Healthcare). All of the plasmids were verified by DNA sequencing. Expression and purification of ΔIBB-importin α5 (20), PB2, MBP-PB2 (32), and a complex of ΔIBB-importin α5 bound to PB2 (18) were performed as previously described. GST-Nup50 was expressed overnight at 20 °C in the Escherichia coli strain BL21; lysed in 100 mm sodium phosphate, pH 7.4, 0.6 m NaCl, and 5 mm β-mercaptoethanol; and purified on glutathione-agarose beads (Pierce). Either the GST-Nup50 was eluted with 15 mm reduced glutathione, or PreScission protease was added to cleave Nup50 from the GST. Both were further purified by gel filtration chromatography on a Superdex 75 column (GE Healthcare) equilibrated in GF buffer, consisting of 1× PBS (3.2 mm Na2HPO4, 0.5 mm KH2PO4, 135 mm NaCl, pH 7.4) and 5 mm β-mercaptoethanol. To form homogeneous complexes of ΔIBB-importin α5 bound to Nup50, two E. coli lysates expressing GST-Nup50 and His-ΔIBB-importin α5 were mixed in ratio 1:2 and immobilized on glutathione-agarose beads (Pierce) as described (27). After washing the beads in GF buffer plus 0.5% Tween 20, PreScission protease was added to the beads to cleave the importin α5-Nup50 complex from GST. The complex was further purified by gel filtration chromatography on a Superdex 75 column (GE Healthcare) pre-equilibrated in GF buffer.
Crystallization and Structure Determination
The ΔIBB-importin α5-Nup50 (residues 1–109) complex was concentrated by ultrafiltration to 7.5 mg/ml and crystallized in the presence of 24% PEG 3350, 100 mm BisTris, pH 6.5, using the hanging drop vapor diffusion technique. The drop was equilibrated at 20 °C, and crystals appeared within 7 days. 27.5% ethylene glycol was added as cryo-protectant before flash-freezing in a nitrogen stream at −170 °C. X-ray data were collected at NSLS Beamline X25 and X6A on ADSC Quanta CCD detectors. The best crystal resulted in a complete data set to 2.7 Å resolution (supplemental Table SI). All data were indexed, integrated, and scaled using HKL-2000 (33). Crystals of ΔIBB-importin α5-Nup50 complex belong to space group P212121 with two complexes in the asymmetric unit. The structure was determined by molecular replacement using PHASER (34) and the structure ΔIBB-importin α5 as a search model (Protein Data Bank code 2JDQ). The initial solution was refined by rigid body refinement, simulated annealing, and isotropic B-factor in PHENIX (35). Nup50 was built in Fo − Fc electron density difference maps using the program Coot (36) followed by positional refinement with PHENIX (37). Further cycles of positional and isotropic B-factor refinement with distinct TLS domains reduced the Rwork/Rfree to ∼21.5/25.5% (supplemental Table SI). The final model includes residues 82–508 for ΔIBB-importin α5 in chain A and B, residues 1–47 of Nup50 in chain C, residues 1–46 in chain D, and 130 water molecules. Four residues from the expression vector (Pro-Leu-Gly-Ser) upstream of Nup50 residue 1 were also included in the final model. The average refined B-factors for ΔIBB-importin α5, Nup50, and solvent are ∼67.7, 102.8, and 41.6 Å2, respectively. The final model has r.m.s.d. for bond and angles of 0.005 Å and 0.795°, respectively, with 92.1% of residues in the most favored regions of the Ramachandran plot and no outliers in disallowed regions (supplemental Table SI). The structure was analyzed using PDBsum (38) and PDBePISA (39); structural figures were made using PyMOL (Delano Scientific). Coordinates for the ΔIBB-importin α5-Nup50 complex have been deposited in the Protein Data Bank with accession code 3TJ3.
Biochemical Techniques
EMSA on agarose was performed as described (20, 41). In the assay, 20 μg of ΔIBB-importin α5, 24 μg of a gel filtration purified complex of ΔIBB-importin α5 bound to either PB2 or Nup50 were separated on a 1.5% agarose gel at room temperature for 1 h. For the Nup50 titration, 24 μg of a gel filtration-purified ΔIBB-importin α5-PB2 complex was incubated with a 0.25–10-fold molar excess of Nup50, for 30 min on ice prior to electrophoretic separation on agarose gel. The pulldown assay was performed on glutathione-agarose beads (Pierce) coupled to GST-Nup50 (residues1–109) as previously described (41). 100 μg of ΔIBB-importin α5 (37 μm in 50 μl) was incubated with 25 μl of beads for 20 min at room temperature. For PB2 displacement, a 4-fold molar excess of PB2 (151 μm in 50 μl) was added to the beads after the addition of ΔIBB-importin α5. Alternately, a molar equivalent (120 μg) of importin α5 precomplexed to PB2 was added to beads and incubated 20 min at room temperature. The beads were washed three times at 20 °C in PBS. After washing, all of the samples were dissolved in SDS loading buffer and analyzed on a 12.5% SDS-PAGE.
Affinity Measurements by Flow Fluorimetry
Solution phase equilibrium dissociation constants (KD) were determined using a KinExA 3000 flow fluorimeter (Sapidyne Instruments). The instrument was configured to sample the free (unbound) concentration of ΔIBB-importin α5 in equilibrated solutions of ΔIBB-importin α5, Nup50, and/or PB2. Sampling occurred in fluorescence flow cells containing a small volume of azlactone beads (Pierce) covalently coupled to either GST-Nup50 or MBP-PB2. The beads were prepared by incubating 200 μg of GST-Nup50 or MBP-PB2 in 500 mm sodium bicarbonate, pH 9.5, at 4 °C overnight; unreacted groups were subsequently blocked using BSA (1 mg/ml) in 1 m Tris, pH 8.0, at 20 °C for 30 min. Equilibrated reaction mixtures were passed through the flow cell, and the amount of FLAG-tagged ΔIBB-importin α5 captured on the beads was detected using a FITC-conjugated anti-FLAG (DYKDDDDK) mouse monoclonal antibody (Genscript, catalogue number A01632). The change in fluorescence signal at 518 nm was linearly proportional to the unbound ΔIBB-importin α5 concentration in solution. All of the reaction mixtures were prepared in a buffer of 10 mm Tris, pH 8.0, 150 mm NaCl, 2 mm DTT, 0.1 mg/ml BSA, and 1 mm PMSF and allowed to come to equilibrium (empirically determined) at 20 °C. To determine the KD for the interaction between ΔIBB-importin α5 and Nup50, Nup50 was titrated into solutions of constant ΔIBB-importin α5 concentration (10, 1, or 0.1 nm), and the reaction mixtures were passed over GST-Nup50-conjugated beads. To determine the KD for the interaction between ΔIBB-importin α5 and PB2, PB2 was titrated into solutions of constant ΔIBB-importin α5 concentration (10, 1, or 0.2 nm), and the reaction mixtures were passed over MBP-PB2-conjugated beads. To determine the effect of PB2 on the affinity of ΔIBB-importin α5 for Nup50, Nup50 was titrated into premixed solutions of 10 nm PB2 and a constant ΔIBB-importin α5 concentration (either 10 or 2 nm), and the reaction mixtures were passed over GST-Nup50-conjugated beads. The complex of ΔIBB-importin α5 and Nup50 failed to bind appreciably to MBP-PB2-conjugated beads even at 80 nm; consequently, the effect of Nup50 on the affinity of ΔIBB-importin α5 for PB2 could not be directly measured. The samples were tested in duplicate, and all of the titrations were performed at least twice. The fluorescence signals were globally fit to a general bimolecular interaction model (manufacturer's software).
RESULTS
Structure of Importin α5 Bound to Nup50
We used x-ray crystallography to determine the molecular basis of the interaction between human importin α5 and Nup50. Crystals of ΔIBB-importin α5 bound to Nup50 (residues 1–109) diffracted to 2.7 Å resolution and have two complexes in the asymmetric unit. Inspection of Fo − Fc electron density difference maps revealed an extended tubular density for Nup50 spanning the C terminus of importin α5 (Fig. 1). Residues 1–47 of Nup50 were modeled in this density, and the C terminus of importin α5 (ARM 10) was entirely rebuilt as compared with the search model (importin α5 solved in complex with PB2 (18)). The final model of ΔIBB-importin α5 bound to Nup50 has been refined to Rwork/Rfree of 21.5 and 25.5% (supplemental Table SI). In the crystal structure, importin α5 reveals a characteristic banana shape formed by 10 ARM repeats stacked against one another (10, 42) (Fig. 1, A and B). Importin α5 structure superimposes well to importin α2 (the mouse homologue of importin α1) previously crystallized with an equivalent fragment of the mouse Nup50 (27) and to yeast Kap60p (10) (r.m.s.d. of 1.85 and 1.22 Å, respectively) but shows dramatic deviations in the C-terminal ARM 10 as compared with the crystallographically observed cargo-bound conformation of importin α5 (18) (r.m.s.d., 2.7 Å). Furthermore, the backbone of Nup50 adopts a fully extended conformation, which binds along the surface of importin α5 like a “clip,” burying ∼4,320 Å2 of solvent-accessible surface area (Fig. 1B). Nup50 lacks any tertiary structure, and a two-turn α-helix between residues 31–37 is the only secondary structure element present in this fragment of nucleoporin. Consistently, the B-factor for Nup50 is higher than in importin α5 (102 versus 67 Å2) and varies throughout its structure with its highest value in the acidic patch between residues 18 and 25, which is poorly resolved.
Structural Determinants for Binding to Nup50
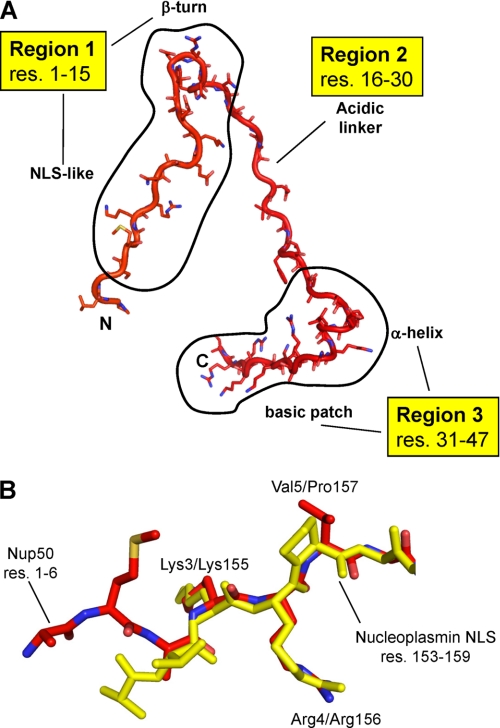
Three molecular determinants contribute to the binding of Nup50 to ARMs 4–10 of importin α5 (Fig. 2A) and account for a total of 22 hydrogen bonds and 273 nonbonded contacts. First, residues 1–15 bind along the minor NLS-binding site, in a direction antiparallel to that of importin α5. The first portion of this moiety, between residues 1 and 6 (1MAKRNA6) is structurally superimposable to the smaller NLS box of the nucleoplasmin bipartite NLS (11) and to the hPLSCR4-NLS (Fig. 2B), which binds exclusively to the minor NLS site (14). This region of Nup50 also largely overlaps with residues 31–37 of Kap60p IBB domain visualized in the context of an export complex, with Cse1p and RanGTP (29). Furthermore, residues 11–15 of Nup50 (11TDRNW15) form a β-turn that packs against the side of importin α5 and inverts the direction of the Nup50 main chain, from being antiparallel to parallel with respect to importin α5. Second, residues 16–30 of Nup50 extend toward the importin α5 C terminus lining on the outer surface of ARM 7–10 (Fig. 1B). This region contains a cluster of five acidic residues (18EDEAEE23), of which only two, Glu20 and Glu22, directly contact importin α5. The other residues are mainly solvent-exposed and have poor electron density. In addition, Met24 and Phe27 of Nup50 make hydrophobic contacts with importin α5 on the outer surface of ARM 9. Third, residues 31–46 of Nup50 account for most of the interactions with ARM 10 of importin α5. Notably, the first part of this moiety, residues 31SEEVMK36, forms two turns of an α-helix tightly packed against the intrarepeat loop connecting ARM 10 helix H2-H3 (Figs. 1B and 2A). Downstream of this short helix, the Nup50 main chain continues in a basic cluster (residues 36KNRAIKKAKRRN47) that stabilizes the acidic surface of ARM 10 helix H3. Finally, residues 48–109 were not visible in the crystal structure, although they were present in the crystal. This region contains 15 glycines and is likely highly flexible. Overall, the binding contacts for Nup50 seen in complex with importin α5 are very similar to those reported for Nup50 bound to mouse importin α2 (27), and the r.m.s.d. between the Nup50s determined in the two crystal structures is ∼0.91 Å.
FIGURE 2.
Extended structure of Nup50. A, regions of Nup50 responsible for interactions with importin α5. B, Nup50 occupies the minor NLS-binding site of importin α5: structural superimposition of residues 1–7 of Nup50 (in red) to residues 153–159 of nucleoplasmin NLS (in yellow) (Protein Data Bank 1EJY). The main determinants that bind to importin α are Lys3/Lys155, Arg4/Arg156, and Val5/Pro157.
Nup50 Stabilizes Closed Conformation of ARM 10 in Importin α5
Perhaps the most striking feature of the importin α5 structure described in this paper lies in the conformation of ARM 10 (Fig. 1B). In our structure, this repeat adopts a canonical, “closed” conformation, with the three ARM helices H1, H2, and H3 stacked on top of each other to generate a hydrophobic core. In contrast, the structure of importin α5 visualized crystallographically in complex with PB2 shows that ARM 10 unfolds as a result of cargo binding (18). Here helices H2 and H3 project by ∼40° and 90°, respectively, from the corresponding position seen in our structure (Fig. 3A). Unfolding of ARM 10 tertiary structure likely stabilizes cargo binding and results in several specific contacts between helix H3 and PB2 (supplemental Fig. S1) (18). Although the overall r.m.s.d. of importin α5 in complex with PB2 and Nup50 is only ∼1.3 Å, the conformation of ARM 10 is drastically different in the two complexes: residue 507 of importin α5 bound to Nup50 is over 40 Å away from its corresponding position in the PB2-bound state (Fig. 3A). Previous biochemical studies have revealed that Tyr476 is critical for ARM 10 swinging (20). This residue is located at the end of the first helix of ARM 10 (helix H1) where it functions like a “Tyr finger” (Fig. 3B). The bulky aromatic side chain projects at the interface of ARM 10 helices H1-H2-H3 to destabilize the interhelical stacking of the three ARM helices, thereby allowing the unfolding of ARM 10 and the extension of helix H3. In our structure, Nup50 stabilizes a closed conformation of ARM 10 that is identical to that seen in importin α1/Kap60p (42, 43), where helices H1-H2-H3 stack onto each other to generate a flat surface. To accommodate the closed conformation of ARM 10, Tyr476 swings 180° in response to Nup50 binding, thus allowing stacking of helix H1-H3 as seen in importin α1, where this residue is replaced by a Gly (20) (Fig. 3B). Thus, the extended conformation of Nup50 that wraps around the C terminus of importin α5 is likely an important structural determinant to stabilize the closed conformation of ARM 10.
FIGURE 3.
Flexibility of ARM 10 of importin α5. A, left panel, the structure of ΔIBB-importin α5 in complex with PB2 (Protein Data Bank 2JDQ) (in blue and yellow, respectively) is superimposed to that of ΔIBB-importin α5 bound to Nup50 (in cyan and red, respectively). Right panel, magnified view of ARM 8–10. Helix H3 in ARM 10 points in opposite directions in the two conformations of importin α5. B, left panel, superimposition of ΔIBB-importin α5 with PB2 and Nup50 as in A but rotated by 180°. Right panel, magnified view of Tyr476 that swings by 180° in response to Nup50 binding, thereby allowing stacking of ARM 10 helices H1–H3.
Importin α5 Can Form a Trimeric Complex with PB2 and Nup50
Both Nup50 and PB2 (18) make wide surface contacts with importin α5, likely essential for nuclear import. The partial overlap between PB2 and Nup50 binding sites in importin α5 (Fig. 3A), as well as the distinct conformation of importin α5 ARM 10 in the two bound conformations led us to investigate whether binding of these two proteins to importin α5 is mutually exclusive. This hypothesis was tested using three independent binding techniques. First, we carried out a pulldown assay on glutathione-agarose beads coupled to GST-Nup50 using ∼40 μm ΔIBB-importin α5 and 150 μm PB2. As shown in Fig. 4A (lanes 5 and 6), GST-Nup50 beads were able to bind ΔIBB-importin α5, whereas no interaction was seen for free PB2. Adding either purified ΔIBB-importin α5 and then PB2 or a gel filtration-purified complex of these two proteins (as in Fig. 4A, lane 4) resulted in three species bound to beads, corresponding to ΔIBB-importin α5, GST-Nup50, and PB2 (Fig. 4A, lanes 7 and 8, respectively). Thus, binding of Nup50 and PB2 to importin α5 is not mutually exclusive, and ΔIBB-importin α5 can form a trimeric complex with PB2 and Nup50. To further explore this hypothesis, we used a native EMSA on agarose gel; in this experiment, increasing concentrations of purified Nup50 were titrated into a preformed importin α5-PB2 complex (Fig. 4B, lanes 3–9). This yielded a trimeric importin α5-Nup50-PB2 complex that migrated more slowly than the importin α5-Nup50 complex (Fig. 4B, lanes 2) and was significantly slowed compared with the importin α5-PB2 complex (Fig. 4B, lane 3). The putative trimeric complex assembled when 1–10 molar equivalents of Nup50 were added to the preformed importin α5-PB2 complex (at ∼20 μm concentration) (Fig. 4B, lanes 6–9). Finally, we characterized the trimeric ΔIBB-importin α5-Nup50-PB2 complex in solution, using analytical size exclusion chromatography on a Superose 12 column. At physiological ionic strength, unliganded ΔIBB-importin α5 migrated as an ∼85-kDa species, slightly larger than the true molecular mass (∼55 kDa), but consistent with the elongated monomer seen in the crystal structure (supplemental Fig. S2) (a similar, larger than expected migration was also observed for importin α1 (44)). A trimeric complex formed using an excess of Nup50 or PB2 added to a gel filtration-purified complex of ΔIBB-importin α5 bound to Nup50 or PB2 contained all three components, although not in exact equimolar ratio (supplemental Fig. S2). Thus, on beads, in agarose and in solution, at concentrations in the low micromolar range ΔIBB-importin α5 can form a tertiary complex with Nup50 and PB2.
FIGURE 4.
Nup50 and PB2 can simultaneously bind to importin α5. A, pulldown assay on glutathione-agarose beads coupled to GST-Nup50 (1–109) (lane 1). GST-Nup50 efficiently pulled down free ΔIBB-importin α5 (lane 6) but not PB2 (lane 5). Adding to the GST-Nup50 beads, either a preformed ΔIBB-importin α5-PB2 complex (lane 8) or free ΔIBB-importin α5 followed by an excess of PB2 (lane 7) results in formation of a trimeric complex. B, EMSA on native agarose gel. Free ΔIBB-importin α5 is in lane 1, and preformed complexes of ΔIBB-importin α5 bound to Nup50 or PB2 are in lanes 2 and 3, respectively. The addition of increasing quantities of Nup50 (from 0.25- to 10-fold molar excess) to a preformed ΔIBB-importin α5-PB2 complex leads to a trimeric species that is retarded as compared with ΔIBB-importin α5 bound to Nup50 (lanes 2) or PB2 (lane 3). Single and double asterisks indicate the position of the dimeric ΔIBB-importin α5-Nup50 and trimeric ΔIBB-importin α5-Nup50-PB2 complex, respectively. The difference in migration between these two species is also emphasized by dashed horizontal lines. M.W., molecular mass.
PB2 Reduces the Binding Affinity of Nup50 for Importin α5
Next, we sought to determine how the binding affinity of importin α5 for Nup50 and PB2 is altered by either factor. To address this question, we used a KinExA 3000 flow fluorimeter. This binding technique uses a solid phase immobilized molecule to probe for free concentration of one interacting partner after allowing sufficient time to reach equilibrium (45). In our experimental set-up (illustrated in supplemental Fig. S3A), equilibrated mixtures of ΔIBB-importin α5 (that also included a short N-terminal epitope FLAG epitope) and Nup50 (residues 1–109) were passed through a fluorescence flow cell containing a small quantity (∼4 μl) of GST-Nup50-conjugated beads. As reaction mixtures were passed through the optical flow cell of the instrument, a small fraction of the total unbound ΔIBB-importin α5 was captured by beads coated with GST-Nup50. The exact amount of captured ΔIBB-importin α5 was quantified using an anti-FLAG-FITC-conjugated mouse monoclonal antibody, and the resulting increase in bead fluorescence (Δf) was directly proportional to the concentration of unbound ΔIBB-importin α5 in the reaction mixture (supplemental Fig. S3B). From full titrations of Nup50 at several ΔIBB-importin α5 concentrations in the low nanomolar to high picomolar range, we have determined the KD between ΔIBB-importin α5 and Nup50 to be <2 pm (Fig. 5A).
FIGURE 5.
Binding affinity of importin α5 for PB2 and Nup50 determined by flow fluorimetry. A, a titration of Nup50 was performed for a range of concentrations of free or PB2-bound ΔIBB-importin α5. The KD values were determined by fitting each set of data to a bimolecular equilibrium binding model (solid lines). Titrations of Nup50 against 100 pm empty ΔIBB-importin α5 gave a KD < 2 pm (left curve); titrations of Nup50 against 2 nm importin α5 prebound to an excess of PB2 resulted in a KD of 815 ± 29 pm (right curve). B, titrations of PB2 against 200 pm ΔIBB-importin α5 gave a KD of 53 ± 2 pm.
Interestingly, when the same experiment was repeated using ΔIBB-importin α5 precomplexed with 10 nm PB2, ΔIBB-importin α5 could still be efficiently captured by the Nup50-conjugated beads in the KinExA flow cell. At this concentration of PB2 (>100-fold above its KD for ΔIBB-importin α5, see below and Fig. 5B), ΔIBB-importin α5 was more than 99% bound to PB2. By titrating Nup50 into pre-equilibrated solutions of the ΔIBB-importin α5-PB2 complex, we determined the apparent KD for Nup50 to be 0.815 ± 0.029 nm (Fig. 5A). Assuming that importin α5 can simultaneously bind Nup50 and PB2 at the concentration used in the KinExA assay (and as also suggested by pulldown, native gel and size exclusion chromatography assays), these data suggest that the presence of bound cargo reduces the binding affinity of importin α5 for Nup50 by >400-fold. Although this affinity is lower than that measured in the absence of PB2, the importin α5-PB2 complex is still captured by Nup50 with robust, nanomolar binding affinity.
In a parallel experiment, we immobilized MBP-PB2 on beads and performed full titrations of PB2 with ΔIBB-importin α5 at concentrations in the low nanomolar to high picomolar range. The results indicated that ΔIBB-importin α5 binds MBP-PB2 beads with a KD = 53 ± 2 pm (Fig. 5B). Interestingly, when a gel filtration-purified complex of ΔIBB-importin α5 prebound to Nup50 (between 2 and 80 nm) was probed for binding of PB2-coated beads, it failed to produce sufficient signal when passed over beads. This lack of capture implies that prebound Nup50 efficiently prevents binding of PB2 to ΔIBB-importin α5. In contrast to the ΔIBB-importin α5-PB2 complex that retains an efficient interaction with Nup50-beads, the ΔIBB-importin α5-Nup50 complex appears to adopt a structure that prevents association with PB2. This argues that at the concentration used in the KinExA assay, the tripartite interaction behaves in a simple competitive manner.
DISCUSSION
Structural Plasticity of ARM 10
α-Karyopherins are composed of ARM repeats and are thought to be predominately rigid. The ARM core generates a cargo-binding surface ideal to function as a static NLS-binding groove (46). This is very different from the HEAT-repeated architecture of β-karyopherins that form superhelical solenoids of remarkable structural flexibility (7, 8, 44). The results presented in this paper, together with previous structural (18) and biochemical (20) reports on importin α5 (Fig. 6), provide compelling evidence that ARM 10 of import α5 functions as a reversible mobile arm. Helix H3 in ARM 10 swings upon binding to the import cargo PB2 and folds back on itself in response to Nup50 binding (Fig. 6). Tyr476 is likely the key residue in importin α5 controlling this switch (20). In the presence of cargo, this Tyr finger folds inward inside the hydrophobic core of ARM 10 (Fig. 3B), destabilizing the interhelical stacking of ARM 10 helices and hence promoting unfolding of ARM 10 (18). Binding of importin α5 to Nup50 causes Tyr476 to swing 180° outward to promote a folded conformation of ARM 10, which is identical to that seen in importin α1/2 (42), where this residue is replaced by a Gly. What is the function of this structural switch in ARM 10 of importin α5? Reanalysis of the importin α5-PB2 crystal structure (18) suggests that the extended conformation of ARM 10 helix H3 stabilizes PB2 binding by forming at least six specific electrostatic contacts between the two proteins, namely, PB2-Lys699 with α5-Gln494, PB2-Arg703 with α5-Thr506/α5-Glu507/α5-Glu509, PB2-Tyr704 with α5-Glu509, and PB2-Lys721 with α5-Glu509 (supplemental Fig. S1). Conversely, the closed conformation of ARM 10 would result in complete loss of these contacts, which likely weakens (but does not fully disrupt) the import complex. Interestingly, Gabriel et al. (47) recently determined that efficient replication of mammalian influenza virus requires the isoform importin α7. This isoform is 81% identical to importin α5 and also presents a Tyr at position 476 (48), supporting the hypothesis that this Tyr finger plays a role in cargo binding by modulating the interconversion of ARM 10 between a closed to an open conformation (Fig. 6). However, PB2 may not be the most representative cargo for importin α5, because it also binds (and can be imported) by other importin α isoforms (32, 47). The nonclassical import cargo STAT1, instead, is strictly dependent on the extended conformation of ARM 10. Phosphorylation at Tyr701 triggers a conformational change in STAT1 that exposes a nonclassical NLS, which binds specifically to import α5 but has negligible affinity for importin α1 (20). Mutagenesis studies revealed that replacing Tyr476 in importin α5 with Gly (as in importin α1) or swapping the C terminus of importin α5 with equivalent residues of importin α1 dramatically disrupts binding to STAT1 (20). Thus, Tyr476 and the structural switch of ARM 10 in importin α5 (as well as in other importin α5-like isoforms like importin α6 and α7 (48)) likely serve the function of stabilizing cargo binding and promoting specificity toward certain specialized cargos.
FIGURE 6.
Model of involvement of Nup50 in importin α5 import cycle. Schematic diagram of the binding interactions of importin α5, Nup50, and PB2. Free importin α5 oscillates between an open (α5O) and closed (α5C) conformation of ARM 10; binding of PB2 or Nup50 to importin α5 stabilizes a given conformation. In all diagrams, the IBB domain of importin α5 is dashed, and for simplicity, importin β is not drawn.
Role of Nup50 in Cargo Displacement and Importin α5 Recycling
Several reports have pointed out a role of Nup50 (or its yeast homologue Nup2p) in destabilizing the importin α1/NLS cargo interaction (25, 27, 31, 43, 49, 50). In this study, we have determined that Nup50 binds importin α5 and stabilizes the closed conformation of ARM 10. Association of Nup50 with importin α5 is compatible with binding to PB2. At concentrations in the range of 5–40 μm (as used in all experiments except for KinExA), a trimeric species formed by importin α5, Nup50, and PB2 can be stably assembled in solution and is sufficiently long-lived to be visualized using conventional biochemical techniques such pulldown assay, native gel electrophoresis, and size exclusion chromatography (Fig. 4 and supplemental Fig. S2). However, the physiological concentration for these proteins is likely to be lower in vivo. For instance, the cellular concentration of all importin α isoforms is estimated at 1 μm (51), whereas the concentration of Nup50 in the nucleus is ∼130 nm (based on 32 copies/NPC (21) and ∼2,770 NPC/cell (52) and a nuclear volume of 1,130 μm3 (52)), and PB2 concentration is likely in the nanomolar/picomolar range (and stoichiometric to that of importin α5 inside the NPC). This range of concentration was explored in the KinExA assay, where we determined that the presence of 10 nm PB2 decreases the KD of importin α5 for Nup50 by >400-fold, likely because of formation of a trimeric complex. Although decreased, the binding affinity of the importin α5-PB2 complex for Nup50 remains very high (KD = ∼0.85 nm), which is likely incompatible with free dissociation of cargo. Furthermore, negligible interaction with the PB2 immobilized on beads was observed when Nup50 was prebound to importin α5, although this may be due to a kinetic effect during the attempted capture of the importin α5-Nup50 complex rather than a lack of trimeric complex formation. These results contrast with the report that, in vitro, Nup50 can actively displace a chimeric cargo consisting of GST-NLS from importin α2 (27). Instead, our observations are consistent with previous reports that a trimeric complex of importin α1, NLS cargo, and Nup50, or its yeast homologue Nup2p, can be isolated from both mammalian (28) and yeast (40, 49) extracts. Furthermore, Nup50 does not bind to the major NLS-binding site of importin α5 or α2 (27), which would be the logical target for a factor with cargo disassembling activity. Together, these direct and indirect observations argue against the idea that the primary function of Nup50 is to actively displace import cargos from importin α5 (Fig. 6, function a). In agreement with this idea, single molecule analysis of importin α/cargo complex dissociation at the NPC found that Nup50 alone is insufficient to promote importin α1/cargo complex dissociation on the time scale of transport (30). Instead, in vivo cargo displacement is mainly exerted by the combined action of CAS and RanGTP (30). RanGTP dissociates the importin α/β complex, thereby freeing up the IBB domain that functions as a potent, intramolecular binder of major (42) and minor (29) NLS-binding sites. In turn, CAS in complex with RanGTP loads empty importin α to recycle the adaptor into the cytoplasm. Given the exceedingly high binding affinity of Nup50 for importin α5 (KD < 2 pm), we propose that Nup50 may have a preferential role in binding empty, as opposed to cargo-bound importin α5. In this scenario, Nup50 would “capture” importin α5 after RanGTP/IBB-mediated displacement of cargo to prevent its rebinding to importin α5 (Fig. 6, function b). Similarly, Nup50 could bind directly to empty importin α5 that has escaped the cytoplasm in the absence of cargo and avoid futile import of unliganded adaptors (Fig. 6, function c).
Nup50 was proposed to serve as a scaffold that binds both the importin α1-NLS cargo and CAS-RanGTP complex, possibly facilitating cargo dissociation by tethering the two complexes together (27). A similar function can be hypothesized for importin α5, with the noticeable difference that in this isoform the conformations competent for binding to cargo (α5O) and CAS (α5C) are likely distinct (Fig. 6). Like Nup50, in fact, CAS binds ARM 10 of Kap60p in a closed conformation (29), suggesting that the primary role of Nup50 may be to function in concert with CAS to fold back ARM 10 in preparation for recycling (Fig. 6). However, if the affinity of Nup50 for importin α5 is so high, how can the CAS-importin α5-RanGTP export complex assemble? The answer to this question likely lies in the modular nature of Nup50 interaction complexes. Nup50 could “transfer” importin α5 in a closed conformation to CAS in a two-step mechanisms. First, the IBB domain (which is stabilized by CAS in an autoinhibited conformation (29)) could compete off the first portion of Nup50 (residues 1–11), weakening the overall affinity for importin α5; this is plausible, because the IBB presents more binding determinants in the minor binding site (29) than Nup50. Second, CAS may bind the C terminus ARM 10 of importin α5, releasing the Nup50 and allowing for re-entry into the cytoplasm. In conclusion, the results presented in this paper expand our understanding of the isoform importin α5 and suggest a role for Nup50 in promoting the disassembly of the importin α5 import complex indirectly by capturing empty importin α5 and preventing cargo rebinding.
Supplementary Material
Acknowledgments
We thank John Pascal and Lan Ho at Thomas Jefferson University for help in data collection and technical assistance and Florence Baudin at EMBL for the importin α5 expression plasmid. We are grateful to Vivian Stojanoff and staff at NSLS Beamlines X6A and X25 for beamtime and beamline assistance. KinExA data were collected at the Kimmel Cancer Center X-ray Crystallography and Molecular Characterization shared resource facility at Thomas Jefferson University.
This work was supported, in whole or in part, by National Institutes of Health Grant GM074846 (to G. C.).

This article contains supplemental text, Table SI, and Figs. S1–S3.
- NPC
- nuclear pore complex
- NLS
- nuclear localization signal
- ARM
- armadillo
- IBB
- importin β-binding
- Nup
- nucleoporin
- MBP
- maltose-binding protein
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Stewart M. (2007) Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8, 195–208 [DOI] [PubMed] [Google Scholar]
- 2. Mosammaparast N., Pemberton L. F. (2004) Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14, 547–556 [DOI] [PubMed] [Google Scholar]
- 3. Marfori M., Mynott A., Ellis J. J., Mehdi A. M., Saunders N. F., Curmi P. M., Forwood J. K., Bodén M., Kobe B. (2011) Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta 1813, 1562–1577 [DOI] [PubMed] [Google Scholar]
- 4. Lott K., Cingolani G. (2011) The importin β binding domain as a master regulator of nucleocytoplasmic transport. Biochim. Biophys. Acta 1813, 1578–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wente S. R., Rout M. P. (2010) The nuclear pore complex and nuclear transport. Cold Spring Harb. Perspect. Biol. 2, a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cingolani G., Petosa C., Weis K., Müller C. W. (1999) Structure of importin-β bound to the IBB domain of importin-α. Nature 399, 221–229 [DOI] [PubMed] [Google Scholar]
- 7. Conti E., Müller C. W., Stewart M. (2006) Karyopherin flexibility in nucleocytoplasmic transport. Curr. Opin. Struct. Biol. 16, 237–244 [DOI] [PubMed] [Google Scholar]
- 8. Forwood J. K., Lange A., Zachariae U., Marfori M., Preast C., Grubmüller H., Stewart M., Corbett A. H., Kobe B. (2010) Quantitative structural analysis of importin-β flexibility: paradigm for solenoid protein structures. Structure 18, 1171–1183 [DOI] [PubMed] [Google Scholar]
- 9. Goldfarb D. S., Corbett A. H., Mason D. A., Harreman M. T., Adam S. A. (2004) Importin α: a multipurpose nuclear-transport receptor. Trends Cell Biol. 14, 505–514 [DOI] [PubMed] [Google Scholar]
- 10. Conti E., Uy M., Leighton L., Blobel G., Kuriyan J. (1998) Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell 94, 193–204 [DOI] [PubMed] [Google Scholar]
- 11. Fontes M. R., Teh T., Kobe B. (2000) Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J. Mol. Biol. 297, 1183–1194 [DOI] [PubMed] [Google Scholar]
- 12. Conti E., Kuriyan J. (2000) Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin α. Structure Fold Des 8, 329–338 [DOI] [PubMed] [Google Scholar]
- 13. Giesecke A., Stewart M. (2010) Novel binding of the mitotic regulator TPX2 (target protein for Xenopus kinesin-like protein 2) to importin-α. J. Biol. Chem. 285, 17628–17635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lott K., Bhardwaj A., Sims P. J., Cingolani G. (2011) A minimal nuclear localization signal (NLS) in human phospholipid scramblase 4 that binds only the minor NLS-binding site of importin α1. J. Biol. Chem. 286, 28160–28169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Köhler M., Speck C., Christiansen M., Bischoff F. R., Prehn S., Haller H., Görlich D., Hartmann E. (1999) Evidence for distinct substrate specificities of importin α family members in nuclear protein import. Mol. Cell. Biol. 19, 7782–7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meyer T., Vinkemeier U. (2004) Nucleocytoplasmic shuttling of STAT transcription factors. Eur. J. Biochem. 271, 4606–4612 [DOI] [PubMed] [Google Scholar]
- 17. Kitamura R., Sekimoto T., Ito S., Harada S., Yamagata H., Masai H., Yoneda Y., Yanagi K. (2006) Nuclear import of Epstein-Barr virus nuclear antigen 1 mediated by NPI-1 (Importin α5) is up- and down-regulated by phosphorylation of the nuclear localization signal for which Lys379 and Arg380 are essential. J. Virol. 80, 1979–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tarendeau F., Boudet J., Guilligay D., Mas P. J., Bougault C. M., Boulo S., Baudin F., Ruigrok R. W., Daigle N., Ellenberg J., Cusack S., Simorre J. P., Hart D. J. (2007) Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat. Struct. Mol. Biol. 14, 229–233 [DOI] [PubMed] [Google Scholar]
- 19. Resa-Infante P., Jorba N., Zamarreño N., Fernández Y., Juárez S., Ortín J. (2008) The host-dependent interaction of α-importins with influenza PB2 polymerase subunit is required for virus RNA replication. PLoS One 3, e3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nardozzi J., Wenta N., Yasuhara N., Vinkemeier U., Cingolani G. (2010) Molecular basis for the recognition of phosphorylated STAT1 by importin α5. J. Mol. Biol. 402, 83–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rabut G., Doye V., Ellenberg J. (2004) Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6, 1114–1121 [DOI] [PubMed] [Google Scholar]
- 22. Guan T., Kehlenbach R. H., Schirmer E. C., Kehlenbach A., Fan F., Clurman B. E., Arnheim N., Gerace L. (2000) Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol. Cell. Biol. 20, 5619–5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smitherman M., Lee K., Swanger J., Kapur R., Clurman B. E. (2000) Characterization and targeted disruption of murine Nup50, a p27(Kip1)-interacting component of the nuclear pore complex. Mol. Cell. Biol. 20, 5631–5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dilworth D. J., Suprapto A., Padovan J. C., Chait B. T., Wozniak R. W., Rout M. P., Aitchison J. D. (2001) Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J. Cell Biol. 153, 1465–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Solsbacher J., Maurer P., Vogel F., Schlenstedt G. (2000) Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin α. Mol. Cell. Biol. 20, 8468–8479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hood J. K., Casolari J. M., Silver P. A. (2000) Nup2p is located on the nuclear side of the nuclear pore complex and coordinates Srp1p/importin-α export. J. Cell Sci. 113, 1471–1480 [DOI] [PubMed] [Google Scholar]
- 27. Matsuura Y., Stewart M. (2005) Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J. 24, 3681–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lindsay M. E., Plafker K., Smith A. E., Clurman B. E., Macara I. G. (2002) Npap60/Nup50 is a tri-stable switch that stimulates importin-α:β-mediated nuclear protein import. Cell 110, 349–360 [DOI] [PubMed] [Google Scholar]
- 29. Matsuura Y., Stewart M. (2004) Structural basis for the assembly of a nuclear export complex. Nature 432, 872–877 [DOI] [PubMed] [Google Scholar]
- 30. Sun C., Yang W., Tu L. C., Musser S. M. (2008) Single-molecule measurements of importin α/cargo complex dissociation at the nuclear pore. Proc. Natl. Acad. Sci. U.S.A. 105, 8613–8618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ogawa Y., Miyamoto Y., Asally M., Oka M., Yasuda Y., Yoneda Y. (2010) Two isoforms of Npap60 (Nup50) differentially regulate nuclear protein import. Mol. Biol. Cell 21, 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boivin S., Hart D. J. (2011) Interaction of the influenza A virus polymerase PB2 C-terminal region with importin α isoforms provides insights into host adaptation and polymerase assembly. J. Biol. Chem. 286, 10439–10448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 34. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 37. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 38. Laskowski R. A., Hutchinson E. G., Michie A. D., Wallace A. C., Jones M. L., Thornton J. M. (1997) PDBsum: a Web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci. 22, 488–490 [DOI] [PubMed] [Google Scholar]
- 39. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 40. Booth J. W., Belanger K. D., Sannella M. I., Davis L. I. (1999) The yeast nucleoporin Nup2p is involved in nuclear export of importin α/Srp1p. J. Biol. Chem. 274, 32360–32367 [DOI] [PubMed] [Google Scholar]
- 41. Mitrousis G., Olia A. S., Walker-Kopp N., Cingolani G. (2008) Molecular basis for the recognition of snurportin 1 by importin β. J. Biol. Chem. 283, 7877–7884 [DOI] [PubMed] [Google Scholar]
- 42. Kobe B. (1999) Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin α. Nat. Struct. Biol. 6, 388–397 [DOI] [PubMed] [Google Scholar]
- 43. Matsuura Y., Lange A., Harreman M. T., Corbett A. H., Stewart M. (2003) Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import. EMBO J. 22, 5358–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cingolani G., Lashuel H. A., Gerace L., Müller C. W. (2000) Nuclear import factors importin α and importin β undergo mutually induced conformational changes upon association. FEBS Lett. 484, 291–298 [DOI] [PubMed] [Google Scholar]
- 45. Darling R. J., Brault P. A. (2004) Kinetic exclusion assay technology: characterization of molecular interactions. Assay Drug Dev. Technol. 2, 647–657 [DOI] [PubMed] [Google Scholar]
- 46. Andrade M. A., Petosa C., O'Donoghue S. I., Müller C. W., Bork P. (2001) Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309, 1–18 [DOI] [PubMed] [Google Scholar]
- 47. Gabriel G., Klingel K., Otte A., Thiele S., Hudjetz B., Arman-Kalcek G., Sauter M., Shmidt T., Rother F., Baumgarte S., Keiner B., Hartmann E., Bader M., Brownlee G. G., Fodor E., Klenk H. D. (2011) Differential use of importin-α isoforms governs cell tropism and host adaptation of influenza virus. Nat. Commun. 2, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nardozzi J. D., Lott K., Cingolani G. (2010) Phosphorylation meets nuclear import: a review. Cell Commun. Signal. 8:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gilchrist D., Rexach M. (2003) Molecular basis for the rapid dissociation of nuclear localization signals from karyopherin α in the nucleoplasm. J. Biol. Chem. 278, 51937–51949 [DOI] [PubMed] [Google Scholar]
- 50. Gilchrist D., Mykytka B., Rexach M. (2002) Accelerating the rate of disassembly of karyopherin.cargo complexes. J. Biol. Chem. 277, 18161–18172 [DOI] [PubMed] [Google Scholar]
- 51. Riddick G., Macara I. G. (2005) A systems analysis of importin-α-β mediated nuclear protein import. J. Cell Biol. 168, 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ribbeck K., Görlich D. (2001) Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20, 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.