Background: Expansion of a polyglutamine repeat in atrophin-1 causes progressive neuronal dysfunction in neurodegenerative dentatorubropallidoluysian atrophy.
Results: Nuclear inclusions of expanded atrophin-1 are more dynamic compared with the cytoplasmic aggregates.
Conclusion: Structural variations of the aggregate core determine these dynamics.
Significance: Probing the molecular properties of the inclusions is crucial for understanding the enhanced cellular toxicity of nuclear aggregates in polyglutamine pathologies.
Keywords: Aggregation, Amyloid, Biophysics, Polyglutamine, Polyglutamine Disease, DRPLA, Atrophin-1, Cross-seeding, Microscopy
Abstract
Preferential accumulation of mutant proteins in the nucleus has been suggested to be the molecular culprit that confers cellular toxicity in the neurodegenerative disorders caused by polyglutamine (polyQ) expansion. Here, we use dynamic imaging approaches, orthogonal cross-seeding, and composition analysis to examine the dynamics and structure of nuclear and cytoplasmic inclusions of atrophin-1, implicated in dentatorubropallidoluysian atrophy, a polyQ-based disease with complex clinical features. Our results reveal a large heterogeneity in the dynamics of the nuclear inclusions compared with the compact and immobile cytoplasmic aggregates. At least two types of inclusions of expanded atrophin-1 with different mobility of the molecular species and ability to exchange with the surrounding monomer pool coexist in the nucleus. Intriguingly, the enrichment of nuclear inclusions with slow dynamics parallels changes in the aggregate core architecture that are dominated by the polyQ stretch. We propose that the observed complexity in the dynamics of the nuclear inclusions provides a molecular explanation for the enhanced cellular toxicity of the nuclear aggregates in polyQ-based neurodegeneration.
Introduction
Expansion of unstable CAG repeat-encoding homopolymeric polyglutamine (polyQ)4 stretch is the only common molecular feature of 10 dominantly inherited polyglutamine diseases (also known as CAG repeat disorders) that include Huntington's disease, dentatorubropallidoluysian atrophy (DRPLA), spinobulbar muscular atrophy, and several spinocerebellar ataxias (1, 2). PolyQ-containing proteins have an inherent ability to aggregate in the nucleus and cytoplasm of affected neurons when the length of the polyQ stretch expands over a critical disease threshold (2, 3). PolyQ expansion confers toxic properties on the host protein (2, 4), and whether aggregation is critical or not, the nucleus is suggested to be the primary site of cellular dysgenesis (5). Some overlapping gene expression changes have been observed in mouse models of Huntington's disease, DRPLA, spinobulbar muscular atrophy, and spinocerebellar ataxia type 7 (6), implying a common mechanism of cellular dysregulation because of polyQ expansion. All described polyQ disorders, however, are characterized by a distinct pathology, cell-type specific aggregation, and cytotoxicity pattern, which in turn cannot be explained by a common polyQ-based aggregation mechanism. Alternatively, each disease protein is characterized by a unique set of transcript alterations (6). Clearly, the expansion of the polyQ stretch per se is necessary but not sufficient to trigger neurodegeneration. Protein domains outside the polyQ tract have an important role in the selective neurotoxicity of the various polyQ disorders.
In vitro, polyQ peptides aggregate in a nucleated growth polymerization with a direct formation of amyloid-like fibrils after structural transition of the polyQ stretch within the monomer (7, 8). By contrast, the aggregation of expanded ataxin-1, ataxin-3, and huntingtin (Htt) exon-1 is a multistep process in which sequences outside the polyQ stretch transiently interact to initiate aggregation (9–12). These initial interactions are polyQ-independent, and expansion of the polyQ tract over the pathological threshold (Q ≥ 35) induces a conformation transition to a more compact toxic β-sheet structure (13, 14) that assembles into highly stable, detergent-insoluble amyloid-like aggregates (9, 10, 15, 16). Regions outside the polyQ stretch modulate the aggregation propensity of the polyQ expansion (17, 18), and variations in the polyQ-flanking sequences induce the following diversity in the aggregate morphology: amorphous bulky aggregates, small inclusions with crisp boundaries, or large aggregates with dense inaccessible core (18, 19).
Many cellular proteins, including transcriptional factors, molecular chaperones, and components of the ubiquitin proteasome, can be stably recruited into the aggregates, thus depleting crucial cellular functionalities (19–26), which is believed to play a major role in the cellular dysgenesis. Although some cellular proteins are irreversibly sequestered in the polyQ aggregates, others retain their flexibility, albeit with impaired functionality (19, 22, 26). Importantly, the ability to interact with cellular proteins varies among the aggregates composed of different disease proteins, which may explain the disease-specific pattern of the polyQ disorders (19). Fluorescent techniques with high time resolution (i.e. fluorescent recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP)) have revealed that cellular aggregates of ataxin-1 and Htt exon-1 are highly heterogeneous with different mobility of the monomers inside them (19, 22, 26). These various aggregate types may expose different parts of the primary protein sequences, thus providing different surfaces for biophysical interactions with the cellular proteins. Therefore, understanding the dynamics of the aggregation process and the molecular architecture of the different aggregate species formed in the cell is critical for understanding the variations in the mechanisms underlying polyQ pathologies.
In an effort to address this question, we sought to characterize the dynamics and structure of the aggregates of atrophin-1 (ATN1), which is implicated in DRPLA pathology. ATN1 contains putative nuclear localization sequence (NLS) and nuclear export (NES) sequence at the N- and C terminus, respectively, and localizes in the nucleus and cytoplasm (27). The role of the aggregate formation in the cellular dysgenesis is still elusive. Whereas ATN1 with a normal and expanded polyQ length show no difference in cellular toxicity in a neuroblastoma cell model, preferential accumulation in the nucleus by removal of the NES sequence confers cellular toxicity (28). The cellular dysfunction and toxic effect might be due to an altered distribution pattern of promyelocytic leukemia protein nuclear bodies by intranuclear aggregates of ATN1 with an expanded polyQ stretch (29). Moreover, some nuclear aggregates of ATN1 with an expanded polyQ tract can sequester the transcriptional coactivator, the cAMP response element-binding protein (30) through interactions with its short polyQ stretch (23). ETO/MTG8, a component of the nuclear receptor corepressor complexes, cofractionates in nuclear matrix preparations from brains of DRPLA patients and colocalizes with nuclear aggregates in a cellular model system (5). Furthermore, sequences flanking the polyglutamine region are involved in interactions with the cellular proteins. SH3 domain of insulin receptor tyrosine kinase substrate protein interacts with the proline-rich region cognate to the polyQ tract in the ATN1 (31). The variety of cellular interaction partners raises the intriguing hypotheses of whether different parts of the parental protein are involved, transiently or irreversibly, in the aggregate core, which may suggest an explanation for the observed variety in the clinical features of DRPLA, i.e. epilepsy versus dementia. Here, we show that the expansion of the polyQ stretch over the pathological threshold changes the dynamics and biophysical properties of nuclear ATN1 aggregates. The polyQ flanking sequences associated independently of the polyQ-length and sequestered ATN1 in highly mobile nuclear inclusions. In a second step, polyQ expansion over the threshold induced the conversion into amyloid-like aggregates with reduced mobility and a core composed of the polyQ stretch. Our findings provide a mechanistic insight into the complexity of ATN1 aggregation to explain the heterogeneity in pathogenesis of DRPLA and other polyQ disorders.
EXPERIMENTAL PROCEDURES
Constructs and Cell Lines
Full-length ATN1 (FL-ATN1) and truncated variant lacking the first 75 amino acids (ΔN-ATN1) with 19Q or 71Q, respectively (FL19Q, FL71Q, ΔNLS19Q, ΔNLS71Q) were kindly provided by Dr. T. Miyashita cloned in pEGFP-C2 vector (32). The promoter of pEGFP-C2 was exchanged with pTREhCMV-1 promoter from pTRE2hyg plasmid (Clontech) using a combined restriction with AflII and NotI. AflII restriction site was introduced by site-directed mutagenesis. To enable controlled protein expression under the Tet-off promoter, we used neuroblastoma N2a cells stably transfected with pTet-Off plasmid (N2a/Tet-Off cells) which were a kind gift from Dr. P Breuer.
pLac-DsRed inducible mammalian expression vector was obtained by subcloning the DsRed coding sequence from the pDsRed2 plasmid (Clontech) into the KpnI/BglII restriction sites of the pLac/MCS plasmid (33). The pLac/MCS plasmid is an engineered pORSVI/MCS plasmid for isopropyl 1-thio-β-d-galactopyranoside-inducible expression in mammalian cells and was kindly provided by Dr. R. Morimoto (Northwestern University, Chicago, IL). The ATN1 fragments comprising different parts of the FL-ATN1 were subcloned in-frame downstream of the DsRed moiety using the BglII/XbaI restriction sites. The stable cell line N2a/Tet-Off/LacI for coexpression of proteins under the Tet-Off was created by transfection of pCMV-LacI-NLS plasmid in N2a/Tet-Off cells. Stably transfected cells were selected with 0.7 mg/ml hygromycin B.
Cell Culture and Protein Expression
N2a/Tet-Off cells were used for expression of FL-ATN1 or ΔN-ATN1 variants. For in vivo cross-seeding experiments, N2a/Tet-Off/LacI cells were transiently cotransfected with the FL and ATN1 fragments at a stoichiometric 1:1 ratio. Transfections were performed using Lipofectamine Plus (Invitrogen) or JetPrime reagent (PeqLab) following the manufacturer's instructions. Cells were grown in DMEM (PAN Biotech) supplemented with 10% Tet system-approved FBS (Clontech), 0.3 mg/ml L-Glu (Invitrogen), 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) at 37 °C in 5% CO2. For maintenance of stably transfected N2a/Tet-Off cells, the medium was supplemented with 0.3 mg/ml G418 (Calbiochem). In the case of N2a/Tet-Off/LacI cells, 0.3 mg/ml hygromycin B (Clontech) was added. The expression of pTREhCMV-1-controlled constructs (FL and ΔN-ATN1) was switched off by addition of 0.3 μg/ml doxycycline (Clontech). The ATN1 fragments that were controlled under the Lac Switch expression system were induced by addition of 15 mm isopropyl 1-thio-β-d-galactopyranoside to the medium.
Microscopy and Cell Imaging
Transfected N2a/Tet-Off cells were grown on poly-L-lysine-precoated cover slides, fixed in 4% paraformaldehyde in PBS permeabilized with 1% Triton X-100 (Sigma) at room temperature and mounted using an anti-fade mounting kit (Invitrogen). DNA was counterstained using DAPI (Sigma). Cells were imaged on an LSM710 Axiovert confocal microscope (Zeiss) equipped with the ×63 oil objective lens, and images were taken at 488 nm, 405 nm, and 561 nm laser wavelengths for GFP, DAPI, and DsRed, respectively. For life-cell imaging, FRAP and iFRAP analyses, transfected N2a/Tet-Off or N2a/Tet-Off/LacI cells were grown in poly-L-lysine-precoated 8-well microslides (Ibidi). For FRAP experiments, an area of 2.98 μm2 was photobleached for 1 s (120 iterations) with a 488-nm laser wavelength at 100% power, and single images were collected before and every 1.5 s after photobleaching within an interval of 8.5 min. For the iFRAP experiments the area of interest was defined using Zen software, and the whole cell outside the defined area was photobleached for 4.5 s (90 iterations) with a 488-nm laser wavelength at 100% power. 300 or 500 z-stacks, each consisting of five slices in 0.7 μm intervals and seven slices in 0.9 μm intervals, were collected for each single image in the FRAP and iFRAP experiments, respectively. The intensity of the images recorded in FRAP was normalized to the relative intensity (RI) as established for in-cell FRAP analysis (22, 26, 34). RIt = i = (Ii/Inbi)/(I0/Ipb), where Ii and Inbi are the integrated intensities of the bleached and non-bleached area at any given time point and I0 and Ipb are the integrated intensities before photobleaching of the bleached and non-bleached area, respectively. The images recorded in the iFRAP experiments were normalized to the relative intensity values RIt = i = (Ii/Inbi). The constitutive photobleaching was measured by collecting control images of at least 10 non-bleached cells. The half-life time was determined as described elsewhere (26), and the standard deviation was calculated from three independent cells.
Fractionation of the Cells
Cells were washed twice in PBS harvested in a lysis buffer (50 mm Tris (pH 7.4) containing 150 mm NaCl, 100 mm MgCl2, protease inhibitor mixture (Roche)), lysed on ice at the lowest blender-homogenizer power, and subsequently treated with BenzonaseTM (Sigma) for 20 min on ice. The lysates were centrifuged for 10 min at low speed (80 × g) at 4 °C, and the pellet fraction was resuspended in the lysis buffer. The supernatant was further separated into five fractions by sucrose density centrifugation (50,000 × g, Ti65-Beckman-Coulter rotor) for 45 min at 4 °C in a discontinuous sucrose density gradient in 50 mm Tris buffer (pH 7.4) containing 150 mm NaCl and protease inhibitor mixture.
For fractionation into nuclear and cytoplasmic fractions, cells were washed twice in PBS, harvested in PBS buffer containing 0.25 m sucrose and protease inhibitor mixture, centrifuged at 300 × g for 3 min, and resuspended in hypotonic buffer (10 mm HEPES (pH 7.4) containing protease inhibitor mixture). After swelling for 10 min, the cells were dounce-homogenized on ice, centrifuged for 10 min at 80 × g at 4 °C to separate the non-disrupted cells and the largest cytoplasmatic, SDS-insoluble inclusions (without precipitating the nuclei), and the nuclear fraction was further pelleted at 600 × g for 10 min at 4 °C. Both the pellet after the first, slow-centrifugation step and the nuclei fraction were resuspended in the hypotonic buffer containing 100 mm MgCl2 to a volume equal to that of the supernatant, subjected to an additional mechanical disruption, and incubated for 15 min with benzonase on ice. Samples were analyzed by immunoblot analysis on a nitrocellulose membrane or filter retardation assay (35) on cellulose acetate membrane (0.2 μm pore size) and detected using anti-GFP (1:1000, Roche) as primary antibodies.
RESULTS
Expansion of the polyQ Stretch Changes the Inclusion Dynamics
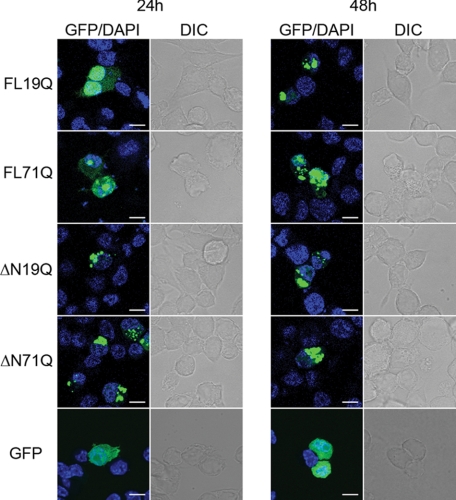
We monitored the in vivo dynamics of full-length ATN1-GFP fusions with a polyQ tract of a non-pathological (FLQ19) or pathological (FLQ71) length in neuroblastoma N2a/Tet-Off cells under the control of a Tet-Off promoter. The pathological variant, FL71Q, localized readily into hyperfluorescent loci in both the cytoplasm and the nucleus, even 24 h post-transfection (Fig. 1 and supplemental Table S1). A fraction of it, however, remained diffusely distributed throughout the cell, even at later times of expression, thus recapitulating the expression pattern of ATN1 with an expanded polyQ tract previously observed in degenerated neurons (28). The diffusive fluorescence was not due to an unspecific cleavage or release of GFP from the fusions. Using Western blot analysis we did not observe a monomeric GFP 24 h or 48 h after transfection (data not shown). At an early time point (24 h), FL19Q exhibited a predominantly diffuse pattern of localization with some rarely detectable, small hyperfluorescent puncta mostly in the nuclei of some cells (Fig. 1 and supplemental Table S1). The fraction of the hyperfluorescent loci increased with the expression time (48 h), with a much higher proportion of the inclusions in the nucleus compared with the cytosol (Fig. 1 and supplemental Table S1). Notably, only the FL71Q variant formed SDS-resistant species (supplemental Table S1). Detergent resistance represents an important pathological hallmark of the polyQ aggregation and is indication of the formation of amyloid structures (protofibrils or mature fibrils) (21).
FIGURE 1.
Cellular localization of the ATN1 variants. GFP-tagged FL-ATN1 variants partitioned between the cytoplasm and the nucleus and ΔN-ATN1 variants were localized in the cytoplasm. The nuclear localization was confirmed with DAPI counterstaining. N2a/Tet-Off cells were transiently transfected with GFP-tagged ATN1 variants and visualized by fluorescence (GFP) and differential interference-contrast (DIC) microscopy after 24 and 48 h. GFP was soluble over the whole expression cycle and is included as a control. Scale bar = 10 μm.
The nucleocytoplasmic shuttling of ATN1 is mediated by the two functional NLS and NES sequences and can be abrogated by a putative caspase cleavage at Asp-109, which alters the localization of the protein solely to the cytoplasm (28). Such N-terminally truncated ATN1 fragments (ΔN-ATN1) have been identified in DRPLA disease model mice and DRPLA patients (30, 37). To compare the cytoplasmic inclusions of FL-ATN1, we expressed the corresponding N-terminally truncated ATN1 variants with a short (ΔN19Q) or an expanded (ΔN71Q) polyQ stretch. Both ΔN19Q and ΔN71Q formed hyperfluorescent loci that were undistinguishable in shape and size (Fig. 1). However, similar to FL-ATN1 proteins, only the variant with the expanded polyQ stretch ΔN71Q) assembled into detergent-resistant aggregates (supplemental Fig. S1).
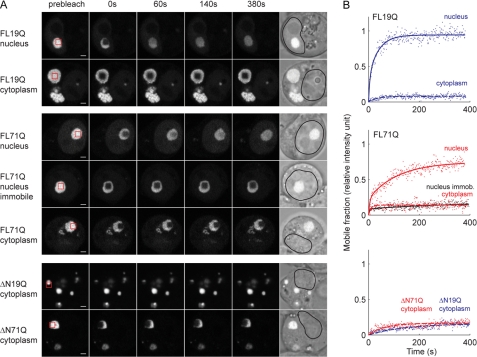
Taken together, the data suggest that the FL-ATN1 and ΔN-ATN1 variants, independently of the polyQ length, formed discernible inclusions. However, polyQ expansion shortened the time of appearance and introduced differences in their detergent solubility. This raised the question as to whether the hyperfluorescent loci of ATN1 with non-pathological and pathological Q- length, even with undistinguishable morphologies on the static microscopic images, have different mobility and dynamic properties. To assess the dynamic properties of the inclusions, we used FRAP, which is usually employed to assess the diffusion and mobility of fluorescently tagged species (34). In the case of the inclusions, a small sector of a hyperfluorescent spot was directly bleached, and the degree of fluorescence recovery within this area indicated the mobility of the fluorescent species within the inclusion. All nuclear FL19Q inclusions were highly mobile, displaying nearly complete recovery within 100 s (Fig. 2, A and B). In contrast, FL71Q formed two types of nuclear inclusions with high and low recovery (two representative examples are included in Fig. 2, A and B). Interestingly, the profile of the high-recovering FL71Q nuclear inclusions showed an initial fast recovery phase followed by a slower, incomplete recovery (Fig. 2B). Mathematical analysis of the FRAP data supported these assertions, as the curve fit of the recovery is consistent with two populations: a mobile fraction that accounted for 69 ± 7% and a less mobile fraction that contributed to 31 ± 4% of the total fluorescent protein within aggregates (Fig. 2B). This recovery pattern strongly differed from the single protein population detected in the aggregates of Htt exon-1 with an expanded polyQ stretch (22, 38). The nuclear, diffusely distributed fractions of both FL71Q and FL19Q were highly mobile and displayed immediate recovery comparable with the GFP recovery. Complete recovery was reached within the first postbleach measurement (at 1.5 s) (data not shown). Cytoplasmic aggregates, whether built of the FL-ATN1 or ΔN-ATN1 variants with pathological and non-pathological Q-lengths, exhibited poor recovery profiles (Fig. 2, A and B and supplemental Table S2) that corresponded to stable aggregate structures containing immobile proteins.
FIGURE 2.
Mobility analysis of ATN-1 aggregates. A, FRAP analysis of FL-ATN1 and ΔN-ATN1 aggregates. Different ATN1 variants were transiently expressed in N2a/Tet-Off cells for 48 h. The red box on the prebleach image marks the area of bleaching. Scale bar = 2 μm. B, quantitative analysis of the FRAP images. Fluorescence intensity in the bleached area of each FRAP image (A) is converted into relative fluorescence intensity. Solid or dashed lines represent a curve fit to a three-parameter single or a five-parameter double exponential function, respectively.
FRAP analyses suggested that two populations with different mobility were present within the nuclear FL71Q inclusions. However, these dynamics may represent variations in the ability of the aggregates to exchange with the external mobile protein pool (outside of the inclusion). Thus, we employed a complementary assay, inverted FRAP (iFRAP), which reports on the exchange of fluorescently labeled species from aggregates or with the neighboring environment or compartment (39). In brief, the whole cell, leaving the inclusion of interest intact, is completely photobleached. If the monomer species can dissociate from the inclusions, we would expect the fluorescence of the inclusions to decrease on the basis of the migration of the monomer to the surrounding environment. The fluorescence of the nucleoplasmic FL19Q inclusions decreased rapidly, with a half-life of 48 min at 24 h of expression (supplemental Table S2), suggesting that the FL19Q monomer was loosely incorporated. Similar to the FRAP results, the nuclear FL71Q inclusions exhibited two different half-lives of decay (supplemental Table S2). The low-recovering FL71Q assemblies in the FRAP experiments decayed with a half-life an order of magnitude longer than that of the high-recovering FL71Q species. Moreover, the fluorescence decay of the high-recovering inclusions better fitted a two-phase exponential decay curve, supporting the presence of at least two distinct protein populations within these inclusions. The fluorescence intensity of the cytoplasmic assemblies of all constructs remained constant, implying that the corresponding monomers are stably associated in the aggregates (supplemental Table S2).
ATN1 with Expanded PolyQ Stretch Form Detergent-resistant Inclusions That Associate with Nuclear Membranes
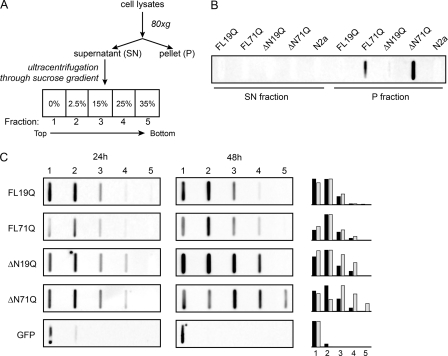
Next, to determine whether the observed differences in the inclusion dynamics correlated with the composition and structural features of the aggregates, we separated the inclusions into nuclear and cytoplasmic and analyzed their detergent-resistance. SDS-insoluble FL71Q aggregates composed of protofibrillar or fibrillar species were bulky structures that sedimented at a very low velocity (supplemental Fig. S2). A small SDS-resistant fraction was also found in the nucleus that most likely represented the low-recovering species in the FRAP experiments (Fig. 2, A and B). Furthermore, the lysates of cells expressing the FL-ATN1 or ΔN-ATN1 variants were fractionated by size and density through sucrose gradient centrifugation (Fig. 3A). The variants with pathological Q-length, FL71Q and ΔN71Q, built bulky SDS-resistant aggregates that sedimented at a very low velocity (Fig. 3B). The supernatant, comprising the SDS-labile, small-sized nuclear and cytoplasmic aggregates (Fig. 3A), was further subjected to sucrose density centrifugation. At an early time point of expression (24 h), SDS-labile aggregates showed a similar pattern for each pair, FL19Q/FL71Q and ΔN19Q/ΔN71Q (Fig. 3C). Although the density of the non-pathological FL19Q and ΔN19Q did not change at later expression time points (48 h), the FL71Q and ΔN71Q aggregates migrated at a higher density (fractions 3 and 4 were enriched with FL71Q and fractions 3–5 of ΔN71Q, Fig. 3C). The separation of the aggregates by density centrifugation is determined by their mass, density, and shape; i.e. the enrichment of FL71Q and ΔN71Q aggregates in the higher-density fractions would account for species with a higher mass, larger density, and/or increased compactness. These changes in the global structure of the FL71Q and ΔN71Q aggregates mirrored the decrease in the mobility observed in the FRAP analyses (supplemental Table S2).
FIGURE 3.
The detergent-labile inclusions of the FL-ATN1 and ΔN-ATN1 with expanded polyQ differ in their density. A, schematic of the fractionation procedure of the inclusions. B, large inclusions pelleted at low centrifugation speed in the P fraction. Only FL71Q and ΔN71Q form detergent-resistant species. The SDS insolubility was monitored by filter retardation assay 48 h after transfection and probed with anti-GFP antibodies. Mock-transfected N2a/Tet-Off cells (N2a) are used as a control. C, fractionation of the inclusions in the supernatant (SN) fraction on a non-continuous sucrose gradient. Two million N2a/Tet-Off cells were transfected with all FL-ATN1 and ΔN-ATN1 variants and lysed after 24 or 48 h. The fractions from the sucrose gradient were loaded onto a nitrocellulose membrane, which retains all proteins, and the GFP-ATN1 positive fractions were visualized by immunostaining with anti-GFP antibodies. The intensity of the GFP-ATN1-positive fractions was related to the spot with the highest intensity within each blot, and the relative intensity is graphically summarized for 24 h (black) and 48 h (gray) for each construct (at the right side of the immunoblot analyses). The numbers over the blots correspond to the fractions from the sucrose gradient in A. Cells expressing GFP protein serve as a control of completely soluble protein that is found in the lightest fraction.
Interestingly, the fraction of the bulky, detergent-resistant FL71Q aggregates that sedimented at a very slow centrifugation speed was strongly lamin B1-positive (supplemental Fig. S2C), even though the majority of those aggregates localized in the cytoplasm, as detected in the FRAP experiments (Fig. 2). As already observed, other proteins with expanded polyQ stretches assemble into perinuclear aggregates that typically associate with the nuclear membrane, forming a characteristic distortion of the nuclear surface (40, 41). Next, we analyzed the state of the nuclear envelope using lamin B1 as a marker for nuclear integrity (42). Immunostaining analysis revealed that small FL71Q inclusions were clearly associated with the nuclear envelope and caused discernible discontinuity in the lamin B1 rim (supplemental Fig. S3). In addition, the cytoplasmic aggregates also associated with the nuclear membrane, forming a local depression (supplemental Fig. S3).
Expansion of the PolyQ Stretch Changes the Aggregate Core
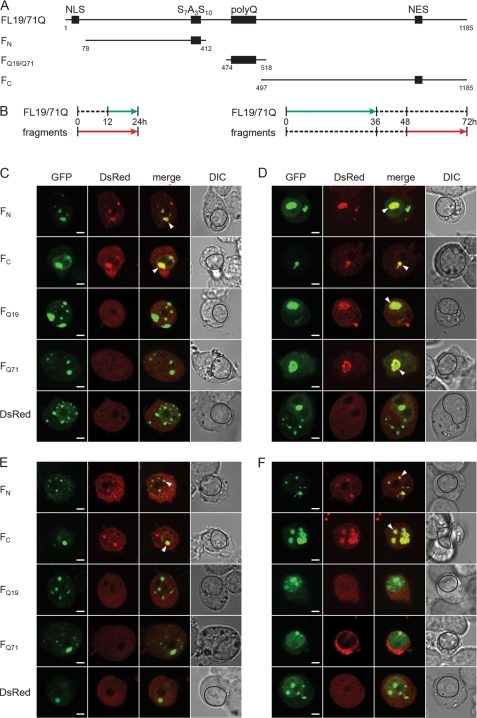
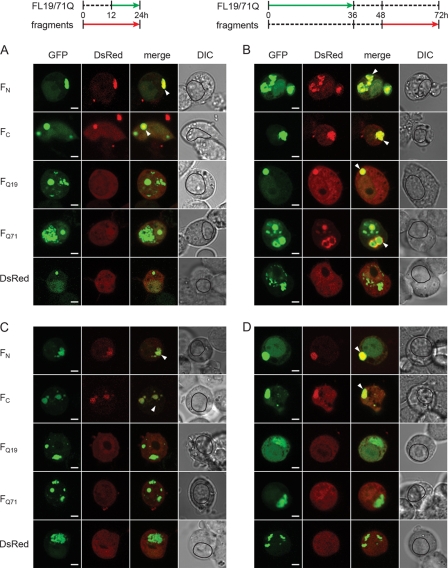
The observed time-dependent evolution in the detergent resistance of FL71Q and ΔN71Q inclusions (supplemental Fig. S1) raised the intriguing question as to whether this is conditioned by changes in the structure of the aggregate core. We next assessed the nature of the aggregate core of the FL19Q and FL71Q inclusions by comparing their recruitment properties using an orthogonal cross-seeding approach (43). Conceptually, this approach is based on the highly discriminating ability of preformed aggregates to recruit monomers with equivalent sequences (44–46). At different times of expression of FL19Q and FL71Q, various fragments comprising parts of the full-length protein were pulse-expressed (Fig. 4, A and B) in a tightly controlled manner under the isopropyl 1-thio-β-d-galactopyranoside-inducible Lac promoter, and their incorporation into the inclusions of FL19Q and FL71Q was monitored by confocal imaging microscopy. We expected the FL19Q or FL71Q aggregates to recruit only the fragment bearing the primary sequence that participates in the aggregate core. A prerequisite for this approach is that the coexpressed fragments remain soluble over the cross-seeding cycle (supplemental Fig. S4). It should be noted that the fragments, despite the lack of NLS, were equally distributed between the nucleoplasm and cytoplasm (supplemental Fig. S4C), probably due to free diffusion into the nucleus as observed for small proteins (47). The uniform distribution of the fragments over the cell volume allowed us to examine the recruiting properties of both nuclear and cytoplasmic inclusions. The N- and C-terminal sequences flanking the polyQ region (FN and FC), but not the polyQ-containing fragments, colocalized with the nuclear FL71Q aggregates formed at an early expression time (Fig. 4C). This implies that at early time points, FL71Q forms inclusions through interactions of the N- and C-terminal regions. At later time points, nuclear FL71Q aggregates also recruited polyQ fragments (FQ19 and FQ71), implying a change in the aggregate core to the polyQ-containing one (Fig. 4D). At the late time point, nuclear and cytoplasmic FL71Q aggregates partitioned between the detergent-resistant and detergent-labile fractions (Fig. 3C and supplemental Fig. S2B), suggesting that the ability to recruit polyQ-fragments paralleled with the appearance of the SDS-insolubility.
FIGURE 4.
Orthogonal cross-seeding in vivo reveals polymorphic interactions of nuclear FL19Q and FL71Q inclusions with the ATN1 fragments. A, schematic of the ATN1 fragments used in the cross-seeding experiments. The numbering is according to the amino acid sequence of FL19Q. B, expression scheme for the coexpression of the fragments and FL-ATN1. Each expression scheme is representative for the image series below it. C and D, recruitment properties of the FL71Q aggregates at early (C) and late (D) expression times. E and F, recruitment ability of FL19Q inclusions at early (E) and late (F) times of expression. N2a/Tet-Off/LacI cells were cotransfected with GFP-tagged FL19Q or FL71Q and a DsRed-tagged fragment thereof (marked on the left of each image row). The degree of colocalization is illustrated by merging the GFP and DsRed images. The recruited fragments in the FL-ATN1 inclusions are marked with an arrow on the merged images. The boundaries of the nuclei are designated with a black line on the differential interference contrast (DIC) images. Note that there are some hyperfluorescent spots on the FQ19 (D) and FC (F) images (visible on the DsRed images) whose origin is unclear. Most likely they are debris from neighboring cells, as they are localized at the edge of the cells. Scale bar = 4 μm.
The cytoplasmic FL71Q aggregates mirrored the recruiting ability of the nuclear inclusions. FN and FC fragments colocalized with the cytoplasmic FL71Q aggregates at early time points (Fig. 5A). At a late time point of expression, the cytoplasmic FL71Q inclusions represented a mixed population of aggregates with different core structures as they recruited all types of fragments (Fig. 5B). The nuclear and cytoplasmic inclusions of the non-pathological variant FL19Q did not evolve over the duration of the expression time and only recruited the FN and FC fragments (Figs. 4, E and F, and 5, C and D).
FIGURE 5.
Orthogonal cross-seeding of the cytoplasmic FL19Q and FL71Q inclusions with the ATN1 fragments. A and B, cytoplasmic FL71Q aggregates recruited FN and FC fragments at early expression times (A) and additionally FQ19 and FQ71 at later expression times (B). The scheme for the coexpression of the fragments and FL-ATN1 is included, and each scheme is representative for the image series below it. C and D, FN and FC fragments colocalized with the cytoplasmic FL19Q inclusions at early (C) and late expression times (D). For details, see the legend of Fig. 4. DIC, differential interference contrast.
Interestingly, the cytoplasmic inclusions of ΔN-ATN1 resembled the phenotype of the cytoplasmic FL-ATN1 inclusions. An evolution in the recruiting ability was observed only for the cytoplasmic ΔN71Q aggregates (supplemental Fig. S5, A and B) but not for the inclusions composed of the non-pathological ΔN19Q variant (supplemental Fig. S5, C and D). These data allow us to conclude that the expansion of the polyQ stretch beyond the pathological threshold induces a change in the aggregate core to polyQ-based.
Expansion of the PolyQ Stretch Alters the Dynamics of Interactions with Cellular Proteins
Immunohistochemical and biochemical analyses established that aggregates of polyQ-containing proteins are associated with a variety of cellular proteins (19–26). Given the different core structure of the FL19Q and FL71Q inclusions observed, we wondered if these structural variations reflected the interactions with cellular proteins. To test this, we fractionated the cells into nucleoplasmic and cytoplasmic fractions, immunoprecipitated the FL19Q and FL71Q inclusions using anti-GFP antibodies (supplemental Fig. S6C), dissolved the aggregates, and analyzed protein composition using two-dimensional gel electrophoresis (36). We compared the interactome of the FL19Q with FL71Q inclusions (supplemental Fig. S6, A and B) and identified only spots that differ between the two proteins using MALDI-TOF MS analysis. For the nuclear inclusions, we mainly identified proteins that are involved in RNA processing and DNA repair (supplemental Table S3). Transcriptional profiles of these proteins have been found to be specifically altered in the DRPLA mouse model (6). Some of the proteins interacting with the nuclear inclusions, including heterologous nuclear protein L and endoplasmin (supplemental Table 3), showed overlapping changes in the microarray profiling common to different polyQ-disease proteins (6). In the nuclear FL71Q species, we observed an intense spot of nucleolin (supplemental Fig. S6A and Table S3), which is a crucial protein involved in the first steps of ribosome biogenesis. Its presence in the FL71Q nuclear aggregates was confirmed by immunoblotting of the fractionated nucleoplasmic FL71Q aggregates with anti-nucleolin antibody (supplemental Fig. S6D).
The difference between the cytoplasmic FL19Q and FL71Q inclusions was a depletion of two proteins from the FL71Q interactome that belong to the group of molecular chaperones (supplemental Table S3). Hsp90β and its murine homolog endoplasmin were depleted in the cytoplasmic FL71Q aggregates as opposed to the inclusions of the non-pathological FL19Q counterpart (supplemental Fig. S6B). A decreased level of the Hsp90β transcript has also been detected in the DRPLA mouse model (6).
DISCUSSION
ATN1 with polyQ length in the non-pathological (FL19Q) or pathological (FL71Q) range localize to nuclear and cytoplasmic hyperfluorescent loci, which on static images obtained from fixed cell studies may support the notion of immobile aggregates. Here, we present a systematic biophysical analysis of these inclusions and identify various aggregates with different dynamic properties and molecular architecture. The pathological FL71Q protein forms at least two types of inclusions with distinct dynamic properties that coexist in the nucleus: 1) inclusions composed of high-mobility species that exchange quickly with the soluble nucleoplasmic pool, and 2) aggregates consisting most likely of detergent-resistant, low-mobility species that exchanged at a lower frequency with the surrounding environment. By contrast, the FL19Q molecular species were also spatially sequestered in inclusions. However, they retain a high mobility within the inclusions and a fast exchange with the surrounding environment. ATN1 and ATN2 are physically associated in vivo, although the functional significance of these interactions is still unknown. ATN2 associates mainly in promyelocytic leukemia protein nuclear bodies and sequesters ATN1 (48). The sequestration may be of functional relevance to ATN1 and the primary reason for the observed spatial confinement of FL19Q. Alternatively, ATN1 may also have an intrinsic propensity to associate, and this initial association is driven by sequence elements outside the polyQ tract, as suggested by the cross-seeding experiments (Figs. 4 and 5 and supplemental Fig. S5). Unlike the nuclear aggregates, the cytoplasmic ATN1 with pathological and non-pathological Q-length assemble into bulky but compact and immobile structures. Spatial sequestration of the aggregates in such bulky cytoplasmic structures may facilitate their clearance (49).
Our in-cell orthogonal cross-seeding analyses suggest that the architecture of the aggregate core of FL71Q inclusions in the nucleus and cytoplasm evolves into polyQ-based, and we propose that the polyQ-stretch expansion over the threshold triggers a conformational switch to facilitate formation of amyloid structures. Our model is consistent with the “conformational conversion” model of yeast prion amyloid formation (50) and the proposed multistep aggregation model for other polyQ disease proteins (10–12) and model proteins (45, 51). The initial aggregation step is triggered by the association of folded domains or secondary structural elements located N-terminally upstream of the polyQ sequence (11, 12, 45, 52). For ATN1, our cross-seeding analyses suggest that two domains, the N- and C-terminal regions, initially associate with highly flexible inclusions and that the polyQ sequence is excluded from the aggregate core.
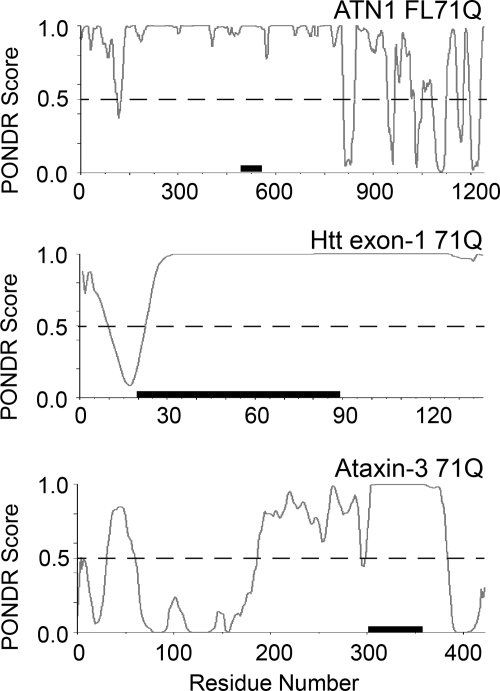
The ATN1 sequence bears some unique features, as revealed by the analysis using the disorder predictor PONDR (53). Both the N- and C-terminal domains show a tendency toward order, and the PONDR score of well below 0.5 median in the C terminus is associated with a higher tendency of being structured (54) (Fig. 6). In contrast, PONDR analysis of the Htt exon-1 sequence predicts some order only in the N-terminal region (Fig. 6) that, as experimentally suggested, probably takes up an α-helical conformation and promotes aggregation by interacting with the polyQ segment (11). The N-terminal segment of ataxin-3 with PONDR scores far below 0.5 median comprises the globular, autonomously folded Josephine domain that imparts an altered aggregation behavior on the polyQ stretch (52). PONDR analysis also suggests a tendency toward order in the C-terminal region (Fig. 6). Its impact on aggregation kinetics of ataxin-3 has not been experimentally addressed yet. Although structural details on the mechanisms by which flanking regions initiate the aggregation of polyQ-disease proteins remain to be clarified, our data and the examples in the literature clearly suggest that structurally stable elements can initially form flexible aggregates, with the polyQ-sequence excluded from the aggregate core. Depending on the sequence features that are unique for each polyQ protein, however, the structured regions can be N- or C-terminally localized.
FIGURE 6.
PONDR analysis of the FL71Q protein, Htt exon-1, and ataxin-3. Segments with a PONDR score between 0 and 0.5 are predicted to be stably folded, and scores near 1.0 are associated with disorder. The polyQ sequence of FL71Q is located between residues 483 and 554 and is flanked by largely disordered regions. The black bars represent the polyQ sequence. For comparison, the polyQ stretches of all three proteins are set to 71Q.
As observed for Htt exon-1 with an expanded polyQ tract (11, 12, 45, 52), the initial step in the FL71Q aggregation is non-nucleated and independent of the polyQ length because FL19Q forms similar inclusions with a mixed N- or C-core, as they recruit both N- and C-terminal fragments of ATN1 (Figs. 4 and 5). At the second stage, the initial, relatively loosely packed FL71Q nuclear inclusions that intensively exchange with the surrounding monomer pool convert into more stable, SDS-resistant amyloid structures with the polyQ-dominated aggregate core. We also detected some alterations in the interaction partners of the FL71Q compared with the non-pathological FL19Q inclusions, which paralleled the changes in the molecular architecture of the aggregates.
The site of cellular toxicity in polyQ diseases has been controversial, but the nucleus is suggested to be more sensitive to aggregation, and nuclear localization contributes to cellular toxicity (28, 55, 56). Our finding that nuclear inclusions are more dynamic provides a mechanistic explanation of the enhanced cellular toxicity of the nuclear ATN1 aggregates (28). This model may apply to some other polyQ diseases, and it resonates with previous observations on ataxin-1. Nuclear inclusions of ataxin-1 with a pathological polyQ length contain both fast- and slow-exchanging components, and the former ones contain high ubiquitin levels (19, 26, 39). By contrast, Htt exon-1 with an expanded polyQ tract forms immobile intranuclear inclusions (19), underscoring the importance of the protein context in polyQ diseases. Furthermore, we observed that intranuclear and cytoplasmic juxtanuclear FL71Q inclusions colocalize with the nuclear envelope component lamin B1 (supplemental Fig. S3). Changes in the lamin B1 rim and mechanical depression in the nuclear membrane are reminiscent of the focal distortion of the nuclear envelope observed for mutant Htt exon-1 (40). Aberrant interactions with the nuclear membrane are relevant to the pathophysiology of some neurological diseases (57). Together with the observation of highly dynamic and structurally heterogeneous nuclear inclusions, our results suggest a mechanistic explanation of why preferential accumulation of mutant ATN1 in the nucleus confers cellular toxicity.
Furthermore, our results contribute to the emerging concept that cellular aggregation is a dynamic multistep process with different sequences of the aggregating protein that is transiently or stably involved in the aggregate core. The idea that in the aggregation time course various surfaces will be accessible for biophysical interactions with cellular proteins goes beyond the role of protein misfolding and may have important implications for a large number of other neurodegenerative pathologies (58).
Supplementary Material
Acknowledgments
We thank Dr. T. Miyashita (National Research Institute of Child Care and Development) for the DRPLA constructs; Dr. R. Morimoto (Northwestern University, Chicago, IL) for the pLac/MCS and pCMV-LacI-NLS plasmids; Dr. P. Breuer (University Bonn) for the N2a/Tet-Off cells; Drs. Otto Baumann, Ralph Gräf, and Bernd Walz (University of Potsdam) for the help with the confocal microscope; and Dr. Jörg Fettke (University of Potsdam) for the mass spectrometry analysis. We also thank Molecular Kinetics for access to PONDR.
This work was supported by Deutsche Forschungsgemeinschaft Grant IG 73/8-1 (to Z. I.).

This article contains supplemental Figs. S1–S6, Tables S1–S3, and materials and methods.
- polyQ
- polyglutamine
- DRPLA
- dentatorubropallidoluysian atrophy
- Htt
- huntingtin
- FRAP
- fluorescent recovery after photobleaching
- FLIP
- fluorescent loss in photobleaching
- NLS
- nuclear localization sequence
- NES
- nuclear export sequence
- FL
- full-length
- iFRAP
- inverted fluorescent recovery after photobleaching.
REFERENCES
- 1. Hands S. L., Wyttenbach A. (2010) Neurotoxic protein oligomerization associated with polyglutamine diseases. Acta Neuropathol. 120, 419–437 [DOI] [PubMed] [Google Scholar]
- 2. Orr H. T., Zoghbi H. Y. (2007) Trinucleotide repeat disorders. Annu. Rev. Neurosci. 30, 575–621 [DOI] [PubMed] [Google Scholar]
- 3. Scherzinger E., Sittler A., Schweiger K., Heiser V., Lurz R., Hasenbank R., Bates G. P., Lehrach H., Wanker E. E. (1999) Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils. Implications for Huntington's disease pathology. Proc. Natl. Acad. Sci. U.S.A. 96, 4604–4609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harjes P., Wanker E. E. (2003) The hunt for huntingtin function. Interaction partners tell many different stories. Trends Biochem. Sci. 28, 425–433 [DOI] [PubMed] [Google Scholar]
- 5. Wood J. D., Nucifora F. C., Jr., Duan K., Zhang C., Wang J., Kim Y., Schilling G., Sacchi N., Liu J. M., Ross C. A. (2000) Atrophin-1, the dentato-rubral and pallido-luysian atrophy gene product, interacts with ETO/MTG8 in the nuclear matrix and represses transcription. J. Cell Biol. 150, 939–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luthi-Carter R., Strand A. D., Hanson S. A., Kooperberg C., Schilling G., La Spada A. R., Merry D. E., Young A. B., Ross C. A., Borchelt D. R., Olson J. M. (2002) Polyglutamine and transcription. Gene expression changes shared by DRPLA and Huntington's disease mouse models reveal context-independent effects. Hum. Mol. Genet. 11, 1927–1937 [DOI] [PubMed] [Google Scholar]
- 7. Bhattacharyya A. M., Thakur A. K., Wetzel R. (2005) Polyglutamine aggregation nucleation. Thermodynamics of a highly unfavorable protein folding reaction. Proc. Natl. Acad. Sci. U.S.A. 102, 15400–15405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen S., Ferrone F. A., Wetzel R. (2002) Huntington's disease age of onset linked to polyglutamine aggregation nucleation. Proc. Natl. Acad. Sci. U.S.A. 99, 11884–11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Chiara C., Menon R. P., Dal Piaz F., Calder L., Pastore A. (2005) Polyglutamine is not all. The functional role of the AXH domain in the ataxin-1 protein. J. Mol. Biol. 354, 883–893 [DOI] [PubMed] [Google Scholar]
- 10. Ellisdon A. M., Thomas B., Bottomley S. P. (2006) The two-stage pathway of ataxin-3 fibrillogenesis involves a polyglutamine-independent step. J. Biol. Chem. 281, 16888–16896 [DOI] [PubMed] [Google Scholar]
- 11. Tam S., Spiess C., Auyeung W., Joachimiak L., Chen B., Poirier M. A., Frydman J. (2009) The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat. Struct. Mol. Biol. 16, 1279–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thakur A. K., Jayaraman M., Mishra R., Thakur M., Chellgren V. M., Byeon I. J., Anjum D. H., Kodali R., Creamer T. P., Conway J. F., Gronenborn A. M., Wetzel R. (2009) Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat. Struct. Mol. Biol. 16, 380–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nagai Y., Inui T., Popiel H. A., Fujikake N., Hasegawa K., Urade Y., Goto Y., Naiki H., Toda T. (2007) A toxic monomeric conformer of the polyglutamine protein. Nat. Struct. Mol. Biol. 14, 332–340 [DOI] [PubMed] [Google Scholar]
- 14. Poirier M. A., Jiang H., Ross C. A. (2005) A structure-based analysis of huntingtin mutant polyglutamine aggregation and toxicity. Evidence for a compact β-sheet structure. Hum. Mol. Genet. 14, 765–774 [DOI] [PubMed] [Google Scholar]
- 15. Poirier M. A., Li H., Macosko J., Cai S., Amzel M., Ross C. A. (2002) Huntingtin spheroids and protofibrils as precursors in polyglutamine fibrilization. J. Biol. Chem. 277, 41032–41037 [DOI] [PubMed] [Google Scholar]
- 16. Wanker E. E. (2000) Protein aggregation and pathogenesis of Huntington's disease. Mechanisms and correlations. Biol. Chem. 381, 937–942 [DOI] [PubMed] [Google Scholar]
- 17. Bhattacharyya A., Thakur A. K., Chellgren V. M., Thiagarajan G., Williams A. D., Chellgren B. W., Creamer T. P., Wetzel R. (2006) Oligoproline effects on polyglutamine conformation and aggregation. J. Mol. Biol. 355, 524–535 [DOI] [PubMed] [Google Scholar]
- 18. Duennwald M. L., Jagadish S., Muchowski P. J., Lindquist S. (2006) Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc. Natl. Acad. Sci. U.S.A. 103, 11045–11050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chai Y., Shao J., Miller V. M., Williams A., Paulson H. L. (2002) Live-cell imaging reveals divergent intracellular dynamics of polyglutamine disease proteins and supports a sequestration model of pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 99, 9310–9315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cummings C. J., Mancini M. A., Antalffy B., DeFranco D. B., Orr H. T., Zoghbi H. Y. (1998) Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat. Genet. 19, 148–154 [DOI] [PubMed] [Google Scholar]
- 21. Kazantsev A., Preisinger E., Dranovsky A., Goldgaber D., Housman D. (1999) Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 96, 11404–11409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim S., Nollen E. A., Kitagawa K., Bindokas V. P., Morimoto R. I. (2002) Polyglutamine protein aggregates are dynamic. Nat. Cell Biol. 4, 826–831 [DOI] [PubMed] [Google Scholar]
- 23. Nucifora F. C., Jr., Sasaki M., Peters M. F., Huang H., Cooper J. K., Yamada M., Takahashi H., Tsuji S., Troncoso J., Dawson V. L., Dawson T. M., Ross C. A. (2001) Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science 291, 2423–2428 [DOI] [PubMed] [Google Scholar]
- 24. Rajan R. S., Illing M. E., Bence N. F., Kopito R. R. (2001) Specificity in intracellular protein aggregation and inclusion body formation. Proc. Natl. Acad. Sci. U.S.A. 98, 13060–13065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steffan J. S., Kazantsev A., Spasic-Boskovic O., Greenwald M., Zhu Y. Z., Gohler H., Wanker E. E., Bates G. P., Housman D. E., Thompson L. M. (2000) The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl. Acad. Sci. U.S.A. 97, 6763–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stenoien D. L., Mielke M., Mancini M. A. (2002) Intranuclear ataxin1 inclusions contain both fast- and slow-exchanging components. Nat. Cell Biol. 4, 806–810 [DOI] [PubMed] [Google Scholar]
- 27. Yazawa I., Nukina N., Hashida H., Goto J., Yamada M., Kanazawa I. (1995) Abnormal gene product identified in hereditary dentatorubral-pallidoluysian atrophy (DRPLA) brain. Nat. Genet. 10, 99–103 [DOI] [PubMed] [Google Scholar]
- 28. Nucifora F. C., Jr., Ellerby L. M., Wellington C. L., Wood J. D., Herring W. J., Sawa A., Hayden M. R., Dawson V. L., Dawson T. M., Ross C. A. (2003) Nuclear localization of a non-caspase truncation product of atrophin-1, with an expanded polyglutamine repeat, increases cellular toxicity. J. Biol. Chem. 278, 13047–13055 [DOI] [PubMed] [Google Scholar]
- 29. Yamada M., Sato T., Shimohata T., Hayashi S., Igarashi S., Tsuji S., Takahashi H. (2001) Interaction between neuronal intranuclear inclusions and promyelocytic leukemia protein nuclear and coiled bodies in CAG repeat diseases. Am. J. Pathol. 159, 1785–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yamada M., Wood J. D., Shimohata T., Hayashi S., Tsuji S., Ross C. A., Takahashi H. (2001) Widespread occurrence of intranuclear atrophin-1 accumulation in the central nervous system neurons of patients with dentatorubral-pallidoluysian atrophy. Ann. Neurol. 49, 14–23 [DOI] [PubMed] [Google Scholar]
- 31. Okamura-Oho Y., Miyashita T., Ohmi K., Yamada M. (1999) Dentatorubral-pallidoluysian atrophy protein interacts through a proline-rich region near polyglutamine with the SH3 domain of an insulin receptor tyrosine kinase substrate. Hum. Mol. Genet. 8, 947–957 [DOI] [PubMed] [Google Scholar]
- 32. Miyashita T., Nagao K., Ohmi K., Yanagisawa H., Okamura-Oho Y., Yamada M. (1998) Intracellular aggregate formation of dentatorubral-pallidoluysian atrophy (DRPLA) protein with the extended polyglutamine. Biochem. Biophys. Res. Commun. 249, 96–102 [DOI] [PubMed] [Google Scholar]
- 33. Matsumoto G., Kim S., Morimoto R. I. (2006) Huntingtin and mutant SOD1 form aggregate structures with distinct molecular properties in human cells. J. Biol. Chem. 281, 4477–4485 [DOI] [PubMed] [Google Scholar]
- 34. Lippincott-Schwartz J., Snapp E., Kenworthy A. (2001) Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2, 444–456 [DOI] [PubMed] [Google Scholar]
- 35. Wanker E. E., Scherzinger E., Heiser V., Sittler A., Eickhoff H., Lehrach H. (1999) Membrane filter assay for detection of amyloid-like polyglutamine-containing protein aggregates. Methods Enzymol. 309, 375–386 [DOI] [PubMed] [Google Scholar]
- 36. Neuhoff V., Arold N., Taube D., Ehrhardt W. (1988) Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9, 255–262 [DOI] [PubMed] [Google Scholar]
- 37. Schilling G., Wood J. D., Duan K., Slunt H. H., Gonzales V., Yamada M., Cooper J. K., Margolis R. L., Jenkins N. A., Copeland N. G., Takahashi H., Tsuji S., Price D. L., Borchelt D. R., Ross C. A. (1999) Nuclear accumulation of truncated atrophin-1 fragments in a transgenic mouse model of DRPLA. Neuron 24, 275–286 [DOI] [PubMed] [Google Scholar]
- 38. Holmberg C. I., Staniszewski K. E., Mensah K. N., Matouschek A., Morimoto R. I. (2004) Inefficient degradation of truncated polyglutamine proteins by the proteasome. EMBO J. 23, 4307–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krol H. A., Krawczyk P. M., Bosch K. S., Aten J. A., Hol E. M., Reits E. A. (2008) Polyglutamine expansion accelerates the dynamics of ataxin-1 and does not result in aggregate formation. PLoS ONE 3, e1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chapple J. P., Bros-Facer V., Butler R., Gallo J. M. (2008) Focal distortion of the nuclear envelope by huntingtin aggregates revealed by lamin immunostaining. Neurosci. Lett. 447, 172–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Waelter S., Boeddrich A., Lurz R., Scherzinger E., Lueder G., Lehrach H., Wanker E. E. (2001) Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell 12, 1393–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lammerding J., Fong L. G., Ji J. Y., Reue K., Stewart C. L., Young S. G., Lee R. T. (2006) Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 281, 25768–25780 [DOI] [PubMed] [Google Scholar]
- 43. Hinz J., Gierasch L. M., Ignatova Z. (2008) Orthogonal cross-seeding. An approach to explore protein aggregates in living cells. Biochemistry 47, 4196–4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chien P., Weissman J. S. (2001) Conformational diversity in a yeast prion dictates its seeding specificity. Nature 410, 223–227 [DOI] [PubMed] [Google Scholar]
- 45. Ignatova Z., Thakur A. K., Wetzel R., Gierasch L. M. (2007) In-cell aggregation of a polyglutamine-containing chimera is a multistep process initiated by the flanking sequence. J. Biol. Chem. 282, 36736–36743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. O'Nuallain B., Williams A. D., Westermark P., Wetzel R. (2004) Seeding specificity in amyloid growth induced by heterologous fibrils. J. Biol. Chem. 279, 17490–17499 [DOI] [PubMed] [Google Scholar]
- 47. Allen T. D., Cronshaw J. M., Bagley S., Kiseleva E., Goldberg M. W. (2000) The nuclear pore complex. Mediator of translocation between nucleus and cytoplasm. J. Cell Sci. 113, 1651–1659 [DOI] [PubMed] [Google Scholar]
- 48. Shen Y., Peterson A. S. (2009) Atrophins' emerging roles in development and neurodegenerative disease. Cell Mol. Life Sci. 66, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kaganovich D., Kopito R., Frydman J. (2008) Misfolded proteins partition between two distinct quality control compartments. Nature 454, 1088–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Serio T. R., Cashikar A. G., Kowal A. S., Sawicki G. J., Moslehi J. J., Serpell L., Arnsdorf M. F., Lindquist S. L. (2000) Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289, 1317–1321 [DOI] [PubMed] [Google Scholar]
- 51. Bulone D., Masino L., Thomas D. J., San Biagio P. L., Pastore A. (2006) The interplay between PolyQ and protein context delays aggregation by forming a reservoir of protofibrils. PLoS ONE 1, e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Masino L., Nicastro G., Menon R. P., Dal Piaz F., Calder L., Pastore A. (2004) Characterization of the structure and the amyloidogenic properties of the Josephin domain of the polyglutamine-containing protein ataxin-3. J. Mol. Biol. 344, 1021–1035 [DOI] [PubMed] [Google Scholar]
- 53. Romero P., Obradovic Z., Dunker A. K. (2004) Natively disordered proteins. Functions and predictions. Appl. Bioinformatics 3, 105–113 [DOI] [PubMed] [Google Scholar]
- 54. Mohan A., Oldfield C. J., Radivojac P., Vacic V., Cortese M. S., Dunker A. K., Uversky V. N. (2006) Analysis of molecular recognition features (MoRFs). J. Mol. Biol. 362, 1043–1059 [DOI] [PubMed] [Google Scholar]
- 55. Klement I. A., Skinner P. J., Kaytor M. D., Yi H., Hersch S. M., Clark H. B., Zoghbi H. Y., Orr H. T. (1998) Ataxin-1 nuclear localization and aggregation. Role in polyglutamine-induced disease in SCA1 transgenic mice. Cell 95, 41–53 [DOI] [PubMed] [Google Scholar]
- 56. Saudou F., Finkbeiner S., Devys D., Greenberg M. E. (1998) Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell 95, 55–66 [DOI] [PubMed] [Google Scholar]
- 57. Iwahashi C. K., Yasui D. H., An H. J., Greco C. M., Tassone F., Nannen K., Babineau B., Lebrilla C. B., Hagerman R. J., Hagerman P. J. (2006) Protein composition of the intranuclear inclusions of FXTAS. Brain 129, 256–271 [DOI] [PubMed] [Google Scholar]
- 58. Rousseau F., Schymkowitz J., Serrano L. (2006) Protein aggregation and amyloidosis. Confusion of the kinds? Curr. Opin. Struct. Biol. 16, 118–126 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.