Background: Human ABH8 is a tRNA-hypermodifying enzyme paralogous to the DNA repair enzyme AlkB.
Results: Crystal structures of the RRM/AlkB domains of ABH8 were determined in conjunction with thermodynamic assays.
Conclusion: Substrate specificity and catalytic activity are modulated by conformational adaptations in and around the active site.
Significance: These results provide insight into the functional expansion of the AlkB enzyme family in higher eukaryotes.
Keywords: Crystal Structure, Dioxygenase, DNA Repair, Enzyme Mechanisms, Enzyme Structure, Multifunctional Enzymes, RNA Modification, RNA-Protein Interaction, AlkB, tRNA Modification
Abstract
Humans express nine paralogs of the bacterial DNA repair enzyme AlkB, an iron/2-oxoglutarate-dependent dioxygenase that reverses alkylation damage to nucleobases. The biochemical and physiological roles of these paralogs remain largely uncharacterized, hampering insight into the evolutionary expansion of the AlkB family. However, AlkB homolog 8 (ABH8), which contains RNA recognition motif (RRM) and methyltransferase domains flanking its AlkB domain, recently was demonstrated to hypermodify the anticodon loops in some tRNAs. To deepen understanding of this activity, we performed physiological and biophysical studies of ABH8. Using GFP fusions, we demonstrate that expression of the Caenorhabditis elegans ABH8 ortholog is widespread in larvae but restricted to a small number of neurons in adults, suggesting that its function becomes more specialized during development. In vitro RNA binding studies on several human ABH8 constructs indicate that binding affinity is enhanced by a basic α-helix at the N terminus of the RRM domain. The 3.0-Å-resolution crystal structure of a construct comprising the RRM and AlkB domains shows disordered loops flanking the active site in the AlkB domain and a unique structural Zn(II)-binding site at its C terminus. Although the catalytic iron center is exposed to solvent, the 2-oxoglutarate co-substrate likely adopts an inactive conformation in the absence of tRNA substrate, which probably inhibits uncoupled free radical generation. A conformational change in the active site coupled to a disorder-to-order transition in the flanking protein segments likely controls ABH8 catalytic activity and tRNA binding specificity. These results provide insight into the functional and structural adaptations underlying evolutionary diversification of AlkB domains.
Introduction
Humans express nine recognized paralogs of the Escherichia coli DNA repair enzyme AlkB, which directly reverses alkylation damage to nucleobases using an iron-catalyzed oxidation reaction. All nine of the human AlkB homologs (ABHs)2 are conserved in vertebrate organisms, while five are also conserved in metazoans (1). Variations in the sequence or expression of several of them have been associated with cancer and obesity (1–3). The expansion of the protein family in metazoans and vertebrates suggests that diversification of AlkB domain function has contributed to the evolution of greater developmental complexity in these organisms. However, although this inference is supported by some experimental observations (4–6), the substrate specificity and physiological function remain unknown for most of them. In this context, many questions remain unresolved concerning the structural and functional diversification of the AlkB domain family.
E. coli AlkB is a member of the iron/2-oxoglutarate (Fe(II)/2OG)-dependent dioxygenase enzyme superfamily. E. coli encodes two recognized representatives of this superfamily (AlkB and TauD), whereas humans encode 24, nine of which belong to the AlkB family (7). The enzymes in this superfamily utilize an Fe(II)-catalyzed reaction mechanism to monohydroxylate substrates using molecular oxygen (O2). Oxidation of the 2OG substrate to succinate functions effectively as a molecular sink for the second oxygen atom in O2.
E. coli AlkB directly repairs SN2 alkylation damage on endocyclic nitrogen atoms in DNA and RNA bases by hydroxylating the covalently bound carbon atom, resulting in spontaneous release of an aldehyde product to regenerate the unmodified nucleobase (8, 9). Human ABH1, ABH2, and ABH3 have been demonstrated to possess similar nucleobase repair activities although with different substrate specificities (6, 10). Extensive structural and enzymological studies performed on E. coli AlkB have demonstrated that it has remarkably broad substrate specificity. It is active in repairing methyl, ethyl, and etheno lesions on adenine and cytosine bases in single- and double-stranded DNA as well as RNA (8, 9, 11–13). The intrinsic flexibility of several loops flanking the active site has been demonstrated to play an important role in mediating promiscuous recognition of substrates of varying molecular structure. Although the nucleotide recognition lid formed by these loops is well ordered in all 2OG- or succinate-bound crystal structures of E. coli AlkB, variations in its conformation enable accommodation of diverse substrates in an efficient catalytic geometry (14).
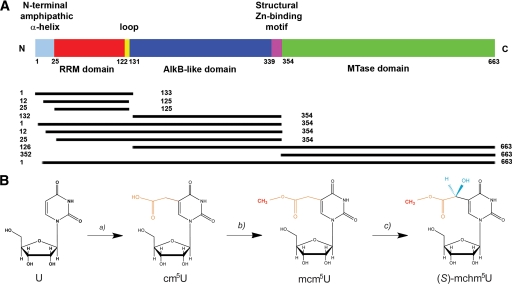
In contrast to the broad substrate specificity that is the hallmark of E. coli AlkB, the human paralog ABH8 has been demonstrated to recognize a unique substrate, a modified nucleobase found at a specific position in several tRNA molecules. This different selectivity is likely to require substantial evolutionary adaptations in the molecular features mediating substrate recognition (15–18). ABH8, which is conserved in metazoans and vertebrates, has a unique tripartite domain organization comprising an N-terminal RNA recognition motif (RRM) domain and a C-terminal methyltransferase (MTase) domain in addition to its central AlkB-like domain (Fig. 1A). Both the AlkB-like and MTase domains have been demonstrated recently to catalyze covalent hypermodifications of the wobble nucleotide base in the anticodon loops of specific tRNAs (Fig. 1B) (15–18). The MTase domain in complex with the accessory protein Trm112 methylates 5-carboxymethyluridine (cm5U) to produce 5-methoxycarbonylmethyluridine (mcm5U) in both yeast and mammalian tRNAs (15, 17, 19). In mammalian tRNA-Gly, the AlkB-like domain hydroxylates mcm5U to generate (S)-5-methoxycarbonylhydroxymethyluridine ((S)-mchm5U) (16, 18). These observations provide the first example of an AlkB-like domain having a biochemical function other than oxidative repair of alkylated nucleotide bases and expand the functional repertoire of the AlkB protein family.
FIGURE 1.
ABH8 domain organization and catalytic activity. A, schematic diagram illustrating ABH8 domain organization and the human protein constructs used in this study. B, an unidentified enzyme or enzymes convert uridine at position 34 (U34) in the anticodon loop of tRNAs into cm5U, which is methylated by the MTase domain of ABH8 in complex with the Trm112 protein to generate mcm5U (15, 17). This product is stereoselectively hydroxylated by the AlkB domain of ABH8 to generate (S)-mchm5U (16, 18).
In addition to providing insight into the functional expansion of the AlkB protein family in higher eukaryotes, analysis of the physiological and biochemical function of ABH8 should contribute to a deeper understanding of tRNA modification and its physiological importance. Over 80 covalent modifications have been detected in tRNAs (20). Although some seem to modulate their folding and stability (21), the modifications in the anticodon loop and particularly in the wobble base have received increasing attention for their influence on protein translation (22–24) and biological phenotype (25–27). Disease-associated single nucleotide polymorphisms have been mapped to tRNA modification enzymes in humans (28), whereas genetic perturbations in these enzymes have been associated with metazoan developmental defects (29, 30). The observation that ABH8 is frequently overexpressed in human bladder cancers provides a link between this tRNA modification enzyme and a biological process critically influenced by DNA repair (1). A more direct link comes from the observation that Trm9, the yeast ortholog of the ABH8 MTase domain, up-regulates translation of a wide variety of proteins involved in DNA repair whose transcripts are enriched in specific codons (19). This connection places ABH8 at the center of questions concerning the evolution of the catalytic activities of AlkB proteins and the relationship between their DNA repair activity and translational regulation of the DNA damage response and of other cellular processes.
To address these questions, we undertook in vivo genetic investigations of the Caenorhabditis elegans ortholog C14B1.10 (35% identical to the human ortholog over all three domains; Fig. 2) in addition to characterizing the RNA binding properties of human ABH8 and the crystal structure of its RRM/AlkB domains. These multidisciplinary studies clarify the relationship between the structural evolution of the AlkB domain and the biological functions of AlkB family enzymes.
FIGURE 2.
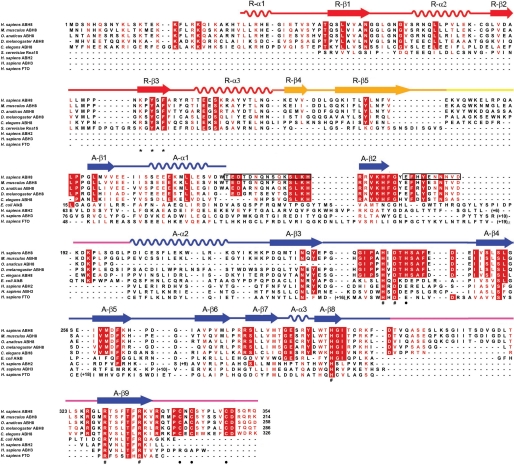
Sequence-structure alignment for the ABH8 protein. A sequence alignment (59) of the RRM/AlkB domains of a set of metazoan ABH8 orthologs is combined with structure-based alignments of E. coli AlkB, human ABH2, human ABH3, the AlkB domain of human FTO, and the RRM domain of S. cerevisiae RNa15. The RNP1 consensus sequence in the RRM domain is indicated by asterisks (*), the invariant residues chelating Mn(II) and 2OG in the AlkB domain are indicated by pound signs (#), and the residues ligating Zn(II) in this domain are indicated by black dots (●). The black boxes indicate disordered regions in the crystal structure of the RRM/AlkB domains of human ABH8 (Table 2 and Fig. 6 below). Conserved and semiconserved residues, grouped according to their biophysical characteristics, are colored white on a red background and red on a white background, respectively. The secondary structural elements found in the crystal structure of the RRM/AlkB domains of human ABH8 are shown schematically above the sequence alignment, with α-helices represented as sine curves and β-strands as arrows. H., Homo; M., Mus; O., Ornithorhynchus; D., Drosophila; C., Caenorhabditis; S., Saccharomyces; E., Escherichia.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Most proteins were expressed in E. coli Rosetta2 cells (Novagen), which were grown in LB at 20 °C until A600 reached 0.6–0.8 and then induced overnight at the same temperature with 1 mm isopropyl 1-thio-β-d-galactopyranoside (0.0075 mm for the constructs in complex with Trm112). For SeMet labeling, the 25–354-His6 construct was expressed in E. coli B834λ(DE3) cells, which are auxotrophic for methionine. An overnight culture from a single colony was grown overnight at 37 °C in LB and diluted 1:100 into M9 minimal medium containing 50 mg/liter dl-SeMet (Sigma-Aldrich) for a final overnight growth at 37 °C. This medium was supplemented with 5 mg/liter tryptophan and tyrosine, 50 mg/liter each remaining amino acid other than methionine, 1% (v/v) Kao and Michayluk Vitamin Solution (Sigma-Aldrich), and 0.005% (w/v) kanamycin. The concentrated tryptophan/tyrosine stock was made in 200 mm HCl, whereas the other amino acids were dissolved in water. The overnight culture in supplemented M9 medium was diluted into the same medium, grown at 37 °C until A600 reached 0.6–0.8, and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 16 h at 20 °C. Cells were washed and then lysed by sonication, and the supernatant from a low speed spin was purified by Ni-NTA chromatography (Qiagen, Valencia, CA). The protein-containing eluate from this column was concentrated in a Centricon (Millipore Inc., Billerica, MA), purified by Sephacryl S200 gel filtration chromatography in storage buffer (typically 100–150 mm NaCl, 5% glycerol, 10 mm Tris-Cl, pH 7.5), concentrated, and snap frozen in liquid N2 for storage in small aliquots at −80 °C. When the hexahistidine tag was cleaved, tobacco etch virus (TEV) protease was added to the Ni-NTA eluate at a 1:30 weight ratio during overnight dialysis at 4 °C to remove the imidazole, and the reaction was passed through a second Ni-NTA column to remove the cleaved tag and hexahistidine-tagged TEV protease (31) prior to concentration for gel filtration chromatography. The buffers used for purification were optimized for each construct (supplemental Table S1). The constructs in complex with Trm112 were purified using the published protocol (17) except for the substitution of 50 mm Tris-Cl, pH 7.5 for phosphate buffer and the addition of a final gel filtration chromatography step.
In Vitro tRNA Transcription
Human tRNA-Gly and tRNA-Glu were transcribed using T7 RNA polymerase (Stratagene, San Diego, CA). Double-stranded DNA coding-sequence templates (Genomic tRNA Database) with added T7 promoter sequences were constructed by PCR using three overlapping primers (supplemental Table S2) (32) and purified by precipitation via addition of 300 mm NaOAc, 2.5 volumes of EtOH, and 0.1 mg/ml glycogen (Fermentas-Thermo Fisher Scientific). In vitro transcription reactions contained 0.2–1.0 μg of precipitated PCR product, 2 μl of 10× transcription buffer, 5 mm NTPs, 10 mm DTT, and 1 μl of T7 polymerase in a total volume of 20 μl. For radioligand binding assays, 1 μl of [α-32P]GTP was added to the reaction mixture. Transcripts were purified on an 8% polyacrylamide gel containing 7 m urea, and bands identified by UV shadowing were excised and extracted overnight with 300 mm NaOAc. Prior to each binding assay, tRNAs were heated at 65 °C for 10 min and then cooled at room temperature to promote proper folding.
Filter Binding Assays
Radiolabeled RNA (5 nm) was mixed with 0.05–4.0 μm protein in 50 μl of RNA binding buffer (150 mm NaCl, 1 mm MgCl2, 20 mm HEPES, pH 7.5) and incubated at room temperature for 30 min. Triplicate reactions were applied to a 96-well Minifold Dot-Blot system (Millipore, Billerica, MA) containing two membranes equilibrated in RNA binding buffer, a protein-binding 0.45-μm Whatman Protran nitrocellulose membrane and a nucleic acid-binding Hybond-N+ nylon membrane (Thermo Fisher Scientific, Pittsburgh, PA). The filters were washed three times with the same buffer and then allowed to dry. Signals from the protein-binding and RNA-binding membranes were quantified using a Storm phosphorimaging system (GE Healthcare). The ratio of protein-bound versus total RNA was analyzed as a function of protein concentration using the curve fitting procedure described in the supplemental Methods.
Fluorescence Anisotropy Assays
Synthetic HPLC-purified RNAs (supplemental Table S2) with 5′-fluorescein labels (Invitrogen) were dissolved at a 200 μm concentration in RNase-free water (Ambion, Austin, TX) and stored in 10 μl aliquots at −80 °C. RNA samples were thawed immediately prior to use and diluted to 60 nm (17-mer stem-loop) or 73 nm (aptamer ABH8-2.2) concentration in 1.1 ml of freshly degassed RNA binding buffer in diethyl pyrocarbonate-treated water (Sigma-Aldrich; 97% NMR grade). Protein titrations were performed in a jacketed cell holder maintained at 25 °C by a water bath with temperature monitored by a Digi-Sense T-type thermocouple thermometer. Fluorescein anisotropy at 523 nm was measured in a T-format QuantaMaster C61 spectrofluorometer (Photon Technology International, Birmingham, NJ) with Glan-Thompson polarizers using 490-nm vertically polarized excitation and 5-nm slits. Photomultipliers were used in digital photon counting mode. Curve fitting was performed as described in the supplemental Methods.
Protein Crystallization
Although lead crystals were obtained for an RRM/AlkB construct containing the intact N terminus (i.e. 1–354), excision of the first 24 N-terminal residues and retention of the C-terminal hexahistidine tag yielded much stronger diffraction. This 25–354-His6 construct was crystallized in 2:1 (protein:precipitant) microbatch reactions under paraffin oil at 20 °C using a 5 mg/ml protein stock solution containing 2.8 mm MnCl2 and 8.6 mm 2OG that was mixed with precipitant containing 28.5–30.5% (w/v) PEG 4000, 15% (v/v) glycerol, 170 mm NH4(CH3COO), 85 mm sodium citrate, pH 5.6. Crystals of the SeMet-labeled construct were obtained in a different space group using the same protocol with a precipitant containing 30% PEG 3350, 0.15 m dl-malic acid, pH 7. Both crystal forms grew to useful dimensions in 4 days and were cryoprotected with 15% (w/v) ethylene glycol.
X-ray Structure Determination
A single wavelength anomalous diffraction data set at 3.2-Å resolution was collected from a SeMet-labeled crystal at 100 K at the anomalous peak wavelength of selenium on the X4C beamline of the National Synchrotron Light Source at Brookhaven National Laboratory. The diffraction images were processed and merged using the HKL package (33). Although the selenium site pattern was clear, attempts to solve the structure using the single wavelength anomalous diffraction method were unsuccessful. The structure was solved from this data set using the molecular replacement method as implemented in the program PHASER (34). A dual search model was used comprising the NMR structure of the RRM domain of human ABH8 (Protein Data Bank code 2CQ2) after removal of the flexible backbone segments (i.e. retaining residues Tyr40–Asp125) and a homology model of the AlkB-like domain covering residues Lys169–Val337 generated by the program PHYRE from the crystal structure of E. coli AlkB (Protein Data Bank code 2FDI (35); 31% identity to human ABH8). The initial refinement of the solution from PHASER using CNS 1.2 (36) resulted in Rwork and Rfree values of 40.1 and 52.4%, respectively. However, the resulting electron density map was of sufficient quality to complete the structure (supplemental Table S1) using iterative cycles of manual rebuilding in XtalView (37) and computational refinement in CNS. This model from the SeMet crystals was used to solve the structure of the non-SeMet-labeled crystals via molecular replacement using the program MOLREP (38) followed by refinement in CNS maintaining strong non-crystallographic symmetry restraints (300 kcal/Å and σB = 2) throughout the RRM domains in all four protomers in the asymmetric unit. The final cycle of refinement for both crystal structures included all structure factors (i.e. without applying any σ cutoff). A diffraction data set collected on the non-SeMet-labeled crystals at the Zn(II) anomalous edge (9671.7618 eV) on the X4A beamline showed 5–6-σ peaks at the Zn(II) position in the C-terminal structural Zn(II)-binding site in the A, B, and D subunits and a 3-σ peak at this position in the C subunit (data not shown).
RESULTS
C. elegans ABH8 Ortholog Displays Developmentally Regulated Expression
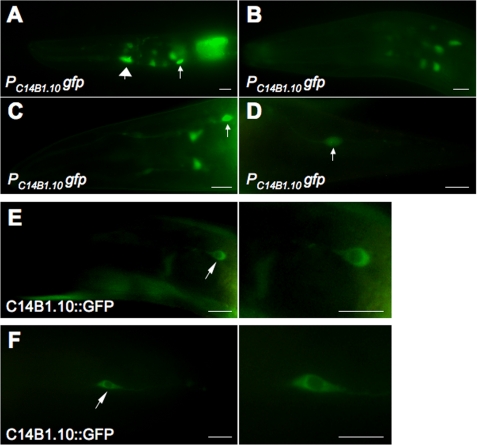
Previous genomic studies demonstrated that knockdown of ABH8 in Drosophila melanogaster produces a fatal defect in cardiac development (30) and that an internal deletion in the gene encoding C. elegans ABH8 (designated C14B1.10) causes embryonic lethality or sterility in animals surviving to adulthood (39). We observed that a PC14B1.10gfp promoter fusion to the GFP protein is expressed broadly in early stage C. elegans larvae but only in a small number of cells, primarily neurons, in adults (Fig. 3, A–D). This progressive restriction in its expression pattern suggests that the physiological function of ABH8 may change and become more specialized during the course of development and that it may play a role in neuronal function in adults (see “Discussion”).
FIGURE 3.
Developmentally regulated expression of C14B1.10, the C. elegans ABH8 ortholog. A–D, PC14B1.10gfp, a fusion of the C14B1.10 promoter to the GFP gene, shows expression in neuronal (smaller arrows) and non-neuronal (large arrow) cells. Expression is higher and more widely distributed at the early larval stage (A), reduced at the middle larvae stage (B), and almost exclusively detected in neuronal cells in the head (C) and tail (D) of adult animals. E and F, C14B1.10::GFP, the GFP protein fused to the C14B1.10 protein under its endogenous promoter control, is exclusively localized to the cytoplasm. Arrows indicate the positions of selected neurons in the head (E) and tail (F) of an adult animal. Scale bars, 5 μm.
A C14B1.10::GFP protein fusion is localized to the cytoplasm of the neurons in which it is expressed in adult worms (Fig. 3, E and F). Notably, similar expression and localization patterns were observed in C. elegans for ELPC-1 and ELPC-3, proteins that are required for the synthesis of mcm5U (i.e. the substrate for the AlkB domain of ABH8); mutations in these enzymes produce neurobehavioral defects (29, 40). ELPC-3 is a radical S-adenosylmethionine (SAM) enzyme homologous to MiaB, an E. coli enzyme that catalyzes a nucleobase modification in tRNA (41). ELPC-1 and ELPC-3 were identified previously as components of “Elongator,” an RNA polymerase II-associated histone acetyltransferase complex (42, 43). In this context, it is noteworthy that the C. elegans ABH8 ortholog is expressed in an operon (WormBase) that includes a subunit of a conserved histone trimethylation complex as well as diphthamide synthetase, an enzyme that covalently modifies eukaryotic translational elongation factor 2. Therefore, multiple connections exist between neurodevelopmental processes, covalent modification of histones, and covalent modification of the translational apparatus in C. elegans.
Protein Constructs for Biophysical Analyses of Human ABH8
To dissect the in vitro biophysical and structural properties of ABH8, we expressed and purified from E. coli a series of protein constructs derived from the human ABH8 ortholog: three RRM single domain constructs (1–133, 12–125, and 25–125), two RRM/AlkB double domain constructs (1–354 and 25–354), an AlkB single domain construct (132–354), an AlkB/MTase double domain construct (126–663), an MTase single domain construct (352–663), and the full-length protein (1–663) (Fig. 1A). Constructs containing the MTase domain were only soluble in the presence of the accessory protein Trm112 as reported previously (15, 17). A basic N-terminal protein segment predicted to form an amphipathic α-helix spanning residues 13–32 in human ABH8 (44) was included in some protein constructs but not others.
Hydrodynamic analyses of the concentrated protein stocks were conducted using analytical gel filtration chromatography monitored by refractive index and light scattering detectors (supplemental Fig. S1), which provide an accurate measurement of mass-averaged molecular weight. Many constructs showed some tendency to aggregate, especially those containing the MTase domain and Trm112. However, all constructs showed a substantial population of monomers during elution from the gel filtration column. The full-length human ABH8-Trm112 complex eluted as a rapidly interconverting monomer and dimer (supplemental Fig. S1E). After His tag cleavage, the 25–354 RRM/AlkB double domain construct eluted as a monodisperse monomer (supplemental Fig. S1A), although the construct retaining the tag had a tendency to form a varying amount of a stable tetramer (∼10–40% in different preparations; supplemental Fig. S1, B–D).
Inductively coupled plasma mass spectrometry (ICP-MS) assays indicated co-purification of 1.5 mol of Zn(II)/mol of the AlkB domain construct. Combined with additional data presented below, this observation supports formation of a structural Zn(II)-binding site by the conserved cysteine-rich sequence at the C terminus of the AlkB domain in ABH8 (Fig. 1A and supplemental Fig. S2A); omission of this motif from constructs containing the AlkB domain produced insoluble protein (data not shown).
The AlkB Domain of Human ABH8 Is Stabilized by a C-terminal Structural Zn(II)-binding Site
Thermal stability assays were used to assess ligand interaction with ABH8 constructs containing the AlkB domain (Fig. 4 and supplemental Fig. S3). Based on the observed behavior of other Fe(II)/2OG dioxygenases, this domain would be expected to bind the Fe(II) cofactor and 2OG co-substrate cooperatively in the active site even in the absence of tRNA substrate. However, the presence of a possible structural Zn(II)-binding site at the C terminus of the AlkB domain complicated efforts to verify divalent cation binding in the active site. To dissect the influence of divalent cation interaction at the two possible binding sites, thermal stability assays were performed on a variety of constructs, including a C341A/C349A double mutant lacking two of the three invariant Cys residues in the putative structural Zn(II)-binding site. Assays were conducted in the absence or presence of excess EDTA (5 mm), which should remove all divalent cations from the AlkB domain, as well as varying concentrations of Mn(II). This divalent cation has been widely used as a catalytically inactive analog of Fe(II) because it generally binds to the active site in Fe(II)/2OG dioxygenases with an affinity similar to Fe(II). Although Mn(II) might also interact with the structural Zn(II)-binding site, it would be expected to do so with substantially lower affinity than Zn(II).
FIGURE 4.
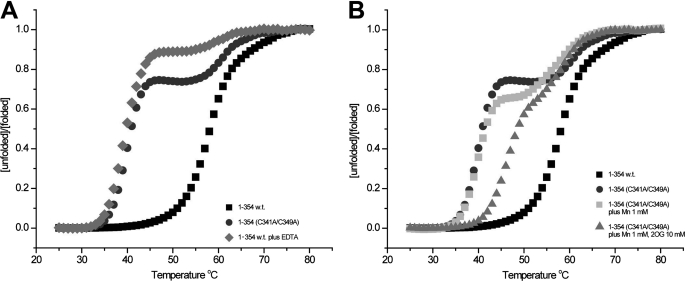
The C-terminal structural Zn(II)-binding site stabilizes the ABH8 RRM/AlkB double domain construct. A and B, thermal denaturation assays monitored using the fluorescent reporter SYPRO Orange demonstrate that the Zn(II)-binding site strongly stabilizes the folding of the AlkB domain. A shows thermal denaturation of the wild-type 1–354 construct in 150 mm NaCl, 50 mm Hepes, pH 7.5 either in the absence (black squares) or in the presence (gray diamonds) of 5 mm EDTA as well as thermal denaturation of the C341A/C349A double mutant of the same construct in the absence of EDTA (dark gray circles). B shows equivalent assays performed on the double mutant in the absence of EDTA with the addition of either 1 mm Mn(II) (light gray squares) or 1 mm Mn(II) plus 10 mm 2OG (gray triangles); the WT and double mutant denaturation curves in the absence of EDTA are shown here again for comparison. The cooperative action of Mn(II)/2OG in stabilizing the first of the two sequential transitions in the double mutant indicates that this transition corresponds to unfolding of the AlkB domain.
Thermal denaturation of AlkB domain-containing constructs of human ABH8 was monitored using the fluorescent reporter dye SYPRO Orange. The 1–354 RRM/AlkB double domain construct exhibits a single unfolding transition with a midpoint (Tm) at ∼58 °C. An equivalent change in thermal denaturation behavior is observed either upon introduction of the C341A/C349A double mutation, which should block the binding of Zn(II) to the C-terminal Cys-rich sequence motif in the AlkB domain, or upon addition of 5 mm EDTA to the wild-type construct. Both of these variations split the single transition with Tm ∼58 °C into two sequential unfolding transitions with midpoints at ∼40 and ∼61 °C (Fig. 4A). The observations that metal chelation and the Cys mutations have the same effect demonstrate that the AlkB domain is destabilized by removal of a Zn(II) ion bound to its C-terminal Cys-rich sequence motif and that this destabilization thermodynamically decouples the unfolding of the AlkB domain from that of the RRM domain.
When 1 mm Mn(II) and 10 mm 2OG co-substrate are added to the RRM/AlkB double domain construct harboring the C341A/C349A mutations, the Tm of the lower unfolding transition shifts up by ∼7 °C to ∼48 °C without changing the Tm of the higher transition (Fig. 4B). (Binding of 2OG to Fe(II)/2OG dioxygenases generally requires binding of the directly interacting metal cofactor.) In contrast, at most, a minimal upshift in the Tm of this transition is observed in the presence of 1 mm Mn(II) in the absence of 2OG. These results demonstrate that the AlkB domain in this construct functionally interacts with the metal cofactor and 2OG co-substrate even without occupancy of its structural Zn(II)-binding site. Moreover, they confirm the inference that the first thermal transition reflects unfolding of the AlkB domain in the double domain construct. The isolated AlkB domain construct (residues 132–354) similarly binds 2OG in an Mn(II)-dependent manner (supplemental Fig. S3).
An N-terminal α-Helix Critically Contributes to RNA Binding by ABH8
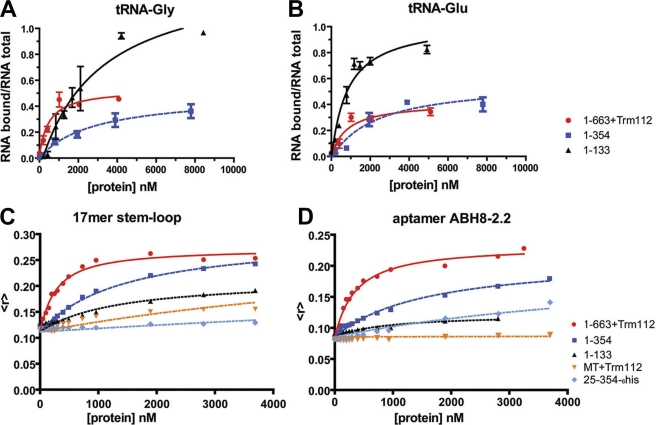
Assays evaluating the binding of different protein constructs to different RNA species were used to dissect the energetic contributions of regions of ABH8 to its affinity and specificity for the tRNA substrate. Filter binding assays were used to analyze the binding of ABH8 constructs to in vitro transcribed tRNAs identified previously as possible ABH8 substrates (15, 17, 18) (Fig. 5, A and B, supplemental Fig. S4B, and Table 1), whereas fluorescence anisotropy assays were used to analyze their binding to shorter synthetic RNA species (Fig. 5, C and D, supplemental Fig. S4B, and Table 1). The synthetic RNAs included a 17-mer matching the anticodon stem-loop of tRNA-Gly (18), a control 17-mer with a randomly chosen sequence, and a 44-base aptamer called ABH8-2.2 selected to bind to the 1–354 construct using systematic evolution of ligands by exponential enrichment (SELEX) (45). Aptamer ABH8-2.2, which had the highest affinity of the aptamers isolated by SELEX, has 41 and 53% identity to the anticodon stem-loops of tRNA-Glu and tRNA-Gly, respectively, including a perfect match in the anticodon loop of tRNA-Gly (supplemental Fig. S4A). The 5′-region of this aptamer is enriched in A bases as was the equivalent region in aptamers of slightly lower affinity (data not shown).
FIGURE 5.
RNA binding properties of ABH8 protein constructs. A and B, filter binding assays in which increasing concentrations of protein were titrated at room temperature onto 5 nm radiolabeled tRNA-Gly (A) or tRNA-Glu (B) in 150 mm NaCl, 1 mm MgCl2, 20 mm HEPES, pH 7.5. The mean and standard deviation of the fraction of protein-bound RNA in triplicate assays are plotted for an RRM domain construct (1–133; black), an RRM/AlkB double domain construct (1–354; blue), and the entirety of ABH8 in complex with the Trm112 protein (1–633; red). C and D, fluorescence anisotropy assays in the same buffer in which increasing concentrations of protein were titrated at 25 °C onto 5′-fluorescein-labeled synthetic 17-mer step-loop matching the anticodon loop of tRNA-Gly (C) or aptamer ABH8-2.2 (D), which was selected in vitro to bind to the 1–354 RRM/AlkB double domain construct. Results from a single titration are plotted for an RRM domain construct (1–133; black), RRM/AlkB domain constructs either with (1–354; dark blue) or without (25–354; light blue) the first 24 N-terminal residues, an MTase domain construct in complex with Trm112 (MT; orange), and the entirety of ABH8 protein in complex with Trm112 (1–633; red).
TABLE 1.
Dissociation constants for binding of RNA species to ABH8 domains
See supplemental Methods and the legend to Fig. 5 for buffer conditions.
| Protein constructs | RNA molecules |
||||
|---|---|---|---|---|---|
| tRNA-Glya | tRNA-Glua | 17-mer stem-loopb | Aptamer ABH8-2.2b | Control 17-merb | |
| 1–133 | 3.0 ± 0.9 μm | 830 ± 330 nm | 1.3 ± 0.4 μm | 3.9 ± 0.2 μm | NDc |
| 1–354 | 2.9 ± 0.8 μm | 2.3 ± 1.8 μm | 1.4 ± 0.2 μm | 2.3 ± 0.4 μmd | 6.3 ± 1.0 μm |
| 1–354(C341A/C349A) | ND | ND | 808 ± 151 nm | 2 ± 0.2 μm | ND |
| 25–354-His6 | ND | ND | 26 ± 5 μm | 4.8 ± 0.3 μm | ND |
| 352–663+Trm112e | ND | ND | 9.1 ± 0.9 μm | 61 ± 6 μm | ND |
| 1–663+Trm112e | 490 ± 290 nm | 830 ± 590 nm | 240 ± 29 nm | 240 ± 50 nmd | 350 ± 20 nm |
a Values calculated from radiolabeled filter binding assays at room temperature.
b Values calculated from fluorescence anisotropy assays at 25 °C.
c ND, not determined.
d Filter binding assays on the same RNA species without a fluorescent label gave equivalent binding affinity for the 1–663+Trm112 construct (350 ± 20 nm) but higher affinity for the 1–354 construct (290 ± 90 nm). Either the fluorescent label reduces aptamer affinity for the 1–354 construct or a conformational change in this construct upon filter binding increases its binding affinity.
e Assays on constructs containing the MTase domain (352–663) displayed reduced binding at the highest protein concentrations, suggesting aggregation, limiting the accuracy of the dissociation constants measured for these constructs.
All of the ABH8 protein constructs containing the basic N-terminal α-helix bind RNA with significant affinity but low sequence specificity (Fig. 5, A–D, and Table 1). The full-length 1–663 construct in complex with Trm112 binds to all assayed RNA species, including the nonspecific control 17-mer, with similar 200–800 nm affinities (Fig. 5, supplemental Fig. S4B, and Table 1). RRM single domain and RRM/AlkB double domain constructs both bind RNA with slightly weaker 1–4 μm affinities and an ∼3-fold preference for the tRNA-related species compared with the nonspecific control 17-mer (Fig. 5, supplemental Fig. S4B, and Table 1). The structural Zn(II)-binding site at the C terminus of the AlkB domain does not contribute to this RNA binding affinity based on the results observed with the C341A/C349A double mutant (supplemental Fig. S2B). However, removal of the basic N-terminal α-helix preceding the RRM domain reduces binding affinity for the tRNA-related species by at least 10-fold (Table 1). Constructs lacking this protein segment retain strong affinity only for the selected ABH8-2.2 aptamer, which is most remote from the physiological tRNA substrate. Moreover, the isolated AlkB domain does not show detectable interaction with any of the RNA species tested (Table 1 and supplemental Fig. S4B). Therefore, fairly strong but mostly nonspecific binding of RNA is mediated by the RRM domain of ABH8 in conjunction with its basic N-terminal α-helix, which makes a critical contribution to the binding energy.
X-ray Crystal Structures Show That RRM and AlkB Domains in ABH8 Form a Continuous Surface Likely to Mediate RNA Interaction
We solved crystals that grew in two different space groups, both containing the 25–354 RRM/AlkB double domain construct with a C-terminal TEV-protease-cleavable hexahistidine tag (Fig. 6A, supplemental Figs. S5–S7, and Table 2). These structures with bound Mn(II) and 2OG were refined to working R-factors of 22 and 21.7% and free R-factors of 27.7 and 28.3% at 3.0 and 3.2 Å, respectively. Other constructs, including the equivalent construct retaining the basic N-terminal α-helix, failed to yield high quality crystals. The diffraction power of the 25–354 crystals declined significantly upon removal of the hexahistidine tag.
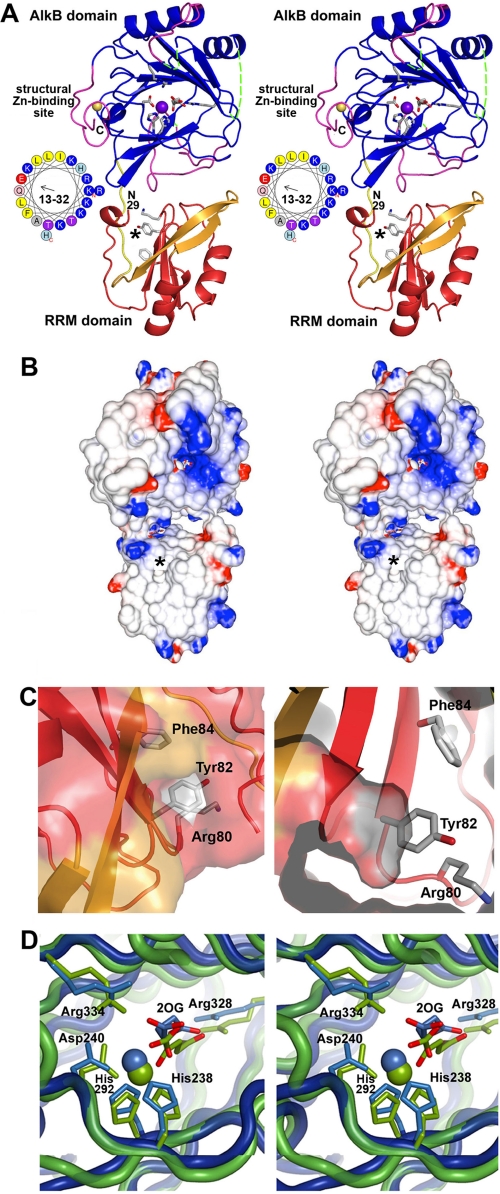
FIGURE 6.
Crystal structure of the RRM/AlkB domains in human ABH8. A, stereo ribbon diagram of the structure in complex with Zn(II) and Mn(II)/2OG. The conserved and non-conserved regions in the RRM domain are colored red and orange, respectively, whereas the conserved and non-conserved regions in the AlkB domain are colored blue and magenta, respectively. The loop connecting the two domains is shown in yellow. Conserved residues in the RRM (Lys80, Tyr82, and Phe84) and AlkB (His238, Asp240, His292, Arg328, and Arg334) domains as well as the 2OG bound to the AlkB domain are shown in stick representation (with carbon and oxygen colored white and red, respectively). The Mn(II) ion bound in the active site and the Zn(II) ion bound in the C-terminal structural Zn(II)-binding site are shown as purple and yellow spheres, respectively. The asterisk in A and B marks the location of a putative pyrimidine-binding pocket in the RRM domain (shown in C). The observed N terminus of the RRM domain at residue 29 and C terminus of the AlkB domain at residue 354 are labeled in black (“N29” and “C”, respectively). Residues 13–32 in ABH8, most of which were deleted to improve crystal quality, are shown as an α-helical wheel with basic amino acids colored blue. Assessment of structural conservation and the numbering of the secondary structural elements (supplemental Fig. S7A) in the AlkB domain are based on comparison with other AlkB family enzymes (48–50) (rather than the Fe(II)/2OG dioxygenase superfamily as done in Yu et al. (35)). Residues 156–174 and 181–192 in the AlkB domain (dotted green lines), which are topologically equivalent to the segments forming the nucleotide-binding lid in E. coli AlkB, are disordered in ABH8. B, stereopair showing the electrostatic potential of the molecular surface of the domains oriented as in A. Fully saturated blue and red colors represent potentials of ±8 kT at 100 mm ionic strength as calculated by GRASP2 (60). C, two views of a putative pyrimidine-binding pocket formed in part by the RNP1 motif in the RRM domain. The entrance to this cavity, which can accommodate a pyrimidine base without steric clash, lies on the surface of the RRM domain below the active site in the AlkB domain. The entrance is marked by a black asterisk in A and B and in supplemental Fig. S7A. Residue Tyr82 is positioned at the base of the cavity (as shown in the right panel) where it could make a stacking interaction with a bound pyrimidine. The residues in the RNP1 motif are shown in stick representation colored according to atomic identity (carbon in gray, oxygen in red, and nitrogen in blue). The molecular surface is colored like the backbone except for the regions formed by the side chains of the residues in RNP1 motif, which are colored according to atomic identity. D, stereopair showing superposition of the active sites in ABH8 (blue) and E. coli AlkB (green; Protein Data Bank code 2FDH) with the invariant residues, the 2OG co-substrates, and the Mn(II) cofactors colored the same as the domains. Mn(II) is widely used in studies of Fe(II)/2OG dioxygenases as a catalytically inactive analog of Fe(II) that preserves active site stereochemistry. Although some high resolution crystal structures of Fe(II)/2OG dioxygenases have a weakly ordered H2O molecule in the final ligation position on the Fe(II), Mn(II), or Co(II) ion occupying the catalytic site, this ligation site is empty in other structures (14, 35, 48–50), as it is in the ABH8 active site shown here. However, other structures generally preserve octahedral coordination geometry for the ligating atoms as shown in D for E. coli AlkB, as opposed to the distorted geometry observed in all of the crystallographically independent views of the ABH8 active site in the structures reported in this study.
TABLE 2.
Data collection and refinement statistics
Standard definitions were used for parameters (61). Entries in parentheses report data for the limiting resolution shell. All observations with I ≥ −3σI were merged and included in the calculation of Rsym, and all resulting structure factors were used in refinement. Data collection and refinement statistics come from SCALEPACK (33) and CNS (36), respectively.
| SeMet | Unlabeled | |
|---|---|---|
| Crystal parameters | ||
| Space group | I222 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 68.3, 81.7, 144.7 | 150.2, 73.3, 149.8 |
| α, β, γ, (°) | 90, 90, 90 | 90, 112.7, 90 |
| Matthews coefficient (Å3/Da) | 2.7 | 2.5 |
| Data quality | ||
| Wavelength (Å) | 0.97885 | 0.97912 |
| Resolution (Å) | 40-3.2 | 40-3.0 |
| Rsym (%) | 15.4 (43.7) | 8.2 (30.4) |
| No. of observations | 94,671 | 105,161 |
| No. of reflections | 12,770 | 29,876 |
| No. of reflections in Rfree set | 1,210 | 2,904 |
| Mean redundancy | 7.4 (6.4) | 3.5 (3.4) |
| Completeness, overall (%) | 99.9 (99.6) | 99.5 (98.6) |
| Mean I/σI | 14.3 (3.7) | 16.4 (3.7) |
| Refinement residuals | ||
| Rfree (%) | 28.3 | 27.7 |
| Rwork (%) | 21.7 | 22.0 |
| Model quality | ||
| r.m.s.d. bond lengths (Å) | 0.009 | 0.009 |
| r.m.s.d. bond angles (°) | 1.1 | 1.2 |
| Ramachandran plot | ||
| Favored (%) | 81.7 | 84.3 |
| Allowed (%) | 17.9 | 15.2 |
| Generously allowed (%) | 0.4 | 0.6 |
| Disallowed (%) | 0 | 0 |
| Mean B-factors (Å2) | ||
| Protein | 44.2 | 48.7 |
| Metals | 35.0 | 39.5 |
| Ligand | 48.9 | 49.7 |
| Model contents | ||
| Protomers in asymmetric unit | 1 | 4 |
| Protein residues | 18–156, 174–181, 192–306, 309–360 | A: 28–157, 174–180, 199–363; B: 28–158, 174–183, 189–304, 308–362; C: 28–158, 174–181, 100–306, 309–356; D: 28–159, 173–181, 200–360 |
| Ligands | 1 | 4 |
| No. of atoms | ||
| Protein | 2,413 | 9,613 |
| Metal ion | 2 | 8 |
| Ligands | 10 | 40 |
| Water molecules | 0 | 100 |
| Protein Data Bank code | 3THP | 3THT |
Both crystal structures show the RRM and AlkB domains connected by a well ordered loop (Fig. 6A and supplemental Fig. S7, A and B). The structural Zn(II)-binding site at the C terminus of the AlkB domain is adjacent to its active site, positioned above a wide groove formed at the interface between the AlkB and RRM domains. (As discussed in more detail below, the well ordered C-terminal tag, omitted in most figures, directly participates in a crystal-packing contact.) Comparing the two crystal structures of the same construct in different space groups shows preservation of the interaction geometry of the two domains with only a small degree of flexibility in the linkage between them (supplemental Fig. S7B). This observation, combined with the observation of a single thermal unfolding transition for the 1–354 construct (Fig. 4), suggests a relatively tight interaction between the RRM and AlkB domains in ABH8. However, there could be more flexibility between the domains in solution than suggested by the comparison of the crystal structures because their interaction geometry in the crystal lattices might be stabilized by a shared tetramer structure that is unlikely to represent a physiological interaction (see below).
The electrostatic potential on the proximal surface of the protein (as shown in Fig. 6A) is strongly basic near the active site in the AlkB domain but otherwise not consistently charged (Fig. 6B). Nonetheless, based on the location of the active site and additional analyses presented in this study, tRNA substrates are likely to bind to this surface and interact simultaneously with the catalytically active AlkB domain and the RRM domain.
A short α-helix observed at the start of the RRM domain likely represents the end of a basic N-terminal α-helix formed by residues 13–32 in ABH8. This α-helix, which is demonstrated above to contribute to nonspecific RNA binding (Fig. 5, supplemental Fig. S4B, and Table 1), was truncated in the 25–354 construct to promote crystallization. When present, it will project over the groove at the interdomain interface between the RRM and AlkB domains, i.e. in an ideal position to interact with tRNAs bound to the putative interaction surface spanning both of these domains (Fig. 6A).
The same tetramer (supplemental Fig. S7C) is observed in both crystal structures of the 25–354 construct with a C-terminal hexahistidine tag. Despite the fact that these crystals grew from different mother liquors and otherwise have different intermolecular packing interactions, this tetramer seems unlikely to have physiological relevance given the observations that the 25–354 construct without the C-terminal tag is a monodisperse monomer and that the full-length ABH8-Trm112 complex equilibrates between monomer and dimer forms in solution (supplemental Fig. S1). The crystallographic tetramer (supplemental Fig. S7C) represents a dimer of dimers. One interprotomer dimer interface, which buries ∼960 Å2 of solvent-accessible surface area per protomer, involves formation of a two-stranded antiparallel β-sheet by the protein segment linking the RRM and AlkB domains together. The other interprotomer dimer interface, which buries ∼990 Å2 of solvent-accessible surface area per protomer, involves contacts with the C-terminal structural Zn(II)-binding site and segments of the uncleaved hexahistidine tag. The crystallographic tetramer therefore could represent the stable oligomer observed to varying extents in analytical gel filtration chromatography of different preparations of the construct retaining the C-terminal hexahistidine tag but not observed after tag cleavage (supplemental Fig. S1).
The Crystal Structure of the RRM Domain of ABH8 Suggests Novel RNA Interaction Mode
The α/β fold (46) of the RRM domain is identical to that in an NMR structure of this single domain deposited previously in the Protein Data Bank (code 2CQ2). The x-ray and the lowest energy NMR structures superimpose with a root mean square deviation (r.m.s.d.) of 0.9 Å, and there is close correspondence between the regions with elevated B-factors in the x-ray structure and significant backbone conformational dispersion in the NMR structure (supplemental Fig. S6). These regions are limited to the short α-helix at the start of the domain and a turn in the β-hairpin formed by the final two β-strands in the domain. This β-hairpin is longer compared with most homologous domains and forms part of the putative tRNA interaction surface in ABH8 (Figs. 6, A and B, and 7A).
FIGURE 7.
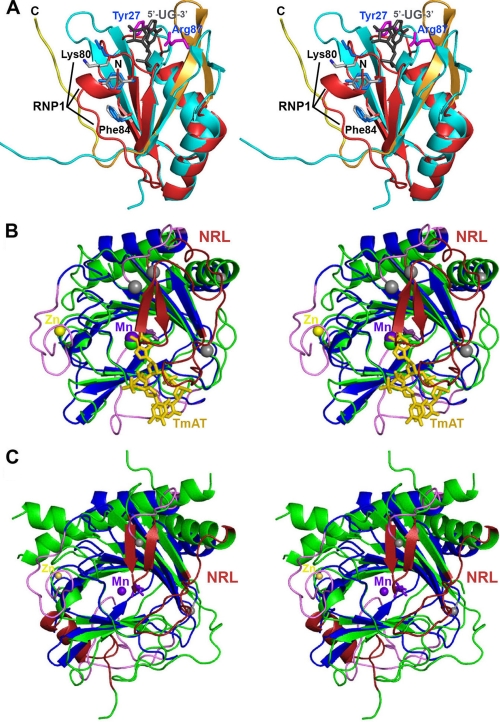
Structural alignments of human ABH8 domains. A, structural alignment (51) of the RRM domains from ABH8 and S. cerevisiae Rna15 (56). The backbone of the ABH8 RRM domain is colored red and orange as in Fig. 6A, and the residues in its RNP1 motif are highlighted and shown in gray in stick representation (with black labels except for Tyr82, which is unlabeled). The N terminus of the ABH8 domain is indicated by the black label “N.” The backbone of Rna15 is colored cyan; its crystallographically observed side chains interacting with RNA (magenta with blue labels), the consensus residues in its RNP1 motif (light blue), and its bound RNA ligand (gray) are all shown in stick representation. Note that this RNA ligand binds to Rna15 on the ridge of the RRM domain proximal to the AlkB domain in the structure of ABH8. The residues in the RNP1 motif are solvent-exposed in Rna15 but partially buried by the N-terminal α-helical segment in ABH8. B, structural alignment of the ABH8 AlkB domain with E. coli AlkB (35). ABH8 is colored as in Fig. 6A with gray spheres added to highlight the termini of its disordered backbone segments. Its bound Mn(II) and Zn(II) cations are shown, respectively, as purple and yellow spheres, whereas its bound 2OG co-substrate is shown in purple in stick representation (adjacent to the Mn(II) cofactor). The core of E. coli AlkB is colored green, its nucleotide-recognition lid (NRL) is colored red, and its bound DNA substrate (TmAT) is colored orange. C, structural alignment of the AlkB domains from ABH8 and human FTO with the latter colored like E. coli AlkB in B (50). The secondary structure elements that differ between ABH8 and its homologs are colored in magenta in B and C.
The RRM domain of ABH8 contains three residues (Lys80, Tyr82, and Phe84) that form the so-called RNP1 consensus sequence motif (Figs. 6A and 7A), one of the signature motifs of canonical RRM domains (46). However, it is missing an additional RNA-interacting aromatic residue often found on the first β-strand of RRM domain (46, 47). In some but not all homologous structures, the RNP1 motif interacts with RNA (supplemental Fig. S8); in these cases, a nucleotide base generally stacks with one of its aromatic residues (46, 47). In ABH8, the residues in this motif are covered by the RRM/AlkB interdomain loop and by the short α-helical segment at the N terminus of the RRM domain. Together, these structures form a cavity that could accommodate without steric clash a pyrimidine nucleotide, which would base stack with Tyr82 in the RNP1 motif. The orifice of this cavity is located on the protein surface hypothesized to bind tRNA, directly below the groove at the interdomain interface. This location places the cavity in an ideal position to contribute to tRNA binding (Fig. 6C). Binding of a nucleotide base into such a sequestered cavity would represent a novel RNA interaction mode for an RRM domain (see “Discussion”).
Unique Conformational Features in AlkB Domain of ABH8
The topology of the AlkB domain in ABH8 is identical to that of other AlkB family enzymes (Fig. 7, B and C, and supplemental Fig. S9). Its core comprises a double-stranded β-helix with eight β-strands organized in a jellyroll fold (35, 48–50). It bears the strongest structural similarity (51) to human ABH3 (supplemental Fig. S9B; Z-score3 of 16 and 2.4-Å r.m.s.d. for alignment of 153 of 194 Cα atoms with 19% sequence identity in Protein Data Bank code 2IUW). It bears a nearly equivalent level of similarity to E. coli AlkB (Fig. 7B; Z-score of 15 and 2.5-Å r.m.s.d. for alignment of 149 of 194 Cα atoms with 23% sequence identity in Protein Data Bank code 2FDH), and it has only slightly lower similarity to the most remote known member of the AlkB family, human FTO (Fig. 7C; Z-score of 12 and 2.6-Å r.m.s.d. for alignment of 147 of 194 Cα atoms with 15% sequence identity in Protein Data Bank code 3LFM).
Three significant structural differences are observed in the AlkB domain of ABH8 compared with other AlkB family members. First, invariant residues Cys341, Cys343, and Cys349 in the 15-residue C-terminal extension unique to ABH8 ligate a single Zn(II) ion, which is also ligated by His242 from the AlkB domain (supplemental Figs. S2 and S5B). This His residue is located between strands A-β3 and A-β4 in the core region of the domain shared by all Fe(II)/2OG dioxygenases. The bridging interaction to this residue made by the Zn(II) ion explains the observed thermodynamic stabilization of the AlkB domain by this C-terminal structural Zn(II)-binding site (Fig. 4).
The second structural adaptation in the AlkB domain of ABH8 is a longer and partially disordered loop between strands A-β8 and A-β9 (residues 299–324). This loop could mediate regulatory interactions remote from the active site.
The third and final structural adaptation occurs in the protein loops corresponding to the nucleotide recognition lid in E. coli AlkB; these loops are completely disordered in both of our crystal structures of the RRM and AlkB domains of ABH8 (Fig. 7B). In E. coli AlkB, these protein segments mediate most contacts to the DNA substrate, and they have been shown by amide 1H/2H exchange mass spectrometry to be more dynamic prior to DNA binding (35). The corresponding loops in ABH8 are somewhat shorter (22 and 21 versus 26 and 28 residues) and more highly charged (six acidic and two basic residues versus two acidic and two basic residues). Based on their close proximity to the active site in ABH8, they are likely to undergo a disorder-to-order transition upon binding specific tRNA substrates.
Given the disordered state of these loops, the active site in ABH8 is completely solvent-exposed (Fig. 6A). Although this structural variation is probably necessary to accommodate the folded macromolecular tRNA substrate, it could potentially promote promiscuous catalysis of 2OG oxidation in the absence of tRNA, resulting in adventitious release of reactive oxygen species. However, ABH8 shows lower basal 2OG oxidation than E. coli AlkB in the absence of the nucleic acid substrate.4 The active site in ABH8 shares the invariant iron-ligating (His238, Asp240, and His292) and 2OG-ligating (Arg328 and Arg334) residues with other members of the Fe(II)/2OG dioxygenase superfamily. However, the bound 2OG co-substrate does not interact with the metal cofactor with proper octahedral coordination geometry in our refined crystal structures of ABH8 (Fig. 6D and supplemental Fig. S10), in contrast to what has been observed in many crystal structures of the Michaelis complexes of E. coli AlkB (14, 35) and other superfamily members (48–50). The electron density in this region of ABH8, including after high temperature simulated annealing omitting the metal cofactors and 2OG co-substrate (supplemental Fig. S5A), suggests that there may be some minor variation in the exact conformation of the bound 2OG. However, its conformation is inconsistent with adoption of the canonical catalytically active conformation. The catalytically essential residue Arg334 also adopts an alternative conformation with its guanidino group failing to make the typical bridging interaction between the Fe(II)-ligating Asp and 2OG groups (Fig. 6D and supplemental Fig. S10). These variations from canonical catalytic geometry likely explain the lower uncoupled turnover of 2OG by the AlkB domain of ABH8. A conformational change in the active site will be needed upon tRNA binding to generate a catalytically active complex; this activation mechanism likely represents an important adaptation to having a solvent-exposed Fe(II)/2OG active site acting on a folded macromolecular substrate.
DISCUSSION
Implications of in Vivo Expression Pattern of ABH8 for Its Physiological Function
In this study, we present data on the evolutionary diversification of AlkB domain function in the multifunctional enzyme ABH8, and we elucidate the structural adaptations underlying this diversification. We demonstrate that, in the nematode C. elegans, the ABH8 ortholog C14B1.10 is expressed in a tissue-specific pattern that changes during the lifespan of the animal, suggesting that its tRNA modification activity may play a role in regulating metazoan development (30). Such a role would be consistent with studies showing substantial differences in ABH8 expression level in different human tissues (4).
An alternative hypothesis for ABH8 function is that it regulates the translation level of proteins involved in DNA damage repair. This hypothesis was based on a study showing that Trm9, a Saccharomyces cerevisiae enzyme that is homologous to the MTase domain in ABH8 and that catalyzes the same tRNA methylation reaction, enhances the translation of a set of DNA repair enzymes encoded by mRNAs enriched in specific codons (19). This hypothesis for ABH8 function is appealing because it ties the altered catalytic activity of the metazoan ABH8 enzyme to the DNA repair activity of the bacterial AlkB enzyme. Consistent with this hypothesis, knockdown of ABH8 expression in human tissue culture cells produces a modest increase in their sensitivity to the DNA-damaging agents methyl methanesulfonate and bleomycin, and bleomycin treatment induces a modest increase in the expression level of ABH8 (17). The widespread expression that we observe for the ABH8 ortholog in C. elegans larvae but not adults (Fig. 3) could potentially reflect a greater need for DNA repair activity in dividing cells. Overexpression of ABH8 in human bladder cancer cells (1) similarly could be related to such an effect, because the genome is often destabilized in cancer cells.
The data presented in this study showing very restricted expression of the ABH8 ortholog in C. elegans adults provide evidence that ABH8 could additionally play a role in the development of this multicellular organism. There are well established examples of proteins mediating basic metabolic processes in unicellular organisms being adapted in metazoans to regulate different processes related to the development of multicellular organisms. For example, translational control of the GCN4 transcription factor regulates the response to amino acid deprivation in yeast, whereas translational control of the murine homolog ATF4 plays a role in regulating long term memory formation (52). The C. elegans expression data presented in this study suggest that ABH8 may similarly have been adapted to play a role in animal development, possibly in neuronal cells, as inferred by the similar expression pattern of the ABH8 ortholog and the Elongator complex in C. elegans. The ELPC-1 and ELPC-3 proteins in this complex are involved in the production of mcm5U at position 34 in tRNA, the substrate for ABH8, and mutations in these proteins impair neural function and development in worms (29). Notably, mutations in the human homologs of these proteins can cause the neurological diseases familial dysautonomia and amyotrophic lateral sclerosis (53, 54). Finally, a C. elegans paralog of the methyltransferase domain in ABH8 (gene C35D10.12) is expressed in the nervous system of both larvae and adults, providing yet another association between tRNA modification and worm neurodevelopment (55). In summary, the available data on the proteins producing the substrate for ABH8, combined with the conservation of ABH8 in all metazoa and our data on ABH8 expression in adult worms (Fig. 3), suggest that ABH8 has a specialized, developmentally regulated physiological function. Further research will be required to elucidate its exact physiological functions, whether these vary in different developmental states, and the molecular mechanism(s) connecting these functions to its tRNA modification activity.
Structural and Functional Adaptations in the RRM Domain of ABH8
Our thermodynamic and crystallographic studies on ABH8 provide insight into the molecular adaptations enabling the AlkB domain in ABH8 to catalyze a highly specific covalent modification of tRNA instead of promiscuously catalyzing repair of alkylation lesions in DNA. The first such adaptation is the fusion of the AlkB domain to an RRM domain. Based on the binding studies reported above, this adaptation provides micromolar level binding affinity for RNA oligomers albeit with little sequence specificity. The RRM domain of ABH8 adopts the α/β fold characteristic of this family and also contains a canonical RNP1 sequence/structure motif (46). However, its RNA binding mode is inferred to be substantially different from that of previously characterized RRM domains. The basic N-terminal α-helix in ABH8 that critically contributes to nonspecific RNA binding affinity (Fig. 5 and supplemental Fig. S4B) is not a typical feature of RRM domains. Moreover, the residues in the RNP1 motif, which are solvent-exposed in other RRM domains, are partially buried in ABH8 by the protein segment connecting the RRM and AlkB domains and by the end of the N-terminal basic α-helix (i.e. its C-terminal segment that is retained and visualized in the crystallized protein construct). These structural interactions create a cavity whose orifice is located just below the interface between the RRM and AlkB domains (Fig. 6, A–C). This cavity could accommodate a pyrimidine base, which would form a base-stacking interaction with Tyr82 from the RNP1 motif. Further research will be required to determine whether such an interaction takes place during tRNA binding to ABH8 and whether it contributes primarily to nonspecific binding energy or specific substrate recognition.
If a pyrimidine base does bind in this cavity in the RRM domain of ABH8, it would provide yet another paradigm for the structural and functional adaptation of RRM domains, which are broadly associated with RNA-binding proteins but display diverse structural interactions (supplemental Fig. S8, A–D). Yeast Rna15, a subunit of cleavage factor 1A involved in mRNA maturation, represents one of the RRM domains with closest structural similarity to ABH8 (Z-score of 11 and 1.4-Å r.m.s.d. for alignment of 75 of 103 Cα carbons with 23% sequence identity in Protein Data Bank code 2X1A) (51, 56). A mononucleotide interacts with the region of Rna15 equivalent to that lining the interdomain groove in ABH8 (Figs. 6A and 7A), a likely region of tRNA interaction in ABH8. In other RRM domains, the RNP1 motif is solvent-exposed and directly binds RNA (46, 47). However, in the RRM domain of the exon-junction complex, this motif is buried in an intersubunit interface and therefore unable to participate directly in binding RNA (supplemental Fig. S8D) (57). The putative pyrimidine-binding cavity in the RRM domain of ABH8 presents a hybrid stereochemical paradigm in which the RNP1 motif is partially buried but still able to participate in RNA binding.
Structural and Functional Adaptations in AlkB Domain of ABH8
Another striking characteristic of human ABH8 is that its AlkB domain has an extremely narrow substrate specificity, contrary to bacterial AlkB DNA repair enzymes whose hallmark is a broad substrate specificity. Bacterial AlkBs efficiently dealkylate 1-methyladenine, 3-methylcytosine, and 1,N6-ethenoadenine in addition to larger alkylation lesions in single-stranded and double-stranded DNA and RNA (12, 13). Joint enzymological and structural studies have shown that the loops forming the nucleotide-binding lid in the bacterial enzymes play a central role in this promiscuous substrate recognition (14). These loops constitute most of the surface of a deep binding slot that can accommodate nucleobases with diverse stereochemistry in the active site. The flexibility of these protein loops enables preservation of their interactions with the polynucleotide backbone while different nucleobases slide into the slot to differing degrees as optimal for catalysis of their oxidation (14). The equivalent structural elements in the AlkB domain in ABH8 are disordered in the absence of the nucleic acid substrate. A disorder-to-order transition in these loops is likely responsible for forming the binding pocket that specifically recognizes the modified nucleobase mcm5U in folded tRNAs. Therefore, the structural properties of these loops represent the second key evolutionary adaptation contributing to the altered catalytic activity of the AlkB domain in ABH8 (with the other being the fusion of the RRM domain as discussed above).
The binding studies presented in this study indicate that the RRM domain of ABH8 contributes primarily to nonspecific RNA binding affinity, which is likely to facilitate the search for correctly modified tRNAs in vivo. However, mcm5U-containing tRNAs probably bind to ABH8 with substantially higher affinity given its observed co-purification with substrate tRNAs (17). Nucleic acid modifications often make critical energetic contributions to substrate binding by enzymes operating on modified RNA and DNA substrates (41). Notably, E. coli AlkB shows ∼50-fold higher affinity for methylated DNA substrates compared with equivalent unmodified DNAs (58).5 The methoxy substituent in mcm5U, which is added by the MTase domain of ABH8 (15, 17), has been inferred to have a similarly strong influence on the binding affinity of the AlkB domain of ABH8 based on the observation that a model anticodon substrate containing mcm5U can be hydroxylated, whereas the equivalent RNA containing cm5U cannot (18). This exquisitely specific recognition of mcm5U-containing tRNAs must be derived at least in part from interactions of the modified nucleobase with the 22- and 21-residue disordered loops that flank the active site in its AlkB domain. These loops are topologically equivalent to the 26- and 28-residue segments that form most of the promiscuous nucleobase-binding cavity in E. coli AlkB. In ABH8, they also likely contribute to maintaining its solvent-exposed Fe(II)/2OG center in a catalytically inactive conformation prior to binding the macromolecular substrate. The dramatic alterations in the catalytic properties of the AlkB domain based primarily on remodeling of these two short loops provide a remarkable example of the structural plasticity underlying protein evolution.
Supplementary Material
Acknowledgments
We thank A. Lauricella, G. DeTitta, J. Everrett, S. N. Tong, and J. Seetharaman for technical assistance and G. Boel, B. Yu, G. T. Montelione, and M. Chalfie for advice and critical review of the manuscript.
This work was supported by National Institutes of Health Grants R01GM077360 (to J. F. H.) and U54GM094597 (to the Northeast Structural Genomics Consortium).

This article contains supplemental Methods, Figs. S1–S10, and Tables S1 and S2.
The atomic coordinates and structure factors (codes 3THP and 3THT) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The Z-score is a standard measure of statistical significance defined in this case as the ratio of the structural similarity score to the standard deviation of that score on randomly related structures. Z-scores above 2 in the program DALI (42) are generally interpreted to indicate significant similarity in backbone fold.
C. Pastore, B. Ergel, and J. F. Hunt, unpublished results.
B. Ergel and J. F. Hunt, manuscript in preparation.
- ABH
- AlkB homolog
- RRM
- RNA recognition motif
- r.m.s.d.
- root mean square deviation
- 2OG
- 2-oxoglutarate
- MTase
- methyltransferase
- cm5U
- 5-carboxymethyluridine
- mcm5U
- 5-methoxycarbonylmethyluridine
- (S)-mchm5U
- (S)-5-methoxycarbonylhydroxymethyluridine
- SeMet
- selenomethionine
- Ni-NTA
- nickel-nitrilotriacetic acid
- TEV
- tobacco etch virus
- SAM
- S-adenosylmethionine
- SELEX
- systematic evolution of ligands by exponential enrichment.
REFERENCES
- 1. Shimada K., Nakamura M., Anai S., De Velasco M., Tanaka M., Tsujikawa K., Ouji Y., Konishi N. (2009) Cancer Res. 69, 3157–3164 [DOI] [PubMed] [Google Scholar]
- 2. Liu B. Q., Wu Y. D., Li P. H., Wei J. X., Zhang T., Liu R. L. (2007) Asian J. Androl. 9, 821–826 [DOI] [PubMed] [Google Scholar]
- 3. Church C., Moir L., McMurray F., Girard C., Banks G. T., Teboul L., Wells S., Brüning J. C., Nolan P. M., Ashcroft F. M., Cox R. D. (2010) Nat. Genet. 42, 1086–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tsujikawa K., Koike K., Kitae K., Shinkawa A., Arima H., Suzuki T., Tsuchiya M., Makino Y., Furukawa T., Konishi N., Yamamoto H. (2007) J. Cell. Mol. Med. 11, 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aas P. A., Otterlei M., Falnes P. O., Vågbø C. B., Skorpen F., Akbari M., Sundheim O., Bjørås M., Slupphaug G., Seeberg E., Krokan H. E. (2003) Nature 421, 859–863 [DOI] [PubMed] [Google Scholar]
- 6. Duncan T., Trewick S. C., Koivisto P., Bates P. A., Lindahl T., Sedgwick B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16660–16665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kurowski M. A., Bhagwat A. S., Papaj G., Bujnicki J. M. (2003) BMC Genomics 4, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Falnes P. ø., Johansen R. F., Seeberg E. (2002) Nature 419, 178–182 [DOI] [PubMed] [Google Scholar]
- 9. Trewick S. C., Henshaw T. F., Hausinger R. P., Lindahl T., Sedgwick B. (2002) Nature 419, 174–178 [DOI] [PubMed] [Google Scholar]
- 10. Westbye M. P., Feyzi E., Aas P. A., Vågbø C. B., Talstad V. A., Kavli B., Hagen L., Sundheim O., Akbari M., Liabakk N. B., Slupphaug G., Otterlei M., Krokan H. E. (2008) J. Biol. Chem. 283, 25046–25056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Falnes P. ø., Klungland A., Alseth I. (2007) Neuroscience 145, 1222–1232 [DOI] [PubMed] [Google Scholar]
- 12. Delaney J. C., Smeester L., Wong C., Frick L. E., Taghizadeh K., Wishnok J. S., Drennan C. L., Samson L. D., Essigmann J. M. (2005) Nat. Struct. Mol. Biol. 12, 855–860 [DOI] [PubMed] [Google Scholar]
- 13. Falnes P. ø., Bjørås M., Aas P. A., Sundheim O., Seeberg E. (2004) Nucleic Acids Res. 32, 3456–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu B., Hunt J. F. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14315–14320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Songe-Møller L., van den Born E., Leihne V., Vågbø C. B., Kristoffersen T., Krokan H. E., Kirpekar F., Falnes P. ø., Klungland A. (2010) Mol. Cell. Biol. 30, 1814–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van den Born E., Vågbø C. B., Songe-Møller L., Leihne V., Lien G. F., Leszczynska G., Malkiewicz A., Krokan H. E., Kirpekar F., Klungland A., Falnes P. ø. (2011) Nat. Commun. 2, 172. [DOI] [PubMed] [Google Scholar]
- 17. Fu D., Brophy J. A., Chan C. T., Atmore K. A., Begley U., Paules R. S., Dedon P. C., Begley T. J., Samson L. D. (2010) Mol. Cell. Biol. 30, 2449–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fu Y., Dai Q., Zhang W., Ren J., Pan T., He C. (2010) Angew. Chem. Int. Ed. Engl. 49, 8885–8888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Begley U., Dyavaiah M., Patil A., Rooney J. P., DiRenzo D., Young C. M., Conklin D. S., Zitomer R. S., Begley T. J. (2007) Mol. Cell 28, 860–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bjork G. R. (1995) in tRNA: Structure, Biosynthesis and Function (Söll D., Rajbhandary U. L., eds) pp. 165–205, ASM Press, Washington, D. C [Google Scholar]
- 21. Motorin Y., Helm M. (2010) Biochemistry 49, 4934–4944 [DOI] [PubMed] [Google Scholar]
- 22. Agris P. F. (2008) EMBO Rep. 9, 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gustilo E. M., Vendeix F. A., Agris P. F. (2008) Curr. Opin. Microbiol. 11, 134–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Y., Zhou H. (2009) Sci. China C Life Sci. 52, 245–252 [DOI] [PubMed] [Google Scholar]
- 25. Suzuki T. (2005) in Fine-Tuning of RNA Functions by Modification and Editing (Grosjean H., ed) pp. 23–69, Springer, Berlin [Google Scholar]
- 26. Chan C. T., Dyavaiah M., DeMott M. S., Taghizadeh K., Dedon P. C., Begley T. J. (2010) PLoS Genet. 6, e1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grosjean H. (2005) Fine-Tuning of RNA Functions by Modification and Editing (Grosjean H., ed) pp. 1–12, Springer, Berlin [Google Scholar]
- 28. Steinthorsdottir V., Thorleifsson G., Reynisdottir I., Benediktsson R., Jonsdottir T., Walters G. B., Styrkarsdottir U., Gretarsdottir S., Emilsson V., Ghosh S., Baker A., Snorradottir S., Bjarnason H., Ng M. C., Hansen T., Bagger Y., Wilensky R. L., Reilly M. P., Adeyemo A., Chen Y., Zhou J., Gudnason V., Chen G., Huang H., Lashley K., Doumatey A., So W. Y., Ma R. C., Andersen G., Borch-Johnsen K., Jorgensen T., van Vliet-Ostaptchouk J. V., Hofker M. H., Wijmenga C., Christiansen C., Rader D. J., Rotimi C., Gurney M., Chan J. C., Pedersen O., Sigurdsson G., Gulcher J. R., Thorsteinsdottir U., Kong A., Stefansson K. (2007) Nat. Genet. 39, 770–775 [DOI] [PubMed] [Google Scholar]
- 29. Chen C., Tuck S., Byström A. S. (2009) PLoS Genet. 5, e1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim Y. O., Park S. J., Balaban R. S., Nirenberg M., Kim Y. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tropea J. E., Cherry S., Waugh D. S. (2009) Methods Mol. Biol. 498, 297–307 [DOI] [PubMed] [Google Scholar]
- 32. Nakamura A., Yao M., Chimnaronk S., Sakai N., Tanaka I. (2006) Science 312, 1954–1958 [DOI] [PubMed] [Google Scholar]
- 33. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 34. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu B., Edstrom W. C., Benach J., Hamuro Y., Weber P. C., Gibney B. R., Hunt J. F. (2006) Nature 439, 879–884 [DOI] [PubMed] [Google Scholar]
- 36. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D. Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 37. McRee D. E. (1999) J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 38. Vagin A. A., Isupov M. N. (2001) Acta Crystallogr. D. Biol. Crystallogr. 57, 1451–1456 [DOI] [PubMed] [Google Scholar]
- 39. Byrne A. B., Weirauch M. T., Wong V., Koeva M., Dixon S. J., Stuart J. M., Roy P. J. (2007) J. Biol. 6, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang B., Johansson M. J., Byström A. S. (2005) RNA 11, 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pierrel F., Douki T., Fontecave M., Atta M. (2004) J. Biol. Chem. 279, 47555–47563 [DOI] [PubMed] [Google Scholar]
- 42. Winkler G. S., Kristjuhan A., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3517–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wittschieben B. O., Otero G., de Bizemont T., Fellows J., Erdjument-Bromage H., Ohba R., Li Y., Allis C. D., Tempst P., Svejstrup J. Q. (1999) Mol. Cell 4, 123–128 [DOI] [PubMed] [Google Scholar]
- 44. Ward J. J., McGuffin L. J., Bryson K., Buxton B. F., Jones D. T. (2004) Bioinformatics 20, 2138–2139 [DOI] [PubMed] [Google Scholar]
- 45. Stoltenburg R., Reinemann C., Strehlitz B. (2007) Biomol. Eng. 24, 381–403 [DOI] [PubMed] [Google Scholar]
- 46. Maris C., Dominguez C., Allain F. H. (2005) FEBS J. 272, 2118–2131 [DOI] [PubMed] [Google Scholar]
- 47. Cléry A., Blatter M., Allain F. H. (2008) Curr. Opin. Struct. Biol. 18, 290–298 [DOI] [PubMed] [Google Scholar]
- 48. Sundheim O., Vågbø C. B., Bjørås M., Sousa M. M., Talstad V., Aas P. A., Drabløs F., Krokan H. E., Tainer J. A., Slupphaug G. (2006) EMBO J. 25, 3389–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang C. G., Yi C., Duguid E. M., Sullivan C. T., Jian X., Rice P. A., He C. (2008) Nature 452, 961–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Han Z., Niu T., Chang J., Lei X., Zhao M., Wang Q., Cheng W., Wang J., Feng Y., Chai J. (2010) Nature 464, 1205–1209 [DOI] [PubMed] [Google Scholar]
- 51. Holm L., Rosenström P. (2010) Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Costa-Mattioli M., Gobert D., Stern E., Gamache K., Colina R., Cuello C., Sossin W., Kaufman R., Pelletier J., Rosenblum K., Krnjevi K., Lacaille J. C., Nader K., Sonenberg N. (2007) Cell 129, 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rubin B. Y., Anderson S. L. (2008) Neuromolecular Med. 10, 148–156 [DOI] [PubMed] [Google Scholar]
- 54. Simpson C. L., Lemmens R., Miskiewicz K., Broom W. J., Hansen V. K., van Vught P. W., Landers J. E., Sapp P., Van Den Bosch L., Knight J., Neale B. M., Turner M. R., Veldink J. H., Ophoff R. A., Tripathi V. B., Beleza A., Shah M. N., Proitsi P., Van Hoecke A., Carmeliet P., Horvitz H. R., Leigh P. N., Shaw C. E., van den Berg L. H., Sham P. C., Powell J. F., Verstreken P., Brown R. H., Jr., Robberecht W., Al-Chalabi A. (2009) Hum. Mol. Genet. 18, 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hunt-Newbury R., Viveiros R., Johnsen R., Mah A., Anastas D., Fang L., Halfnight E., Lee D., Lin J., Lorch A., McKay S., Okada H. M., Pan J., Schulz A. K., Tu D., Wong K., Zhao Z., Alexeyenko A., Burglin T., Sonnhammer E., Schnabel R., Jones S. J., Marra M. A., Baillie D. L., Moerman D. G. (2007) PLoS Biol. 5, e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pancevac C., Goldstone D. C., Ramos A., Taylor I. A. (2010) Nucleic Acids Res. 38, 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bono F., Ebert J., Lorentzen E., Conti E. (2006) Cell 126, 713–725 [DOI] [PubMed] [Google Scholar]
- 58. Holland P. J., Hollis T. (2010) PLoS One 5, e8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 60. Petrey D., Honig B. (2003) Methods Enzymol. 374, 492–509 [DOI] [PubMed] [Google Scholar]
- 61. Drenth J. (1994) Principles of Protein X-ray Crystallography, Springer, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.