Background: Familial hemiplegic migraine type II (FHM2) is caused by mutations in the Na+,K+-ATPase α2-isoform.
Results: Several FHM2 mutations inhibit phosphorylation or dephosphorylation.
Conclusion: These mutations cause FHM2 by local and long range effects on the catalytic site and not by reducing the affinity for external K+.
Significance: Insights into the pathophysiological mechanism of FHM2 and the molecular mechanism of the Na+,K+-ATPase have been obtained.
Keywords: ATPases; Membrane Transport; Mutagenesis Site-specific; Mutant; Na,K-ATPase; Phosphorylation; P-type ATPase; Migraine; Neurological Disease; Vanadate
Abstract
The neurological disorder familial hemiplegic migraine type II (FHM2) is caused by mutations in the α2-isoform of the Na+,K+-ATPase. We have studied the partial reaction steps of the Na+,K+-pump cycle in nine FHM2 mutants retaining overall activity at a level still compatible with cell growth. Although it is believed that the pathophysiology of FHM2 results from reduced extracellular K+ clearance and/or changes in Na+ gradient-dependent transport processes in neuroglia, a reduced affinity for K+ or Na+ is not a general finding with the FHM2 mutants. Six of the FHM2 mutations markedly affect the maximal rate of phosphorylation from ATP leading to inhibition by intracellular K+, thereby likely compromising pump function under physiological conditions. In mutants R593W, V628M, and M731T, the defective phosphorylation is caused by local perturbations within the Rossmann fold, possibly interfering with the bending of the P-domain during phosphoryl transfer. In mutants V138A, T345A, and R834Q, long range effects reaching from as far away as the M2 transmembrane helix perturb the function of the catalytic site. Mutant E700K exhibits a reduced rate of E2P dephosphorylation without effect on phosphorylation from ATP. An extremely reduced vanadate affinity of this mutant indicates that the slow dephosphorylation reflects a destabilization of the phosphoryl transition state. This seems to be caused by insertion of the lysine between two other positively charged residues of the Rossmann fold. In mutants R202Q and T263M, effects on the A-domain structure are responsible for a reduced rate of the E1P to E2P transition.
Introduction
Hemiplegic migraine is a severe subtype of migraine with aura associated with transient motor weakness and sensory as well as speech difficulties. It is autosomal dominantly inherited (familial hemiplegic migraine). Besides mutations in neuronal voltage-gated Ca2+ and Na+ channels (FHM1 and FHM3), familial hemiplegic migraine has been associated with mutations in the α2-isoform of the Na+,K+-ATPase (FHM2) (1). The Na+ and K+ gradients created across the cell membrane by the Na+,K+-ATPase are of vital importance for cellular function and activities, including generation of action potentials and secondary active transport of ions, nutrients, and neurotransmitters. In the mammalian brain, three different isoforms of the catalytic α-subunit are expressed, α1, α2, and α3. The α2-isoform is mainly distributed in glial cells, whereas the α3-isoform is found in neurons and is absent in glial cells (2). So far, we know of more than 50 Na+,K+-ATPase α2 mutations associated with hemiplegic migraine. It has been suggested that the pathophysiology results from an impaired clearance of extracellular K+, producing a wide cortical depolarization (3, 4). Moreover, the gradients of both Na+ and K+ are important for the reuptake of the excitatory transmitter glutamate from the synaptic cleft via the glutamate transporter, and impaired Na+ transport would also lead to a rise of intracellular Ca2+ via the Na+/Ca2+ exchanger with secondary effects on Ca2+ signaling (1, 5, 6). Cell survival studies of transfected cells, in which the wild type has been inhibited by ouabain, have demonstrated the inability of certain FHM2 mutants to sustain cell growth in accordance with the theory of haploinsufficiency, i.e. only the wild type allele encodes a functional enzyme (1, 7). Other FHM2 mutants have nevertheless been found to retain transport function (4, 8–11), and more functional mutants exist, as indicated by the present results. A crucial question is therefore whether and how their function deviates from that of the wild type. One study has reported a 2-fold reduced apparent K+ affinity for activation of the ATPase reaction of mutant T345A, which was suggested to give rise to a reduced rate of external K+ clearance in vivo (4).
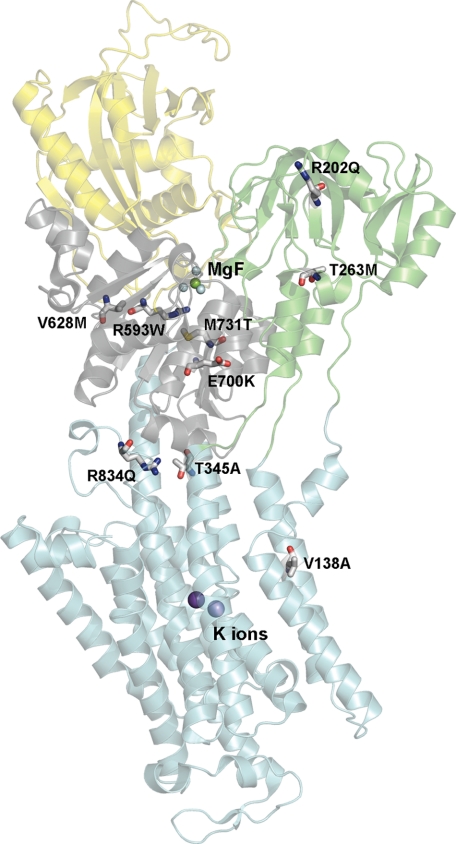
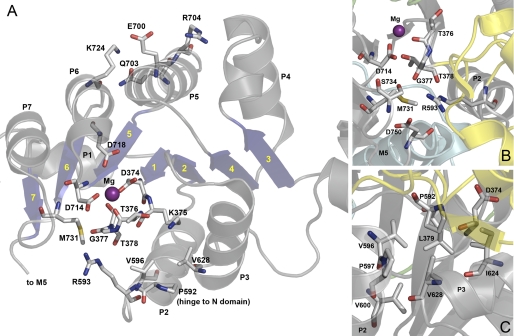
To fully understand the functional implications of the FHM2 mutations and the structural basis, it is necessary to reveal the mutational effects on the individual partial reaction steps of the pump cycle and the interaction with Na+ and K+ and to try to relate the observed effects to known structural features. We present here the functional consequences of nine Na+,K+-ATPase α2 mutations, including T345A, which all have been found in patients exhibiting the characteristic symptoms of hemiplegic migraine (12–17). The α-subunit carrying the mutations consists of 10 transmembrane helices M1–M10 (harboring the Na+- and K+-binding sites) and a cytoplasmic “head” made up of three subdomains as follows: A (“actuator”), N (“nucleotide binding”), and P (“phosphorylation”) (18, 19). Generally, FHM2 mutations are widely scattered over the Na+,K+-ATPase protein, but with a preference for the P-domain (7). Four of the mutations investigated here are located in the P-domain (R593W, V628M, E700K, and M731T), two are in the A-domain (R202Q and T263M), one is in M2 (V138A), another is in M4 near the interface to the P-domain (T345A), and the last is in the L6–7 loop between M6 and M7 close to the P-domain (R834Q) (Fig. 1). We assessed their functional impact in a panel of analyses of the partial reaction steps (cf. Scheme 1), using both steady-state and transient kinetic measurements, the latter allowing determination of the rates of the rapid phosphorylation and dephosphorylation reactions. The observed effects are analyzed in the context of information derived from the recently determined crystal structures of the Na+,K+-ATPase (18, 19).
FIGURE 1.
Na+,K+-ATPase structure with indication of the FHM2 mutations studied. The structure shown has Protein Data Bank code 2ZXE (E2[K2] with bound MgF42− as phosphate analog (19)). Color codes for P-, A-, N-, and M-domain are gray, green, yellow, and cyan, respectively. The bound K+ ions are shown as purple spheres and MgF42− (MgF) in the catalytic site as one green sphere with four cyan spheres. The mutated residues (numbered according to the human α2-isoform) are highlighted as sticks colored according to the elements (carbon, gray; oxygen, red; nitrogen, blue; sulfur, yellow).
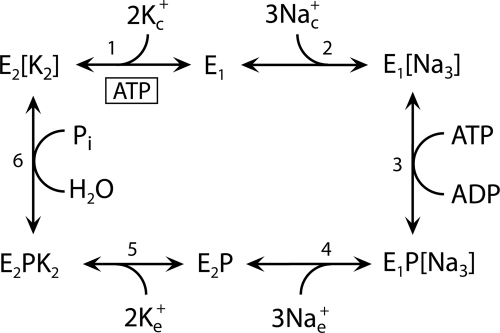
SCHEME 1.
Model of the reaction cycle of Na+,K+-ATPase (“Post-Albers scheme”). Ligand binding and dissociation steps and conformational changes between E1/E1P and E2/E2P forms are indicated. Na+ binds from the cytoplasmic side to the E1 form and activates phosphorylation from ATP bound with high affinity to E1. The E1P form is ADP-sensitive, i.e. able to donate the phosphoryl group back to ADP, forming ATP. The E2P form is K+-sensitive, i.e. hydrolysis of E2P is activated by binding of K+ from the extracellular side. Occluded ions are shown in brackets. Subscripts c and e indicate the cytoplasmic and extracellular sides, respectively. The ATP molecule binding with low affinity to E2 is shown boxed. Because ATP binds with higher affinity to E1 than to E2, an increase in the apparent affinity for ATP upon mutation reflects a shift of the E1-E2 distribution toward the E1 form, whereas a decrease in the affinity for ATP reflects a shift toward the E2 form.
EXPERIMENTAL PROCEDURES
FHM2 mutations were introduced into full-length cDNA encoding an ouabain-resistant version of the human α2-isoform (4). Mutants and wild type were expressed in COS-1 cells under ouabain selection pressure (20). The plasma membrane fraction was isolated and made leaky to allow access of incubation media from both sides of the membrane, and Na+,K+-ATPase activity was studied at 37 °C by following the liberation of Pi (20). The catalytic turnover rate was calculated by relating the ATPase activity to the active site concentration determined by phosphorylation at 0 °C in the presence of 150 mm Na+ and oligomycin (21, 22). Measurements of phosphorylation and dephosphorylation were performed using either a manual mixing technique at 0 °C or a QFM-5 quench-flow module (Bio-Logic Instruments, Claix, France), allowing transient kinetic studies at 25 °C (21, 22). To eliminate the contribution of the endogenous Na+,K+-ATPase, ouabain was included in the reaction media (21). Data processing was performed using SigmaPlot (SPSS, Inc.) for linear regression analysis (21), and the results are reported as average values ± S.E. (shown by error bars in figures when larger than the size of the symbols). The number of independent determinations is indicated by n in Tables 1 and 2. Structural figures were prepared using PyMOL.
TABLE 1.
ATPase activity parameters
| Ouabain-affinitya | Turnover rateb | K0.5(K+)c | K0.5(VO43−)c | K0.5(ATP)c | |
|---|---|---|---|---|---|
| μm | min−1 | μm | μm | μm | |
| Wild type α2 | 297 ± 29 | 6843 ± 380 | 716 ± 19 | 27 ± 2 | 104 ± 6 |
| (n = 3) | (n = 6) | (n = 5) | (n = 6) | (n = 8) | |
| V138A | 693 ± 38 | 3768 ± 157 | 763 ± 18 | 290 ± 18 | 47 ± 2 |
| (n = 3) | (n = 6) | (n = 4) | (n = 4) | (n = 7) | |
| R202Q | 288 ± 40 | 5420 ± 277 | 748 ± 33 | 113 ± 6 | 67 ± 4 |
| (n = 3) | (n = 10) | (n = 5) | (n = 8) | (n = 4) | |
| T263M | 1323 ± 95 | 3233 ± 168 | 465 ± 19 | 73 ± 4 | 21 ± 1 |
| (n = 3) | (n = 6) | (n = 5) | (n = 8) | (n = 4) | |
| T345A | 657 ± 48 | 3419 ± 50 | 747 ± 20 | 283 ± 21 | 31 ± 2 |
| (n = 4) | (n = 6) | (n = 4) | (n = 5) | (n = 5) | |
| R593W | 2409 ± 518 | 1029 ± 56 | 263 ± 35 | >1000 | 19 ± 1 |
| (n = 4) | (n = 6) | (n = 7) | (n = 6) | (n = 6) | |
| V628M | 1282 ± 206 | 1885 ± 63 | 273 ± 15 | >1000 | 71 ± 6 |
| (n = 3) | (n = 6) | (n = 4) | (n = 5) | (n = 5) | |
| E700K | 146 ± 23 | 3669 ± 229 | 1145 ± 36 | >1000 | 28 ± 1 |
| (n = 3) | (n = 7) | (n = 3) | (n = 6) | (n = 3) | |
| M731T | 2708 ± 597 | 1238 ± 84 | 216 ± 9 | 778 ± 14 | 26 ± 1 |
| (n = 3) | (n = 6) | (n = 5) | (n = 3) | (n = 3) | |
| R834Q | 2134 ± 534 | 1236 ± 46 | 332 ± 21 | >1000 | 24 ± 2 |
| (n = 3) | (n = 6) | (n = 5) | (n = 4) | (n = 5) |
a The rate of ATP hydrolysis was determined at 37 °C in the presence of 130 mm NaCl, 20 mm KCl, 3 mm ATP, 3 mm MgCl2, 30 mm histidine buffer (pH 7.4), 1 mm EGTA, and various concentrations of ouabain. The data points were fitted using a function with the ouabain-inhibited enzyme represented by the sum of two hyperbolic components corresponding to the exogenous enzyme (affinity constant K1) and the endogenous COS cell enzyme (affinity constant K2 ≤1 μm), respectively (24): V/Vmax = 1 − a1[ouabain]/(K1 + [ouabain]) − a2[ouabain]/(K2 + [ouabain]). The values determined for K1 are indicated in the table.
b The Na+,K+-ATPase activity was determined at 37 °C in the presence of 30 mm histidine buffer (pH 7.4), 130 mm NaCl, 3 mm ATP, 3 mm MgCl2, 1 mm EGTA, ouabain to inhibit the endogenous enzyme, and 20 mm KCl. The catalytic turnover rate was calculated as the ratio between the Na+,K+-ATPase activity and the active site concentration (maximum phosphorylation from [γ-32P]ATP measured at 0 °C in the presence of 150 mm NaCl and oligomycin) (21, 22).
TABLE 2.
Phosphorylation parameters
| K0.5(Na+)a | EP/EPoligob | kphos, 2 μmATPc | Vmaxc | Kmc | E1Pd | Rate of E2P dephosphorylatione |
||
|---|---|---|---|---|---|---|---|---|
| 1 mm K+ | 20 mm K+ | |||||||
| μm | % | s−1 | s−1 | μm | % | s−1 | s−1 | |
| Wild type α2 | 538 ± 15 | 76 ± 2 | 22 ± 1 | 83 ± 10 | 6.7 ± 0.8 | 44 ± 4 | 43 ± 2 | 132 ± 5 |
| (n = 8) | (n = 5) | (n = 3) | (n = 8) | (n = 8) | (n = 9) | (n = 6) | (n = 3) | |
| V138A | 807 ± 25 | 56 ± 3 | 12 ± 1 | 45 ± 8 | 5.1 ± 1.0 | 48 ± 9 | 32 ± 2 | 108 ± 4 |
| (n = 4) | (n = 6) | (n = 5) | (n = 13) | (n = 13) | (n = 5) | (n = 3) | (n = 3) | |
| R202Q | 541 ± 13 | 76 ± 3 | 17 ± 1 | ND | ND | 61 ± 2 | 59 ± 4 | NDb |
| (n = 7) | (n = 5) | (n = 3) | (n = 6) | (n = 3) | ||||
| T263M | 633 ± 20 | 85 ± 5 | 23 ± 1 | ND | ND | 96 ± 2 | NDa | NDb |
| (n = 6) | (n = 6) | (n = 3) | (n = 4) | |||||
| T345A | 814 ± 34 | 59 ± 3 | 13 ± 1 | 39 ± 2 | 4.5 ± 0.2 | 33 ± 14 | 43 ± 2 | NDb |
| (n = 7) | (n = 3) | (n = 3) | (n = 9) | (n = 9) | (n = 4) | (n = 3) | ||
| R593W | 694 ± 23 | 16 ± 2 | 10 ± 1 | 28 ± 10 | 5.1 ± 2.0 | 71 ± 6 | 36 ± 2 | NDb |
| (n = 7) | (n = 3) | (n = 3) | (n = 10) | (n = 10) | (n = 10) | (n = 3) | ||
| V628M | 890 ± 29 | 18 ± 2 | 12 ± 1 | 45 ± 11 | 5.9 ± 1.5 | 74 ± 6 | 41 ± 2 | NDb |
| (n = 7) | (n = 4) | (n = 3) | (n = 7) | (n = 7) | (n = 4) | (n = 3) | ||
| E700K | 621 ± 19 | 85 ± 2 | 20 ± 1 | 69 ± 3 | 5.3 ± 0.2 | 59 ± 5 | 17 ± 1 | 42 ± 3 |
| (n = 6) | (n = 4) | (n = 2) | (n = 7) | (n = 7) | (n = 5) | (n = 3) | (n = 3) | |
| M731T | 218 ± 10 | 30 ± 1 | 5 ± 1 | 14 ± 3 | 3.0 ± 0.7 | 66 ± 4 | 41 ± 2 | NDb |
| (n = 8) | (n = 5) | (n = 4) | (n = 11) | (n = 11) | (n = 5) | (n = 5) | ||
| R834Q | 2496 ± 89 | 21 ± 4 | 15 ± 1 | 35 ± 13 | 5.9 ± 2.3 | 56 ± 4 | 41 ± 2 | NDb |
| (n = 7) | (n = 5) | (n = 3) | (n = 9) | (n = 9) | (n = 6) | (n = 6) | ||
a Extracted from the data in Fig. 4.
b Ratio between phosphorylation levels without (EP) and with oligomycin (EPoligo). Phosphorylation was carried out for 10 s at 0 °C in 20 mm Tris (pH 7.5), 3 mm MgCl2, 2 μm [γ-32P]ATP, 10 μm ouabain, 150 mm NaCl, without and with 20 μg of oligomycin/ml.
c Extracted from the data in Figs. 5 and 6. The kinetic parameters were not determined (ND) for R202Q and T263M, because the phosphorylation rate at 2 μm ATP was close to wild type.
d Extracted from the data in Fig. 7.
e Extracted from the data in Fig. 3. For T263M, the E2P dephosphorylation rate constant was not determined (ND), because the high extent of accumulation of E1P, even at 25 °C, precluded such a determination. The dephosphorylation rate at 20 mm K+ was determined only when the dephosphorylation at 1 mm K+ was slower than that of the wild type.
RESULTS
Overall Function
The nine FHM2 mutations were introduced into the ouabain-resistant version of the human α2-isoform, and the mutants were expressed in mammalian COS-1 cells and subjected to ouabain-selection pressure (20), taking advantage of the >100-fold difference between the ouabain affinities of the exogenous Na+,K+-ATPase and the endogenous COS cell enzyme. Like the wild type, all mutants were able to confer ouabain resistance in growth medium containing 5 μm ouabain, indicating that the rate of transport of Na+ and K+ is sufficiently high to sustain cell growth. Determination of the ouabain dependence of Na+,K+-ATPase activity showed 2–9-fold reduction of the apparent affinity for ouabain for mutants V138A, T263M, T345A, R593W, V628M, M731T, and R834Q, wild type-like apparent affinity for R202Q, and 2-fold increased apparent affinity for E700K (Table 1). Hence, all the functional studies described below were carried out in the presence of ouabain to selectively inhibit the endogenous Na+,K+-ATPase (K0.5 ≤ 1 μm).
The catalytic turnover rate determined in the presence of 130 mm Na+, 20 mm K+, and a saturating concentration of MgATP was found reduced in all mutants, most severely for R593W, V628M, M731T, and R834Q, displaying less than one-third the turnover rate of the wild type. T263M, T345A, E700K, and V138A displayed a turnover rate around 50%, whereas R202Q showed ∼20% reduction (Table 1).
K+ Interaction
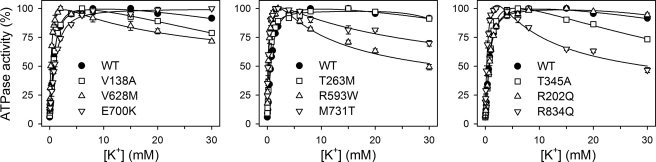
A critical issue is whether the affinity for external K+ is lowered, thereby resulting in reduced ability to clear K+ from the extracellular space. The interaction with K+ was investigated by determining the K+ dependence of ATPase activity (Fig. 2). In the wild type, K+ at a submillimolar concentration activates ATP hydrolysis by binding at extracellularly facing sites of the E2P form, thereby stimulating dephosphorylation (cf. Scheme 1, Reaction 6). The majority of the mutants displayed a K0.5 value for K+ activation similar to or lower (i.e. corresponding to higher affinity) than that determined for the wild type, with E700K being the only exception, exhibiting a slight 1.6-fold increase in the K0.5 value for K+ activation relative to wild type (Fig. 2 and Table 1). K+ concentrations above 15 mm inhibited the wild type somewhat, which is explained by K+ binding in competition with Na+ at the cytoplasmically facing sites of the enzyme in E1 conformation, leading to conversion of E1 back to the K+-occluded E2 state (cf. Scheme 1, Reaction 1) (23, 24). Mutants V138A, T345A, R593W, V628M, M731T, and R834Q exhibited a more distinct inhibitory phase than the wild type, whereas no inhibition was observed for E700K at the K+ concentrations studied (Fig. 2).
FIGURE 2.
K+ dependence of Na+,K+-ATPase activity. ATPase activity was measured at 37 °C in 40 mm NaCl, 3 mm ATP, 3 mm MgCl2, 30 mm histidine (pH 7.4), 1 mm EGTA, 10 μm ouabain, and K+ concentrations as indicated. K0.5 values for K+ activation are listed in Table 1.
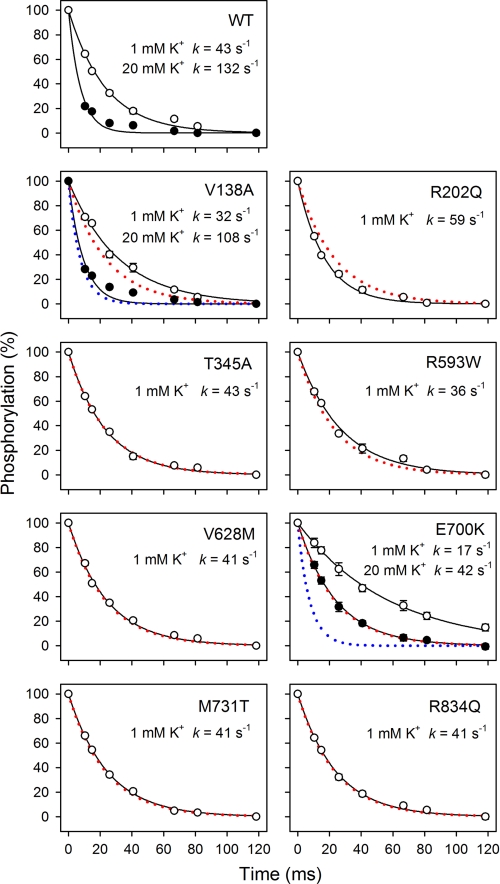
Because the variable inhibitory phase in the K+ dependence of ATPase activity inevitably will influence the apparent affinity for K+ activation corresponding to the rising phase, the activation by extracellular K+ was also examined more directly by determining the rate of E2P dephosphorylation (Fig. 3 and Table 2). At a nonsaturating K+ concentration of 1 mm (Fig. 3, open symbols), T345A, R593W, V628M, M731T, and R834Q displayed a dephosphorylation rate constant almost identical to that of the wild type, indicating that these mutations did not weaken the interaction with extracellular K+. R202Q exhibited a higher rate constant of dephosphorylation than wild type. By contrast, the dephosphorylation rate constant was reduced for V138A and E700K relative to wild type (1.3- and 2.5-fold, respectively). To determine whether this was related to a reduction in affinity for external K+, similar experiments were performed at a saturating K+ concentration of 20 mm (Fig. 3, filled symbols). The calculated ratios between the dephosphorylation rate constants at 1 and 20 mm K+ are 0.29 ± 0.02, 0.42 ± 0.04, and 0.32 ± 0.02 for V138A, E700K, and wild type, respectively. Hence, at 1 mm K+, the saturation of the external sites is similar to wild type for V138A and is actually higher for E700K, indicating that the reduced dephosphorylation rate is caused by a lower Vmax of phosphoenzyme hydrolysis rather than a lower affinity for K+.
FIGURE 3.
Rapid kinetic measurements of K+-induced E2P dephosphorylation. Phosphorylation was performed for 5 s at 25 °C in the presence of 2 μm [γ-32P]ATP in 20 mm Tris (pH 7.5), 20 mm NaCl, 3 mm MgCl2, 1 mm EGTA, 130 mm choline chloride, and 10 μm ouabain, followed by dephosphorylation for the indicated times in the same medium containing in addition 1 mm nonradioactive ATP and 1 mm K+ (open symbols) or 20 mm K+ (filled symbols). Each line represents the best fit of a mono-exponential decay function, giving the indicated rate constants (also listed in Table 2 with statistics). The wild type data are represented in each panel by dotted red and blue lines.
Na+ Interaction
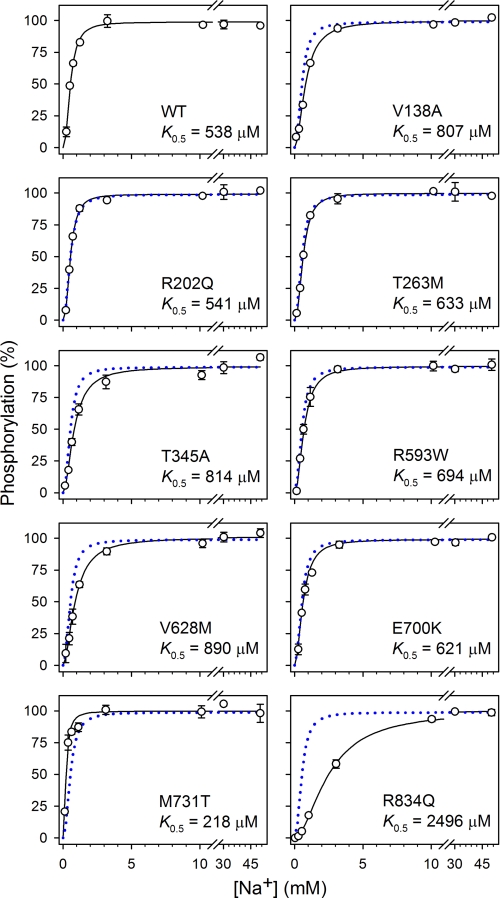
Phosphorylation from ATP is triggered when three Na+ ions have bound to intracellularly facing high affinity sites of the E1 form and become occluded in the E1[Na3] form (Scheme 1, Reaction 2) (25). To assess the Na+ affinity of the E1 form of the mutants, we studied the Na+ dependence of phosphorylation from ATP in the absence of K+ and presence of oligomycin to support occlusion (Fig. 4). The mutants displayed affinities similar to that obtained for the wild type; only R834Q exhibited a more than 2-fold reduced apparent affinity for Na+ relative to wild type.
FIGURE 4.
Na+ dependence of phosphorylation. Phosphorylation was carried out for 10 s at 0 °C in 20 mm Tris (pH 7.5), 3 mm MgCl2, 2 μm [γ-32P]ATP, 10 μm ouabain, 20 μg of oligomycin/ml, NaCl to obtain the indicated concentrations of Na+, and various concentrations of N-methyl-d-glucamine to maintain the ionic strength. Oligomycin was added to optimize phosphorylation and prevent dephosphorylation, thereby minimizing effects on apparent Na+ affinity of variation of the phosphorylation and dephosphorylation rates. Each line represents the best fit of the equation EP = EPmax·[Na+]n/(K0.5n + [Na+]n), giving the indicated K0.5 values (also listed in Table 2 with statistics). The data corresponding to wild type are represented in each panel by a dotted blue line.
Rate of Phosphorylation
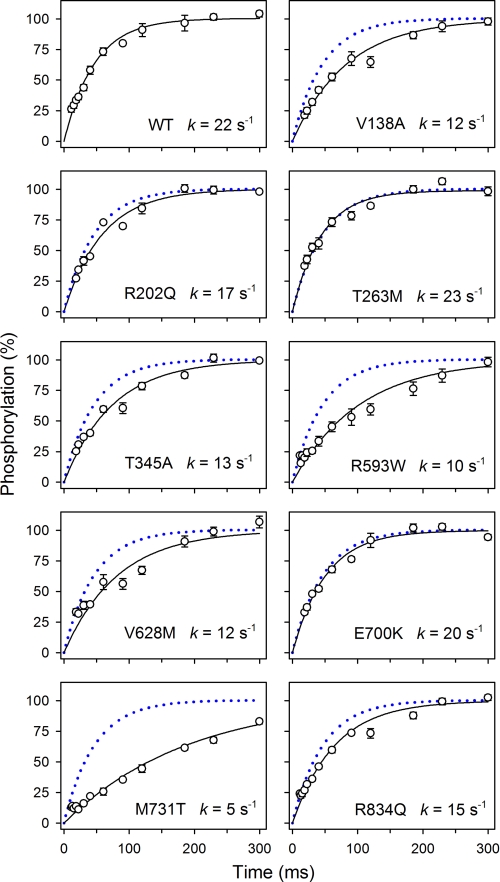
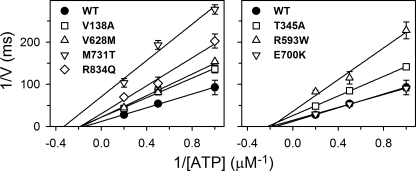
Because oligomycin stabilizes the Na+-occluded E1[Na3] form, thereby promoting phosphorylation (25, 26) and blocking the E1P-E2P transition (Scheme 1, Reaction 4), the presence of oligomycin enables the phosphoenzyme to build up a maximum level. Table 2 lists the level of phosphoenzyme accumulated at 150 mm NaCl in the absence of oligomycin relative to the maximum level obtained in the presence of oligomycin (EP/EPoligo). Mutants R593W, V628M, M731T, and R834Q displayed a markedly reduced EP/EPoligo ratio, and V138A and T345A showed a slight reduction relative to wild type, whereas T263M and E700K displayed higher EP/EPoligo ratios than the wild type. Such effects can be due to changes in either the rate of phosphorylation or the rate of dephosphorylation. In the presence of oligomycin, the level of phosphoenzyme built up was sufficiently high to allow rapid kinetic studies of the rate of phosphorylation at a millisecond time scale at 25 °C, and thus the time course of phosphorylation in the presence of 2 μm ATP was determined (Fig. 5). Mutants V138A, T345A, R593W, V628M, M731T, and R834Q all displayed a reduced rate constant relative to wild type, most pronounced for M731T, suggesting that either the phosphorylation reaction or the binding of ATP is affected in these mutants. To determine the maximal rate of phosphorylation and the apparent affinity of these mutants for ATP we performed the analysis at varying ATP concentrations. E700K was included, because the dephosphorylation studies (Fig. 3) indicated that the catalytic site might be defective in this mutant. In Fig. 6, the results are depicted as double-reciprocal plots of the initial phosphorylation rate per ATPase molecule as a function of the concentration of ATP. The linear dependence allows extraction of the maximal phosphorylation rate per ATPase molecule, Vmax, and the apparent affinity for ATP (the Michaelis constant Km). M731T exhibited a most severe 6-fold reduction of Vmax, relative to wild type. Mutants V138A, T345A, R593W, V628M, and R834Q showed 2–3-fold reduction, whereas E700K was wild type-like (Table 2). None of the mutations increased the Km value. A significant reduction of Km is noted for M731T (Fig. 6 and Table 2), likely reflecting the marked reduction of Vmax, which leads to increased accumulation of nonphosphorylated enzyme with ATP bound. According to the equation Km = (k−1 + k2)/k1, where k1 and k−1 are the respective rate constants for binding and dissociation of ATP, and k2 equals Vmax (measured in units of rate per ATPase molecule), the Km value will decrease with k2, unless k−1 is much higher than the k2 value, which is not the case here, because Na+,K+-ATPase binds the nucleotide rather tightly (27). In an analogous way, the accumulation of E1 explains the increased apparent affinity for Na+ in M731T (Table 2).
FIGURE 5.
Rapid kinetic measurements of phosphorylation rate. Phosphorylation was performed for the indicated times at 25 °C in the presence of 2 μm [γ-32P]ATP, 100 mm NaCl, 40 mm Tris (pH 7.5), 3 mm MgCl2, 1 mm EGTA, 10 μm ouabain, and 20 μg/ml oligomycin. Each line represents the best fit of a mono-exponential “rise to max function,” giving the indicated rate constants. The wild type data are represented in each panel by a dotted blue line.
FIGURE 6.
ATP dependence of phosphorylation rate. Initial rates of phosphorylation, determined as in Fig. 5 at 1, 2, and 5 μm [γ-32P]ATP, are shown as a function of ATP concentration in double-reciprocal plots. For Vmax and Km values, see Table 2.
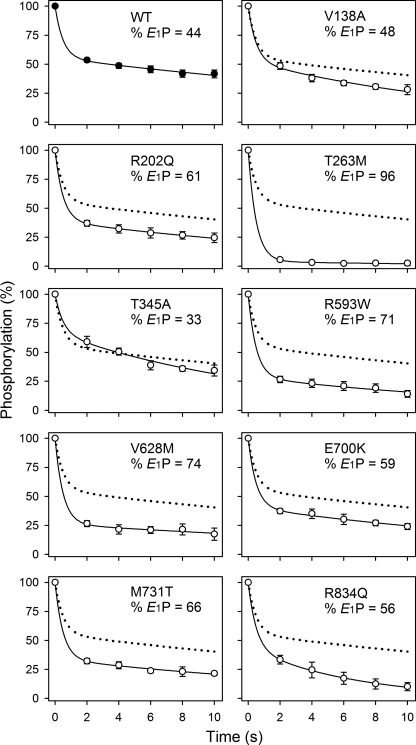
E1P-E2P Distribution
The phosphorylated form of the Na+,K+-ATPase exists in E1P and E2P states that can be distinguished by their different sensitivities to K+ and ADP (Scheme 1). To estimate the extent of each phosphoenzyme pool present at steady state, the dephosphorylation time course was examined following addition of ADP to the phosphoenzyme. A bi-exponential function could be fitted to the data, permitting extraction of a rapid and a slow decay component, reflecting the initial amounts of E1P and E2P, respectively (Fig. 7). Table 2 lists the E1P fraction. Notably, mutant T263M displayed a marked increase in the level of E1P (to 96% versus 44% for the wild type). The other mutants displayed a less pronounced shift of the E1P-E2P distribution toward the E1P form or were wild type-like.
FIGURE 7.
Distribution of the phosphoenzyme between E1P and E2P. Phosphorylation was carried out for 10 s at 0 °C in the presence of 2 μm [γ-32P]ATP, 20 mm NaCl, 130 mm choline chloride, 20 mm Tris (pH 7.5), 3 mm MgCl2, 1 mm EGTA, and 10 μm ouabain. Dephosphorylation was followed by addition of a chase solution producing final concentrations of 2.5 mm ADP and 1 mm unlabeled ATP. The dephosphorylation reaction was terminated by acid quenching after the indicated time intervals. A bi-exponential decay function was fitted to the data. The rate constant corresponding to the rapid phase, reflecting the ADP reaction with E1P, was set to 2 s−1. The fraction of E1P phosphoenzyme obtained from this analysis is indicated in the panel (also listed in Table 2 with statistics). The wild type data are represented in each panel by a dotted line.
Vanadate and ATP Affinities
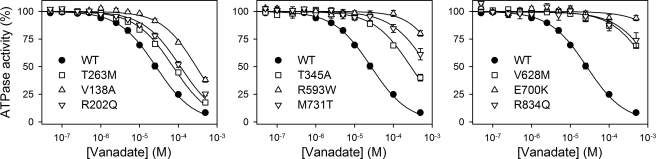
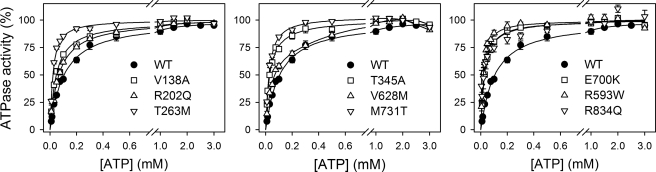
Vanadate is an inhibitor that acts as analog of the phosphoryl group in the transition state during E2P hydrolysis and binds exclusively to the enzyme in the E2 conformation (28). The properties of the E2 state were investigated by determining the apparent affinity for vanadate inhibition of ATP hydrolysis (Fig. 8). All the mutants exhibited a reduced apparent affinity for vanadate (Table 1). For R593W, V628M, E700K, M731T, and R834Q, the affinity was extremely low (≥30-fold reduced relative to wild type); in fact, no significant inhibition was seen for E700K within the vanadate concentration range examined (Fig. 8, right panel). A reduced sensitivity to vanadate inhibition can in principle arise from either a lowering of the intrinsic binding affinity of E2 (possibly reflecting destabilization of the transition state of E2P hydrolysis) or from a shift of the E1-E2 distribution away from the vanadate binding E2 form in favor of E1. To examine the mutational effects on the E1-E2 distribution, the ATP dependence of ATPase activity was also studied (Fig. 9). Because ATP binds with higher affinity to E1 than to E2, an increase in the apparent affinity for ATP upon mutation reflects a shift of the E1-E2 distribution toward the E1 form, whereas a decrease in the affinity for ATP reflects a shift toward the E2 form. The results summarized in Table 1 show a significant increase of the apparent ATP affinity for all the mutants (2–5-fold), indicating a shift toward the E1 form. However, for R593W, V628M, E700K, M731T, and R834Q, the effect on ATP affinity was much less spectacular than the reduction of vanadate affinity, indicating that the latter arises not only from a shift in E1-E2 distribution but also from a change in the intrinsic affinity of the E2 form for vanadate.
FIGURE 8.
Vanadate dependence of Na+,K+-ATPase activity. ATPase activity was measured at 37 °C in 130 mm NaCl, 20 mm KCl, 3 mm MgCl2, 3 mm ATP, 30 mm histidine (pH 7.4), 1 mm EGTA, 10 μm ouabain, and vanadate concentrations as indicated. Each line represents the best fit of the equation V = Vmax·(1 − [vanadate]n/(K0.5n + [vanadate]n)), with n ranging between 0.9 and 1.1, giving the K0.5 values indicated in Table 1.
FIGURE 9.
ATP dependence of Na+,K+-ATPase activity. ATPase activity was measured at 37 °C in the presence of 130 mm NaCl, 20 mm KCl, 3 mm MgCl2, 30 mm histidine (pH 7.4), 1 mm EGTA, 10 μm ouabain, and the indicated concentrations of ATP. K0.5 values for activation are listed in Table 1.
DISCUSSION
The nine FHM2 mutations studied here, which are found in various regions of the Na+,K+-ATPase α2-isoform, lead to a functionally altered but active enzyme capable of sustaining cell viability. Our results do not lend support to the hypothesis that a reduced affinity for external K+, causing a selective disturbance of K+ clearance, is an obligatory part of the mechanism underlying the disease (4). Neither can a defective Na+ interaction generally account for the pathophysiology of the disease, as only R834Q exhibited a significant reduction of the Na+ affinity. This is contrary to the neurological disorder rapid-onset dystonia parkinsonism, caused by mutation of the α3-isoform of Na+,K+-ATPase, where all mutants characterized so far display markedly reduced Na+ affinity (21, 29, 30). At saturating Na+ and K+ concentrations, the FHM2 mutants studied here display a reduced catalytic turnover rate relative to wild type. A reduced turnover rate has previously been reported for mutants T263M, T345A, M731T, and R834Q, but the responsible change(s) in partial reaction step(s) were not elucidated (4, 9, 10). Our rapid kinetic studies of the phosphorylation from ATP identify a reduced Vmax of phosphorylation as a major factor contributing to the reduction of the catalytic turnover rate of mutants V138A, T345A, R593W, V628M, M731T, and R834Q. These mutants displayed decreases in the Vmax of phosphorylation of 2–6-fold (Fig. 6 and Table 2). To be able to carry out transient kinetic measurements, oligomycin was added to promote phosphorylation by stabilizing the Na+-occluded E1[Na3] form and blocking the E1P[Na3] → E2P transition. In the absence of oligomycin, the relative level of phosphoenzyme at steady state was in fact very low for R593W, V628M, M731T, and R834Q (Table 2, “EP/EPoligo”), suggesting that under physiological conditions without oligomycin the phosphorylation rate is even more dramatically affected than judged from the kinetic data obtained with oligomycin.
The low phosphorylation rate also accounts for the enhanced K+ inhibition of ATPase activity at high K+ concentrations seen particularly for R593W, V628M, M731T, and R834Q (Fig. 2). Hence, the low phosphorylation rate leads to increased availability of E1 at steady state and consequently to enhanced K+ competition with Na+ at the intracellular E1 sites. In the living cell with cytoplasmic concentrations of ∼150 mm K+ and only ∼10 mm Na+, the enhanced K+ competition with Na+ may be highly relevant as a factor contributing to compromise pump function.
For R593W, V628M, E700K, M731T, and R834Q, the apparent affinity for vanadate was lowered to an extent that could not be accounted for by the observed shift of the conformational equilibrium toward E1 (Fig. 8 and Table 1). Because vanadate binds at the phosphorylation site of the E2 form as an analog of the phosphoryl group in the transition state between E2 and E2P, the extraordinary low affinity for vanadate might be related to a perturbation of the phosphorylation site in E2 and/or the transition state. For the mutants showing both a reduced Vmax of phosphorylation from ATP in E1 and an extraordinary low affinity for vanadate (R593W, V628M, M731T, and R834Q), the underlying structural changes in E1 and E2 forms might be similar. By contrast E700K did not show a reduced phosphorylation rate with ATP, and for this mutant the phosphorylation site therefore seems not to be perturbed in E1. Besides its extraordinary lack of sensitivity to vanadate, E700K was characterized by a relatively low maximal rate of dephosphorylation of E2P (42 s−1 versus 132 s−1 for wild type), which probably causes the reduced catalytic turnover rate of E700K. Both the disruption of vanadate binding and the reduced dephosphorylation rate can be explained by assuming that the transition state of E2P dephosphorylation is destabilized. It is furthermore of note that E700K exhibited an increased apparent affinity for ouabain, and even though E700K did not show reduced affinity for K+ activation of dephosphorylation (Fig. 3 and Table 2), a slight 1.6-fold reduction of apparent K+ affinity in activation of ATPase activity was noted (Fig. 2 and Table 1). Both of these effects might result from accumulation at steady state of the E2P[Na2] phosphoenzyme form, which is an intermediate in the E1P[Na3] → E2P transition (31) exhibiting a particularly high affinity for cardiotonic steroids (“E*P” cf. Refs 32 and 33). Thus, the evidence indicates that mutation E700K is most disturbing in the part of the pump cycle involving E2P and E2P-like states.
From the crystal structure it is known how the residues studied here are positioned in the E2[K2] state with bound MgF42− as a phosphate analog (18, 19), and although crystal structures are static snapshots without the flexibility of the native enzyme, they are useful as a basis for analysis of the mechanisms underlying the observed mutational effects. Arg593, Val628, Glu700, and Met731 are located in the Rossmann fold, which is a central feature of the P-domain, consisting of a seven-stranded parallel β-sheet associated with seven short α-helices (P1–P7) as indicated in Fig. 10. Asp374 (phosphorylation site) and residues involved in Mg2+ binding, Thr376, Asp714, and Asp718, are located centrally in the Rossmann fold (Fig. 10). The phosphorylation of Asp374 is triggered by an approach between the N- and P-domains associated with a bending of the P-domain. This conformational change is the result of transmission of the effect of Mg2+ binding to the P7 helix of the Rossmann fold (34, 35), meaning that the loop between β6 and P7, where Met731 is located (Figs. 10 and 11), has to be strained when Mg2+ binds to Asp714. The role of Met731 may thus be to reduce the freedom of movement of this loop, thereby ensuring the strain. In fact the side chain of Met731 is flanked on one side by Asp714, Thr378, and Arg593 (Fig. 10B), and the introduction of the threonine side chain in M731T likely pushes Asp714, thus preventing a proper bending of the P-domain. It is interesting to note that M731T and R593W are the mutations giving the largest reduction of phosphorylation rate observed in this study (cf. Table 2). Considering that the Met731 and Arg593 side chains are located within a distance of only 4–5 Å from each other in the crystal structure (Fig. 10, A and B), it is possible that the bulky tryptophan in R593W actually interferes by disturbing Met731. Furthermore, Arg593 is within hydrogen bonding distance of the backbone carbonyls of Gly377 and Thr378, which are located in the same loop as Asp374 and Thr376 (Fig. 10, A and B). The clash resulting from insertion of the tryptophan might therefore also disturb phosphorylation by shifting the positions of Asp374 and Thr376. It is noteworthy that mutations T376M and T378N have been found in familial hemiplegic migraine patients and were reported as loss of function mutations based on cell survival studies (14, 36–38).
FIGURE 10.
Location of the FHM2 mutations with relation to the Rossmann fold in the Na+,K+-ATPase structure. The structure used is the same as in Fig. 1. Side chains corresponding to mutated residues and interaction partners (numbered according to the human α2-isoform) are highlighted as sticks, as are key residues in the catalytic site as follows: Thr376, Asp714, and Asp718 (Mg2+-binding residues), and Asp374 (phosphorylation site). Atoms are colored according to the elements (carbon, gray; oxygen, red; nitrogen, blue; sulfur, yellow). The catalytic Mg2+ ion is shown as a purple sphere. A, Rossmann fold of the P-domain viewed from the cytosol. P1–P7 helices with interposed β-strands (blue arrows with yellow numbers 1–7) are indicated. Note the proximity of Arg593 and Met731 to Gly377 and Thr378, and the position of Glu700 between the two positively charged side chains of Arg704 and Lys724. B, close up of interaction network around Arg593 and Met731. C, close up of hydrophobic/van der Waals interaction network around Val628 (Pro592 and Val596 also indicated in A).
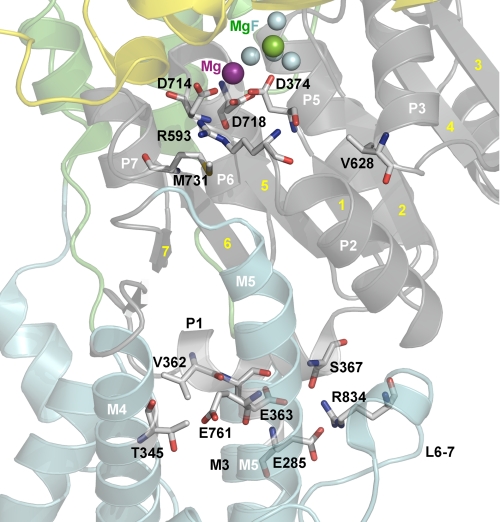
FIGURE 11.
Side view of the membrane interface of the P-domain. The structure used is the same as in Fig. 1 with the same color codes for P-, A-, N-, and M-domain, and the phosphate analog MgF42− (MgF) shown as one green and four cyan spheres. Side chains corresponding to mutated residues and interaction partners (numbered according to the human α2-isoform) are highlighted as sticks, as are key residues in the catalytic site. Atoms are colored according to the elements (carbon, gray; oxygen, red; nitrogen, blue; sulfur, yellow). P-domain helices P1–P3 and P5–P7 as well as the interposed β-strands (yellow numbers) and transmembrane helices M3–M5 are indicated. Note the location of the P1 helix with Val362 interacting with Thr345, and Glu363 and Ser367 interacting with Arg834. Glu761 (M5) may also interact with Thr345, depending on the actual rotational state of Thr345. In addition, Glu285 (M3) interacts with Arg834.
The valine replaced in mutant V628M is positioned at the end of the P3 helix in the Rossmann fold. It is part of a hydrophobic/van der Waals interaction network (Fig. 10, A and C), and the interaction with Pro592 and Val596 at the N-terminal end of the P2 helix may contribute to stabilize the positions of helices P2 and P3. The V628M mutation might lead to a shift of the position of Pro592, thereby disturbing the interactions of the adjacent Arg593 with Gly377 and Thr378 mentioned above.
The glutamate replaced in E700K is located in the P5 helix of the Rossmann fold. In the crystal structure of the Na+,K+-ATPase E2[K2] state with bound MgF42−, Glu700 is positioned between two positively charged residues, Arg704 (at the end of the P5 helix) and Lys724 (P6 helix), with a distance of 4.6 Å to each (Fig. 10A). Moreover, Lys724 is within hydrogen bonding distance to Gln703 in the P5 helix (2.7 Å). It can be imagined that the extra positive charge introduced between Arg704 and Lys724 by E700K leads to a repulsion between the three positive charges disturbing the positioning of P5 and P6. Such a disturbance could lead to a shift of β5 and the loop between β5 and P6 containing Asp714 and Asp718 critical to Mg2+ binding (Fig. 10A). We wondered whether Glu700 and either Lys724 or Arg704 might actually be closer to each other in the vanadate-bound E2 state than the 4.6 Å in the E2[K2] crystal structure. The Na+,K+-ATPase residues Glu700, Lys724, and Gln703 are conserved as Glu689, Lys713, and Gln692 in the Ca2+-ATPase, which has been crystallized not only in the MgF42−-bound E2 form similar to the Na+,K+-ATPase crystal form but also in several other E2 states, including the AlF4−-bound form. The latter is considered the state closest to the E2 form with vanadate bound, as AlF4− and vanadate are both believed to mimic the trigonal bipyramidal structure of the penta-coordinated phosphate in the transition state of E2P dephosphorylation (39, 40). Interestingly, among the AlF4−-bound E2 crystal structures of the Ca2+-ATPase, 4 out of 7 have a distance between Glu689 and Lys713 <4 Å, i.e. short enough to indicate the presence of a salt link between these residues in the E2P transition state. In contrast, in the MgF42−-bound Ca2+-ATPase structures, the corresponding distance is 4–5 Å, consistent with the distance in the MgF42−-bound form of Na+,K+-ATPase (cf. supplemental Table S1). If this scenario for the E2P transition state were extrapolated to the Na+,K+-ATPase, it would explain the very strong destabilization of the vanadate-bound state of E700K (Fig. 8). If in addition Glu700 and Lys724 were further apart in the E1[Na3] state, the lack of effect of the mutation on the phosphorylation from ATP would be understandable. The Ca2+-ATPase E1 structures do not provide a clear answer to this question, but it is noteworthy that 4 out of 8 E1 structures show a distance larger than 4 Å (cf. supplemental Table S1).
T345A in the cytoplasmic extension of M4 and R834Q in the L6–7 loop are both located at the boundary between the P-domain and the transmembrane region, and the reason that these mutations also reduce the phosphorylation rate significantly may be the participation of Thr345 and Arg834 in interaction networks involving helices connected with the Rossmann fold (Fig. 11). Thr345 might participate in van der Waals interactions with Val362 at the N-terminal end of the P-domain helix P1, from which the central β-strand of the Rossmann fold (β-strand “1” in Fig. 11) leads to the phosphorylation site with Asp374. Interestingly, mutation V362E has also been found in patients with FHM2 (41). Depending on the actual rotational state of the Thr345 side chain, the hydroxyl group might form a hydrogen bond with Glu761 of M5. Mutation R834Q seems to disrupt bonds from P1-helix residues Glu363 and Ser367 (C-terminal end of the P1-helix) to the L6–7 loop (19). R834Q furthermore disrupts the bond between Arg834 and Glu285 of M3 (Fig. 11), which may explain the reduction of Na+ affinity caused by R834Q, since mutation of Glu285 has been shown to reduce Na+ affinity, possibly a consequence of the involvement of Glu285 in control of the cytoplasmic entrance pathway for Na+ (22).
Surprisingly, the mutation V138A also had significant impact on the phosphorylation rate, even though Val138 is positioned in the transmembrane segment M2 far from the phosphorylation site, thus indicating a long range effect, which might be exerted through interference with the hydrophobic/van der Waals interactions between M1 and M2 (cf. Fig. 12A). These interactions seem to allow M1 to close the Na+ binding pocket through contact with the ion binding Glu332 of M4 (42), a conformational change believed to be propagated to the phosphorylation site in a yet undefined way and result in phosphoryl transfer (35). Indeed, a reduced rate of phosphorylation was previously observed following replacement of M1 residues Leu94 and Gly97 (42, 43).
FIGURE 12.
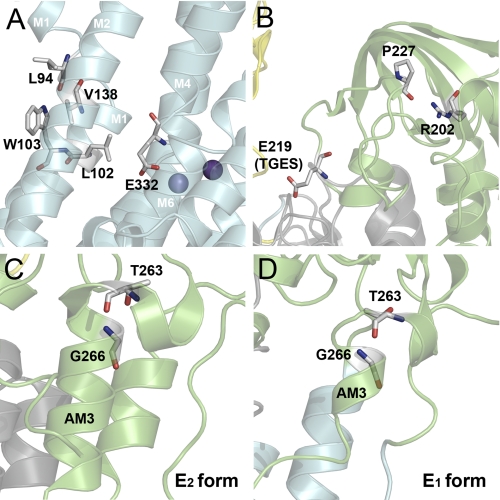
Structural relationships of residues altered in mutants V138A, R202Q, and T263M. The structure used in A–C is the same as in Fig. 1 with the same color codes for P-, A-, N-, and M-domain. Side chains corresponding to mutated residues and interaction partners (numbered according to the human α2-isoform) are highlighted as sticks. Atoms are colored according to the elements (carbon, gray; oxygen, red; nitrogen, blue). A, Val138 in M2 interacts with M1 residues Leu94 and Trp103. B, interaction between Arg202 and Pro227 may stabilize the loop containing the conserved TGES motif with Glu219. C, Thr263 is located in the AM3 linker segment connecting the A-domain to the M3 transmembrane segment (cf. Fig. 1). In the E2 and E2P conformations, part of the AM3 linker is coiled up, forming an α-helical arrangement, which is partially unwound at Thr263, with the two parts of the α-helix being almost orthogonal to each other. The side chain hydroxyl group of the threonine is within hydrogen bonding distance to the backbone amide nitrogen of Gly266. D, homology model of the Na+,K+-ATPase in E1 based on the structure of the closely related Ca2+-ATPase in E1 (Protein Data Bank code 1T5T). The helical structure of the AM3 linker is much more unwound than in E2 conformations of Na+,K+-ATPase and Ca2+-ATPase, with less strict requirement for the bond between Thr263 and Gly266.
For R202Q and T263M, the phosphorylation rate was wild type-like, and the reason for the reduced catalytic turnover rate seems to be a slow conversion of E1P to E2P, in particular T263M showed a conspicuous accumulation of E1P. Furthermore, both R202Q and T263M appeared to shift the E1-E2 distribution of the dephosphoenzyme in favor of E1. These mutations are both associated with the A-domain, which undergoes drastic structural rearrangements during the E1-E2 and E1P-E2P transitions (35, 44). Arg202 is positioned in a β-strand of the A-domain, where it appears to form a hydrogen bond with the backbone carbonyl oxygen of Pro227 (2.7 Å distance, Fig. 12B). This bond could be important for stabilization of the loop containing the conserved TGES motif, and hence for interaction of TGES with the P-domain in E2 and E2P. Thr263 is located in the AM3 linker segment connecting the A-domain to M3 and appears to be involved in stabilization of the kinked α-helix present in the AM3 linker in E2, but not in E1 (see Fig. 12, C and D). Hence, in E2 the side chain hydroxyl group of the threonine seems to form a hydrogen bond with the backbone amide nitrogen of Gly266 (2.8 Å distance). This structural arrangement is likely destabilized by the T263M substitution due to the bulkiness of the methionine side chain. The ability of the E1 conformation to better accommodate the methionine, due to a looser structure compared with the helical arrangement in E2 (Fig. 12D), explains that the conformational equilibrium is shifted toward E1 in the mutant. In line with this interpretation, mutation of Gly266 to alanine, disturbing the kinked helix arrangement, was previously shown to cause accumulation of E1 (45).
To summarize, if disturbance of K+ clearance by glial cells is the reason for development of FHM2, this disturbance must be attributed to a low maximum turnover rate of the Na+,K+-ATPase and not to a reduced affinity for external K+. In several of the FHM2 mutants studied here, the function of the catalytic site in phosphorylation (V138A, T345A, R593W, V628M, M731T, and R834Q) or dephosphorylation (E700K) is affected, involving local effects on the catalytic assembly as well as helices connected with the Rossmann fold, and long range effects transmitted from as far away as the membrane domain. The last two mutations (R202Q and T263M) affect the maximum turnover rate by destabilizing the A-domain in the E2/E2P conformations.
Supplementary Material
Acknowledgments
We thank Kirsten Lykke Pedersen, Janne Petersen, and Nina Juste for expert technical assistance. We also thank Professor C. Toyoshima, University of Tokyo, for discussion and helpful suggestions.
Footnotes
This work was supported in part by grants from the Danish Medical Research Council, the Novo Nordisk Foundation (Fabrikant Vilhelm Pedersen og Hustrus Legat), and the Lundbeck Foundation.

This article contains supplemental Table S1.
REFERENCES
- 1. De Fusco M., Marconi R., Silvestri L., Atorino L., Rampoldi L., Morgante L., Ballabio A., Aridon P., Casari G. (2003) Haploinsufficiency of ATP1A2 encoding the Na+/K+ pump α2 subunit associated with familial hemiplegic migraine type 2. Nat. Genet. 33, 192–196 [DOI] [PubMed] [Google Scholar]
- 2. McGrail K. M., Phillips J. M., Sweadner K. J. (1991) Immunofluorescent localization of three Na,K-ATPase isozymes in the rat central nervous system: both neurons and glia can express more than one Na,K-ATPase. J. Neurosci. 11, 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Somjen G. G. (2001) Mechanisms of spreading depression and hypoxic spreading depression-like depolarization. Physiol. Rev. 81, 1065–1096 [DOI] [PubMed] [Google Scholar]
- 4. Segall L., Scanzano R., Kaunisto M. A., Wessman M., Palotie A., Gargus J. J., Blostein R. (2004) Kinetic alterations due to a missense mutation in the Na,K-ATPase α2 subunit cause familial hemiplegic migraine type 2. J. Biol. Chem. 279, 43692–43696 [DOI] [PubMed] [Google Scholar]
- 5. Lencesova L., O'Neill A., Resneck W. G., Bloch R. J., Blaustein M. P. (2004) Plasma membrane-cytoskeleton-endoplasmic reticulum complexes in neurons and astrocytes. J. Biol. Chem. 279, 2885–2893 [DOI] [PubMed] [Google Scholar]
- 6. Rose E. M., Koo J. C., Antflick J. E., Ahmed S. M., Angers S., Hampson D. R. (2009) Glutamate transporter coupling to Na,K-ATPase. J. Neurosci. 29, 8143–8155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jen J. C., Klein A., Boltshauser E., Cartwright M. S., Roach E. S., Mamsa H., Baloh R. W. (2007) Prolonged hemiplegic episodes in children due to mutations in ATP1A2. J. Neurol. Neurosurg. Psychiatry 78, 523–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Todt U., Dichgans M., Jurkat-Rott K., Heinze A., Zifarelli G., Koenderink J. B., Goebel I., Zumbroich V., Stiller A., Ramirez A., Friedrich T., Göbel H., Kubisch C. (2005) Rare missense variants in ATP1A2 in families with clustering of common forms of migraine. Hum. Mutat. 26, 315–321 [DOI] [PubMed] [Google Scholar]
- 9. Segall L., Mezzetti A., Scanzano R., Gargus J. J., Purisima E., Blostein R. (2005) Alterations in the α2 isoform of Na,K-ATPase associated with familial hemiplegic migraine type 2. Proc. Natl. Acad. Sci. U.S.A. 102, 11106–11111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tavraz N. N., Friedrich T., Dürr K. L., Koenderink J. B., Bamberg E., Freilinger T., Dichgans M. (2008) Diverse functional consequences of mutations in the Na+/K+-ATPase α2-subunit causing familial hemiplegic migraine type 2. J. Biol. Chem. 283, 31097–31106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vanmolkot K. R., Stam A. H., Raman A., Koenderink J. B., de Vries B., van den Boogerd E. H., van Vark J., van den Heuvel J. J., Bajaj N., Terwindt G. M., Haan J., Frants R. R., Ferrari M. D., van den Maagdenberg A. M. (2007) First case of compound heterozygosity in Na,K-ATPase gene ATP1A2 in familial hemiplegic migraine. Eur. J. Hum. Genet. 15, 884–888 [DOI] [PubMed] [Google Scholar]
- 12. Vanmolkot K. R., Kors E. E., Hottenga J. J., Terwindt G. M., Haan J., Hoefnagels W. A., Black D. F., Sandkuijl L. A., Frants R. R., Ferrari M. D., van den Maagdenberg A. M. (2003) Novel mutations in the Na+, K+-ATPase pump gene ATP1A2 associated with familial hemiplegic migraine and benign familial infantile convulsions. Ann. Neurol. 54, 360–366 [DOI] [PubMed] [Google Scholar]
- 13. Kaunisto M. A., Harno H., Vanmolkot K. R., Gargus J. J., Sun G., Hämäläinen E., Liukkonen E., Kallela M., van den Maagdenberg A. M., Frants R. R., Färkkilä M., Palotie A., Wessman M. (2004) A novel missense ATP1A2 mutation in a Finnish family with familial hemiplegic migraine type 2. Neurogenetics 5, 141–146 [DOI] [PubMed] [Google Scholar]
- 14. Riant F., De Fusco M., Aridon P., Ducros A., Ploton C., Marchelli F., Maciazek J., Bousser M. G., Casari G., Tournier-Lasserve E. (2005) ATP1A2 mutations in 11 families with familial hemiplegic migraine. Hum. Mutat. 26, 281. [DOI] [PubMed] [Google Scholar]
- 15. Pierelli F., Grieco G. S., Pauri F., Pirro C., Fiermonte G., Ambrosini A., Costa A., Buzzi M. G., Valoppi M., Caltagirone C., Nappi G., Santorelli F. M. (2006) A novel ATP1A2 mutation in a family with FHM type II. Cephalalgia 26, 324–328 [DOI] [PubMed] [Google Scholar]
- 16. Vanmolkot K. R., Kors E. E., Turk U., Turkdogan D., Keyser A., Broos L. A., Kia S. K., van den Heuvel J. J., Black D. F., Haan J., Frants R. R., Barone V., Ferrari M. D., Casari G., Koenderink J. B., van den Maagdenberg A. M. (2006) Two de novo mutations in the Na,K-ATPase gene ATP1A2 associated with pure familial hemiplegic migraine. Eur. J. Hum. Genet. 14, 555–560 [DOI] [PubMed] [Google Scholar]
- 17. Thomsen L. L., Kirchmann M., Bjornsson A., Stefansson H., Jensen R. M., Fasquel A. C., Petursson H., Stefansson M., Frigge M. L., Kong A., Gulcher J., Stefansson K., Olesen J. (2007) The genetic spectrum of a population-based sample of familial hemiplegic migraine. Brain 130, 346–356 [DOI] [PubMed] [Google Scholar]
- 18. Morth J. P., Pedersen B. P., Toustrup-Jensen M. S., Sørensen T. L., Petersen J., Andersen J. P., Vilsen B., Nissen P. (2007) Crystal structure of the sodium-potassium pump. Nature 450, 1043–1049 [DOI] [PubMed] [Google Scholar]
- 19. Shinoda T., Ogawa H., Cornelius F., Toyoshima C. (2009) Crystal structure of the sodium-potassium pump at 2.4 A resolution. Nature 459, 446–450 [DOI] [PubMed] [Google Scholar]
- 20. Vilsen B. (1992) Functional consequences of alterations to Pro-328 and Leu-332 located in the 4th transmembrane segment of the α-subunit of the rat kidney Na+,K+-ATPase. FEBS Lett. 314, 301–307 [DOI] [PubMed] [Google Scholar]
- 21. Rodacker V., Toustrup-Jensen M., Vilsen B. (2006) Mutations Phe785Leu and Thr618Met in Na+,K+-ATPase, associated with familial rapid-onset dystonia parkinsonism, interfere with Na+ interaction by distinct mechanisms. J. Biol. Chem. 281, 18539–18548 [DOI] [PubMed] [Google Scholar]
- 22. Toustrup-Jensen M., Vilsen B. (2002) Importance of Glu(282) in transmembrane segment M3 of the Na+,K+-ATPase for control of cation interaction and conformational changes. J. Biol. Chem. 277, 38607–38617 [DOI] [PubMed] [Google Scholar]
- 23. Skou J. C. (1957) The influence of some cations on an adenosine triphosphatase from peripheral nerves. Biochim. Biophys. Acta 23, 394–401 [DOI] [PubMed] [Google Scholar]
- 24. Vilsen B. (1999) Mutant Phe788 → Leu of the Na+,K+-ATPase is inhibited by micromolar concentrations of potassium and exhibits high Na+-ATPase activity at low sodium concentrations. Biochemistry 38, 11389–11400 [DOI] [PubMed] [Google Scholar]
- 25. Esmann M., Skou J. C. (1985) Occlusion of Na+ by the Na,K-ATPase in the presence of oligomycin. Biochem. Biophys. Res. Commun. 127, 857–863 [DOI] [PubMed] [Google Scholar]
- 26. Skou J. C. (1991) in The Sodium Pump: Recent Developments (Kaplan J. H., De Weer P., eds) pp. 317–319, The Rockefeller University Press, New York [Google Scholar]
- 27. Fedosova N. U., Champeil P., Esmann M. (2003) Rapid filtration analysis of nucleotide binding to Na,K-ATPase. Biochemistry 42, 3536–3543 [DOI] [PubMed] [Google Scholar]
- 28. Cantley L. C., Jr., Cantley L. G., Josephson L. (1978) A characterization of vanadate interactions with the Na,K-ATPase. Mechanistic and regulatory implications. J. Biol. Chem. 253, 7361–7368 [PubMed] [Google Scholar]
- 29. Blanco-Arias P., Einholm A. P., Mamsa H., Concheiro C., Gutiérrez-de-Terán H., Romero J., Toustrup-Jensen M. S., Carracedo A., Jen J. C., Vilsen B., Sobrido M. J. (2009) A C-terminal mutation of ATP1A3 underscores the crucial role of sodium affinity in the pathophysiology of rapid-onset dystonia-parkinsonism. Hum. Mol. Genet. 18, 2370–2377 [DOI] [PubMed] [Google Scholar]
- 30. Einholm A. P., Toustrup-Jensen M. S., Holm R., Andersen J. P., Vilsen B. (2010) The rapid-onset dystonia parkinsonism mutation D923N of the Na+,K+-ATPase α3 isoform disrupts Na+ interaction at the third Na+ site. J. Biol. Chem. 285, 26245–26254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jorgensen P. L. (1991) in The Sodium Pump: Structure, Mechanism, and Regulation (Kaplan J. H., De Weer P., eds) pp. 189–200, The Rockefeller University Press, New York [Google Scholar]
- 32. Yoda A., Yoda S. (1987) Two different phosphorylation-dephosphorylation cycles of Na,K-ATPase proteoliposomes accompanying Na+ transport in the absence of K+. J. Biol. Chem. 262, 110–115 [PubMed] [Google Scholar]
- 33. Yoda S., Yoda A. (1987) Phosphorylated intermediates of Na,K-ATPase proteoliposomes controlled by bilayer cholesterol. Interaction with cardiac steroid. J. Biol. Chem. 262, 103–109 [PubMed] [Google Scholar]
- 34. Toyoshima C., Mizutani T. (2004) Crystal structure of the calcium pump with a bound ATP analogue. Nature 430, 529–535 [DOI] [PubMed] [Google Scholar]
- 35. Toyoshima C. (2009) How Ca2+-ATPase pumps ions across the sarcoplasmic reticulum membrane. Biochim. Biophys. Acta 1793, 941–946 [DOI] [PubMed] [Google Scholar]
- 36. Castro M. J., Stam A. H., Lemos C., Barros J., Gouveia R. G., Martins I. P., Koenderink J. B., Vanmolkot K. R., Mendes A. P., Frants R. R., Ferrari M. D., Sequeiros J., Pereira-Monteiro J. M., van den Maagdenberg A. M. (2007) Recurrent ATP1A2 mutations in Portuguese families with familial hemiplegic migraine. J. Hum. Genet. 52, 990–998 [DOI] [PubMed] [Google Scholar]
- 37. Bassi M. T., Bresolin N., Tonelli A., Nazos K., Crippa F., Baschirotto C., Zucca C., Bersano A., Dolcetta D., Boneschi F. M., Barone V., Casari G. (2004) A novel mutation in the ATP1A2 gene causes alternating hemiplegia of childhood. J. Med. Genet. 41, 621–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Swoboda K. J., Kanavakis E., Xaidara A., Johnson J. E., Leppert M. F., Schlesinger-Massart M. B., Ptacek L. J., Silver K., Youroukos S. (2004) Alternating hemiplegia of childhood or familial hemiplegic migraine? A novel ATP1A2 mutation. Ann. Neurol. 55, 884–887 [DOI] [PubMed] [Google Scholar]
- 39. Danko S., Yamasaki K., Daiho T., Suzuki H. (2004) Distinct natures of beryllium fluoride-bound, aluminum fluoride-bound, and magnesium fluoride-bound stable analogues of an ADP-insensitive phosphoenzyme intermediate of sarcoplasmic reticulum Ca2+-ATPase: changes in catalytic and transport sites during phosphoenzyme hydrolysis. J. Biol. Chem. 279, 14991–14998 [DOI] [PubMed] [Google Scholar]
- 40. Cornelius F., Mahmmoud Y. A., Toyoshima C. (2011) Metal fluoride complexes of Na,K-ATPase: characterization of fluoride-stabilized phosphoenzyme analogues and their interaction with cardiotonic steroids. J. Biol. Chem. 286, 29882–29892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Castro M. J., Nunes B., de Vries B., Lemos C., Vanmolkot K. R., van den Heuvel J. J., Temudo T., Barros J., Sequeiros J., Frants R. R., Koenderink J. B., Pereira-Monteiro J. M., van den Maagdenberg A. M. (2008) Two novel functional mutations in the Na+,K+-ATPase α2-subunit ATP1A2 gene in patients with familial hemiplegic migraine and associated neurological phenotypes. Clin. Genet. 73, 37–43 [DOI] [PubMed] [Google Scholar]
- 42. Einholm A. P., Andersen J. P., Vilsen B. (2007) Importance of Leu99 in transmembrane segment M1 of the Na+, K+-ATPase in the binding and occlusion of K+. J. Biol. Chem. 282, 23854–23866 [DOI] [PubMed] [Google Scholar]
- 43. Einholm A. P., Toustrup-Jensen M., Andersen J. P., Vilsen B. (2005) Mutation of Gly-94 in transmembrane segment M1 of Na+,K+-ATPase interferes with Na+ and K+ binding in E2P conformation. Proc. Natl. Acad. Sci. U.S.A. 102, 11254–11259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Patchornik G., Goldshleger R., Karlish S. J. (2000) The complex ATP-Fe2+ serves as a specific affinity cleavage reagent in ATP-Mg2+ sites of Na,K-ATPase: altered ligation of Fe2+ Mg2+ ions accompanies the E1 → E2 conformational change. Proc. Natl. Acad. Sci. U.S.A. 97, 11954–11959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Toustrup-Jensen M., Hauge M., Vilsen B. (2001) Mutational effects on conformational changes of the dephospho- and phospho-forms of the Na+,K+-ATPase. Biochemistry 40, 5521–5532 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.