Background: Trophoblast cells, the functional components of the placenta, are derived from multipotent trophoblast stem (TS) cells.
Results: SATB homeobox proteins regulate the TS cell stem state through up-regulation of a stem-specific transcription factor, EOMES, and inhibition of trophoblast differentiation.
Conclusion: SATB proteins regulate TS cell development.
Significance: Understanding TS cell biology is crucial to determining processes underlying placental development.
Keywords: Development, Differentiation, Placenta, Stem Cells, Trophoblast, CDX2, EOMES, SATB Proteins
Abstract
The morphogenesis of the hemochorial placenta is dependent upon the precise expansion and differentiation of trophoblast stem (TS) cells. SATB homeobox 1 (SATB1) and SATB2 are related proteins that have been implicated as regulators of some stem cell populations. SATB1 is highly expressed in TS cells, which prompted an investigation of SATB1 and the related SATB2 as regulators of TS cells. SATB1 and SATB2 were highly expressed in rat TS cells maintained in the stem state and rapidly declined following induction of differentiation. SATB proteins were also present within the rat placenta during early stages of its morphogenesis and disappeared as gestation advanced. Silencing Satb1 or Satb2 expression decreased TS cell self-renewal and increased differentiation, whereas ectopic expression of SATB proteins promoted TS cell expansion and blunted differentiation. Eomes, a key transcriptional regulator of TS cells, was identified as a target for SATB proteins. SATB knockdown decreased Eomes transcript levels and promoter activity, whereas SATB ectopic expression increased Eomes transcript levels and promoter activity. Electrophoretic mobility shift assay as well as chromatin immunoprecipitation analyses demonstrated that SATB proteins physically associate with a regulatory site within the Eomes promoter. We conclude that SATB proteins promote TS cell renewal and inhibit differentiation. These actions are mediated in part by regulating the expression of the TS cell stem-associated transcription factor, EOMES.
Introduction
The early mammalian embryo is the source of at least three different stem cell populations that can be propagated in vitro (1). They include embryonic stem (ES)5 cells (2–4), extraembryonic endoderm stem cells (5), and trophoblast stem (TS) cells (6, 7). Each has become a powerful model system for elucidating regulatory mechanisms controlling cell fate and differentiation decisions.
TS cells are the antecedents of all trophoblast lineages that comprise the mature placenta (8, 9). Culture conditions have been established that promote TS cell stemness or facilitate differentiation (6, 10). The involvement of several transcription factors as regulators of trophoblast cell lineage determination, maintenance of the stem state, or differentiation has been demonstrated (11–13). Efforts have also been initiated to understand the integration of these gene regulators in TS cells (14). It is evident that there is a higher order orchestration, extending beyond individual transcription factors, responsible for trophoblast development that includes epigenetic regulators (15–18).
Chromatin reorganization plays a fundamental role in regulating gene expression during stem cell renewal and lineage-specific differentiation (19, 20). Among the myriad of proteins that possess instructive actions on chromatin are two structurally related proteins termed SATB homeobox 1 (SATB1) (21) and SATB2 (22, 23). These proteins facilitate assembly of chromatin remodeling proteins and transcription factors, thereby modulating chromatin architecture to facilitate binding of transcription factors to active promoter regions leading to gene activation or repression (22, 23, 24–26). SATB proteins bind to AT-rich elements in matrix attachment regions of actively transcribed genes (21, 23, 27–31). The actions of SATB1 and SATB2 are cell lineage-specific. SATB1 acts as a genome organizer and gene regulator essential for T cell differentiation (24, 26, 28, 29, 32–34), erythroid development (35), and mammary epithelial transformation (36) and also facilitates gene silencing by modulating the expression of Xist in embryonic cells (37). SATB2 is linked to craniofacial patterning and osteoblast differentiation (38), as well as development of cortical neurons (23, 25, 39, 40). SATB1 and SATB2 possess reciprocal actions in regulating ES cell differentiation (41). Thus SATB1 and SATB2 are crucial determinants of cellular differentiation in a number of systems.
A few connections between SATB proteins, especially SATB1, and trophoblast development have been reported. Activation of diapaused mouse embryos is associated with trophectoderm-specific up-regulation of Satb1 expression (42). An increase in Satb1 expression is also detected during reprogramming of mouse ES cells to a TS cell fate (43). Moreover, Satb1 is abundantly expressed in TS cells and dramatically down-regulated in differentiated trophoblast cells (44). These observations, although correlative, are intriguing and along with the known actions of SATB proteins in other cell-types constitute the basis for this report on their functional roles in TS cells.
EXPERIMENTAL PROCEDURES
Animals and Tissue Collection
Holtzman Sprague-Dawley rats were obtained from Harlan Laboratories (Indianapolis, IN) and housed in an environmentally controlled facility with free access to food and water. For timed pregnancies, sperm in the vaginal lavage was defined as gestation day (E) 0.5. Rat placental tissues were collected on E9.5, E11.5, E13.5, E15.5, and E18.5. Placentation site dissections were performed as previously described (45). Tissues for histological analysis were frozen in dry ice-cooled heptane and stored at −80 °C. Tissue samples for RNA and protein extraction were frozen in liquid nitrogen and stored at −80 °C. The University of Kansas Animal Care and Use Committee approved all protocols for the care and use of animals.
Cell Culture
Blastocyst-derived rat TS cells (7) and Rcho-1 TS cells (46) were maintained in the stem state or differentiated by culturing in appropriate culture conditions (7, 47). Rat TS cells were grown in TS cell basal medium (Cellgro RPMI 1640 (Mediatech, Manassas, VA), 20% FBS (Atlanta Biologicals, Norcross, GA), 1 mm sodium pyruvate (Mediatech), 100 μm 2-mercaptoethanol (Sigma), 100 μm penicillin, and 100 units/ml streptomycin (Mediatech)), supplemented with FGF4 (37.5 ng/ml; Sigma), heparin (1.5 μg/ml; Sigma), and rat embryonic fibroblast-conditioned medium (80% of final volume). The culture medium was replaced every day, and rat TS cells were subcultured once they achieved >70% confluence. Differentiation was induced by removal of FGF4, heparin, and rat embryonic fibroblast-conditioned medium. Rcho-1 TS cells were maintained in TS cell basal medium. Differentiation was induced by growing cells to near confluence and then replacing the TS cell basal medium with differentiation medium (NCTC-135 medium supplemented with 1% horse serum (Atlanta Biologicals), 10 mm HEPES, 1 mm sodium pyruvate, and 50 μm 2-mercaptoethanol). High cell density and the absence of mitogenic factors (removal of FBS) facilitate Rcho-1 TS cell differentiation (47).
Mouse TS cells (obtained from Dr. Janet Rossant, Hospital for Sick Children, Toronto, Canada) were maintained in FGF4/heparin supplemented TS culture medium (containing 30% TS basal medium, 70% mouse embryonic fibroblast-conditioned medium, 25 ng/ml FGF4, and 1 μg/ml heparin) as previously described (6). Differentiation of the cells was induced by removal of FGF4, heparin, and mouse embryonic fibroblast-conditioned medium (6).
RT-PCR
Transcript levels were estimated by RT-PCR. Two μg of total RNA and 100 ng of random primers were used for reverse transcription by Superscript II (Invitrogen) in a 20-μl reaction. cDNAs were diluted 1:10 and subjected to conventional PCR or quantitative PCR (qPCR) analysis with specific primers (supplemental Tables S1–S3). Conventional PCR was performed for 25 cycles (denature, 94 °C for 30 s; anneal, 55 °C for 30 s; extension, 72 °C for 45 s), and amplified products were resolved by electrophoresis in 1.5% agarose gels and ethidium bromide staining. qPCR was carried out in 25-μl reactions containing SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). Amplification and fluorescence detection were carried out using an ABI Prism 7500 real time PCR system (Applied Biosystems). Cycling conditions included an initial hold step (95 °C for 10 min) and 40 cycles of a two-step PCR (92 °C for 15 s and then 60 °C for 1 min), followed by a dissociation step (95 °C for 15 s, 60 °C for 15 s, and then a sequential increase to 95 °C). Relative mRNA expression was calculated by the comparative ΔΔCt method, using 18S rRNA as a reference RNA.
Western Blotting
Cellular protein expression was assessed by Western blot analysis. The cell lysates were prepared in radioimmune precipitation assay buffer (150 mm NaCl, 25 mm Tris-HCl pH 7.5, 1 mm EDTA, 0.1% SDS, 0.5% Triton X-100, 0.5% Igepal CA-630, 1% sodium deoxycholate, 1 mm PMSF, and 1% protease inhibitor mixture (Sigma)). Protein concentrations were determined using the Bio-Rad protein assay. Approximately 30 μg of total proteins were separated by SDS-PAGE and transferred to PVDF membranes. SATB proteins were detected using mouse monoclonal antibodies (SATB1 antibody, BD Biosciences, San Jose, CA; 611182; and SATB2 antibody, Abcam, Cambridge, MA; ab51502) at a dilution of 1:2000. CDX2 was detected using a rabbit monoclonal antibody (Epitomics, Burlingame, CA; EPR2764Y) at a dilution of 1:3000, and EOMES was detected using a rabbit polyclonal antibody (Abcam; ab23345) at a dilution of 1:2500. Mouse monoclonal antibodies to GAPDH (Millipore Corporation, Billerica, MA; MAB374, dilution 1:5000) or ACTB (Sigma; A5441, dilution 1:5000) were used as controls to monitor loading. Following incubations with peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies (Sigma) at a dilution of 1:5000 (GAPDH and ACTB: 1:10000), immunoreactive proteins were visualized using Luminata Crescendo Western HRP substrate (Millipore).
Immunofluorescence and Histological Analyses
Localization of cellular proteins in trophoblast cells was accomplished using immunofluorescence staining of histological sections. The analyses were performed on rat placental cryosections (10 μm). Briefly, following fixation in freshly prepared cold 4% formaldehyde for 15 min and blocking in 10% normal goat serum for 30 min at room temperature, incubations were performed overnight at 4 °C with rabbit anti-SATB1 monoclonal (Epitomics; EPR3951) or mouse anti-SATB2 monoclonal (Abcam; ab51502) antibodies (dilution 1:250) in 10% normal goat serum. Washed sections were subsequently incubated with cyanine 3-labeled goat anti-rabbit IgG (Cy3; Jackson ImmunoResearch Laboratories, West Grove, PA; dilution 1:250) or goat anti-mouse IgG tagged with Alexa 568 (Invitrogen; dilution 1:400) secondary antibodies for 30 min at room temperature. Negative controls were performed with normal serum and did not exhibit positive reactivity. Tissue sections were counterstained with DAPI (Molecular Probes, Carlsbad, CA) to visualize nuclei. The images were captured using a Leica DMI 4000 microscope equipped with a Leica CCD camera (Leica Microsystems, Welzlar, Germany).
SATB Knockdown and Ectopic Expression
We established Rcho-1 TS cells as well as mouse TS cells with stable knockdown of Satb1 or Satb2. shRNAs targeted to Satb1 or Satb2 were delivered by transducing the cells with lentivirus carrying shRNA-encoding pLKO.1-puro vectors (Addgene, Cambridge, MA) (48). Selection in puromycin (2.5 μg/ml) was used to obtain stably transduced cells. Knockdown efficiencies were evaluated using RT-qPCR and Western blotting. For each target gene, multiple shRNAs with common target sites in rat and mouse were tested, and the two most effective shRNAs (Satb1 shRNA 9 and 12; Satb2 shRNA 7 and 10) were selected for further analysis. An shRNA to green fluorescent protein (Control shRNA G) and an shRNA with no known mammalian gene targets (Control shRNA S) were used as controls (Addgene; 49). shRNA target sites and sequences are shown in supplemental Table S4.
We also stably transfected Rcho-1 TS cells for ectopic expression of SATB1 or SATB2. Full-length mouse Satb1 and Satb2 cDNAs with HA tag constructs (41) were each subcloned into modified pcDNA3 mammalian expression vectors (Invitrogen). HA-tagged SATB1 and SATB2 proteins were used to distinguish ectopic from endogenous SATB proteins by Western blotting with mouse monoclonal anti-HA antibody (Sigma). Stably transfected Rcho-1 TS cells were selected with 250 μg/ml of geneticin (G418; Invitrogen). After stable transfection and colony screening, we expanded three clones with SATB1 ectopic expression, three with SATB2 ectopic expression, and another three with the empty vector for further experiments. In preliminary experiments, we confirmed that clones for each construct behaved similarly. Results from representative clones are presented.
Assessment of Cell Proliferation
Control, Satb knockdown, and Satb ectopically expressed Rcho-1 TS cells were monitored for cell proliferation by direct cell counting using a hemocytometer. The cells were plated into 24-well plates (10000 cells/well) and cultured under stem conditions for 3 days or differentiating conditions for 6 days. The cell counts were normalized to the number of cells counted on day 1 (24 h after plating) to adjust for variations in plating.
Analysis of Endoreduplication
Following induction of differentiation, Rcho-1 TS cells undergo endoreduplication and form trophoblast giant cells (44). DNA content was estimated by flow cytometry as previously described (7, 44). The cells were detached with trypsin, fixed with cold 70% ethanol, stained with propidium iodide, and analyzed by flow cytometry using BD LSR II (FACSDiva software) or BD FACSCalibur (CellQuest Pro software; BD Biosciences, San Jose, CA). Propidium iodide staining was expressed in a logarithmic scale. Because Rcho-1 TS cells are tetraploid (47), flow cytometry histogram peaks reflected cell populations with 4n, 8n, 16n, and >16n DNA contents. Cells with ≤8n DNA (4n cells in the G0/G1 phase, 4n-8n cells in the S phase, and 8n cells in the G2/M phase) are defined as being in the stem state, whereas cells with >8n DNA correspond to differentiated trophoblasts.
Assessment of TS Cell Differentiation State-associated Gene Expression
To assess the effects of SATB loss-of-function or gain-of-function on Rcho-1 TS cell differentiation state, Eomes, Cdx2, Esrrb, and Id2 (stem associated genes) and Prl3d1, Prl3b1, Gcm1, and Tpbpa (differentiation associated genes) were assessed by RT-qPCR analysis (7, 44). RNA was collected using TRIzol (Invitrogen) according to the manufacturer's instructions. Rcho-1 TS cells expressing control, Satb1, or Satb2 shRNAs were examined in stem conditions, whereas cells transfected with control, HA-SATB1, or HA-SATB2 expression vectors were examined during the course of differentiation. Stem- and differentiation-associated gene expression was also examined in mouse TS cells following SATB protein knockdown.
Promoter-Reporter Analyses
A potential Satb-binding site with characteristic AT-rich DNA sequences (31, 50) was identified in the upstream sequence flanking mouse and rat Eomes genes using rVista 2.0 (51). The SATB-binding site (TCCAATTAATAAATACATAATTGAATTAGTG) 125 bp upstream of the predicted rat Eomes transcription start site was found conserved between mouse and rat. The sequence was also checked for prediction of matrix attachment regions using the MAR-WIZ software. A 2.8-kbp (−2150 to +650) segment of the Eomes regulatory region was PCR-amplified and cloned into the pGL2 luciferase vector between KpnI and XhoI sites. Cloned promoter sequences were confirmed by DNA sequencing. Potential regulatory roles for SATB proteins on Eomes promoter-reporter activity were evaluated in Rcho-1 TS cells, as well as in mouse TS cells. The cells were transiently transfected with 300 ng of the reporter construct, 100 ng of the shRNA (Satb1 or Satb2) or HA-tagged SATB (SATB1 or SATB2) expression vectors, and 50 ng of pCDNA3.1-His-LacZ in each well of a 24-well plate using Lipofectamine 2000 (Invitrogen). The cells were maintained in stem conditions, and cell lysates were collected 72 h after transfection. Luciferase activities were measured with the luciferase assay system (Promega, Madison, WI), and β-galactosidase activities were measured using the Luminescent β-galactosidase detection kit 2 (Clontech).
To determine the specificity of SATB binding, the −125 SATB-binding site in the Eomes promoter construct was mutated (TCCCTTCGTAAATACTGAACTGAATTAGTG) by site-directed mutagenesis. Sequences of the oligonucleotide primers for introducing site-directed mutagenesis are shown in supplemental Table S5. Briefly, mutagenic primers were annealed to the denatured template of Eomes promoter-reporter construct and PCR-amplified using Pfu ultra high fidelity DNA polymerase (Stratagene, La Jolla, CA). Then the template reporter plasmids were digested with DpnI (New England Biolabs, Ipswich, MA), and amplified products were used to transform XL1-Blue competent cells (Stratagene). From 10 positive colonies of the transformed bacteria, plasmids were purified, and introduction of the desired mutation was confirmed by DNA sequencing. To avoid any PCR-introduced error in the luciferase vector backbone, mutated Eomes promoter from a sequenced plasmid was reintroduced into the pGL2 luciferase vector between KpnI and XhoI sites. Subsequently, activity of the Eomes promoter construct with the mutated SATB-binding site was evaluated after knockdown or ectopic expression of SATB proteins in Rcho-1 TS cells, as described above.
Electrophoretic Mobility Shift Assay
EMSA was performed with the SATB-binding oligonucleotide probe and nuclear proteins isolated from Rcho-1 TS cells. The SATB-binding oligonucleotide probe corresponded to the putative SATB-binding element located 125 bp upstream of the Eomes transcription start site (supplemental Fig. S1). Synthesized oligonucleotides (supplemental Table S6) were annealed and labeled at the 3′ end with digoxigenin-ddUTP by terminal transferase using the DIG gel shift kit (Roche Applied Science). Nuclear proteins from Rcho-1 TS cells were extracted as described earlier (21, 52). Briefly, Rcho-1 TS cell pellets were swelled and lysed in hypotonic cell lysis buffer (10 mm HEPES-KOH, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm DTT, 0.625% Nonidet P-40, 0.5 mm PMSF). After centrifugation, the nuclear pellets were lysed with nuclear lysis buffer (20 mm HEPES-KOH, pH 7.9, 400 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm DTT, 1 mm PMSF). Protein concentrations in nuclear extracts were measured by a Bio-Rad protein assay. Rcho-1 TS cell nuclear extract (5 μg) was incubated with 0.4 ng of digoxigenin-labeled oligonucleotides and 1 μg of poly(dI-dC) (Roche Applied Science) in binding buffer (10 mm HEPES-KOH, pH 7.9, 50 mm KCl, 2.5 mm MgCl2, 10% glycerol, 1 mm DTT) for 20 min at room temperature. Following binding, the reactions were resolved on 4% native polyacrylamide gels and transferred to a Hybond N+ nylon membranes (GE Healthcare). Membrane cross-linking was performed at 120 mJ, and digoxigenin-labeled oligonucleotides were reacted with anti-digoxigenin-AP antibody (Roche Applied Science; 1:5000 dilution). Subsequently, signals of the digoxigenin-labeled EMSA bands were detected by using CSPD chemiluminescent substrate following manufacturer's protocol (Roche Applied Science). The specificity of DNA-protein interactions was investigated by conducting the binding reactions in the presence of a molar excess of annealed unlabeled wild or mutant oligonucleotide competitors (supplemental Table S6). To determine whether SATB1 or SATB2 were involved in the protein-nucleotide complexes, either anti-Satb1 (BD Biosciences; 611182) or anti-Satb2 (Abcam; ab34735) antibodies were added to the EMSA probe-nuclear protein incubation mixture. An anti-SRF antibody (Santa Cruz Biotechnology, Santa Cruz, CA; sc-335) was included in the supershift assay as a control.
Chromatin Immunoprecipitation
To examine Satb1 and Satb2 binding to potential sites in the Eomes regulatory region, chromatin fragments from Rcho-1 TS cells stably expressing HA-Satb1 or HA-Satb2 were immunoprecipitated with an anti-HA antibody (Sigma; mouse monoclonal clone HA-7, H9658). ChIP assay procedures were performed as described (22). Briefly, Rcho-1 TS cells were fixed with 0.4% formaldehyde added directly to the culture medium and neutralized with 125 mm glycine. Harvested cells were lysed with nuclear lysis buffer, and nuclear lysates were sonicated with a Bioruptor sonicator (Wolfe Laboratories, York, UK) to prepare chromatin fragments at a size of ∼500 bp. Antibodies (either anti-HA, Sigma, H9658; or anti-acetyl-histone H3K9, Millipore, 06-599) were added to precleared chromatin samples at a concentration of 1 μg/106 cells and incubated overnight with protein G Plus-Agarose (Santa Cruz Biotechnology). Mouse or rabbit IgG (BD Biosciences) was used as a control for nonspecific binding. Immunoprecipitated chromatin was eluted from washed agarose beads, and DNA-protein interactions were reverse cross-linked with 300 mm NaCl at 65 °C for 4 h. After RNase A and proteinase K digestion at 42 °C, DNA fragments were extracted with phenol-chloroform and precipitated with ethanol. qPCR analysis was performed with SYBR Green Master Mix (Applied Biosystems) using Eomes promoter-specific primers (supplemental Table S7 and Fig. S1).
Statistical Analyses
All of the experimental procedures were repeated at least three times. Statistical comparisons between two means were performed using Student's t test. Comparisons of multiple groups were evaluated using analysis of variance. The source of variation from significant F-ratios was determined using Tukey's HSD multiple comparison test. All of the analyses were performed using the SPSS Statistical Package (IBM, Armonk, NY).
RESULTS
Satb1 and Satb2 Are Abundantly Expressed in TS Cell Stem State and Down-regulated during Differentiation
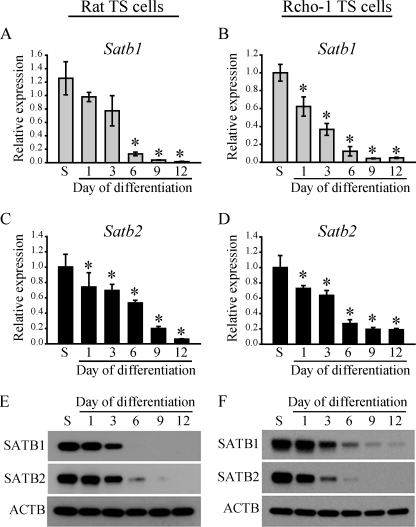
Transcriptome profiling of Rcho-1 TS cells indicated that Satb1 was a transcript with robust expression in the stem state and minimal expression following induction of differentiation (44). RT-qPCR and Western blot analyses were used to verify the differential expression pattern for SATB1 and the related SATB2 in Rcho-1 TS cells and blastocyst-derived rat TS cells in stem and differentiated states. As expected, SATB1 and SATB2 transcripts and proteins were high in the stem state and decreased following differentiation (Fig. 1). Similar to rat TS cell models, mouse TS cells also expressed high levels of SATB proteins in the stem state, which were markedly down-regulated after induction of differentiation (supplemental Fig. S2).
FIGURE 1.
Expression of SATB1 and SATB2 in rat TS cells and Rcho-1 TS cells. Rat TS cells derived from blastocysts (A, C, and E) and Rcho-1 TS cells (B, D, and F) were assessed for expression of SATB1 and SATB2. RT-qPCR (A–D) and Western blotting (E and F) of Satb1 and Satb2 in the stem state (S) and following induction of differentiation. The RT-qPCR data are expressed as the means ± S.D. Comparisons of stem state versus differentiated. *, p < 0.05 (n = 3). ACTB, β-actin.
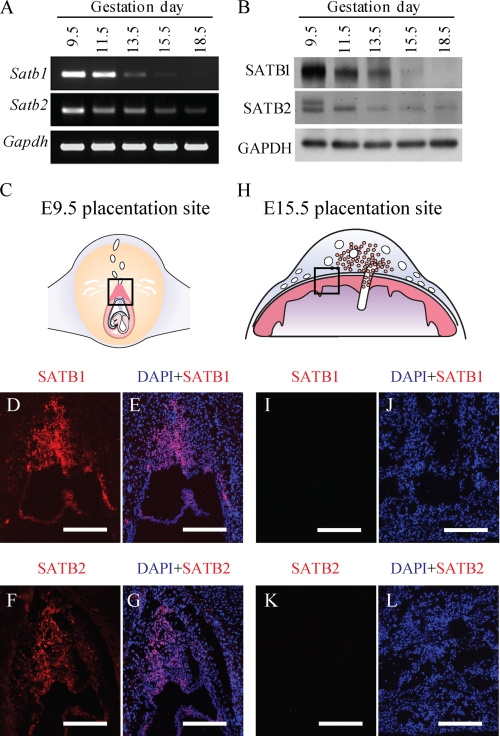
We next examined the expression of SATB1 and SATB2 in trophoblast cells within the placenta. SATB transcripts and proteins were readily detected during early stages of placental development and became undetectable as gestation progressed (Fig. 2). SATB1 and SATB2 proteins were immunolocalized to nuclei of trophoblast cells present in the E9.5 ectoplacental cone (primordial placenta) (Fig. 2, C–G) but not in placentas from later in gestation (Fig. 2, H–L). These observations indicate that SATB1 and SATB2 expression is restricted to the TS cell stem state.
FIGURE 2.
Satb1 and Satb2 are down-regulated in placenta as gestation progresses. RT-PCR (A) and Western blot (B) analyses of SATB1 and SATB2 expression in the rat placenta during various stages of gestation. In addition, immunofluorescence staining for SATB1 and SATB2 in rat placentation sites at E9.5 (C–G) and E15.5 (H–L) was performed. Schematic representations of E9.5 and E15.5 placentation sites are shown in C and H, respectively. Bar, 0.25 mm (n = 3).
Satb1 and Satb2 Promote TS Cell Stem State and Inhibit Differentiation
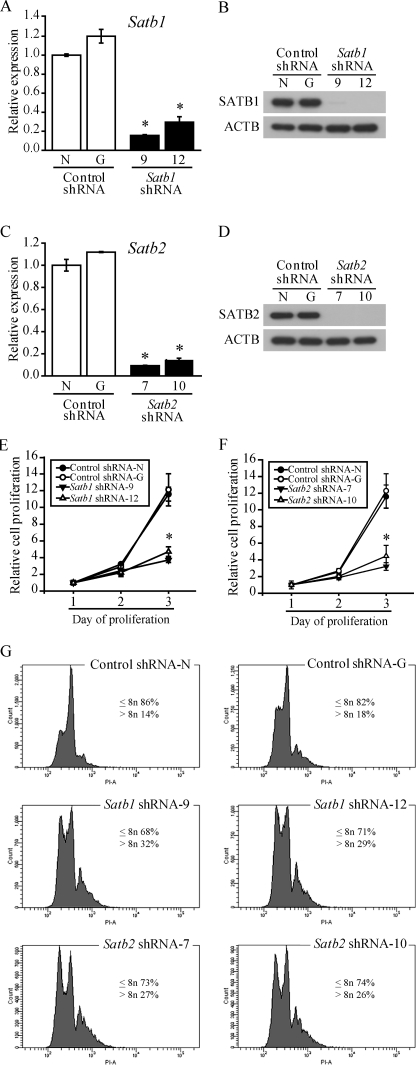
The developmental pattern of SATB1 and SATB2 expression was suggestive of potential regulatory roles for the proteins and prompted us to analyze cellular phenotypes following their knockdown and ectopic expression in TS cells. Rcho-1 TS cells as well as mouse TS cells were stably transduced with lentiviral vectors encoding control, Satb1, or Satb2 shRNAs. Satb1 shRNAs (targeted to regions in exons 9 or 12 of Satb1; Fig. 3, A and B, and supplemental Fig. S3, A and B) and Satb2 shRNAs (targeted to regions in exons 7 or 10 of Satb2; Fig. 3, C and D, and supplemental Fig. S3, C and D) exhibited knockdown efficiencies of ∼80%.
FIGURE 3.
Knockdown of Satb1 or Satb2 in Rcho-1 TS cells decreases proliferation and induces endoreduplication. Rcho-1 TS cells were transduced with pLKO.1-puro lentiviral vectors encoding the control or Satb shRNAs and selected for puromycin resistance. RT-qPCR and Western blot analyses were performed for Satb1 (A and B) and Satb2 expression (C and D) in Rcho-1 TS cells transduced with control shRNA (N and G) or shRNA specifically targeted to Satb1 (9 and 12) or Satb2 (7 and 10). In addition, assessment of Rcho-1 TS cell proliferation was done following knockdown of Satb1 (E) or Satb2 (F). The data are expressed as the means ± S.D. Comparisons of control versus Satb shRNAs samples (n ≥ 3). *, p < 0.05. Because in vitro TS cell differentiation results in endoreduplication, flow cytometric evaluation of ploidy (G) was done in Rcho-1 TS cells transduced with control (shRNA-N and -G), Satb1 (shRNA-9 and -12), or Satb2 (shRNA-7 and -10) shRNAs. Knockdown of SATB1 or SATB2 increased the number of differentiated cells with >8n. ACTB, β-actin.
Under culture conditions that promote proliferation, silencing of either Satb1 or Satb2 in Rcho-1 TS cells slowed cell proliferation (Fig. 3, E and F) and enhanced the number of differentiated cells (trophoblast giant cells) present in the cultures. The latter observation was verified by flow cytometry. Knockdown of either Satb1 or Satb2 increased the number of polyploid cells (>8n; Fig. 3G).
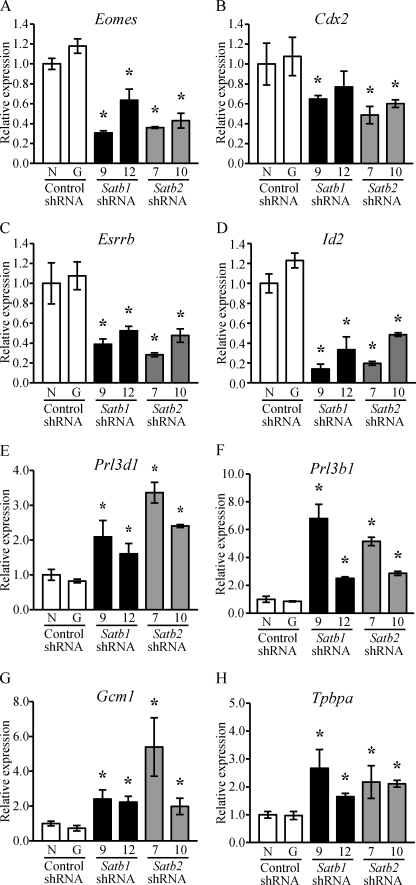
SATB1 or SATB2 knockdown in Rcho-1 TS cells was also associated with a decrease in transcripts indicative of the stem state (Eomes, Cdx2, Esrrb, and Id2) and an increase in transcripts reflective of trophoblast differentiation (Prl3d1, Prl3b1, Gcm1, and Tpbpa) (Fig. 4 and supplemental Fig. S4). Similarly, knockdown of SATB proteins in mouse TS cells also decreased the expression of stem-associated genes and increased the expression of differentiation-associated genes (supplemental Fig. S5).
FIGURE 4.
Knockdown of Satb1 or Satb2 in Rcho-1 TS cells decreases expression of genes associated with the stem state and increases expression of genes associated with differentiation. Rcho-1 TS cells were transduced with pLKO.1-puro lentiviral vectors encoding the shRNAs and selected for puromycin resistance. Subsequently, RT-qPCR analyses were performed for Eomes (A), Cdx2 (B), Esrrb (C), Id2 (D), Prl3d1 (E), Prl3b1 (F), Gcm1 (G), and Tpbpa (H) in Rcho-1 TS cells transduced with control shRNA (N and -G) or shRNA specifically targeted to Satb1 (9 and 12) or Satb2 (7 and 10). RT-qPCR data are expressed as the means ± S.D. Comparisons of control versus Satb shRNAs samples. *, p < 0.05 (n = 3).
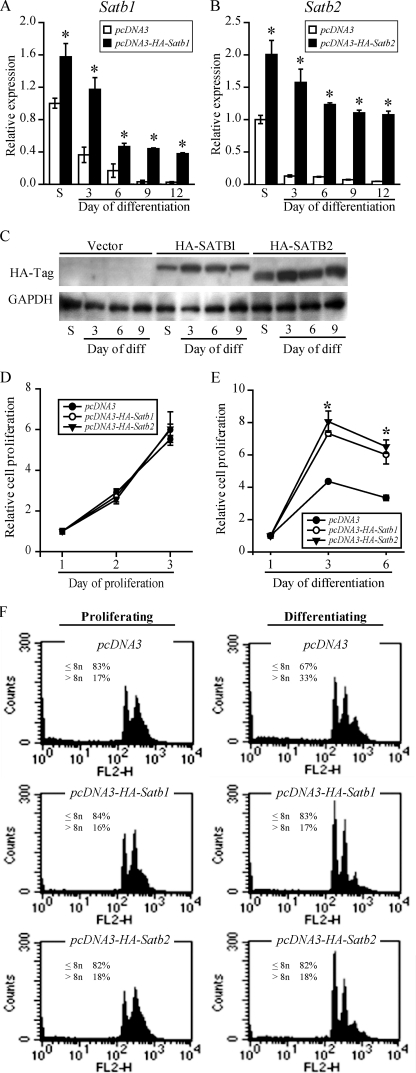
The reciprocal experiments were also performed. Rcho-1 TS cells were stably transfected with vectors expressing HA-tagged SATB1 or SATB2 (Fig. 5). Cells transfected with empty pCDNA3 vectors were used as controls. In proliferating culture conditions, ectopic expression of HA-SATB1 or HA-SATB2 did not significantly affect cell proliferation (Fig. 5D) or endoreduplication (Fig. 5F, left panels). However, in Rcho-1 TS cells induced to differentiate (via mitogen removal), ectopic expression of either HA-SATB1 or HA-SATB2 increased cell numbers (Fig. 5E) and inhibited endoreduplication (reduction in cells with >8n; Fig. 5F, right panels).
FIGURE 5.
Ectopic expression of Satb1 or Satb2 in Rcho-1 TS cells sustains proliferation and inhibits endoreduplication. Rcho-1 TS cells were stably transfected with pcDNA3, pcDNA3-HA-Satb1, or pcDNA3-HA-Satb2 expression vectors and selected for G418 resistance. A–C, following selection and screening, Satb1 (A) and Satb2 (B) RT-qPCR and Western blot analyses (C) were performed on representative clones of transfected Rcho-1 TS cells in stem (S) and differentiating states. An anti-HA tag antibody was used to detect ectopic SATB protein expression. D and E, assessment of cell proliferation in transfected Rcho-1 TS cells cultured in conditions that promote proliferation (D) or differentiation (E). The data are expressed as the means ± S.D. Comparisons of control versus HA-SATB ectopic expression samples. *, p < 0.05 (n = 3). To evaluate endoreduplication, flow cytometric analyses of DNA content (F) were performed in transfected Rcho-1 TS cells cultured in proliferative conditions (left panels) or conditions that promote differentiation (right panels). Ectopic expression of SATB1 or SATB2 did not alter the ploidy profile in proliferative cells (left panels), but it decreased endoreduplication in differentiating cells (right panels) (n = 3).
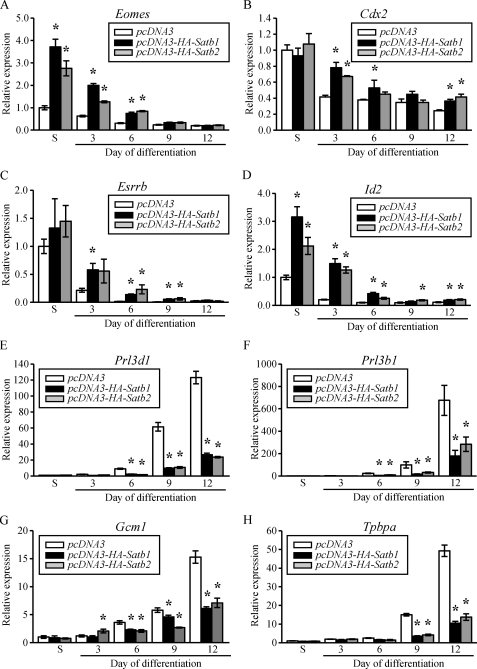
Ectopic expression of HA-SATB1 or HA-SATB2 also significantly increased transcript levels for Eomes, Cdx2, Esrrb, and Id2 and decreased transcript levels for Prl3d1, Prl3b1, Gcm1, and Tpbpa (Fig. 6). The up-regulation of stem-associated genes following ectopic expression of HA-tagged SATB proteins differed. Eomes and Id2 showed a robust up-regulation, which was most prominent in the stem state (Fig. 6, A and D), whereas the Cdx2 and Esrrb increase was not detected in the stem state and was modest following induction of differentiation (Fig. 6, B and C). Inhibition of Prl3d1, Prl3b1, Gcm1, and Tpbpa was dramatic and sustained and not dependent upon an up-regulation of Eomes or other stem-associated genes (Fig. 6, E–H). Collectively, these results indicate that SATB proteins promote stemness and inhibit differentiation of TS cells.
FIGURE 6.
Ectopic expression of Satb1 or Satb2 in Rcho-1 TS cells increases expression of genes associated with the stem state and decreases expression of genes associated with differentiation. Rcho-1 TS cells were stably transfected with pcDNA3, pcDNA3-HA-Satb1, or pcDNA3-HA-Satb2 expression vectors and selected for G418 resistance. Transcript levels were monitored in the stem state (S) and in culture conditions that promote differentiation. RT-qPCR analyses for Eomes (A), Cdx2 (B), Esrrb (C), Id2 (D), Prl3d1 (E), Prl3b1 (F), Gcm1 (G), and Tpbpa (H) showing significant up-regulation of stem-associated genes and down-regulation of differentiation-associated genes. RT-qPCR data are expressed as the means ± S.D. Comparisons of control versus HA-SATB ectopic expression samples. *, p < 0.05 (n = 3).
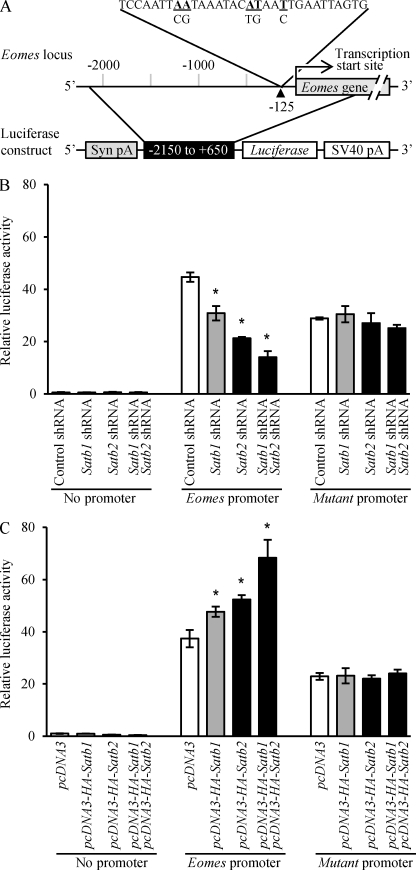
SATB Proteins Regulate Eomes Promoter Activity
Based on the above analyses, Eomes appeared to be a potential target for the regulatory function of SATB proteins. Consequently, the Eomes regulatory region was studied for responsiveness to manipulation of SATB1 or SATB2 expression. A 2.8-kbp Eomes promoter region (−2150 to +650), which included the potential SATB-binding site at −125 (Fig. 7A), was subcloned into a luciferase reporter vector and evaluated in Rcho-1 TS cells and mouse TS cells. Eomes promoter activity was affected by manipulation of SATB protein levels. Knockdown of Satb1 and/or Satb2 inhibited Eomes promoter activity (Fig. 7B and supplemental Fig. S6B), whereas ectopic expression of HA-Satb1 and/or HA-Satb2 significantly increased Eomes promoter activity (Fig. 7C and supplemental Fig. S6C). As expected, mutation of the −125 SATB-binding site in the Eomes promoter construct rendered it insensitive to manipulation of SATB protein levels in Rcho-1 TS cells (Fig. 7). These findings support a role for SATB proteins in the transcriptional regulation of Eomes gene expression.
FIGURE 7.
SATB proteins regulate Eomes promoter activity in Rcho-1 TS cells. A, bioinformatic analysis identified a potential SATB-binding site at 125 bp upstream of the Eomes transcription start site (−125). Accordingly, a 2.8-kbp segment (−2150 to +650) of the Eomes proximal promoter including the putative SATB-binding site was cloned into pGL2 luciferase vector. Subsequently, Rcho-1 TS cells maintained in stem conditions were transiently transfected with a promoterless luciferase construct (No promoter) or the Eomes promoter-luciferase reporter construct (Eomes promoter). The SATB-binding site in the Eomes promoter was mutated (Mutant promoter, as shown in A, underlined bases in bold type were mutated to the bases indicated below) to examine the specificity of regulation by SATB proteins. A β-galactosidase reporter construct was used as an internal control. SATB1 or SATB2 levels were manipulated by transient transfection with control versus Satb shRNAs (Satb1 shRNA-9 or Satb2 shRNA-7) (B) or control versus pcDNA3-HA-Satb expression vectors (C). Relative luciferase activity data are expressed as the means ± S.D. Comparisons of control versus Satb shRNA or HA-SATB ectopic expression samples. *, p < 0.05 (n = 3).
The Eomes Regulatory Locus Is a Direct Target for SATB Proteins
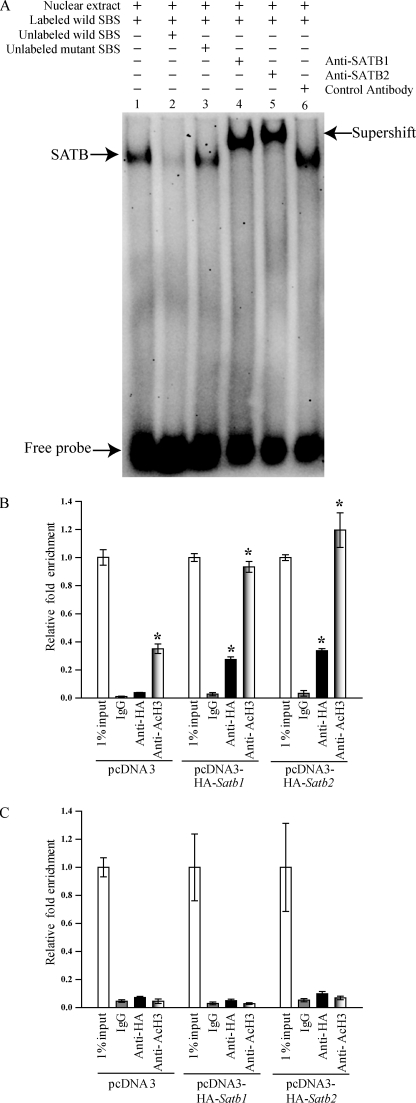
We next evaluated whether SATB proteins physically associate with the putative SATB-binding element at the Eomes locus. Using EMSA, we observed binding of Rcho-1 TS nuclear extracts to an oligonucleotide probe containing the SATB-binding element located 125 bp upstream of the Eomes transcription start site (Fig. 8A, lane 1). Specificity of the interaction was determined by competition with unlabeled wild-type and mutant oligonucleotides (Fig. 8A, lanes 2 and 3). The identity of the proteins binding to the oligonucleotide probe was assessed by SATB antibody-specific supershift analysis. Co-incubation with either anti-Satb1 or anti-Satb2 antibodies slowed the mobility of the protein-oligonucleotide probe complexes, indicating that both SATB1 and SATB2 were capable of binding the SATB-binding element located within the regulatory region of the Eomes promoter (Fig. 8A, lanes 4 and 5).
FIGURE 8.
SATB proteins bind to a region within the Eomes promoter. A, EMSA demonstrating binding of SATB1 and SATB2 to an oligonucleotide containing a predicted SATB-binding element located 125 bp upstream of the Eomes transcription start site (−125). Labeled SATB probes were incubated with Rcho-1 TS cell nuclear lysates as indicated above the lanes. Lane 1, without any competitor oligonucleotide; lane 2, with unlabeled wild-type competitor; lane 3, with unlabeled mutant competitor; lane 4, with SATB1 antibody; lane 5, with SATB2 antibody; lane 6, with a control antibody. B and C, Rcho-1 TS cells stably transfected with pcDNA3, pcDNA3-HA-Satb1, or pcDNA3-HA-Satb2 and maintained in stem culture conditions were used for ChIP assays. Antibodies to the HA tag (Anti-HA) or acetylated histone H3K9 (Anti-AcH3) were used in the analyses. IgG was used as a control for nonspecific immunoprecipitation. qPCR analyses were performed on immunoprecipitates using primers amplifying the segment (−172 to −63) including the SATB-binding element at −125 (B) and a control Eomes upstream region (−5541 to −5428) devoid of any putative SATB-binding element (C). Relative fold enrichment data are expressed as the means ± S.D. Comparisons of IgG control versus anti-HA or anti-AcH3 samples. *, p < 0.05 (n = 3). SBS, SATB binding site.
Given that SATB proteins had the potential to bind to the AT-rich site at −125 within the Eomes promoter, we investigated whether SATB proteins actually occupied the putative SATB-binding element in situ using a ChIP assay. Rcho-1 TS cells stably transfected with HA-SATB1 or HA-SATB2 constructs were used for the ChIP analysis. Occupancy was measured from HA immunoprecipitates at the −125 SATB-binding site using qPCR. Both SATB1 and SATB2 were bound to the region encompassing the −125 SATB-binding site (Fig. 8B). This region was also enriched for histone H3K9 acetylation, indicating a transcriptionally activated chromatin domain. In contrast, no enrichment was detected at an Eomes upstream region that did not contain potential SATB-binding sites (−5541 to −5428; Fig. 8C). The results indicate that SATB proteins directly interact with the Eomes promoter and positively regulate Eomes transcription.
DISCUSSION
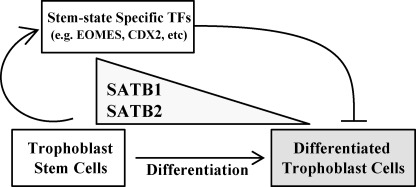
The growth and development of the fetus are dependent upon the in utero environment created by the placenta. Regulatory processes controlling TS cell decisions to expand or to differentiate dictate the functional capacity of the placenta. In this report, we have demonstrated that SATB proteins contribute to the regulation of TS cell stem state. SATB1 and SATB2 act similarly to promote stemness and inhibit TS cell differentiation (Fig. 9).
FIGURE 9.
SATB proteins regulate TS cells by up-regulating stem-associated transcription factors. Schematic presentation showing that high levels of SATB1 and SATB2 expressed in TS cells up-regulate stem-associated transcription factors (TFs), including EOMES and CDX2, which facilitate self-renewal and inhibit induction of trophoblast cell differentiation.
Several proteins have been implicated in the regulation of TS cell developmental states. Some proteins possess TS cell lineage determining actions; others maintain the TS cell stem state, whereas others are required for trophoblast differentiation (11–13, 53). SATB1 and SATB2 do not promote trophoblast lineage commitment (41), nor do they facilitate trophoblast cell differentiation (present study). SATB proteins share the responsibility of maintaining the stem state with several other proteins, including EOMES, CDX2, TCFAP2C, SMARCA4, ESRRB, ETS2, SOX2, TEAD4, and ELF5 (14, 15, 54–65). SATB1 and SATB2 possess a similar TS cell stem state-specific expression pattern with EOMES, CDX2, ESRRB, SOX2, and ELF5 (7). CDX2, EOMES, and ELF5 serve essential roles specifically in the TS cell lineage (14, 15, 54, 55), whereas ESRRB, SOX2, and SATB proteins have broader roles and contribute to regulating the stem state of both ES cells and TS cells (41, 62, 58, 59, 66–68).
A link exists between SATB proteins and Eomes gene regulation. Both proteins maintain the TS cell stem state. SATB1 and SATB2 specifically bind to the Eomes promoter and regulate Eomes transcription. EOMES is a transcription factor targeting several genes essential for maintaining the TS cell stem state, including Tcfap2c, Ets2, Tead4, Elf5, and Eomes genes (14). Our expectation is that SATB proteins coordinate a spectrum of gene targets in addition to Eomes that would be essential for the maintenance of the TS cell stem state. These targets may include stimulatory effects on pro-stem state genes (e.g. Eomes) and inhibitory effects on genes promoting and/or characteristic of differentiation. Whether the gene targets for SATB1 and SATB2 in TS cells are identical is unknown; however, it is expected that at least a set of core gene targets required for TS cell maintenance would be shared.
At this juncture, upstream factors directing stem state-specific expression of Satb1 and Satb2 are unknown. Some transcription factors regulating the TS cell stem state co-regulate one another (14, 15). EOMES regulates TS cells via actions on numerous targets, many of which are shared with TCFAP2C and SMARCA4 (14). Based on global promoter binding analysis, EOMES does not appear to associate with either Satb1 or Satb2 promoters, nor do TCFAP2C, SMARCA4, and ETS2 (14). Elf5 is activated in TS cells as a result of demethylation of CpG dinucleotides in its promoter (15, 69). This mechanism of gene activation does not appear to be used for activation of either Satb1 or Satb2 (69). Although SOX2 has been implicated in the regulation of mouse TS cells (58, 59), SOX2 is not expressed in rat TS cells, eliminating it from consideration as a potential regulator of Satb1 and Satb2 expression (7). CDX2, ELF5, and ESRRB are co-expressed with SATB1 and SATB2 in rat TS cells and represent candidate upstream regulators.
Cellular context is critical in determining the nature of SATB protein actions on stem cell populations. SATB1 and SATB2 have been investigated individually in several developmental systems. SATB1 promotes T cell and erythroid differentiation (24, 26, 28, 29, 32–35), whereas SATB2 stimulates bone differentiation and neuronal specification (23, 25, 38–40). SATB1 and SATB2 activities have been studied together in ES cells, where they have opposing functions (41). SATB2 promotes Nanog expression and ES cell pluripotency, whereas SATB1 antagonizes the actions of SATB2 and stimulates ES cell differentiation (41). In contrast, as described in this report, SATB1 and SATB2 act cooperatively to maintain the TS cell stem state. This overlapping function in regulating TS cells may also help explain the lack of placental phenotypes following disruption of either Satb1 or Satb2 loci (32, 38) and also point to a fundamental difference in the consequences of Satb gene disruptions occurring before or after trophoblast cell lineage commitment. As demonstrated here, sufficient nuclear SATB protein concentrations following trophoblast lineage commitment are critical for maintenance of TS cell stemness. Potential involvement of SATB proteins in other stem populations, including blastocyst-derived extraembryonic endoderm stem cell development has not been reported.
It is apparent that SATB proteins do not have a universal function in regulating stem cell populations. Their activities are cell lineage-dependent and include participation in the maintenance of the stem state or induction of differentiation. SATB proteins are contributors to the regulation of the TS cell ground state. Future efforts will be required to place SATB1 and SATB2 in their appropriate positions within the hierarchy of TS cell regulation.
Supplementary Material
Acknowledgments
We thank Dr. Janet Rossant (Hospital for Sick Children, Toronto, Canada) for providing the mouse TS cells and Dr. Rudolf Grosschedl (Max-Planck Institute of Immunobiology, Freiburg, Germany) for generously providing Satb1 and Satb2 cDNAs used for the construction of expression vectors.
This work was supported, in whole or in part, by National Institutes of Health Grant HD20676.

This article contains supplemental Tables S1–S7 and Figs. S1–S6.
- ES
- embryonic stem
- TS
- trophoblast stem
- En
- gestation day n
- qPCR
- quantitative PCR.
REFERENCES
- 1. Ralston A., Rossant J. (2005) Genetic regulation of stem cell origins in the mouse embryo. Clin. Genet. 68, 106–112 [DOI] [PubMed] [Google Scholar]
- 2. Evans M. J., Kaufman M. H. (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 [DOI] [PubMed] [Google Scholar]
- 3. Martin G. R. (1981) Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U.S.A. 78, 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thomson J. A., Kalishman J., Golos T. G., Durning M., Harris C. P., Becker R. A., Hearn J. P. (1995) Isolation of a primate embryonic stem cell line. Proc. Natl. Acad. Sci. U.S.A. 92, 7844–7848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kunath T., Arnaud D., Uy G. D., Okamoto I., Chureau C., Yamanaka Y., Heard E., Gardner R. L., Avner P., Rossant J. (2005) Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development 132, 1649–1661 [DOI] [PubMed] [Google Scholar]
- 6. Tanaka S., Kunath T., Hadjantonakis A. K., Nagy A., Rossant J. (1998) Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072–2075 [DOI] [PubMed] [Google Scholar]
- 7. Asanoma K., Rumi M. A., Kent L. N., Chakraborty D., Renaud S. J., Wake N., Lee D. S., Kubota K., Soares M. J. (2011) FGF4-dependent stem cells derived from rat blastocysts differentiate along the trophoblast lineage. Dev. Biol. 351, 110–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rielland M., Hue I., Renard J. P., Alice J. (2008) Trophoblast stem cell derivation, cross-species comparison and use of nuclear transfer: new tools to study trophoblast growth and differentiation. Dev. Biol. 322, 1–10 [DOI] [PubMed] [Google Scholar]
- 9. Roberts R. M., Fisher S. J. (2011) Trophoblast stem cells. Biol. Reprod. 84, 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Erlebacher A., Price K. A., Glimcher L. H. (2004) Maintenance of mouse trophoblast stem cell proliferation by TGF-β/activin. Dev. Biol. 275, 158–169 [DOI] [PubMed] [Google Scholar]
- 11. Maltepe E., Bakardjiev A. I., Fisher S. J. (2010) The placenta: transcriptional, epigenetic, and physiological integration during development. J. Clin. Invest. 120, 1016–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sasaki H. (2010) Mechanisms of trophectoderm fate specification in preimplantation mouse development. Dev. Growth Differ. 52, 263–273 [DOI] [PubMed] [Google Scholar]
- 13. Senner C. E., Hemberger M. (2010) Regulation of early trophoblast differentiation: lessons from the mouse. Placenta 31, 944–950 [DOI] [PubMed] [Google Scholar]
- 14. Kidder B. L., Palmer S. (2010) Examination of transcriptional networks reveals an important role for TCFAP2C, SMARCA4, and EOMES in trophoblast stem cell maintenance. Genome Res. 20, 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ng R. K., Dean W., Dawson C., Lucifero D., Madeja Z., Reik W., Hemberger M. (2008) Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat. Cell Biol. 10, 1280–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hemberger M. (2010) Genetic-epigenetic intersection in trophoblast differentiation. Epigenetics 5, 24–29 [DOI] [PubMed] [Google Scholar]
- 17. Rugg-Gunn P. J., Cox B. J., Ralston A., Rossant J. (2010) Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc. Natl. Acad. Sci. U.S.A. 107, 10783–10790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Santos J., Pereira C. F., Di-Gregorio A., Spruce T., Alder O., Rodriguez T., Azuara V., Merkenschlager M., Fisher A. G. (2010) Differences in the epigenetic and reprogramming properties of pluripotent and extra-embryonic stem cells implicate chromatin remodelling as an important early event in the developing mouse embryo. Epigenetics Chromatin 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ho L., Crabtree G. R. (2010) Chromatin remodelling during development. Nature 463, 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lessard J. A., Crabtree G. R. (2010) Chromatin regulatory mechanisms in pluripotency. Annu. Rev. Cell Dev. Biol. 26, 503–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dickinson L. A., Joh T., Kohwi Y., Kohwi-Shigematsu T. (1992) A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell 70, 631–645 [DOI] [PubMed] [Google Scholar]
- 22. Dobreva G., Dambacher J., Grosschedl R. (2003) SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev. 17, 3048–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Britanova O., Akopov S., Lukyanov S., Gruss P., Tarabykin V. (2005) Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur. J. Neurosci. 21, 658–668 [DOI] [PubMed] [Google Scholar]
- 24. Yasui D., Miyano M., Cai S., Varga-Weisz P., Kohwi-Shigematsu T. (2002) SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419, 641–645 [DOI] [PubMed] [Google Scholar]
- 25. Gyorgy A. B., Szemes M., de Juan Romero C., Tarabykin V., Agoston D. V. (2008) SATB2 interacts with chromatin-remodeling molecules in differentiating cortical neurons. Eur. J. Neurosci. 27, 865–873 [DOI] [PubMed] [Google Scholar]
- 26. Notani D., Gottimukkala K. P., Jayani R. S., Limaye A. S., Damle M. V., Mehta S., Purbey P. K., Joseph J., Galande S. (2010) Global regulator SATB1 recruits β-catenin and regulates T(H)2 differentiation in Wnt-dependent manner. PLoS Biol. 8, e1000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dickinson L. A., Dickinson C. D., Kohwi-Shigematsu T. (1997) An atypical homeodomain in SATB1 promotes specific recognition of the key structural element in a matrix attachment region. J. Biol. Chem. 272, 11463–11470 [DOI] [PubMed] [Google Scholar]
- 28. Cai S., Han H. J., Kohwi-Shigematsu T. (2003) Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 34, 42–51 [DOI] [PubMed] [Google Scholar]
- 29. Cai S., Lee C. C., Kohwi-Shigematsu T. (2006) SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 38, 1278–1288 [DOI] [PubMed] [Google Scholar]
- 30. Szemes M., Gyorgy A., Paweletz C., Dobi A., Agoston D. V. (2006) Isolation and characterization of SATB2, a novel AT-rich DNA binding protein expressed in development- and cell-specific manner in the rat brain. Neurochem. Res. 31, 237–246 [DOI] [PubMed] [Google Scholar]
- 31. Yamasaki K., Akiba T., Yamasaki T., Harata K. (2007) Structural basis for recognition of the matrix attachment region of DNA by transcription factor SATB1. Nucleic Acids Res. 35, 5073–5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alvarez J. D., Yasui D. H., Niida H., Joh T., Loh D. Y., Kohwi-Shigematsu T. (2000) The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 14, 521–535 [PMC free article] [PubMed] [Google Scholar]
- 33. Nakayama Y., Mian I. S., Kohwi-Shigematsu T., Ogawa T. (2005) A nuclear targeting determinant for SATB1, a genome organizer in the T cell lineage. Cell Cycle 4, 1099–1106 [PubMed] [Google Scholar]
- 34. Ahlfors H., Limaye A., Elo L. L., Tuomela S., Burute M., Gottimukkala K. V., Notani D., Rasool O., Galande S., Lahesmaa R. (2010) SATB1 dictates expression of multiple genes including IL-5 involved in human T helper cell differentiation. Blood 116, 1443–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wen J., Huang S., Rogers H., Dickinson L. A., Kohwi-Shigematsu T., Noguchi C. T. (2005) SATB1 family protein expressed during early erythroid differentiation modifies globin gene expression. Blood 105, 3330–3339 [DOI] [PubMed] [Google Scholar]
- 36. Han H. J., Russo J., Kohwi Y., Kohwi-Shigematsu T. (2008) SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature 452, 187–193 [DOI] [PubMed] [Google Scholar]
- 37. Agrelo R., Souabni A., Novatchkova M., Haslinger C., Leeb M., Komnenovic V., Kishimoto H., Gresh L., Kohwi-Shigematsu T., Kenner L., Wutz A. (2009) SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev. Cell 16, 507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dobreva G., Chahrour M., Dautzenberg M., Chirivella L., Kanzler B., Fariñas I., Karsenty G., Grosschedl R. (2006) SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell 125, 971–986 [DOI] [PubMed] [Google Scholar]
- 39. Alcamo E. A., Chirivella L., Dautzenberg M., Dobreva G., Fariñas I., Grosschedl R., McConnell S. K. (2008) Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron 57, 364–377 [DOI] [PubMed] [Google Scholar]
- 40. Britanova O., de Juan Romero C., Cheung A., Kwan K. Y., Schwark M., Gyorgy A., Vogel T., Akopov S., Mitkovski M., Agoston D., Sestan N., Molnár Z., Tarabykin V. (2008) Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron 57, 378–392 [DOI] [PubMed] [Google Scholar]
- 41. Savarese F., Dávila A., Nechanitzky R., De La Rosa-Velazquez I., Pereira C. F., Engelke R., Takahashi K., Jenuwein T., Kohwi-Shigematsu T., Fisher A. G., Grosschedl R. (2009) Satb1 and Satb2 regulate embryonic stem cell differentiation and Nanog expression. Genes Dev. 23, 2625–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hamatani T., Daikoku T., Wang H., Matsumoto H., Carter M. G., Ko M. S., Dey S. K. (2004) Global gene expression analysis identifies molecular pathways distinguishing blastocyst dormancy and activation. Proc. Natl. Acad. Sci. U.S.A. 101, 10326–10331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ralston A., Cox B. J., Nishioka N., Sasaki H., Chea E., Rugg-Gunn P., Guo G., Robson P., Draper J. S., Rossant J. (2010) Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137, 395–403 [DOI] [PubMed] [Google Scholar]
- 44. Kent L. N., Konno T., Soares M. J. (2010) Phosphatidylinositol 3 kinase modulation of trophoblast cell differentiation. BMC Dev. Biol. 10, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ain R., Konno T., Canham L. N., Soares M. J. (2006) Phenotypic analysis of the rat placenta. Methods Mol. Med. 121, 295–313 [DOI] [PubMed] [Google Scholar]
- 46. Faria T. N., Soares M. J. (1991) Trophoblast cell differentiation: establishment, characterization, and modulation of a rat trophoblast cell line expressing members of the placental prolactin family. Endocrinology 129, 2895–2906 [DOI] [PubMed] [Google Scholar]
- 47. Sahgal N., Canham L. N., Canham B., Soares M. J. (2006) Rcho-1 trophoblast stem cells: a model system for studying trophoblast cell differentiation. Methods Mol. Med. 121, 159–178 [PubMed] [Google Scholar]
- 48. Stewart S. A., Dykxhoorn D. M., Palliser D., Mizuno H., Yu E. Y., An D. S., Sabatini D. M., Chen I. S., Hahn W. C., Sharp P. A., Weinberg R. A., Novina C. D. (2003) Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 50. Purbey P. K., Singh S., Kumar P. P., Mehta S., Ganesh K. N., Mitra D., Galande S. (2008) PDZ domain-mediated dimerization and homeodomain-directed specificity are required for high-affinity DNA binding by SATB1. Nucleic Acids Res. 36, 2107–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Loots G. G., Ovcharenko I. (2004) rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 32, W217-W221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hoffmann A., Levchenko A., Scott M. L., Baltimore D. (2002) The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science 298, 1241–1245 [DOI] [PubMed] [Google Scholar]
- 53. Simmons D. G., Cross J. C. (2005) Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev. Biol. 284, 12–24 [DOI] [PubMed] [Google Scholar]
- 54. Strumpf D., Mao C. A., Yamanaka Y., Ralston A., Chawengsaksophak K., Beck F., Rossant J. (2005) Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093–2102 [DOI] [PubMed] [Google Scholar]
- 55. Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R., Rossant J. (2005) Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123, 917–929 [DOI] [PubMed] [Google Scholar]
- 56. Nishioka N., Yamamoto S., Kiyonari H., Sato H., Sawada A., Ota M., Nakao K., Sasaki H. (2008) Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech. Dev. 125, 270–283 [DOI] [PubMed] [Google Scholar]
- 57. Nishioka N., Inoue K., Adachi K., Kiyonari H., Ota M., Ralston A., Yabuta N., Hirahara S., Stephenson R. O., Ogonuki N., Makita R., Kurihara H., Morin-Kensicki E. M., Nojima H., Rossant J., Nakao K., Niwa H., Sasaki H. (2009) The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398–410 [DOI] [PubMed] [Google Scholar]
- 58. Avilion A. A., Nicolis S. K., Pevny L. H., Perez L., Vivian N., Lovell-Badge R. (2003) Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Keramari M., Razavi J., Ingman K. A., Patsch C., Edenhofer F., Ward C. M., Kimber S. J. (2010) Sox2 is essential for formation of trophectoderm in the preimplantation embryo. PLoS One 5, e13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Russ A. P., Wattler S., Colledge W. H., Aparicio S. A., Carlton M. B., Pearce J. J., Barton S. C., Surani M. A., Ryan K., Nehls M. C., Wilson V., Evans M. J. (2000) Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 404, 95–99 [DOI] [PubMed] [Google Scholar]
- 61. Wen F., Tynan J. A., Cecena G., Williams R., Múnera J., Mavrothalassitis G., Oshima R. G. (2007) Ets2 is required for trophoblast stem cell self-renewal. Dev. Biol. 312, 284–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tremblay G. B., Kunath T., Bergeron D., Lapointe L., Champigny C., Bader J. A., Rossant J., Giguère V. (2001) Diethylstilbestrol regulates trophoblast stem cell differentiation as a ligand of orphan nuclear receptor ERRβ. Genes Dev. 15, 833–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Georgiades P., Rossant J. (2006) Ets2 is necessary in trophoblast for normal embryonic anteroposterior axis development. Development 133, 1059–1068 [DOI] [PubMed] [Google Scholar]
- 64. Odiatis C., Georgiades P. (2010) New insights for Ets2 function in trophoblast using lentivirus-mediated gene knockdown in trophoblast stem cells. Placenta 31, 630–640 [DOI] [PubMed] [Google Scholar]
- 65. Kuckenberg P., Buhl S., Woynecki T., van Fürden B., Tolkunova E., Seiffe F., Moser M., Tomilin A., Winterhager E., Schorle H. (2010) The transcription factor TCFAP2C/AP-2gamma cooperates with CDX2 to maintain trophectoderm formation. Mol. Cell Biol. 30, 3310–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhou Q., Chipperfield H., Melton D. A., Wong W. H. (2007) A gene regulatory network in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 104, 16438–16443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J., Loh Y. H., Yeo H. C., Yeo Z. X., Narang V., Govindarajan K. R., Leong B., Shahab A., Ruan Y., Bourque G., Sung W. K., Clarke N. D., Wei C. L., Ng H. H. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 [DOI] [PubMed] [Google Scholar]
- 68. Zhang X., Zhang J., Wang T., Esteban M. A., Pei D. (2008) Esrrb activates Oct4 transcription and sustains self-renewal and pluripotency in embryonic stem cells. J. Biol. Chem. 283, 35825–35833 [DOI] [PubMed] [Google Scholar]
- 69. Farthing C. R., Ficz G., Ng R. K., Chan C. F., Andrews S., Dean W., Hemberger M., Reik W. (2008) Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 4, e1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.