Abstract
The role of ArfGAP1 in COPI vesicle biogenesis has been controversial. In work using isolated Golgi membranes, ArfGAP1 was found to promote COPI vesicle formation. In contrast, in studies using large unilamellar vesicles (LUVs) as model membranes, ArfGAP1 functioned as an uncoating factor inhibiting COPI vesicle formation. We set out to discriminate between these models. First, we reexamined the effect of ArfGAP1 on LUVs. We found that ArfGAP1 increased the efficiency of coatomer-induced deformation of LUVs. Second, ArfGAP1 and peptides from cargo facilitated self-assembly of coatomer into spherical structures in the absence of membranes, reminiscent of clathrin self-assembly. Third, in vivo, ArfGAP1 overexpression induced the accumulation of vesicles and allowed normal trafficking of a COPI cargo. Taken together, these data support the model in which ArfGAP1 promotes COPI vesicle formation and membrane traffic and identify a function for ArfGAP1 in the assembly of coatomer into COPI.
Key words: ADP-ribosylation factor, ArfGTPase-activating protein, coatomer, membrane traffic, Golgi apparatus
Introduction
The ArfGAPs are a family of proteins encoded by 31 genes in humans. They have been implicated as regulators of membrane traffic and actin remodeling.1–3 Five ArfGAPs associate with the Golgi apparatus.2,4–7 Three, ArfGAP1, 2 and 3, regulate traffic between the Golgi apparatus and the endoplasmic reticulum (ER). The function of ArfGAP1, 2 and 3 is, at least in part, related to interaction with the substrate protein Arf1•GTP, vesicle coat protein and the cargo proteins that are being transported.8–15
The molecular basis for retrograde traffic between the Golgi cisternae or from the Golgi apparatus to the ER has been extensively studied.2,4–7,16 In these pathways, cargo is transferred from trans-side cisternae to the more cis-side, or from the Golgi apparatus to the endoplasmic reticulum (ER). Cargo proteins include p24 family proteins (p23, p24, p25 and p26) and the KDEL receptor. Two key components to make the transport intermediates are Arf1 and the coatomer vesicle coat protein. Arf1 has two states, Arf1•GTP and Arf1•GDP. Arf1•GTP binds simultaneously to the Golgi membranes and to coatomer, acting as a bridge. The conversion of Arf1•GDP to Arf1•GTP results in the recruitment of coatomer to membranes containing cargo.4,17,18 Coatomer binds to cargo and then polymerizes into COPI, which deforms the membrane into a bud. The coated bud undergoes fission to form a coated vesicle. The coat must be removed for the vesicles to fuse to an acceptor organelle. Uncoating is thought to be triggered by GTP hydrolysis converting Arf1•GTP to Arf1•GDP. In this model, Arf and coatomer are necessary and sufficient for the formation of coated vesicles.
While the model of Arf-dependent COPI vesicle formation was being developed, GTP hydrolysis by Arf was found to be necessary for efficient incorporation of cargo into vesicles.19–23 The finding that interfering with GTP hydrolysis by Arf resulted in production of transport vesicles that lacked cargo was the basis for the suggestion that GTP hydrolysis by Arf1 facilitates cargo sorting into coated vesicles.23–25 The model for Arf-dependent COPI vesicle formation was modified to accommodate the result. ArfGAP1 catalytic activity was thought to be inhibited by cargo.25,26 If cargo were not present, ArfGAP1 would rapidly convert Arf1•GTP to Arf1•GDP, thereby preventing coatomer recruitment and formation of an empty vesicle. With cargo present, Arf1•GTP is stable, allowing recruitment and polymerization of coatomer into COPI with consequent formation of a vesicle. On the other hand, for uncoating, two mechanisms have been envisioned for GTP hydrolysis. In one, ArfGAP1 is selectively recruited to the highly curved vesicle through ArfGAP lipid packing sensor motifs (ALPS).27 In a second proposed mechanism, while ArfGAP1 functions in cargo sorting, ArfGAP2 and ArfGAP3 are the GAPs that mediate uncoating and are recruited to the vesicle by binding to coatomer.28 In these models, ArfGAP1 is thought to be a negative regulator of Arf, either preventing formation of COPI vesicles or accelerating uncoating. Either activity of ArfGAP1 would lead to the prediction that ArfGAP1 reduces accumulation of COPI vesicles.11,12,15,18,29–31
Other studies of ArfGAP1 have not consistently supported the model of ArfGAP1 as a negative regulator of Arf1 in COPI vesicle formation. For instance, reexamination of the kinetics of GTP hydrolysis that included titrations of coatomer, cargo and the substrate Arf1•GTP revealed that both coatomer and cargo stimulate catalytic activity of ArfGAP1.10,13 The previously observed results of the inhibition of GAP activity by cargo could be due to high concentration of cargo peptide. At lower concentrations of cargo peptide, GAP activity is stimulated when coatomer is present. Moreover, the regulation of ArfGAP1 and ArfGAP2 were similar, at least qualitatively. These results are not consistent with ArfGAPs triggering uncoating, given that cargo and coat associate prior to completed formation of a vesicle nor are they consistent with ArfGAP1 and ArfGAP2 functioning at different sites during the process of forming vesicles. Yeast genetic studies identified ArfGAPs, including the yeast ortholog of ArfGAP1, as suppressors of Arf insufficiency, which is difficult to reconcile with a purely negative regulatory role.32,33 Evidence for a positive role in vesicle formation includes the finding that the yeast ArfGAP Glo3 is required for vesicle formation.31 Direct evidence from a mammalian model was the finding that ArfGAP1 induces vesicle formation from isolated Golgi membranes in the presence of coatomer and Arf1•GTP.34–36 Note that this latter result directly contradicts results taken to support the model of ArfGAP1 as a negative regulator of Arf1•GTP, including experiments with isolated Golgi in which different methods were used to detect coated vesicles. Possible explanations have been offered for the discrepancies.17,34,37,38
Here, we describe the results of experiments designed to discriminate between ArfGAP1 functioning (1) to prevent vesicle formation and (2) to promote vesicle formation. Using LUVs containing a peptide from p25, a cargo that binds to ArfGAP1, as a model membrane, we found ArfGAP1 facilitated coatomerand Arf-dependent membrane deformation. Using purified coatomer in solution, we found that ArfGAP1 also facilitated coat polymerization. In in vivo experiments, overexpression of ArfGAP1 generated vesicles that supported retrograde traffic from the Golgi to the ER. Taken together, our results support the idea that ArfGAP1 has a direct role in generating COPI vesicles.
Results
ArfGAPs facilitate coatomer-dependent deformation of large unilamellar vesicles (LUVs).
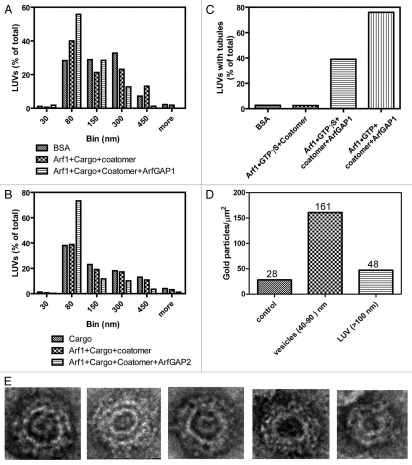
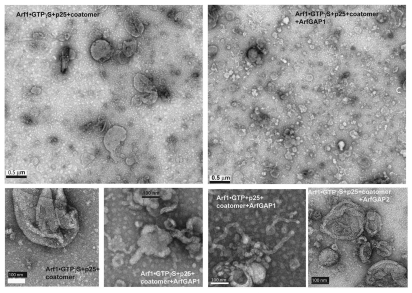
In initial work to discriminate between the model of ArfGAP1 as a negative regulator of COPI vesicle formation and ArfGAP1 as a promoter of COPI vesicle formation, we determined the effect of coatomer, Arf1•GTP⎳S and ArfGAP1 on the size distribution of structures formed from LUVs during incubation, which were examined using negative stained EM. We incorporated a palmitoylated peptide from the cargo p25, which binds to ArfGAP136 and, therefore, could recruit ArfGAP1 to the LUV. First, the size distribution of LUVs incubated with bovine serum albumin was compared with LUVs incubated with myrArf1•GTP⎳S and coat or with myrArf1•GTP⎳S, coatomer and ArfGAP1. The proportion of LUVs between 30 and 80 nm in diameter was greatest in the presence of the three proteins and peptide from cargo (Fig. 1A and 2). The proportion of large LUVs (300–450 nm) was reduced in the presence of the three proteins and cargo pep-tide compared with either BSA alone or myrArf1, coatomer and cargo peptide. The micrographs also revealed that ArfGAP1 with coatomer and myrArf1 induced tubulation of the LUVs that was not observed under other conditions (Fig. 1C and 2; see Fig. S1 for high magnification images of LUVs under control conditions). The effect of ArfGAP2 was also examined. LUVs containing cargo peptide with myrArf1•GTP⎳S+coatomer and myrArf1•GTP⎳S+ArfGAP2+coatomer (Fig. 1B) and myrArf1•GTP⎳S+ArfGAP2 (not shown) were compared. The proportion of 30–80 nm diameter vesicles was greatest and the proportion of 300–450 nm vesicles was least when the three proteins were present together. ArfGAP2 did not affect the proportion without coatomer present. The geometry of many of the 30–80 nm vesicles was consistent with a coated vesicle (Fig. 1E). Immuno-gold labeling indicated the 30–80 nm spherical structures contained the highest concentration of β-COP, a subunit of coatomer (Fig. 1D).
Figure 1.
Effect of ArfGAP1 and ArfGAP2 on LUVs. LUVs were formed by extrusion through membrane with 1.0 µm pore size as described75 and consisted of 40% phosphatidylcholine (PC), 25% phosphatidylethanolamine (PE), 15% phosphatidylserine (PS), 9% phosphatidylinositol (PI), and 10% cholesterol and 1% phosphatidylinositol 4-phosphate (PI4P). LUVs were incubated with the proteins and peptide indicated in the figure at the following concentrations: bovine serum albumin (BSA), 5 µM; myrArf1•GTP⎳S, 0.1 µM; Arf1•GTP, 0.1 µM; ArfGAP1, 0.1 µM; ArfGAP2, 0.1 µM; coatomer, 0.124 µM; palmitoylated p25 peptide, 5 µM. LUVs were imaged by negative stained EM. (A) Effect of ArfGAP1 on LUV size distribution. LUVs were incubated with BSA, p25+Arf1•GTP⎳S+coatomer or p25+Arf1•GTP⎳S+coatomer+ArfGAP1. Diameters of the structures were determined using ImageJ for one of two experiments that were performed. At least 100 LUVs under each condition were examined. (B) Effect of ArfGAP2 on LUV size. The experiment was performed as described in (A) but with the indicated additions. The quantification shown is for one of two experiments. (C) ArfGAP1 induced tubulation of LUVs. The fraction of LUVs containing 2 or more tubules 20 nm in diameter and more than 50 nm in length was determined from the negative stained images. At least 100 LUVs were examined for each condition. (D) Association of coatomer with vesicles. LUVs were immunostained. Coatomer was detected using secondary antibody conjugated to 12 nm gold particles. Gold particles associated with the indicated structures was determined by examination of the negative stained images. The total number of particles associated with each structure is indicated over the bars. (E) Examples of 30–80 nm particles. LUVs were incubated with p25, Arf1•GTP⎳S, Coatomer and ArfGAP2.
Figure 2.
ArfGAP1 and ArfGAP2 facilitate deformation of LUVs containing myrArf1•GTP, coatomer and p25 cargo peptide. LUVs were formed and incubated with the proteins and peptide as described in Figure 1. The combination of ArfGAP, myrArf1•GTP⎳S, palmitoylated p25 and coatomer are indicated on the figure. Concentrations of each are the same as in Figure 1. LUVs were stained with uranyl acetate and examined by transmission electron microscopy (TEM).
Direct effects of ArfGAP1 on coatomer.
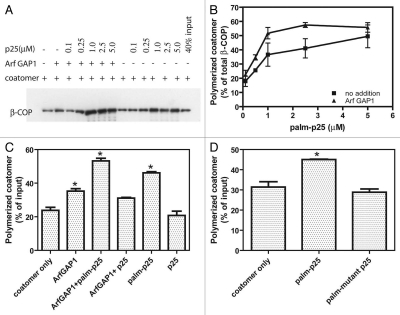
We considered the possibility that ArfGAP1 and 2 may directly influence coatomer polymerization leading to COPI formation. To test this idea, we examined coatomer polymerization in solution. First, we reassessed the effect of p25 peptide. The amount of coatomer sedimented by high speed centrifugation was taken as a measure of polymerization. As previously reported,39 the peptide increased the amount of purified coatomer that sedimented (Fig. 3A–D). In our experiments, we used palmitoylated peptide, which induced polymerization when present at 1–5 µM. The nonpalmitoylated peptide was not effective at this concentration (Fig. 3C). The palmitoylated peptide induced polymerization at concentrations 5–10-fold lower than reported for dimerized peptide.39 Palmitoylated peptide with changes in the amino acids critical to coatomer binding did not induce polymerization (Fig. 3D). ArfGAP1 induced polymerization to a small extent and reduced the amount of palmitoylated peptide required to induce polymerization (Fig. 3A–C).
Figure 3.
Effect of ArfGAP 1 and peptide from cargo cytoplasmic tail on coatomer polymerization. (A) Coatomer polymerization assay. Palmitoylated p25 cargo peptide was titrated into a reaction mixture containing purified coatomer and ArfGAP1, where indicated. Polymerized coatomer was sedimented by centrifugation. An example immunoblot is shown. (B) Quantification of sedimented coatomer. The sedimented proteins were fractionated using SDS-PAGE and detected using immunoblotting as in (A). The immunoblot data were quantified by scanning films for three experiments. Average ± SEM is presented. (C) Comparison of the effect of palmitoylated and unmodified peptide. Coatomer polymerization was determined as described in A in the presence of ArfGAP1 and either 1 µM palmitoylated p25 cargo peptide or 1 µM of the unmodified peptide. Data were analyzed by one way ANOVA followed by the Bonferoni multiple comparisons test for differences from coatomer only. * indicates p < 0.05. (D) Effect of mutant p25 on coatomer polymerization. The experiment was performed as described in (A) using palmitoylated p25 peptide or palmitoylated p25 with the phenylalanines changed to alanine and the two lysines changed to serine. Statistical analysis was performed as described in (C).
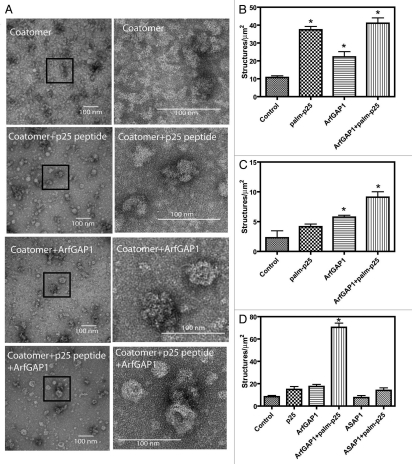
Coatomer incubated with palmitoylated p25 peptide and ArfGAP1 without LUVs was stained with uranyl acetate and examined by TEM (Fig. 4A). Without peptide or ArfGAP1 present, rare spherical structures indicative of self assembly of COPI were present with an average diameter of 25 ± 5 nm (average of 18 particles). Spherical structures approximately 29 ± 7 nm (average of 44 particles) in diameter were observed when coatomer was treated with the peptide alone. When ArfGAP1 was included in the incubations, 31 ± 7 nm (average of 77 particles) nm spherical structures were apparent in the presence of peptide and 34 ± 8 nm (average of 37 particles) in the absence of peptide. Although there were quantitative differences between experiments, consistently more structures were present with both ArfGAP1 and peptide present than with either alone (Fig. 4B–D are the quantification from 3 independent experiments). Another ArfGAP ASAP1, which does not function at the Golgi, had no effect (Fig. 4D). These results are consistent with the idea that ArfGAP1 and 2 can facilitate coatomer polymerization directly.
Figure 4.
Effects of p25 peptide and ArfGAP1 on structure of coatomer. Coatomer incubated with the indicated proteins and peptides as described in Figure 3 was visualized by staining with uranyl acetate and examination by TEM as described in “Materials and Methods.” (A) shows representative images of coatomer incubated under the indicated conditions. The images in the right column are magnifications of the indicated areas in the images in the left column. (B–D) are from three experiments in which the number of spherical structures approximately 30 nm in diameter was determined. In (D), the effect of 0.5 µM [325–724]ASAP 1, an irrelevant ArfGAP, is determined. The average for 5 to 10 random fields ± sem is presented. Data were analyzed by one way ANOVA followed by the Bonferoni multiple comparisons test for differences from control. * indicates p < 0.05.
Effect of ArfGAP1 overexpression on the structure of intracellular organelles.
We used electron microscopy to examine the effect of ArfGAP1 overexpression on endomembranes of cells. We focused our in vivo studies on ArfGAP1, the most extensively studied among ArfGAP1, 2 and 3. The proposed function of ArfGAP is the prevention of COPI vesicle formation.
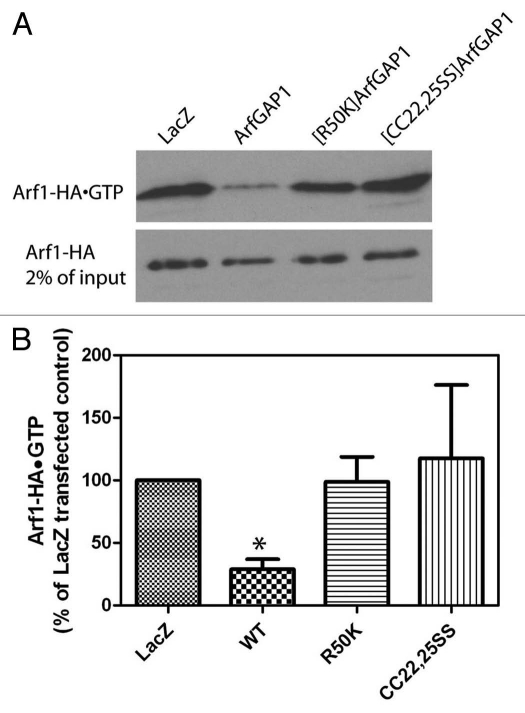
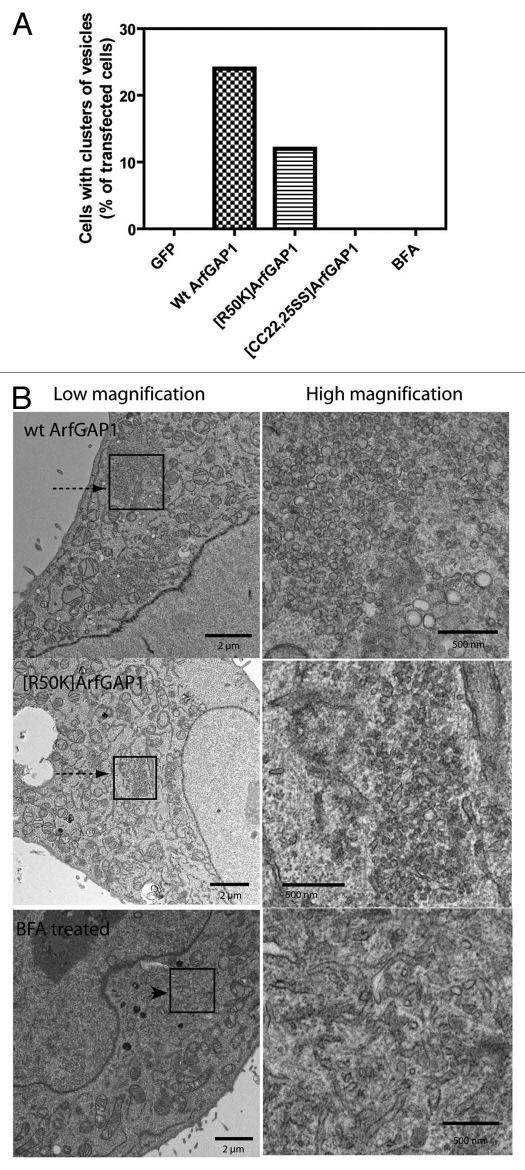
In these experiments we expressed two mutants in addition to wild-type protein. [R50K]ArfGAP1 lacks GAP activity because the catalytic arginine is changed to a lysine. [CC22,25SS] ArfGAP1 lacks GAP activity because binding of the zinc atom essential for activity is disrupted. To confirm the lack of activity in vivo, cells were cotransfected with plasmids directing expression of each of the ArfGAPs together with a plasmid for the expression of Arf1-HA. Cells were lysed and Arf1-HA•GTP levels were determined by a pull-down assay (Fig. 5).40 As anticipated, expression of wild-type ArfGAP1 reduced cellular Arf1-HA•GTP levels whereas expression of the mutants had little or no effect on Arf1-HA•GTP levels as compared with cells expressing LacZ (Fig. 5A and B). Overexpression of ArfGAP1 is known to result in a loss of typical Golgi (see refs. 41 and 67 and Fig. 8). In cells expressing wild-type ArfGAP1 in which Golgi were found, cisternae were dilated as compared with the GFP transfected controls (Fig. S2). We found that in 24% of cells expressing wild-type ArfGAP1, Golgi were difficult to detect but there were patches of 40–70 nm vesicles (Fig. 6A and C). The accumulations of vesicles were distinct from the small tubular vesicular clusters observed in cells treated with Brefeldin A (BFA) (Fig. 6), an inhibitor of Arf guanine nucleotide exchange factors that reduce cellular levels of Arf1•GTP. BFA treated cells did not have patches containing more than 50 vesicles in 4 mm2 square area (Fig. 6A). Less frequently than with wild-type ArfGAP1, vesicle accumulation was also observed with expression of [R50K] ArfGAP1 (12% of cells, Fig. 6A). In contrast, accumulations of vesicles were not observed in cells expressing [CC22,25SS] ArfGAP1, although the Golgi cisternae were dilated (Fig. 6 and S2). Based on these results, we conclude that overexpression of ArfGAP1 facilitates vesicle formation, which is consistent with the results that ArfGAP1 facilitates coatomer polymerization in vitro. Furthermore, given that [R50K]ArfGAP1 expression did not induce accumulation of vesicles to the same extent as did wild-type ArfGAP1, GAP catalytic activity appears to contribute to efficient vesicle formation.
Figure 5.
Arf1•GTP levels in cells overexpressing ArfGAP1 and mutant ArfGAP1. Cells were cotransfected with plasmids directing expression of Arf1-HA and the indicated ArfGAP1-myc. A part of the lysates was taken to determine Arf1-HA expression levels and the remaining lysates were used to determine Arf1-HA•GTP levels.40 (A) Representative experiment. (B) Quantification of 3 experiments. Films were digitized and the signal intensities determined using ImageJ. Normalized signals from 3 experiments were averaged. Data were analyzed by one way ANOVA followed by the Bonferoni multiple comparisons test for differences from control. * indicates p < 0.05.
Figure 8.
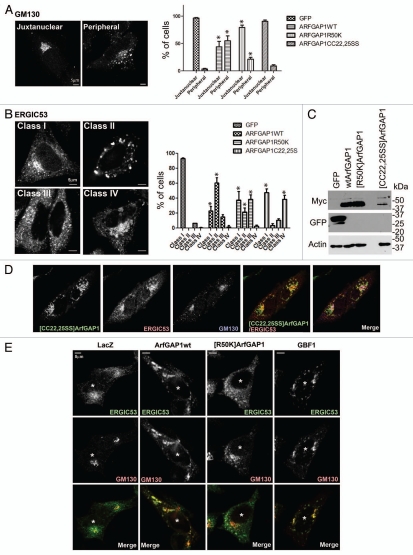
Effect of overexpressed wild-type and mutant ArfGAP1 on organellar morphology. (A) Golgi. Golgi morphology was analyzed using GM130 antibody in HeLa cells. Golgi morphology was classified into 2 classes depicted in the left panel, and more than 50 randomly chosen transfected cells were classified. The data are presented in the right panel, which is the summary of 3 independent experiments. Data were analyzed by two way ANOVA followed by the Bonferoni multiple comparisons test. * indicates p < 0.05 compared with GFP controls. (B) ERGIC. ERGIC morphology, analyzed using ERGIC53 antibody in HeLa cells, was grouped into 4 classes, depicted in the left panel, and quantified as in (A). The summary of the data is presented in the right panel. Statistical analysis was performed as descript in (A). (C) Expression level of ArfGAP1 and mutants. Protein expression was examined by immunoblotting lysates of HeLa cells. Wild-type and mutant ArfGAP1 were detected by anti-myc antibody. The non-specific band is indicated by an asterisk. (D) Relative localization of Golgi and ERGIC proteins in cells expressing [CC22,25SS]ArfGAP1. Cells expressing [CC22,25SS]ArfGAP1 were stained with antibodies against myc (for [CC22,25SS]ArfGAP1), GM130 and ERGIC53. (E) Effect of GBF1 on the Golgi and ERGIC. ArfGAP1 expression affected cells similarly to cells overexpressing GBF1. HeLa cells were transfected as indicated, and the cells were triple stained with anti-myc (not shown), anti-GM130 (green), and anti-ERGIC53 (red). The transfected cells were indicated by an asterisk.
Figure 6.
Effect of ArfGAP1 and mutants on cell ultrastructure. HeLa cells were cotransfected with plasmids directing the expression of the indicated ArfGAP1 and with a plasmid for the expression of Lac Z. The cells were prepared for TEM as described in “Materials and Methods.” Random sections were examined and cells containing crystallized produced of the LacZ reaction, indicating transfection with LacZ, were analyzed. The % of cells with patches of vesicles was determined and is presented in (A). At least 24 cells were examined under each condition. Representative images of vesicle accumulation are shown in (B). Patches of vesicles are indicated by arrows. Tubular-vesicular accumulations are indicated by an arrowhead. High magnification images of the boxed areas are shown in the panels to the right.
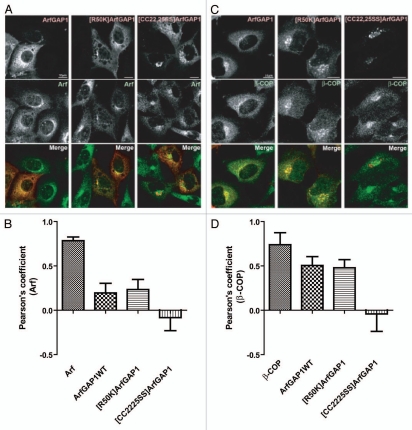
ArfGAP1 overexpression has been found to cause disruption of the Golgi apparatus.41,67 This effect has been attributed to the reduction of Arf1•GTP levels and considered similar to the effect of lowering Arf1•GTP levels by other mechanisms such as expression of dominant negative Arf ([T31N]Arf1) or treatment with Brefeldin A (BFA), which inhibits nucleotide exchange factors. However, we found by examination of thin sections by EM that cells overexpressing ArfGAP1 and treated with BFA had different phenotypes, with ArfGAP1, but not BFA, causing the accumulation of numerous vesicles. These results support the idea that ArfGAP1 and BFA disrupt the Golgi by different mechanisms and are not consistent with the idea that ArfGAP1 inhibits vesicle formation. These considerations motivated us to reexamine the effect of ArfGAP1 on the morphology of the Golgi by immunofluorescence. We started by examining the colocalization of wild-type and mutant ArfGAP1, expressed at low levels, with Arf and β-COP, a subunit of coatomer. Wild-type ArfGAP1 and [R50K]ArfGAP1 colocalized with Arf and β-COP near the nucleus (Fig. 7). However, cells expressing ArfGAP1 at higher levels had more diffuse Arf and β-COP staining and less juxtanuclear staining than control cells (Fig. 7A and B and data not shown). [CC22,25SS]ArfGAP1 was also juxtanuclear, but did not colocalize with Arf or β-COP (Fig. 7).†
Figure 7.
Localization of ArfGAP1 and ArfGAP1 mutants when expressed at low levels. (A) Cellular distribution of exogenous ArfGAP1 with Arf. NIH 3T3 cells were transfected with plasmids directing expression of myc-tagged ArfGAP1 and mutants, as indicated, and stained with anti-Arf (red) and anti-myc (green). (B) Quantitative assessment of colocalization with Arf. Pearson's coefficients were determined for signals from Arf and ArfGAP1 and mutant ArfGAP1. The positive control was Arf1 stained with the same primary but secondary antibodies with different fluors. (C) Localization of exogenous ArfGAP1 relative to coatomer. Cells were treated as in A, then stained with anti-β-COP (green) and anti-myc (red) antibody. (D) Quantitative assessment of colocalization with β-COP. The experiment was performed as described in (C). The positive control used one primary for β-COP and two different secondary antibodies.
To analyze the effect of ArfGAP1 on Golgi morphology, we examined the cis-Golgi marker GM130 in cells overexpressing ArfGAP1 and mutants (Fig. 8A). Relative expression levels were determined by immunoblotting as shown in Figure 8C. We found the Golgi morphology was disrupted in cells overexpressing wild-type ArfGAP1. The “Juxtanuclear” phenotype is a normal distribution of GM130 in the juxtanuclear region. In the “Peripheral” phenotype, GM130 is redistributed into peripheral puncta. Overexpression of wild-type ArfGAP1 caused redistribution of GM130 with a “Peripheral” phenotype in 55% of transfected cells (Fig. 8A), consistent with the previous report of Golgi disruption detected using the Golgi marker, giantin.41 [R50K]ArfGAP1 also caused redistribution of GM130, although it affected a smaller fraction, 21% of cells. [CC22,25SS]ArfGAP1 induced redistribution of GM130 in 9% of cells, and most of the Golgi appeared in the perinuclear region. Giantin localization was also affected by overexpression of wild-type ArfGAP1 and [R50K]ArfGAP1, but not by [CC22,25SS]ArfGAP1 (data not shown). These results are consistent with the previous results that wild-type ArfGAP1 affected giantin, but [C22A]ArfGAP1 did not.41
Next, the morphology of the ER-Golgi intermediate compartment (ERGIC), using ERGIC53 as a marker, was examined (Fig. 8B). Four phenotypes of cells overexpressing ArfGAP1 proteins were identified: class I was no effect, with ERGIC53 concentrated in the juxtanuclear region as well as peripheral dots; in class II, ERGIC53 was in large structures distributed throughout the cell; in class III, ERGIC53 was less concentrated in the juxtanuclear region than class I, but the large structures found in class II were not present, and; in class IV, ERGIC53 accumulated in an enlarged, round structure in the juxtanuclear region. The round structure was bigger than the juxtanuclear dots of class I, but was not distributed in the periphery as in class II. Cells expressing wild-type ArfGAP1 primarily had ERGIC53 in a class II distribution (61% of cells), and fewer cells with a class III phenotype (15%). [R50K]ArfGAP1, deficient in GAP activity, induced similar changes as wild-type ArfGAP1, but less efficiently. [R50K] ArfGAP1 expression resulted in a class II phenotype in 21% of cells and a class III in 38% of cells, with class III appearing to be a weaker phenotype. [CC22,25SS]ArfGAP1 expression resulted in a class IV phenotype in 38% of cells. In the class IV cells, [CC22,25SS]ArfGAP1 colocalized with ERGIC53 but not GM130 (Fig. 8D). In summary, ArfGAP1 causes a redistribution of Golgi and ERGIC proteins, which was at least in part dependent on GAP activity.
Given that (1) ArfGAP1 facilitates COPI assembly in vitro and (2) ArfGAP1 induced vesicle accumulation (detected by EM), we considered that the Golgi fragmentation in wild-type ArfGAP1 expression may be the result of ArfGAP1 promoting excessive vesicle formation. If the fragmentation of the Golgi were due to ArfGAP1 promoting COPI vesicles formation, we expected a similar phenotype when Arf1 was activated to excess at the Golgi apparatus. We tested this prediction by examining the Golgi and ERGIC in cells overexpressing GBF1, a Golgi associated Arf exchange factor.42–45 The overexpression of GBF1 was similar to the effect of wild-type ArfGAP1. At lower expression levels, GBF1 was juxtanuclear, associated with the Golgi apparatus. At higher concentrations, GBF1 induced fragmentation of the Golgi, detected using GM130 as a marker. The ERGIC, detected with an antibody to ERGIC53, was enlarged and distributed in peripheral puncta, partly colocalizing with GM130 as was observed in cells overexpressing ArfGAP1 (Fig. 8E). These results are consistent with the idea that the Golgi fragmentation induced by ArfGAP1 results from acceleration of transport vesicle formation.
Retrograde traffic of a model cargo is intact in cells overexpressing wild-type ArfGAP1.
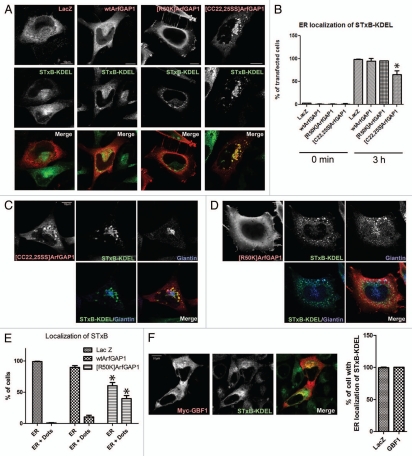
We used a functional assay of retrograde traffic as an additional means of discriminating between the two models of ArfGAP1 function (Fig. 9). If ArfGAP1 functions to prevent vesicle formation, then its overexpression should inhibit transport of a COPI cargo. If ArfGAP1 promoted vesicle formation, overexpression should either not affect or accelerate transport of the cargo. The effect of ArfGAP1 on retrograde traffic was determined using fluorescently tagged Shiga toxin subunit B fused to KDEL (STxB-KDEL). Cells expressing ArfGAP1 were incubated with STxB-KDEL for 3 min at 37°C. Unbound toxin was washed from the cells and the cells were shifted to 37°C for 3 h. After internalization, STxB-KDEL was transported to the Golgi, then to the ER in a COPI dependent manner.46,47 Localization after 3 h was determined by confocal microscopy. In control LacZ-expressing cells and cells expressing wild-type ArfGAP1 or [R50K]ArfGAP1, STxB-KDEL reached the nuclear membrane and a reticular compartment (Fig. 9A and B). In cells expressing [CC22,25SS]ArfGAP1, STxB-KDEL was trapped in the perinuclear region in globular structures. STxB-KDEL colocalized partially with giantin in addition to a structure connected to the giantin-containing structure (Fig. 9A–C). We examined the endosomal markers, EEA1 and transferrin receptor, in cells overexpressing [CC22,25SS]ArfGAP1. We found no colocalization of STxB-KDEL with EEA1 and limited colocalization with transferrin receptor (data not shown). The results indicate most of STxB-KDEL reached the Golgi. STxB-KDEL localization is consistent with association with the dilated structures observed by EM. STxB-KDEL colocalized with [CC22,25SS] ArfGAP1 in this structure. In cells expressing [R50K]ArfGAP1, we noticed that STxB-KDEL was found in punctate structures distributed around the nucleus as well as on the nuclear membrane (Fig. 9D). The punctate structures, which contained either giantin (Fig. 9D) or ERGIC53 (data not shown), were more evident in cells overexpressing [R50K]ArfGAP1 than in wild-type ArfGAP1 (Fig. 9E), suggesting some STxB-KDEL was retained in residual Golgi or ERGIC elements. Overexpression of GBF1 caused similar fragmentation of the ERGIC and Golgi as did wild-type ArfGAP1, which we took as support of the idea that both GBF1 and ArfGAP1 accelerate transport vesicle formation. Based on this interpretation, retrograde transport should occur in the cell overexpressing GBF1 despite a fragmented Golgi apparatus. As predicted, we found that STxB-KDEL transport is intact in GBF1 overexpressing cells as well as wild-type ArfGAP1 overexpressing cells (Fig. 9F). We conclude that COPI traffic is unimpeded in cells overexpressing wild-type ArfGAP1.
Figure 9.
Effect of recombinant ArfGAP1 on COPI dependent retrograde transport. COPI dependent transport was measured using STxB-KDEL conjugated with Alexa 488. (A) Relative distribution of STxB-KDEL and ArfGAP1-myc. STxB-KDEL conjugated with Alexa 488 fluorescence was internalized for 3hr in HeLa cells transfected with indicated constructs. After fixation, cells were stained with anti-myc antibody (red). Representative images are shown. (B) Quantitation of STxB-KDEL targeting to the ER. The percentage of transfected cells with a reticular distribution consistent with an ER localization of STxB-KDEL (referred to as ER localization) was determined. More than 50 cells were counted, and the summary of 3 independent experiments is shown. Data for the 3 h time point were analyzed by one way ANOVA followed by the Bonferoni multiple comparisons test. * indicates p < 0.05 compared with LacZ. (C) Distribution of STxB-KDEL in cells expressing [CC22,25SS]ArfGAP1. After internalization of STxB-KDEL (green) for 3 h, cells expressing [CC22,25SS]ArfGAP1 were stained for the Golgi marker giantin (blue) and [CC22,25SS]ArfGAP1 (red). STxB-KDEL localized at the perinuclear region, partially colocalizing with giantin but also was present in a structure connected to the Golgi. [CC22,25SS] ArfGAP1 colocalized with STxB-KDEL in the latter structure. (D) Distribution of STxB-KDEL in cells expressing [R50K]ArfGAP1. Cells expressing [R50K]ArfGAP1 were treated as described in C. The punctuate structure was also stained by giantin. (E) Quantification of relative reticular and punctate distributions illustrated in (D). The cells with STxB-KDEL localization that was reticular (presumed to be ER) or ER and puncta (dots) were shown as the percentage of transfected cells. Summary of 3 independent experiments is shown. Data were analyzed by two way ANOVA followed by the Bonferoni multiple comparisons test. * indicates p < 0.05 compared with Lac Z control. (F) Relative distribution of STxB-KDEL and GBF1-myc, and quantification of STxB-KDEL localization. STxB-KDEL was internalized as above, and ER localization of STxB-KDEL is shown as the percentage of transfected cells.
Discussion
ArfGAP1 is recognized to be involved in membrane traffic mediated by COPI vesicles but the molecular basis for its function remains controversial. Two opposing models are currently debated.4,17,18,34,37,38,48 In one model, ArfGAP1 acts as a negative regulator of the GTP-binding protein Arf, preventing formation of vesicles and/or triggering coat dissociation from vesicles. In a second model, ArfGAP1 has a positive role for the formation of vesicles. We set out to discriminate between these two models. In our results, ArfGAP1 facilitated (1) deformation of model membranes (large unilamellar vesicles containing a peptide from the cytoplasmic tail of the cargo protein p25) induced by Arf1•GTP and coatomer and (2) polymerization of coatomer. In vivo, ArfGAP1 induced the accumulation of vesicles, visualized by electron microscopy, and induced the redistribution of the Golgi apparatus. The redistribution was similar to that observed with the Arf exchange factor GBF1. Despite the redistribution of the Golgi apparatus, the model cargo Shiga toxin subunit B fused to KDEL was efficiently transported through the Golgi, indicating active formation of functional COPI transport intermediates. Taken together, these data support the model in which ArfGAP1 has a positive role in generating COPI transport intermediates and identify a specific function for ArfGAP1 in facilitating assembly of coatomer into COPI.
Role of ArfGAP in coat formation and membrane deformation in vitro.
The assembly of COPI is an important property of coat proteins.49 Self-assembly was first described for clathrin. Cages could be assembled by altering pH and salt from physiological conditions. Adaptor proteins were found to facilitate assembly under physiologic conditions. Assembly did not require association with a membrane.49,50 COPII has also been found to assemble independent of membrane binding.51–53 The function of ArfGAP1 to facilitate assembly of COPI independent of membranes is analogous to that of clathrin adaptors AP-1 and AP-2 and the catalytic function is analogous to that of Sec23 in COPII, which is thought to be analogous to clathrin adaptors.54–57 Thus, our results are consistent with previous work identifying ArfGAP1 as a coat component.35,36 Furthermore, the in vitro assembly will provide a basis for additional studies of the molecular determinants and regulation of COPI vesicle formation.
While our results contrast with previously reported results18,28,29,34,35,39,58 in which it was reported that Arf1 and coatomer were sufficient for vesicle formation, they do not necessarily contradict these studies. The population of LUVs generated by the extrusion method, which is a frequently used method that we used in this work, contains many vesicles with diameters between 30 and 150 nm. If there were an increase due to Arf+coatomer, it may have been missed due to background signal. Indeed, in some experiments we did observe a small increase in vesicles in the size range of 30–80 nm due to coatomer and Arf. Other factors may account for additional differences. The composition of the liposomes used could account for the differences. In some work, dioleoyl lipids were used, with the most efficient effects observed with dioleoyl-phosphatidic acid.58 The high content of unsaturated lipid increases the elasticity of the bilayer59,60 and, consequently, the membranes are easier to deform. Phosphatidic acid, used in as least two reports58,61 induces membrane curvature,62–64 which would favor vesicle budding. In other work, vesicles contained a peptide from the cargo protein p23, which does not bind ArfGAP1,34,36 10% phosphatidylinositol 4,5-bisphosphate, which does not occur in the Golgi apparatus, and 30% phosphatidylserine, for the EM studies.65 In contrast, we used a peptide from the cargo protein p25, which does bind to ArfGAP1,36 linked to palmitic acid to anchor it to vesicles. The vesicles contained 1% phosphatidylinositol 4-phosphate, a phospholipid normally found in the Golgi apparatus. The vesicles also contained 15% phosphatidylserine, which is higher than the lipid composition of the total Golgi,58 but may more accurately reflect the content of the cytoplasmic leaflet. Regardless of the quantitative differences in vesicle formation due to Arf and coatomer between studies, the important conclusion of our work is that the ArfGAPs facilitate the processs.
Role of ArfGAP on vesicle formation in vivo.
Our results support the idea that ArfGAP1 facilitates vesicle formation. The accumulation of vesicles seen by EM could be explained as either accelerated vesicle formation or slowed vesicle consumption. The accumulation of vesicles seen by EM could be explained as either accelerated vesicle formation or slowed vesicle consumption. Also, by itself, the in vivo result of intact STxB-KDEL transport to the ER with wild-type ArfGAP1 overexpression is not evidence that vesicle formation was promoted (although it is evidence against the idea that ArfGAP1 prevents vesicle formation). However, the in vivo results of (1) intact STxB-KDEL transport with wild-type ArfGAP1 overexpression, (2) impeded transport with expression of mutant ArfGAP1 and (3) vesicle accumulation with ArfGAP1 overexpression, taken together with the in vitro results are evidence for the proposal that vesicle formation is supported by ArfGAP1. Acceleration of vesicle for mation from the Golgi can also explain the Golgi dispersion in cells overexpressing wild-type ArfGAP1 that has been reported by us and others.41,66,67 Previously, the Golgi dispersion has been explained as inhibition of vesicle formation. However, given our -results that (1) ArfGAP1 overexpression leads to the accumulation of vesicles and (2) the phenotype of ArfGAP1 overexpression is similar to GBF1 overexpression, Golgi disruption in cells over-expressing ArfGAP1 may be better explained by acceleration of transport; the contents of newly formed Golgi in the periphery are rapidly recycled to the ER, giving insufficient time for the Golgi to assume its usual juxtanuclear localization.
Our results are also consistent with a role for GTP hydrolysis in vesicle formation. We found that although [R50K]ArfGAP1 expression results in a phenotype similar to wild-type ArfGAP1, there were quantitative and qualitative differences between the effects of wild-type and [R50K]ArfGAP1. [R50K]ArfGAP1 was less effective than wild-type ArfGAP1 for morphological changes (21% Golgi fragmentation in [R50K]ArfGAP1 vs. 55% in wild-type ArfGAP1) and vesicle accumulation. In addition, in cells expressing [R50K]ArfGAP1, retrograde cargo accumulated in dispersed Golgi more frequently (40%) than in cells expressing wild-type ArfGAP1 (10%). These results could indicate that retrograde cargo had a longer Golgi residence time in the cells expressing [R50K]ArfGAP1 than in cells overexpressing wild-type ArfGAP1, consistent with the idea that the role of GTP hydrolysis is related to cargo sorting.19,20,22,23 In vitro, we found that both Arf1•GTP⎳S and Arf1•GTP can induce membrane deformation with cargo, coatomer, and ArfGAP1 in LUVs. However, LUVs were more extensively tubulated and deformed with Arf1•GTP than Arf1•GTP⎳S (Figs. 1C and 2). It is possible that Arf1•GTP⎳S could slow coatomer polymerization, implying a mechanism in which release of Arf1•GTP is required to ensure that cargo associates with coatomer prior to coatomer polymerization. This interpretation is consistent with the results reported by Hsu and colleagues35,36,68 in which they found that the [R50K]ArfGAP1 did not efficiently generate COPI vesicles.
An intact Zn-binding finger in the ArfGAP domain, together with GAP activity, may contribute to transport. The mutation introduced into [CC22,25SS]ArfGAP1 disrupts the ability of the protein to bind to Zn, raising the concern that the effects could result from denaturation of the ArfGAP, in which case the results with [CC22,25SS]ArfGAP1 would be difficult to interpret. However, several results support the idea that the effects of [CC22,25SS]ArfGAP1 are specific and not nonspecific effects of protein aggregation. The expression level of [CC22,25SS] ArfGAP1 was very low, but it colocalized with ERGIC53 and affected the morphology of ERGIC53 in a way not observed with control LacZ or wild-type or [R50K]ArfGAP1. Furthermore, it partly colocalized with ERGIC53, with giantin and, in a separate compartment, with STxB-KDEL indicating association with membrane-delimited structures and not non-specific aggregates of these proteins, in which multiple proteins might colocalize. In addition, expression of [CC22,25SS]ArfGAP1 caused specific dilation of Golgi cisternae. We also cannot exclude the possibility that the defect in GAP activity is more complete in the [CC22,25SS]ArfGAP1 than in [R50K]ArfGAP1, leading to a more extreme phenotype, but we consider this unlikely given the large effect of mutating the catalytic arginine.
In summary, ArfGAP1, which binds both coatomer and cargo proteins, promotes transport vesicle formation. Future studies will determine if the interaction of ArfGAP1 with Arf1 and cargo affects vesicle formation and whether other ArfGAPs, such as ArfGAP2 and 3 and AGAPs, that bind to coat proteins and cargo12,15,69–71 function by a mechanism analogous to ArfGAP1.
Experimental Procedures
Reagents.
Plasmids for expression of C-terminally Myc and His-tagged rat Arf GAP1 were kindly provided by Victor Hsu (Boston, MA). A plasmid for expressing HA-tagged Arf1 was a gift from Kazuhisa Nakayama (Kyoto, Japan). Constructs encoding [T31N]Arf1-HA and [Q71L]Arf1-HA were a gift from Julie Donaldson (NIH). Myc-tagged LacZ control plasmid was purchased from Invitrogen. [R50K] Arf GAP1 and [CC22,25SS] Arf GAP1 were generated using the Quick-change site-directed mutagenesis kit (Stratagene). C-terminally Myc and His-tagged human GBF1 have been described.44 The following antibodies were used; mouse monoclonal antibody against Myc epitope (9E10, Santa Cruz), rabbit polyclonal antibody against Myc epitope (Santa Cruz), goat polyclonal antibody against GM130 (Santa Cruz), mouse monoclonal antibody against ERGIC53 (G1/93, Alexis Biochemicals), mouse monoclonal antibody against GM130 (BD), rabbit polyclonal antibody against ERGIC53 (Sigma). Polyclonal anti-giantin antibodies were purchased from Covance and rat monoclonal anti-HA antibodies were from Roche Applied Science. Alexa Fluor dye conjugated secondary antibodies (Invitrogen). Egg phosphatidylcholine (PC), porcine brain phosphatidylserine (PS), bovine liver phosphatidylethanolamine (PE), bovine liver phosphatidylinositol (PI), porcine brain phosphatidylinositol 4-phosphate (PIP), and cholesterol were from Avanti Polar Lipids. The purification of GST-GGA3 VHS-GAT was as described.40 The plasmid for bacterial expression of KDEL conjugated Shiga Toxin B-subunit (STxB-KDEL) was kindly provided from Ludger Johannes (Paris, France).
Protein purification.
Preparation of proteins used for this work have been described.10,13,47,72–74 Plasmids for the expression of Arf1, and N-myristoyltransferase and [1–415]ArfGAP1-His6 and ArfGAP2-His6 in bacteria have been described previously.10,13 p25 cytoplasmic tail peptide (WRMRHLKSFFEAKKLV) and the mutant WRMRHLKSAAEASSLV, covalently linked to palmitate at the N-terminus where indicated, were synthesized as described.13 Myristoylated Arf1 (myrArf1) was expressed in and purified from bacteria as described.10,72,73 [1–415]ArfGAP1-His6 was expressed and purified from insect cell as described,10 [1–521]ArfGAP2-His6 was expressed in E. coli and purified from inclusion bodies.13 Coatomer was purified from rat liver as described.74
STxB-KDEL was purified and labeled as described.47 Briefly, the protein expression was induced by heat shock at 42°C for 4 h. Bacteria were washed 3 times with 10 mM TRIS-HCl, pH 8.0 and washed once in sucrose buffer (25% sucrose, 1 mM EDTA, 10 mM TRIS-HCl, pH8.0). Bacterial pellets were resuspended in H2O with protease inhibitors for 10 min at 4°C. The protein extract was collected and the pH adjusted with TRIS-HCl to 8.0. The protein was loaded on a HiTrap Q HP column (GE Healthcare) and eluted with a linear gradient of 50–600 mM NaCl in 20 mM TRIS-HCl, pH 8.0. The protein elution was confirmed by monoclonal antibody against STX-1 (Santa Cruz). The STxB-KDEL was conjugated with Alexa 488 fluorescence using Alexa Fluor 488 labeling kit (Invitrogen). The functionality of the fluorescently labeled STxB-KDEL was confirmed by determining whether the protein associated with the Golgi after internalization for 45 min in HeLa cells.
Coatomer polymerization.
Fifteen nanomolar coatomer, 0.5 mM ArfGAP1 and palmitoylated p25 cargo peptide at concentrations from 0.1 mM to 5.0 mM were incubated at 30°C for 10 min, and centrifuged at 100,000xg for 15 min. The pellet was resuspend in SDS-PAGE sample buffer and precipitated coatomer was quantified by immunoblot with an anti-βCOP rabbit polyclonal antibody (Thermo Scientific).
Determining Arf1-HA•GTP levels in cells.
Two × 106 HeLa cells were double transfected with Arf1-HA and other constructs as indicated in the figure legends using Lipofectamine™ 2000 (Invitrogen). After 20 h, Arf1-HA•GTP levels were determined as described.40
Immunofluorescence.
HeLa and NIH3T3 cells were maintained in DMEM (Invitrogen) supplemented with 10% FBS and 200 U of penicillin-streptomycin (Invitrogen). Cells grown on coverslips were transfected with FuGene6 (Roche) for HeLa cells. Lipofectamine™ 2000 was used for NIH3T3 cells. After 18–22 h, cells were washed with PBS and fixed with 4% paraformaldehyde (PFA) for 15 min and quenched with 50 mM NH4Cl. Cells were incubated with blocking solution A (0.04% Saponin, 0.2% BSA, in PBS) for 30 min. Primary and secondary antibodies were diluted with blocking solution A and incubated with cells for 30 min each. Slides were prepared using mounting solution (DakoCytomation). For polyclonal ERGIC53 staining, cells were fixed with methanol at −20°C for 5 min, washed with PBS, and incubated with blocking solution B (10% FBS in PBS). Primary and secondary antibodies were diluted in PBS and cells were processed as for PFA fixed cells. For retrograde transport, Alexa 488 conjugated STxB-KDEL was internalized at 37°C for 3 min, washed twice with DMEM, incubated further at 37°C for 3 h then fixed with 4% PFA. Zeiss 510 meta confocal microscope was used to capture the images. To score the organelle morphology and localization of STxB-KDEL, fields were randomly chosen and all transfected cells in the field were classified. More than 50 transfected cells were counted. The experiment was repeated 3 times.
Immunofluorescence in experiments using [Q71L]Arf1-HA and [T31N]Arf1-HA have been described.44 Cells 16 h post-transfection were washed in phosphate-buffered saline (PBS), fixed in 3% paraformaldehyde for 10 min and quenched with 10 mM ammonium chloride. Cells were permeabilized with 0.1% Triton-X-100 in PBS. The coverslips were then washed with PBS and blocked in PBS, 2.5% goat serum, 0.2% Tween-20 for 5 min followed by blocking in PBS, 0.4% fish skin gelatin and 0.2% Tween-20. Cells were incubated with primary antibody for 1 h at room temperature. Coverslips were washed with PBS, 0.2% Tween-20 and incubated with secondary antibodies for 45 min. Coverslips were washed as described above and mounted on slides in 9:1 glycerol/PBS with 0.1% p-phenylenediamine (Sigma-Aldrich). Fluorescence patterns were visualized with a Leitz Orthoplan microscope with epifluorescence and Hoffman Modulation Contrast optics from Chroma Technology. Optical sections were captured with a CCD high-resolution camera from Roper Scientific equipped with a camera/computer interface. Images were analyzed with a power Mac using IPLab Spectrum software (Scanalytics).
Electron microscopy (EM).
Imaging of LUVs by negative staining was performed as previously described.75 LUVs were prepared as described10,13,75 and incubated at a total phospholipid concentration of 1 mM with the indicated combinations of 0.1 µM myrArf1•GTP⎳S or myrArf1•GTP, 0.1 µM ArfGAP1 or ArfGAP2, 124 nM coatomer and 5 µM palmitoylated p25 peptide in a total volume of 25 µl for 10 min at 30°C.
For coat polymerized in vitro without LUVs, 100 µl of 0.1 µM coatomer was incubated with 0.5 µM ArfGAP1 and 2.5 µM palmitoylated-p25 cargo cytoplasmic tail peptide in a buffer containing 20 mM HEPES, pH 7.4, 100 mM NaCl, 1 mM EDTA and 2 mM MgCl2 at 30°C for 10 min and precipitated by centrifugation at 100,000xg for 15 min as described.39 The precipitate was suspended in 20 µl of 20 mM HEPES, pH 7.4, 100 mM NaCl, 1 mM EDTA and 2 mM MgCl2. Nine microliters of the suspension was applied to carbon coated grids and the grids were stained with 2% uranyl acetate and examined in a TEM (H7600, Hitachi) and images recorded with an AMT CCD camera (Advanced Microscopy Techniques).
For analysis of the ultrastructure of cells overexpressing ArfGAP1 proteins, cultured cells on 6-well plates were processed, embedded in situ, and thin-sections prepared for EM analysis, as previously described.76 Briefly, the cells were fixed overlaying 2% glutaraldehyde77,78 in sodium cacodylate buffer (0.1 M, pH 7.0) for 1 h, replaced with same buffer, and post-fixed in 1% osmium tetroxide (Electron Microscopy Sciences). The cells were en bloc stained in 0.5% uranyl acetate in acetate buffer (0.1 M pH 4.5) for 1 h. The cells were rinsed in acetate buffer and dehydrated in a grade of ethanol (e.g., 35%, 50%, 75%, 95% and 100%). After the final changes in 100% ethanol, cells were washed three changes in 100% epoxy resin (EMbed 812, Electron Microscopy Sciences) infiltrated overnight at room temperature, embedded in a new epoxy resin, and cured in 55°C oven for 48 h. The cured resin was removed from the plate and thin-sections (80 to 90 nm) were made in parallel direction to the growth of cells. The thin-sections were stained in uranyl acetate and lead citrate and examined in the EM (H7600, Hitachi). Images were captured with a CCD camera (AMT). Transfected cells were identified by transfecting cells simultaneously with plasmids for expression of LacZ and wild-type or mutant ArfGAP1. More than 80% of transfected cells were double transfected. After X-gal treatment, LacZ transfected cells develop crystals in cells, and which are easily observed by EM.
Acknowledgments
We thank Richard Kahn, Victor Hsu and Julie Donaldson for helpful discussions. The work was supported by NIH Intramural Program (PAR and JEH) and the National Science Foundation, award NSF 0744471 (ES).
Footnotes
Disruption of the Zn-finger by mutating cysteine to serine could lead to denatured and potentially aggregated protein; however, as described in the discussion, the colocalization with organelle markers in addition to other results in the paper support the idea that [CC22,25SS]ArfGAP1 had specific effects.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Disclaimer
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Supplementary Material
References
- 1.Kahn RA, Bruford E, Inoue H, Logsdon JM, Nie Z, Premont RT, et al. Consensus nomenclature for the human ArfGAP domain containing proteins. J Cell Biol. 2008;182:1039–1044. doi: 10.1083/jcb.200806041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gillingham AK, Munro S. The small G proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- 3.Inoue H, Randazzo PA. Arf GAPs and their interacting proteins. Traffic. 2007;8:1465–1475. doi: 10.1111/j.1600-0854.2007.00624.x. [DOI] [PubMed] [Google Scholar]
- 4.Nie Z, Randazzo PA. Arf GAPs and membrane traffic. J Cell Sci. 2006;119:1203–1211. doi: 10.1242/jcs.02924. [DOI] [PubMed] [Google Scholar]
- 5.Rothman JE. The machinery and principles of vesicle transport in the cell. Nat Med. 2002;8:1059–1062. doi: 10.1038/nm770. [DOI] [PubMed] [Google Scholar]
- 6.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/S0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 7.D'Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 8.Styers ML, O'Connor AK, Grabski R, Cormet-Boyaka E, Sztul E. Depletion of beta-COP reveals a role for COP-I in compartmentalization of secretory compartments and in biosynthetic transport of caveolin-1. Am J Physiol Cell Physiol. 2008;294:C1485–C1498. doi: 10.1152/ajpcell.00010.2008. [DOI] [PubMed] [Google Scholar]
- 9.Saitoh A, Shin HW, Yamada A, Waguri S, Nakayama K. Three homologous ArfGAPs participate in coat protein I-mediated transport. J Biol Chem. 2009;284:13948–13957. doi: 10.1074/jbc. M900749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo R, Randazzo PA. Kinetic analysis of Arf GAP1 indicates a regulatory role for coatomer. J Biol Chem. 2008;283:21965–21977. doi: 10.1074/jbc.M802268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frigerio G, Grimsey N, Dale M, Majoul I, Duden R. Two human ARFGAPs associated with COP-I-coated vesicles. Traffic. 2007;8:1644–1655. doi: 10.1111/j.1600-0854.2007.00631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weimer C, Beck R, Eckert P, Reckmann I, Moelleken J, Brugger B, et al. Differential roles of ArfGAP1, ArfGAP2, and ArfGAP3 in COPI trafficking. J Cell Biol. 2008;183:725–735. doi: 10.1083/jcb.200806140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo R, Ha VL, Hayashi R, Randazzo PA. Arf GAP2 is positively regulated by coatomer and cargo. Cell Signal. 2009;21:1169–1179. doi: 10.1016/j.cellsig.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schindler C, Rodriguez F, Poon PP, Singer RA, Johnston GC, Spang A. The GAP Domain and the SNARE, Coatomer and Cargo Interaction Region of the ArfGAP2/3 Glo3 are Sufficient for Glo3 Function. Traffic. 2009;10:1362–1375. doi: 10.1111/j.1600-0854.2009.00952.x. [DOI] [PubMed] [Google Scholar]
- 15.Kartberg F, Asp L, Dejgaard SY, Smedh M, Fernandez-Rodriguez J, Nilsson T, et al. ArfgGAP2 and ArfGAP3 are essential for COPI coat assembly on the Golgi membrane of living cells. J Biol Chem. 2010;285:36709–36720. doi: 10.1074/jbc. M110.180380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rabouille C, Klumperman J. The maturing role of COPI vesicles in intra-Golgi transport. Nat Rev Mol Cell Biol. 2005;6:812–817. doi: 10.1038/nrm1735. [DOI] [PubMed] [Google Scholar]
- 17.Beck R, Brugger B, Wieland F. GAPs in the context of COPI. Cell Logist. 2011;1:52–54. doi: 10.4161/cl.1.2.15174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beck R, Ravet M, Wieland FT, Cassel D. The COPI system: Molecular mechanisms and function. FEBS Lett. 2009;583:2701–2709. doi: 10.1016/j.febslet.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 19.Pepperkok R, Whitney JA, Gomez M, Kreis TE. COPI vesicles accumulating in the presence of a GTP restricted arf1 mutant are depleted of anterograde and retrograde cargo. J Cell Sci. 2000;113:135–144. doi: 10.1242/jcs.113.1.135. [DOI] [PubMed] [Google Scholar]
- 20.Lanoix J, Ouwendijk J, Lin CC, Stark A, Love HD, Ostermann J, et al. GTP hydrolysis by arf-1 mediates sorting and concentration of Golgi resident enzymes into functional COPI vesicles. EMBO J. 1999;18:4935–4948. doi: 10.1093/emboj/18.18.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanoix J, Ouwendijk J, Stark A, Lin CC, Ostermann J, Nilsson T. GTP gamma S inhibits selective incorporation of resident Golgi enzymes into functional COP-1-dependent vesicular carriers. Mol Biol Cell. 1998;9:578–590. [Google Scholar]
- 22.Nickel W, Malsam J, Gorgas K, Ravazzola M, Jenne N, Helms JB, et al. Uptake by COPI-coated vesicles of both anterograde and retrograde cargo is inhibited by GTP gamma S in vitro. J Cell Sci. 1998;111:3081–3090. doi: 10.1242/jcs.111.20.3081. [DOI] [PubMed] [Google Scholar]
- 23.Lanoix J, Ouwendijk J, Stark A, Szafer S, Cassel D, Dejgaard K, et al. Sorting of Golgi resident proteins into different subpopulations of COPI vesicles: a role for ArfGAP1. J Cell Biol. 2001;155:1199–1212. doi: 10.1083/jcb.200108017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss M, Nilsson T. Sorting proteins into COPI vesicles: What kinetic proofreading can do for you. Mol Biol Cell. 2002;13:1499–1500. [Google Scholar]
- 25.Weiss M, Nilsson T. A kinetic proof-reading mechanism for protein sorting. Traffic. 2003;4:65–73. doi: 10.1034/j.1600-0854.2003.40202.x. [DOI] [PubMed] [Google Scholar]
- 26.Goldberg J. Decoding of sorting signals by coatomer through a GTPase switch in the COPI coat complex. Cell. 2000;100:671–679. doi: 10.1016/S0092-8674(00)80703-5. [DOI] [PubMed] [Google Scholar]
- 27.Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic alphα-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 28.Beck R, Adolf F, Weimer C, Bruegger B, Wieland FT. ArfGAP1 Activity and COPI Vesicle Biogenesis. Traffic. 2009;10:307–315. doi: 10.1111/j.1600-0854.2008.00865.x. [DOI] [PubMed] [Google Scholar]
- 29.Reinhard C, Schweikert M, Wieland FT, Nickel W. Functional reconstitution of COPI coat assembly and disassembly using chemically defined components. Proc Natl Acad Sci USA. 2003;100:8253–8257. doi: 10.1073/pnas.1432391100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanigawa G, Orci L, Amherdt M, Ravazzola M, Helms JB, Rothman JE. Hydrolysis of Bound Gtp by Arf Protein Triggers Uncoating of Golgi-Derived Cop-Coated Vesicles. J Cell Biol. 1993;123:1365–1371. doi: 10.1083/jcb.123.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis SM, Poon PP, Singer RA, Johnston GC, Spang A. The ArfGAP Glo3 is required for the generation of COPI vesicles. Mol Biol Cell. 2004;15:4064–4072. doi: 10.1091/mbc. E04-04-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang CJ, Bowzard JB, Anido A, Kahn RA. Four ARF GAPs in Saccharomyces cerevisiae have both overlapping and distinct functions. Yeast. 2003;20:315–330. doi: 10.1002/yea.966. [DOI] [PubMed] [Google Scholar]
- 33.Zhang CJ, Cavenagh MM, Kahn RA. A family of Arf effectors defined as suppressors of the loss of Arf function in the yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:19792–19796. doi: 10.1074/jbc.273.31.19792. [DOI] [PubMed] [Google Scholar]
- 34.Hsu VW. Role of ARFGAP1 in COPI vesicle biogenesis. Cell Logist. 2011;1:55–56. doi: 10.4161/cl.1.2.15175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang JS, Lee SY, Gao M, Bourgoin S, Randazzo PA, Premont RT, et al. ARFGAP1 promotes the formation of COPI vesicles, suggesting function as a component of the coat. J Cell Biol. 2002;159:69–78. doi: 10.1083/jcb.200206015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SY, Yang JS, Hong WJ, Premont RT, Hsu VW. ARFGAP1 plays a central role in coupling COPI cargo sorting with vesicle formation. J Cell Biol. 2005;168:281–290. doi: 10.1083/jcb.200404008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.East MP, Kahn RA. Models for the functions of Arf GAPs. Semin Cell Dev Biol. 2011;22:3–9. doi: 10.1016/j.semcdb.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kahn RA. GAPs: Terminator versus effector functions and the role(s) of ArfGAP1 in vesicle biogenesis. Cell Logist. 2011;1:49–51. doi: 10.4161/cl.1.2.15153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reinhard C, Harter C, Bremser M, Brügger B, Sohn K, Helms JB, et al. Receptor-induced polymerization of coatomer. Proc Natl Acad Sci USA. 1999;96:1224–1228. doi: 10.1073/pnas.96.4.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon HY, Bonifacino JS, Randazzo PA. In vitro assays of Arf1 interaction with GGA proteins. Methods Enzymol. 2005;404:316–332. doi: 10.1016/S0076-6879(05)04028-0. [DOI] [PubMed] [Google Scholar]
- 41.Huber I, Cukierman E, Rotman M, Aoe T, Hsu VW, Cassel D. Requirement for both the amino-terminal catalytic domain and a noncatalytic domain for in vivo activity of ADP-ribosylation factor GTPase-activating protein. J Biol Chem. 1998;273:24786–24791. doi: 10.1074/jbc.273.38.24786. [DOI] [PubMed] [Google Scholar]
- 42.Claude A, Zhao BP, Kuziemsky CE, Dahan S, Berger SJ, Yan JP. GBF1: A novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5. J Cell Biol. 1999;146:71–84. [PMC free article] [PubMed] [Google Scholar]
- 43.García-Mata R, Sztul E. The membrane-tethering protein p115 interacts with GBF1, an ARF guanine-nucleotide-exchange factor. EMBO Rep. 2003;4:320–325. doi: 10.1038/sj.embor.embor762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szul T, Grabski R, Lyons S, Morohashi Y, Shestopal S, Lowe M, et al. Dissecting the role of the ARF guanine nucleotide exchange factor GBF1 in Golgi biogenesis and protein trafficking. J Cell Sci. 2007;120:3929–3940. doi: 10.1242/jcs.010769. [DOI] [PubMed] [Google Scholar]
- 45.Alvarez C, Garcia-Mata R, Brandon E, Sztul E. COPI recruitment is modulated by a Rab1b-dependent mechanism. Mol Biol Cell. 2003;14:2116–2127. doi: 10.1091/mbc. E02-09-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Girod A, Storrie B, Simpson JC, Johannes L, Goud B, Roberts LM, et al. Evidence for a COP-I-independent transport route from the Golgi complex to the endoplasmic reticulum. Nat Cell Biol. 1999;1:423–430. doi: 10.1038/15658. [DOI] [PubMed] [Google Scholar]
- 47.Johannes L, Tenza D, Antony C, Goud B. Retrograde transport of KDEL-bearing B-fragment of Shiga toxin. J Biol Chem. 1997;272:19554–19561. doi: 10.1074/jbc.272.31.19554. [DOI] [PubMed] [Google Scholar]
- 48.Spang A, Shiba Y, Randazzo PA. Arf GAPs: Gatekeepers of vesicle generation. FEBS Lett. 2010;584:2646–2651. doi: 10.1016/j.febs-let.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keen JH. Clathrin and associated assembly and disassembly proteins. Annu Rev Biochem. 1990;59:415–438. doi: 10.1146/annurev.bi.59.070190.002215. [DOI] [PubMed] [Google Scholar]
- 50.Zaremba S, Keen JH. Assembly polypeptides from coated vesicles mediate reassembly of unique clathrin coats. J Cell Biol. 1983;97:1339–1347. doi: 10.1083/jcb.97.5.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stagg SM, Gurkan C, Fowler DM, LaPointe P, Foss TR, Potter CS, et al. Structure of the Sec13/31 COPII coat cage. Nature. 2006;439:234–238. doi: 10.1038/nature04339. [DOI] [PubMed] [Google Scholar]
- 52.Stagg SM, LaPointe P, Balch WE. Structural design of cage and coat scaffolds that direct membrane traffic. Curr Opin Struct Biol. 2007;17:221–228. doi: 10.1016/j.sbi.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 53.Stagg SM, LaPointe P, Razvi A, Gurkan C, Potter CS, Carragher B, et al. Structural basis for cargo regulation of COPII coat assembly. Cell. 2008;134:474–484. doi: 10.1016/j.cell.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bi X, Corpina RA, Goldberg J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 2002;419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- 55.Fath S, Mancias JD, Bi XP, Goldberg J. Structure and organization of coat proteins in the COPII cage. Cell. 2007;129:1325–1336. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 56.Bi X, Mancias JD, Goldberg J. Insights into COPII coat nucleation from the structure of Sec23 center dot Sar1 complexed with the active fragment of sec31. Dev Cell. 2007;13:635–645. doi: 10.1016/j.devcel.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshihisa T, Barlowe C, Schekman R. Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic-reticulum. Science. 1993;259:1466–1468. doi: 10.1126/science.8451644. [DOI] [PubMed] [Google Scholar]
- 58.Spang A, Matsuoka K, Hamamoto S, Schekman R, Orci L. Coatomer, Arf1p, and nucleotide are required to bud coat protein complex I-coated vesicles from large synthetic liposomes. Proc Natl Acad Sci USA. 1998;95:11199–11204. doi: 10.1073/pnas.95.19.11199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olbrich K, Rawicz W, Needham D, Evans E. Water permeability and mechanical strength of polyun-saturated lipid bilayers. Biophys J. 2000;79:321–327. doi: 10.1016/S0006-3495(00)76294-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rawicz W, Olbrich KC, McIntosh T, Needham D, Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ktistakis NT, Brown HA, Waters MG, Sternweis PC, Roth MG. Evidence that phospholipase D mediates ADP ribosylation factor-dependent formation Golgi coated vesicles. J Cell Biol. 1996;134:295–306. doi: 10.1083/jcb.134.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kooijman EE, Chupin V, Fuller NL, Kozlov MM, de Kruijff B, Burger KNJ, et al. Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry. 2005;44:2097–2102. doi: 10.1021/bi0478502. [DOI] [PubMed] [Google Scholar]
- 63.Kooijman EE, Chupin V, de Kruijff B, Burger KNJ. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic. 2003;4:162–174. doi: 10.1034/j.1600-0854.2003.00086.x. [DOI] [PubMed] [Google Scholar]
- 64.Kooijman EE, Burger KNJ. Biophysics and function of phosphatidic acid: A molecular perspective. Biochim Biophys Acta. 2009;1791:881–888. doi: 10.1016/j.bbalip.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 65.Bremser M, Nickel W, Schweikert M, Ravazzola M, Amherdt M, Hughes CA, et al. Coupling of coat assembly and vesicle budding to packaging of putative cargo receptors. Cell. 1999;96:495–506. doi: 10.1016/S0092-8674(00)80654-6. [DOI] [PubMed] [Google Scholar]
- 66.Aoe T, Cukierman E, Lee A, Cassel D, Peters PJ, Hsu VW. The KDEL receptor, ERD2, regulates intracellular traffic by recruiting a GTPase-activating protein for ARF1. EMBO J. 1997;16:7305–7316. doi: 10.1093/emboj/16.24.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cukierman E, Huber I, Rotman M, Cassel D. The ARF1 GTPase-activating protein - Zinc-finger motif and Golgi complex localization. Science. 1995;270:1999–2002. doi: 10.1126/science.270.5244.1999. [DOI] [PubMed] [Google Scholar]
- 68.Hsu VW, Lee SY, Yang JS. The evolving understanding of COPI vesicle formation. Nat Rev Mol Cell Biol. 2009;10:360–364. doi: 10.1038/nrm2663. [DOI] [PubMed] [Google Scholar]
- 69.Bendor J, Lizardi-Ortiz JE, Westphalen RI, Brandstetter M, Hemmings HC, Sulzer D, et al. AGAP1/AP-3-dependent endocytic recycling of M-5 muscarinic receptors promotes dopamine release. EMBO J. 2010;29:2813–2826. doi: 10.1038/emboj.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nie Z, Boehm M, Boja E, Vass W, Bonifacino J, Fales H, et al. Specific Regulation of the Adaptor Protein Complex AP-3 by the Arf GAP AGAP1. Dev Cell. 2003;5:513–521. doi: 10.1016/S1534-5807(03)00234-X. [DOI] [PubMed] [Google Scholar]
- 71.Nie Z, Fei J, Premont RT, Randazzo PA. The Arf GAPs AGAP1 and AGAP2 distinguish between the adaptor protein complexes AP-1 and AP-3. J Cell Sci. 2005;118:3555–3566. doi: 10.1242/jcs.02486. [DOI] [PubMed] [Google Scholar]
- 72.Ha VL, Thomas GMH, Stauffer S, Randazzo PA. Preparation of myristoylated Arf1 and Arf6. Methods Enzymol. 2005;404:164–174. doi: 10.1016/S0076-6879(05)04016-4. [DOI] [PubMed] [Google Scholar]
- 73.Randazzo PA, Fales HM. Preparation of myristoylated Arf1 and Arf6 proteins. In: Manser E, Leung T, editors. GTPase Protocols: The Ras Superfamily. Vol. 189. Totowa: Humana; 2002. pp. 169–180. [DOI] [PubMed] [Google Scholar]
- 74.Pavel J, Harter C, Weiland FT. Reversible dissociation of coatomer: Functional characterization of a beta/delta-coat protein subcomplex. Proc Natl Acad Sci USA. 1998;95:2140–2145. doi: 10.1073/pnas.95.5.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nie Z, Hirsch DS, Luo R, Jian X, Stauffer S, Cremesti A, et al. A BAR domain in the N Terminus of the Arf GAP ASAP1 affects membrane structure and trafficking of Epidermal Growth Factor Receptor. Curr Biol. 2006;16:130–139. doi: 10.1016/j.cub.2005.11.069. [DOI] [PubMed] [Google Scholar]
- 76.Gonda MA, Aaronson SA, Ellmore N, Zeve VH, Nagashima K. Ultrastructural studies of surface features of human normal and tumor-cells in tissue-culture by scanning and transmission electron-microscopy. J Natl Cancer Inst. 1976;56:245–263. doi: 10.1093/jnci/56.2.245. [DOI] [PubMed] [Google Scholar]
- 77.Tousimis AJ, Hilleman MR. Size and Shape of Adenovirus (Ri-Apc-Ard) Type-4 - Electron Microscopy of Purified and Intracellular Virus. J Appl Phys. 1956;27:1392. [Google Scholar]
- 78.Tousimis AJ. Intracellular Rickettsia-Tsutsugamushi in Tissue Culture Cells As Studied by Ultrathin Sectioning and Electron Microscopy. J Appl Phys. 1956;27:1392–1393. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.