Analysis of the lipid composition of influenza virus–infected cells provides support for the membrane raft-based biogenesis model.
Abstract
The influenza virus (IFV) acquires its envelope by budding from host cell plasma membranes. Using quantitative shotgun mass spectrometry, we determined the lipidomes of the host Madin–Darby canine kidney cell, its apical membrane, and the IFV budding from it. We found the apical membrane to be enriched in sphingolipids (SPs) and cholesterol, whereas glycerophospholipids were reduced, and storage lipids were depleted compared with the whole-cell membranes. The virus membrane exhibited a further enrichment of SPs and cholesterol compared with the donor membrane at the expense of phosphatidylcholines. Our data are consistent with and extend existing models of membrane raft-based biogenesis of the apical membrane and IFV envelope.
Introduction
Influenza viruses (IFVs) are spherical or filamentous RNA viruses (Fujiyoshi et al., 1994) responsible for annual epidemics and rare, but often severe, pandemics (Forrest and Webster, 2010). The life cycle of IFV and many other enveloped viruses is comprised of three fundamental steps: virus entry, replication, and budding. Virus assembly at the plasma membrane (PM) is thought to be initiated by clustering of the two major viral spike proteins, HA (Hess et al., 2005) and neuraminidase, in a process postulated to involve membrane rafts (Scheiffele et al., 1999; Zhang et al., 2000; Schmitt and Lamb, 2005). A third viral protein, the M2 protein, is located at the rim of the budding domain and is involved in fission of the virus during exit from the cell (Schroeder et al., 2005; Rossman et al., 2010). IFV particles are released exclusively from the apical side of polarized epithelial cells (Rodriguez Boulan and Sabatini, 1978), which are characterized by apical and basolateral PM domains with specific protein and lipid compositions (van Meer and Simons, 1986). The unusual robustness of the apical membrane is largely attributable to its special lipid composition rich in glycosphingolipids (GSPs; Kawai et al., 1974; Brasitus and Schachter, 1980; Simons and van Meer, 1988). However, whether this is also true for cultured epithelial cells, such as MDCK cells, widely used as hosts for IFV, is not yet known.
The evidence for raft involvement in IFV assembly is manifold (Fiedler et al., 1993; Harder et al., 1998; Scheiffele et al., 1999; Zhang et al., 2000; Shvartsman et al., 2003; Chen et al., 2008). If membrane rafts were to serve as assembly points where viral components concentrate and host cell proteins are depleted (Veit and Thaa, 2011), this would lead to a viral envelope that should be enriched in raft lipids. Previous studies of the lipid composition of purified IFV are consistent with this prediction (Zhang et al., 2000; Blom et al., 2001). However, so far no comprehensive lipid analysis has been performed using the novel sophisticated mass spectrometric methods available today, and the PM from the virus producer cells has not yet been included in the analysis.
In this paper, we tackle this issue by purifying the apical membrane from MDCK cells and IFV produced in these cells. Our data show that the apical membrane compared with total MDCK membranes is low in lipids with a glycerol backbone (GPLs [glycerophospholipids and the glycerolipid DAG]) but high in cholesterol and sphingolipids (SPs), thus reflecting the general trends of lipid composition of apical membranes in epithelial tissues (Kawai et al., 1974; Brasitus and Schachter, 1980; Simons and van Meer, 1988). The IFV is further depleted in GPLs and enriched in cholesterol and SPs compared with the apical PM, consistent with the virus budding from apical raft platforms (Scheiffele et al., 1999; Zhang et al., 2000).
Results and discussion
Purification of apical membranes
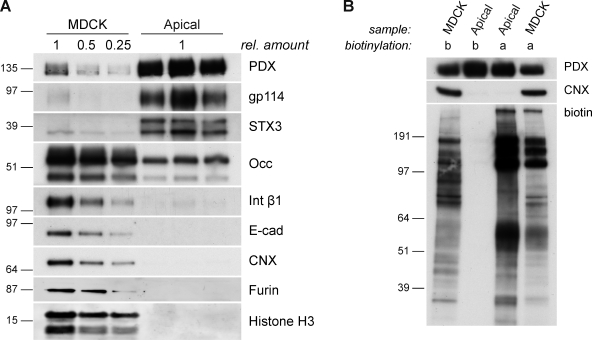
To purify the apical membrane, we adapted a peeling protocol that was originally used to perforate MDCK cells to study intracellular transport and exocytic transport vesicles (Simons and Virta, 1987; Bennett et al., 1989; Wandinger-Ness et al., 1990) and is similar to a recently published method (Fong-ngern et al., 2009). Western blot analysis of this preparation showed strong enrichment of the apical markers podocalyxin, gp114, and syntaxin 3 compared with total cell lysates (Fig. 1 A), whereas basolateral markers, such as integrin β1 and E-cadherin, were depleted. Importantly, markers for organelles like histone H3 for nuclei, furin for the TGN, and calnexin for the ER were absent in the apical membrane preparation (AP). In a rigorous test, we labeled all surface proteins on either the apical or the basolateral surface with biotin and subsequently prepared the apical membrane from these cells (Fig. 1 B). We found the protein pattern of apically biotinylated proteins enriched in the AP, whereas basolaterally biotinylated proteins were not detectable. These data show that we prepared an apical membrane of high purity, which is suitable for lipid analysis.
Figure 1.
Biochemical analysis of the AP. (A) Western blot analysis of apical membrane proteins (Apical) that were recovered from NC membranes peeled from polarized MDCK cells. Total cells (MDCK) serve as a reference. Podocalyxin (PDX), gp114, and syntaxin 3 (STX3) were used as markers for the apical membrane, Occludin (Occ) was used for tight junctions, integrin β1 (Int β1)/E-cadherin (E-cad) were used for the basolateral membrane, calnexin (CNX) was used for the ER, furin was used for the TGN, and histone H3 was used for the nucleus. Samples were standardized to total protein amount. (B) Polarized MDCK cells were either biotinylated apically (a) or basolaterally (b). Apical membrane proteins (Apical) and total cells (MDCK) were prepared as in A. The degree of biotinylation was visualized by streptavidin-HRP. rel. amount, relative amount. Molecular markers are given in kilodaltons.
The lipidome of the apical membrane
We used a shotgun lipidomics approach that combines high mass accuracy and resolution mass spectrometry (MS) and a two-step extraction procedure for optimal recovery of polar lipids with the use of internal standard mixtures for absolute quantification of 18 lipid classes and 419 lipid species (Table S1 and Table S2), recently developed in our laboratories (Ejsing et al., 2009; Sampaio et al., 2011). The data were analyzed by LipidXplorer, a software developed in house (Herzog et al., 2011), and interpreted with a database-based software solution (see Materials and methods), suitable for screening and evaluating large lipidomic datasets.
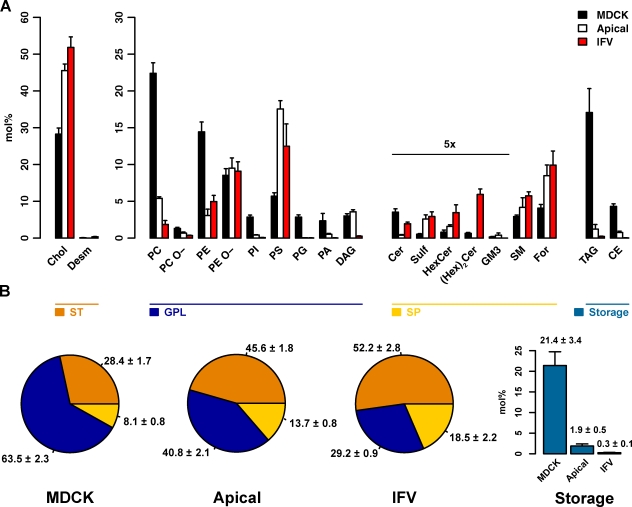
How do the lipids of the AP differ from total cells? For a general overview, we defined functional lipid categories (Fig. 2 B), which are similar to the LIPID MAPS (LIPID Metabolites and Pathways Strategy) categories (Fahy et al., 2005, 2009) but differentiate between membrane and storage lipids. As expected, storage lipids, such as triacylglycerol (TAG) and cholesterol esters (CEs), known to reside in lipid bodies were depleted from the apical membrane (Fig. 2, A and B). Glycerophospholipids and the glycerolipid DAG (combined as GPLs) were reduced to <65% of their total cell amount in the AP, with the mitochondrial lipid PG (Fig. 2) practically absent. Likewise, PC, PI, PA, and PE were reduced to <25% of their whole-cell values, whereas 1.2× DAG and 1.1× PE O- were slightly increased. Interestingly, the only GPL class found to be strongly increased is PS, which was three times more abundant in the AP.
Figure 2.
Lipid class composition and functional categories. (A and B) The content of individual lipid classes (A) and functional lipid categories (B) of total cells (MDCK), the AP (Apical), and influenza virus (IFV) was determined by summing up absolute abundances of all identified species. Values are standardized to mole percentage of all membrane lipids within the sample without storage lipids. Indicated values have been multiplied by five for visibility. Sterols (ST): cholesterol (Chol) and desmosterol (Desm). GPLs: DAG, phosphatidic acid (PA), phosphatidylcholine/-ethanolamine/-glycerol/-inositol/-serine (PC/PE/PG/PI/PS), and ether linked PC/PE (PC O-/PE O-). Sphingolipids (SP): ceramide (Cer), Forssman glycolipid (For), hexosylceramide (HexCer), dihexosylceramide ((Hex)2Cer), sphingomyelin (SM), and sulfatide (Sulf). Storage: cholesterol esters (CE) and triacylglycerol (TAG). Error bars correspond to SDs (n = 3).
Although GPLs were in general reduced, SPs were enriched in the AP, contributing 13.7 ± 0.8 mol% to its composition. The globoside derivative Forssman glycolipid, which is the most abundant SP in polarized MDCK cells, doubled its concentration to ∼8.5 mol% of the AP. Ceramide (Cer) and dihexosyl-Cer ((Hex)2Cer) were the only SPs reduced with respect to total cells (Fig. 2 A).
As expected, we found a high cholesterol content in the AP; cholesterol comprising ∼45 mol% of all apical lipids was the most abundant lipid species. Consequently, the lipid composition of the AP is in agreement with the expected cellular lipid distribution (van Meer et al., 2008): besides the strong depletion of storage lipids, we found most GPLs to be reduced but SPs and sterols to be increased. Furthermore, these data confirm a remarkable co-segregation of PS with cholesterol and SPs found in a recent study (Fairn et al., 2011).
The lipid class composition of apical intestinal brush border membranes has been analyzed earlier (Forstner et al., 1968; Kawai et al., 1974; Brasitus and Schachter, 1980) and is similar to that of the MDCK apical membrane except that the GSP concentration was even higher, namely 37%, in the brush border membrane (Simons and van Meer, 1988). The apical membrane forms the barrier to the external world and therefore has to be constructed to serve this purpose. Intestinal apical membranes need to be especially robust (Simons and van Meer, 1988; Danielsen and Hansen, 2003). However, MDCK cells are derived from canine kidney, and previous analysis of apical membranes from the kidney tubule cells (Carmel et al., 1985) demonstrated that these brush border membranes had less GSPs and instead more sphingomyelin (SM) in line with our findings.
We recently analyzed the lipidomic changes occurring during polarization of MDCK cells (Sampaio et al., 2011). We observed changes in the lipid composition in line with what would be expected when an apical membrane is introduced during polarization. The pronounced remodeling of the SP class and species composition is also in agreement with the generation of a robust impermeable raft barrier (Simons and van Meer, 1988), possibly maintained by a lectin-GSP circuit set up by the most abundant GSP in MDCK cells, the Forssman glycolipid, and by galectin-9 (Mishra et al., 2010).
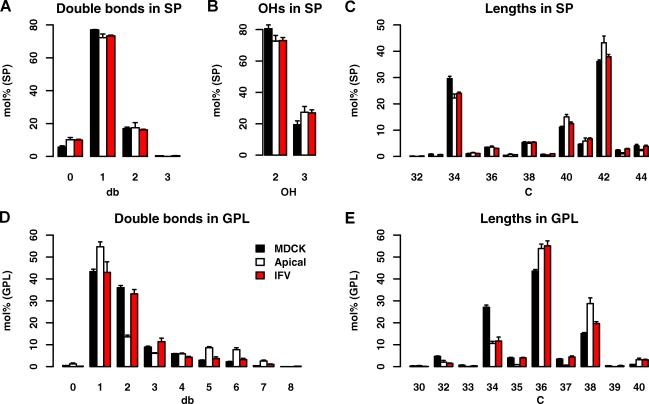
A closer look at the molecular structural details of SP species in the AP versus total MDCK cells shows that SPs in the AP are slightly longer, more saturated, and more hydroxylated (Fig. 3, A–C). Similar, but more pronounced, changes were found in apical GPLs (Fig. 3, D and E): First, they are more saturated, with especially monounsaturated species comprising more than half of all species. Second, they are longer, with the acyl chains below a sum of 36 carbon atoms decreased and species above or equal to a sum of 36 carbon atoms more abundant. The major changes in this respect are a reduction of 34:1 GPL species, whereas 36:1 species are more than doubled (Fig. S2 B). How does this change come about? The species profiles of the main GPL classes show little changes between total cells and the AP (Fig. S2 A). However, the profiles are distinct with PC being rich in short chain species and PS mainly consisting of PS 36:1. As PC and diacyl-PE class levels are high in total MDCK cells, and PS and PE O- levels dominate the AP, the class level changes influence the overall GPL profile: e.g., the significant drop of PC levels in the AP is responsible for the drop in 34:1 species, whereas the enrichment of PS is the main reason for more abundant 36:1 species. Therefore, the differences in length and saturations are mostly caused by a change of lipid class levels and their typical species profiles, rather than a change in the species profile within the classes themselves. In summary, we find the apical membrane to be enriched in SPs and cholesterol, depleted in storage lipids, and reduced in the majority of GPLs.
Figure 3.
Lipid features of total cells, the apical membrane, and IFV. (A–E) Changes in SP saturation as number of double bonds (db) in the sum of long chain base and fatty acid (A), SP hydroxylation (B), SP chain length as number of carbon atoms in the sum of long chain base and fatty acid (C), GPL saturation as number of double bonds in the sum of fatty acid moieties (D), and GPL chain length as number of carbon atoms in the sum of fatty acid moieties (E). Values are standardized to mole percentage of all GPLs or SPs as indicated. Features were calculated from individual quantities of lipid molecular species. Error bars correspond to SDs (n = 3).
The lipidome of IFV particles
Before purifying virus from MDCK cells, we compared the lipidome of total MDCK cells 24 h after infection with that of uninfected control cells. We found no effect of influenza infection on the functional lipid categories (Fig. S1 A, inset); however, we did find effects on some lipid classes: a reduction in PC and an increase in PI were the most prominent changes (Fig. S1 A). However, as described in the second part of this section (Fig. 2 A), these changes are not in line with the differences observed by comparing the AP to the IFV envelope lipids.
Interestingly, we found a sixfold increase in dihydro-SM (34:0:2) as a result of viral infection (Fig. S1 B). This lipid has also been found enriched in retroviruses (Brügger et al., 2006; Chan et al., 2008), vesicular stomatitis virus (VSV) and Semliki Forest virus (SFV; Kalvodova et al., 2009) compared with uninfected cells. It remains to be seen whether in these cases also, viral infection promoted dihydro-SM synthesis.
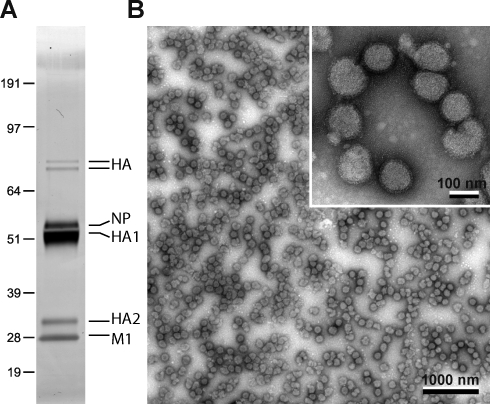
We then infected polarized MDCK monolayers with IFV, collected the apical supernatant after 24 h, and purified virus particles to homogeneity. Silver gel analysis showed only viral protein bands without host cell contamination (Fig. 4 A), and the preparation consisted of spherical viral particles 100 nm in diameter (Fig. 4 B).
Figure 4.
IFV purification. Polarized MDCK cells were infected with IFV, and virus particles were collected after 24 h. (A) A pure virus preparation was obtained by gradient centrifugations and was analyzed by SDS-PAGE and silver staining. (B) Electron micrograph of the virus preparation with negative stain. The inset shows a higher magnification of the same preparation. Molecular markers are given in kilodaltons. NP, influenza nucleoprotein.
The lipidome of the viral particle was significantly different from the AP. Storage lipids are reduced to 0.3 mol% and hence a negligible component of the viral lipidome. GPLs make up 29.2 ± 0.9 mol% compared with 40.8 ± 2.1 mol% in the AP (Fig. 2 B), and viral GPLs consisted mostly of the inner leaflet lipids PE, PE O-, and PS (Fig. 2 A). PC was reduced threefold, with only <2 mol% left in the viral membrane. All other GPL classes contributed only marginally to the lipidome. Interestingly, we found PE O- to be the only GPL class not to be reduced in the virus compared with the AP. PE O- has been consistently found in PM preparations (Chan et al., 2008; Kalvodova et al., 2009), viruses (Brügger et al., 2006), and raft preparations (Pike et al., 2002; Zech et al., 2009). Moreover, dialkylglycerophospholipid membranes have been shown to exhibit higher phase transition temperatures, smaller surface areas, and reduced permeability compared with their diacylglycerophospholipid counterparts (Paltauf, 1994).
SPs represented 18.5 mol% of the viral envelope, ∼1.4 times their amount in the AP (Fig. 2 B). This was caused by enrichment of nearly all SP classes in the virus particle compared with the AP (Fig. S3). GM3 was the only exception, as it was absent from the viral envelope. (Hex)2Cer, however, was highly abundant in the virus, whereas not detectable in the AP. The straightforward explanation is the presence of neuraminidase in the virus envelope. Cleaving neuraminic acid from the ganglioside GM3 results in lactosyl-Cer, which is measured as (Hex)2Cer.
The molecular SP features change little from the AP toward IFV (Fig. 3, A–C). However, in the GPLs, we saw that species with two and three double bonds became more abundant in IFV over AP, whereas polyunsaturated species with 38 carbon atoms (mostly PE O-) were reduced (Fig. 3 D).
Interestingly, the already high sterol levels in the AP are further increased in the viral particle to ∼52 mol%. With IFV leaflet asymmetry maintained (Rothman et al., 1976), the outer IFV envelope leaflet should consist almost entirely of SPs and cholesterol.
All these features of the IFV lipidome confirm the notion that the viral envelope buds from a raft domain in the host membrane and imply that the virus does not bud passively from the lipid bilayer but is selective. Clusters containing self-associated HA could serve as nucleation sites for a polymerizing M1 protein (Rossman et al., 2010; Veit and Thaa, 2011).
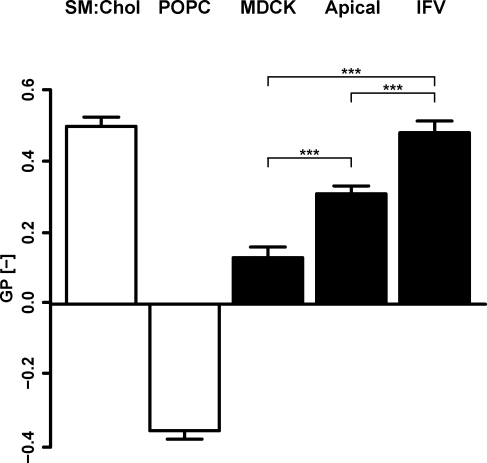
Laurdan spectroscopy shows increased membrane order in the apical membrane and IFV
We used C-laurdan spectroscopy to measure membrane order in the different membrane preparations (Kaiser et al., 2009). Floated membranes of total cells were already found to be quite ordered (general polarization [GP] = 0.13 ± 0.03) compared with the standard mixture of egg POPC liposomes (GP = −0.35 ± 0.02; Fig. 5). This is not surprising, as total MDCK cells already contain 28 mol% cholesterol and 8 mol% SPs. Unfortunately, we could not use our peeled-off APs because the filter disturbed the spectrophotometric C-laurdan measurements, but a floated AP showed a significantly higher GP value (GP = 0.31 ± 0.02) compared with total cells. The GP value of the virus was very high (0.48 ± 0.03) and virtually identical to the value of liposomes prepared with the bona fide liquid-ordered mixture of SM/cholesterol = 1:1 (GP = 0.50 ± 0.03). Similar results have been recorded with diphenylhexatriene fluorescence (Scheiffele et al., 1999). It has to be pointed out that the complete virus particle was measured; the membrane of which is asymmetric, concentrating SPs on one side and packed with spike proteins that may induce additional order (Lorizate et al., 2009; Niemelä et al., 2010).
Figure 5.
Membrane order measurements by C-laurdan spectroscopy. Generalized polarization (GP) values of liposomes with defined liquid-ordered (SM/cholesterol [Chol] = 1:1) and liquid-disordered (POPC) compositions (open bars) compared with GP values of floated total MDCK membranes (MDCK), floated apical membranes (Apical), and influenza virus (IFV; shaded bars). Measurements were performed at 23°C. The p-values have been estimated with one-way analysis of variance and posthoc test with Bonferroni correction (***, P < 0.001). Error bars correspond to SDs (n = 3).
Do viruses in general sort lipids or is this a special feature of “raft viruses?” Comparison of the raft virus HIV lipidome with its host cell PM demonstrated enrichment of cholesterol, the ganglioside GM3, and Cer in the virus envelope (Chan et al., 2008). The lipidomes of the SFV and VSV, on the other hand, highly resembled that of the host PM from which they bud (Kalvodova et al., 2009) with a minor but measurable specificity, probably imposed by geometric restraints originating from lipid asymmetry and high curvature. We observed similar species selectivity (Fig. S3). The VSV lipidome has also been directly compared with IFV budding from the same host cell (Blom et al., 2001). In this study, the different viruses selected distinct lipids from the same PM. IFV strongly enriched GSPs compared with VSV. In conclusion, HIV and IFV are clearly enriched in membrane raft components, suggesting that the viral proteins assemble specific membrane raft platforms in the host PM during budding, whereas other viruses (SFV and VSV) more passively accommodate lipids from the host PM into their envelopes.
We have investigated two lipid-sorting processes in which SPs and cholesterol are enriched. The first process leads to a resilient and durable apical PM but still has to provide enough fluidity, such that membrane function and communication with the outer world is secured (Schuck and Simons, 2004; Meder et al., 2006). The second process leads to the formation of a robust capsule of raft lipids (and proteins) with little other functionality than to withstand damage after exit from the host cell and during airborne transmission (Polozov et al., 2008) and to support virus fusion in the endosome (Mercer et al., 2010).
Materials and methods
Reagents
All common chemicals were purchased from Sigma-Aldrich and were of the best analytical grade. Water for MS, MeOH (both LiChrosolv grade), chloroform (liquid chromatography grade), and silver nitrate were obtained from Merck. Synthetic lipid standards were purchased from Avanti Polar Lipids, Inc. PI 17:0-17:0 was provided by C. Thiele (Life and Medical Sciences Institute, University of Bonn, Bonn, Germany). C-laurdan (6-dodecanoyl-2-N-methyl-N-carboxymethyl-amino-naphthalene) was a gift from B.-R. Cho (Korea University, Seoul, South Korea). The streptavidin-HRP conjugate was obtained from GE Healthcare. Antibodies used were as follows: anti-gp135/podocalyxin (G. Ojakian, State University of New York Downstate Medical Center, Brooklyn, NY; Ojakian and Schwimmer, 1988), mouse anti-gp114 (Simons laboratory, Max-Planck-Institut für molekulare Zellbiologie und Genetik, Dresden, Germany; Balcarova-Ständer et al., 1984), anti–syntaxin 3 (V. Olkkonen, National Institute for Health and Welfare, Helsinki, Finland; Riento et al., 1998), rabbit (rb) anti-Occludin (Invitrogen), rb anti-calnexin (Stressgen), mouse anti–integrin β1 (BD), rb anti–histone H3 (ab1791; Abcam), rb anti-furin (Thermo Fisher Scientific), and mouse anti–E-cadherin (Simons laboratory; Gumbiner and Simons, 1986).
MDCK cell culture
MDCK cell culture was performed as in Sampaio et al. (2011). In short, MDCK strain II cells were maintained in MEM (with Earle’s salts; PAA Laboratories) supplemented with 5% fetal calf serum, 2 mM l-glutamine, and 100 U/ml penicillin/streptomycin (all obtained from Invitrogen) on polystyrene tissue-culture flasks. For subculturing, confluent cells were detached by incubation with trypsin/EDTA (Invitrogen). For polarized growth, cells were plated on Transwell polycarbonate filters (0.4-µm pore size) obtained from Costar.
Cell surface biotinylation
Cells were washed 3× with cold PBS+ (containing Mg2+ and Ca2+), and apical or basolateral surfaces were biotinylated with 2 mg/ml Sulfo-NHS-LC-biotin (Thermo Fisher Scientific) in PBS+ on ice for 20 min. Cold quenching solution (0.1 M glycine in PBS+) was applied on the respective other side of the monolayer. Unreacted biotin was quenched with cold quenching solution 2× for 5 min.
AP with nitrocellulose (NC) membranes
APs were modified from Simons and Virta (1987) and Huber and Simons (1994). NC membranes (85 mm; Immobilon-NC transfer membrane; Millipore) were sequentially preextracted with chloroform/MeOH at 10:1 and 2:1 solvent mixtures and soaked in KOAc buffer (25 mM Hepes, pH 7.4, 115 mM potassium acetate, and 2.5 mM MgCl2). 106 MDCK cells (passage between 3 and 10) were grown for 7 d on 75-mm Transwell polycarbonate filter supports (0.4-µm pore size), washed twice with cold KOAc buffer, and overlaid with Whatman paper predrained NC membranes. Excess moisture was removed by overlaying twice with filter paper (3MM; Whatman) and smoothing with a bent Pasteur pipette. After binding to the cells for 75 s, the NC membrane was wetted with KOAc buffer and carefully peeled away from the cells. NC membranes were washed 3× for 10 min with 150 mM ammonium acetate, pH 7.4, 2× for 5 min with 0.5 M urea, and 3× with water and taken up in 150 mM ammonium bicarbonate. To obtain protein samples, the NC membrane pieces were incubated with lysis buffer (2% NP-40 [Fluka] and 0.2% SDS [Serva] in PBS) in spin columns (Mobicols; MoBiTec GmbH), and the samples were recovered by centrifugation.
Virus production and purification
Avian IFV A/fowl plague virus/Rostock/34 (H7N1) was obtained from H.-D. Klenk (Institut fuer Virologie, Universitaet Giessen, Giessen, Germany). 107 MDCK cells (passage 3–10) were fully polarized on eight 75-mm Transwell polycarbonate filter supports (0.4-µm pore size). All virus work was performed in an S3 laboratory. Cells were infected with IFV at an MOI of 0.005 for 1 h at 37°C and 5% CO2. The virus-containing medium was removed, and cells were incubated for 24 h at 37°C and 5% CO2 in MEM containing 20 mM Hepes, penicillin-streptomycin, and glutamate.
The apical medium was clarified by centrifugation at 3,000 rpm for 30 min. The supernatant was transferred to ultraclear centrifuge tubes (SW28; Beckman Coulter), and the virus was concentrated for 1 h at 25 krpm. The resulting pellet was resuspended overnight in 100 µl TBS (10 mM Tris-HCl and 150 mM NaCl, pH 7.4), subjected to a 30/60% (wt/wt) sucrose step gradient, and centrifuged for 1.5 h at 27 krpm in a rotor (SW40 Ti; Beckman Coulter). The 30/60% interface was harvested, diluted in TBS, and centrifuged at 25 krpm for 1 h in a rotor (SW28). The resulting pellet was resuspended overnight in 100 µl TBS, layered on top of a 20–50% (wt/wt) continuous sucrose gradient with a 60% (wt/wt) sucrose cushion, and centrifugated for 1 h at 25 krpm in a rotor (SW40 Ti). The virus band was collected with a needle, diluted in TBS, and centrifuged for 1.5 h at 27 krpm in a SW28 rotor. The virus pellet was resuspended overnight in 100 µl 150 mM ammonium bicarbonate and washed in the same buffer using a rotor (TLA-55; Beckman Coulter) for 45 min at 50 krpm. The final virus pellet was resuspended in 100 µl ammonium bicarbonate buffer.
EM
For EM, a 2-µl drop of sample was deposited on glow-discharged, carbon-coated EM grids for 1 min and blotted. Viruses were inactivated on the grid with a drop of 1% glutaraldehyde (in water). After blotting of fixative, the samples were negatively stained with 2% uranyl-acetate in water and blotted again. After drying, the samples were examined at room temperature in a transmission EM (Tecnai 12 BioTWIN; FEI) fitted with a 2,000 × 2,000–pixel digital camera (F214; Tietz Video and Image Processing Systems) and EM-MENU 4 software package (Tietz Video and Image Processing Systems). Contrast was adjusted using Fiji Software.
Lipid analysis
Lipid extraction of MDCK cells (Sampaio et al., 2011) and IFV (Kalvodova et al., 2009) was performed as previously described. In brief, 10 µg protein of cell lysates and virus preparation was dissolved in 200 µl of 150 mM ammonium bicarbonate, and 10 µl of an internal standard mixture (see following paragraph for details) was added to the extract. We then added 1 ml of a mixture of chloroform/MeOH (10:1) and shook the extracts for 2 h. After that, we centrifuged the extract for 2 min at 1 krpm and collected the organic phase. We then added 1 ml of a mixture of chloroform/MeOH (2:1) and shook the extracts for 1 h. After that, we centrifuged the extract for 2 min at 1 krpm and collected the organic phase. Lipid extraction of apical rip-offs was performed as follows: 10 µl of an internal standard mixture was added to ∼10-cm2 NC membrane pieces and resuspended in 1 ml of 150 mM ammonium bicarbonate. We then extracted the suspension with 5 ml chloroform/MeOH (10:1) for 2 h. The lower organic phase was collected, and the aqueous phase containing the NC membrane pieces was reextracted with 5 ml chloroform/MeOH (2:1) for 1 h. Then, all organic phases were evaporated in a vacuum desiccator (VWR International; Merck) at 4°C overnight. Lipid extracts were dissolved in 100 µl chloroform/MeOH (1:2 [vol/vol]) and subjected to quantitative lipid analysis as previously described elsewhere (Sampaio et al., 2011). In brief, for positive ion mode, 10 µl extract was mixed with 13 µl of 13 mM ammonium acetate solution in propanol before infusion. For negative ion mode, 10 µl extract was mixed with 10 µl of 0.1% methylamine before infusion. Samples were infused with a robotic nanoflow ion source (TriVersa NanoMate; Advion Biosciences, Inc.) into a mass spectrometer instrument (LTQ Orbitrap; Thermo Fisher Scientific). PA, PS, PE, PE O-, PI, Cer, HexCer, GM3, sulfatide, and PG species were quantified by negative ion mode Fourier transform MS at Rm/z=400 = 100,000; PC, PC O-, SM, DAG, CE, TAG, and Forssman glycolipid were quantified by positive ion mode Fourier transform MS at Rm/z=400 = 100,000. Sterols were quantified as previously described (Sandhoff et al., 1999). In brief, the extracts were dried in the speedvac (Jouan RC1022; Thermo Fisher Scientific), and 20 µl of freshly prepared sulfur trioxide pyridine complex solution (dissolving 25 mg sulfur trioxide pyridine complex in 5 ml absolute pyridine) was added followed by sonication for ∼1 min. The reaction was allowed to take place for ≥30 min at room temperature. Then, 2.1 µl barium acetate solution was added, and the sample was sonicated (1 min). Hydrolysis and precipitation of barium sulfate was allowed to take place for 10 min at room temperature and 60 min at 4°C. Then, 80 µl MeOH was added to the reaction mixture, and the sample was centrifuged at 14 krpm for 15 min at 4°C in a bench-top centrifuge (MiniSpin; Eppendorf). For mass spectrometric analysis, 20 µl of the sample was used and infused as described earlier in this section.
Lipid standards
In every lipid extraction, we added a lipid standard mixture for absolute quantification consisting of 12.7 pmol PA 17:0-17:0, 42.5 pmol PE 17:0-17:0, 9.5 pmol PG 17:0-17:0, 56 pmol PS 17:0-17:0, 39 pmol PC 18:3-18:3, 10 pmol PI 17:0-17:0, 78 pmol cholesterol-d7, 40 pmol SM 18:1;2-17:0;0, 20 pmol lactosyl-Cer 18:1;2-12:0;0; 20 pmol Cer 18:1;2-17:0;0, 20 pmol galactosyl-Cer 18:1;2-12:0;0, 20 pmol DAG 17:0-17:0, 90 pmol CE 17:0, 10 pmol TAG 12:0-12:0-12:0, 10 pmol sulfatide 18:1;2-12:0;0, 120 pmol GM3 18:1;2-12:0;0, and 44 pmol For-d3 standard extract. Internal lipid standards for GM3 were acquired from S. Sonnino (University of Milan, Milan, Italy). Forssman glycolipid (Papirmesiter and Mallette, 1955; Siddiqui and Hakomori, 1971) was purified from sheep erythrocytes by solvent extraction and silica gel chromatography and stored dissolved in chloroform/MeOH (2:1 [vol/vol]) at −20°C. De–N-acetylated glycolipid was prepared as described previously (Lingwood and Nutikka, 1994). In brief, 0.5 mg (330 nmol) purified glycolipid was dried in a glass tube under N2 gas and then resuspended with gentle sonication in 1.5 ml of 1-N NaOH in water. The tube was capped tightly, and the solution was heated at 100°C for 3.5 h, cooled on ice, and then neutralized with HCl. The glycolipid was desalted by solvent partition (CHCl3/CH3OH/H2O; 2:1:0.6 [vol/vol/vol]) and dried under N2 gas. The de–N-acetylation reaction was essentially complete, as determined by MS.
50 nmol de–N-acetyl-Forssman suspended in 1 ml of 0.5-M NaHCO3 was reacted with 0.5 mmol d6 acetic anhydride (99 atom % D; Sigma-Aldrich) at ambient temperature for 1 h. The reacetylated glycolipid was desalted by solvent partition, and the reaction completion was verified by MS.
Software
Automated processing of acquired mass spectra, identification, and quantification of detected molecular lipid species were performed by LipidXplorer software (Herzog et al., 2011) developed in house. Lipidomic datasets were stored in a database, which could be queried to create profiles of lipid-specific features (acyl length, double bonds …) within specific subsets of the lipidome (total sample, lipid class …) combined with flexible standardization (to total sample, lipid class …). Results were immediately and adjustably visualized via a commercially available graphical user interface (Vortex; Dotmatics Ltd.). For statistical analysis and graph plotting, the R software package was used (R Development Core Team, 2010). Digital images were processed using Photoshop and Illustrator (Adobe).
Free-floating AP
Free-floating APs were modified from Fong-ngern et al. (2009). In short, polarized MDCK cells on 75-mm Transwell permeable supports were overlaid with semidry paper (3-MM; Whatman) for 10 min. Whatman papers were rehydrated for 30 min in 150 mM ammonium bicarbonate, and the surface was scraped with a cell scraper. Supernatants containing membranes and floating paper pieces were underlaid with a 60% iodixanol (wt/vol; OptiPrep; Progen Biotechnik) cushion and spun for 3 h at 160,000 g in a rotor (SW40 Ti). The membrane band at the 60% iodixanol interphase was transferred into a tube (SW60), adjusted to 40% iodixanol, and overlayed with 30% iodixanol in ammonium bicarbonate and, finally, with pure ammonium bicarbonate. After centrifugation for 90 min at 280,000 g, the membranes were harvested at the 30% iodixanol interphase. For floated total cell membranes, filter-grown polarized MDCK cells were scraped, homogenized with a 25-gauge needle, and floated identically to apical membranes described earlier in this paragraph.
Formation of liposomes and C-laurdan spectroscopy
Liposomes were created, and C-laurdan spectroscopy was performed as previously described in Kaiser et al. (2009). For liposomes, lipids in chloroform/MeOH were dried under vacuum and recovered in HBS (10 mM Hepes, 150 mM NaCl, and 0.2 mM EDTA, pH 7.25) at 68°C. After 10 freeze-thaw cycles, liposomes were extruded at 70°C using 100-nm polyvinylidene fluoride membranes. Liposome, cellular, and virus membrane amounts were standardized via scattering fluorescence emission at 425 nm (λex = 385 nm) to 30,000 intensity units stained with 100 µM C-laurdan for 15 min at room temperature. C-laurdan was then excited at 385 nm. The GP value was calculated from the following emission bands: Ch1 (400–460 nm) and Ch2 (470–530 nm). GP = (ICh1 − ICh2)/(ICh1 + ICh2), in which I is the fluorescence intensity, from spectra at a 1-nm resolution recorded on a fluorescence spectrometer (FluoroMax-3; Horiba) with a thermostat (SC 100-A5B; Thermo Fisher Scientific) at 23°C.
SDS-PAGE
SDS-PAGE was performed in Novex 4–12% Bis-Tris gels (NuPAGE; Invitrogen) using MOPS or MES SDS running buffer (NuPAGE) and prestained standard (SeeBlue Plus2; Invitrogen). Differences between the known and the apparent molecular weight were observed for some proteins.
Online supplemental material
Fig. S1 shows lipid class composition of MDCK cells infected with IFV. Fig. S2 shows hydrocarbon chains of total cells, the apical membrane, and IFV. Fig. S3 shows enrichment of lipid classes in the apical membrane and IFV. Table S1 shows individual lipid species of total MDCK cells, apical membrane, and IFV lipidomes. Table S2 shows individual lipid species of total MDCK cells and IFV-infected MDCK cell lipidomes. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201108175/DC1.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft Schwerpunktprogramm1175 (grants SI459/2-1 and LI621/4-2), Deutsche Forschungsgemeinschaft Transregio 83 (grant TRR83 TP02), European Science Foundation LIPIDPROD (Lipid–protein interactions in membrane organization; grant SI459/3-1), Bundesministerium für Bildung und Forschung ForMaT (Forschung für den Markt im Team; grant 03FO121), and the Klaus Tschira Foundation. A. Shevchenko was supported by Virtual Liver (code/0315757) grant from Bundesministerium für Bildung und Forschung.
Footnotes
Abbreviations used in this paper:
- AP
- apical membrane preparation
- CE
- cholesterol ester
- Cer
- ceramide
- GP
- general polarization
- GSP
- glyco-SP
- (Hex)2Cer
- dihexosyl-Cer
- IFV
- influenza virus
- MS
- mass spectrometry
- NC
- nitrocellulose
- PA
- phosphatidic acid
- PM
- plasma membrane
- rb
- rabbit
- SFV
- Semliki Forest virus
- SM
- sphingomyelin
- SP
- sphingolipid
- TAG
- triacylglycerol
- VSV
- vesicular stomatitis virus
References
- Balcarova-Ständer J., Pfeiffer S.E., Fuller S.D., Simons K. 1984. Development of cell surface polarity in the epithelial Madin-Darby canine kidney (MDCK) cell line. EMBO J. 3:2687–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.K., Wandinger-Ness A., De Curtis I., Antony C., Simons K., Kartenbeck J. 1989. Perforated cells for studying intracellular membrane transport. Methods Cell Biol. 31:103–126 10.1016/S0091-679X(08)61604-0 [DOI] [PubMed] [Google Scholar]
- Blom T.S., Koivusalo M., Kuismanen E., Kostiainen R., Somerharju P., Ikonen E. 2001. Mass spectrometric analysis reveals an increase in plasma membrane polyunsaturated phospholipid species upon cellular cholesterol loading. Biochemistry. 40:14635–14644 10.1021/bi0156714 [DOI] [PubMed] [Google Scholar]
- Brasitus T.A., Schachter D. 1980. Lipid dynamics and lipid-protein interactions in rat enterocyte basolateral and microvillus membranes. Biochemistry. 19:2763–2769 10.1021/bi00553a035 [DOI] [PubMed] [Google Scholar]
- Brügger B., Glass B., Haberkant P., Leibrecht I., Wieland F.T., Kräusslich H.G. 2006. The HIV lipidome: a raft with an unusual composition. Proc. Natl. Acad. Sci. USA. 103:2641–2646 10.1073/pnas.0511136103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel G., Rodrigue F., Carrière S., Le Grimellec C. 1985. Composition and physical properties of lipids from plasma membranes of dog kidney. Biochim. Biophys. Acta. 818:149–157 10.1016/0005-2736(85)90557-7 [DOI] [PubMed] [Google Scholar]
- Chan R., Uchil P.D., Jin J., Shui G., Ott D.E., Mothes W., Wenk M.R. 2008. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J. Virol. 82:11228–11238 10.1128/JVI.00981-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.J., Leser G.P., Jackson D., Lamb R.A. 2008. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 82:10059–10070 10.1128/JVI.01184-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsen E.M., Hansen G.H. 2003. Lipid rafts in epithelial brush borders: atypical membrane microdomains with specialized functions. Biochim. Biophys. Acta. 1617:1–9 10.1016/j.bbamem.2003.09.005 [DOI] [PubMed] [Google Scholar]
- Ejsing C.S., Sampaio J.L., Surendranath V., Duchoslav E., Ekroos K., Klemm R.W., Simons K., Shevchenko A. 2009. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA. 106:2136–2141 10.1073/pnas.0811700106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy E., Subramaniam S., Brown H.A., Glass C.K., Merrill A.H., Jr, Murphy R.C., Raetz C.R., Russell D.W., Seyama Y., Shaw W., et al. 2005. A comprehensive classification system for lipids. J. Lipid Res. 46:839–861 10.1194/jlr.E400004-JLR200 [DOI] [PubMed] [Google Scholar]
- Fahy E., Subramaniam S., Murphy R.C., Nishijima M., Raetz C.R., Shimizu T., Spener F., van Meer G., Wakelam M.J., Dennis E.A. 2009. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 50(Suppl.):S9–S14 10.1194/jlr.R800095-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairn G.D., Schieber N.L., Ariotti N., Murphy S., Kuerschner L., Webb R.I., Grinstein S., Parton R.G. 2011. High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J. Cell Biol. 194:257–275 10.1083/jcb.201012028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler K., Kobayashi T., Kurzchalia T.V., Simons K. 1993. Glycosphingolipid-enriched, detergent-insoluble complexes in protein sorting in epithelial cells. Biochemistry. 32:6365–6373 10.1021/bi00076a009 [DOI] [PubMed] [Google Scholar]
- Fong-ngern K., Chiangjong W., Thongboonkerd V. 2009. Peeling as a novel, simple, and effective method for isolation of apical membrane from intact polarized epithelial cells. Anal. Biochem. 395:25–32 10.1016/j.ab.2009.08.007 [DOI] [PubMed] [Google Scholar]
- Forrest H.L., Webster R.G. 2010. Perspectives on influenza evolution and the role of research. Anim. Health Res. Rev. 11:3–18 10.1017/S1466252310000071 [DOI] [PubMed] [Google Scholar]
- Forstner G.G., Tanaka K., Isselbacher K.J. 1968. Lipid composition of the isolated rat intestinal microvillus membrane. Biochem. J. 109:51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyoshi Y., Kume N.P., Sakata K., Sato S.B. 1994. Fine structure of influenza A virus observed by electron cryo-microscopy. EMBO J. 13:318–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner B., Simons K. 1986. A functional assay for proteins involved in establishing an epithelial occluding barrier: identification of a uvomorulin-like polypeptide. J. Cell Biol. 102:457–468 10.1083/jcb.102.2.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder T., Scheiffele P., Verkade P., Simons K. 1998. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol. 141:929–942 10.1083/jcb.141.4.929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog R., Schwudke D., Schuhmann K., Sampaio J.L., Bornstein S.R., Schroeder M., Shevchenko A. 2011. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 12:R8 10.1186/gb-2011-12-1-r8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess S.T., Kumar M., Verma A., Farrington J., Kenworthy A., Zimmerberg J. 2005. Quantitative electron microscopy and fluorescence spectroscopy of the membrane distribution of influenza hemagglutinin. J. Cell Biol. 169:965–976 10.1083/jcb.200412058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, L., and K. Simons. 1994. Preparing and purification of post-Golgi transport vesicles from perforated Madin-Darby kidney cells. In Cell Biology: A Laboratory Handbook. Vol. 1. J.E. Celis, editor. Academic Press, San Diego, CA. 517–524.
- Kaiser H.J., Lingwood D., Levental I., Sampaio J.L., Kalvodova L., Rajendran L., Simons K. 2009. Order of lipid phases in model and plasma membranes. Proc. Natl. Acad. Sci. USA. 106:16645–16650 10.1073/pnas.0908987106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvodova L., Sampaio J.L., Cordo S., Ejsing C.S., Shevchenko A., Simons K. 2009. The lipidomes of vesicular stomatitis virus, semliki forest virus, and the host plasma membrane analyzed by quantitative shotgun mass spectrometry. J. Virol. 83:7996–8003 10.1128/JVI.00635-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai K., Fujita M., Nakao M. 1974. Lipid components of two different regions of an intestinal epithelial cell membrane of mouse. Biochim. Biophys. Acta. 369:222–233 [PubMed] [Google Scholar]
- Lingwood C.A., Nutikka A. 1994. A novel chemical procedure for the selective removal of nonreducing terminal N-acetyl hexosamine residues from glycolipids. Anal. Biochem. 217:119–123 10.1006/abio.1994.1091 [DOI] [PubMed] [Google Scholar]
- Lorizate M., Brügger B., Akiyama H., Glass B., Müller B., Anderluh G., Wieland F.T., Kräusslich H.G. 2009. Probing HIV-1 membrane liquid order by Laurdan staining reveals producer cell-dependent differences. J. Biol. Chem. 284:22238–22247 10.1074/jbc.M109.029256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meder D., Moreno M.J., Verkade P., Vaz W.L., Simons K. 2006. Phase coexistence and connectivity in the apical membrane of polarized epithelial cells. Proc. Natl. Acad. Sci. USA. 103:329–334 10.1073/pnas.0509885103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J., Schelhaas M., Helenius A. 2010. Virus entry by endocytosis. Annu. Rev. Biochem. 79:803–833 10.1146/annurev-biochem-060208-104626 [DOI] [PubMed] [Google Scholar]
- Mishra R., Grzybek M., Niki T., Hirashima M., Simons K. 2010. Galectin-9 trafficking regulates apical-basal polarity in Madin-Darby canine kidney epithelial cells. Proc. Natl. Acad. Sci. USA. 107:17633–17638 10.1073/pnas.1012424107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemelä P.S., Miettinen M.S., Monticelli L., Hammaren H., Bjelkmar P., Murtola T., Lindahl E., Vattulainen I. 2010. Membrane proteins diffuse as dynamic complexes with lipids. J. Am. Chem. Soc. 132:7574–7575 10.1021/ja101481b [DOI] [PubMed] [Google Scholar]
- Ojakian G.K., Schwimmer R. 1988. The polarized distribution of an apical cell surface glycoprotein is maintained by interactions with the cytoskeleton of Madin-Darby canine kidney cells. J. Cell Biol. 107:2377–2387 10.1083/jcb.107.6.2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paltauf F. 1994. Ether lipids in biomembranes. Chem. Phys. Lipids. 74:101–139 10.1016/0009-3084(94)90054-X [DOI] [PubMed] [Google Scholar]
- Papirmesiter B., Mallette M.F. 1955. The isolation and some properties of the Forssman hapten from sheep erythrocytes. Arch. Biochem. Biophys. 57:94–105 10.1016/0003-9861(55)90181-8 [DOI] [PubMed] [Google Scholar]
- Pike L.J., Han X., Chung K.N., Gross R.W. 2002. Lipid rafts are enriched in arachidonic acid and plasmenylethanolamine and their composition is independent of caveolin-1 expression: a quantitative electrospray ionization/mass spectrometric analysis. Biochemistry. 41:2075–2088 10.1021/bi0156557 [DOI] [PubMed] [Google Scholar]
- Polozov I.V., Bezrukov L., Gawrisch K., Zimmerberg J. 2008. Progressive ordering with decreasing temperature of the phospholipids of influenza virus. Nat. Chem. Biol. 4:248–255 10.1038/nchembio.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, editors. 2010. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.r-project.org (accessed December 21, 2011)
- Riento K., Galli T., Jansson S., Ehnholm C., Lehtonen E., Olkkonen V.M. 1998. Interaction of Munc-18-2 with syntaxin 3 controls the association of apical SNAREs in epithelial cells. J. Cell Sci. 111:2681–2688 [DOI] [PubMed] [Google Scholar]
- Rodriguez Boulan E., Sabatini D.D. 1978. Asymmetric budding of viruses in epithelial monlayers: a model system for study of epithelial polarity. Proc. Natl. Acad. Sci. USA. 75:5071–5075 10.1073/pnas.75.10.5071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman J.S., Jing X., Leser G.P., Lamb R.A. 2010. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 142:902–913 10.1016/j.cell.2010.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J.E., Tsai D.K., Dawidowicz E.A., Lenard J. 1976. Transbilayer phospholipid asymmetry and its maintenance in the membrane of influenza virus. Biochemistry. 15:2361–2370 10.1021/bi00656a018 [DOI] [PubMed] [Google Scholar]
- Sampaio J.L., Gerl M.J., Klose C., Ejsing C.S., Beug H., Simons K., Shevchenko A. 2011. Membrane lipidome of an epithelial cell line. Proc. Natl. Acad. Sci. USA. 108:1903–1907 10.1073/pnas.1019267108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhoff R., Brügger B., Jeckel D., Lehmann W.D., Wieland F.T. 1999. Determination of cholesterol at the low picomole level by nano-electrospray ionization tandem mass spectrometry. J. Lipid Res. 40:126–132 [PubMed] [Google Scholar]
- Scheiffele P., Rietveld A., Wilk T., Simons K. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274:2038–2044 10.1074/jbc.274.4.2038 [DOI] [PubMed] [Google Scholar]
- Schmitt A.P., Lamb R.A. 2005. Influenza virus assembly and budding at the viral budozone. Adv. Virus Res. 64:383–416 10.1016/S0065-3527(05)64012-2 [DOI] [PubMed] [Google Scholar]
- Schroeder C., Heider H., Möncke-Buchner E., Lin T.I. 2005. The influenza virus ion channel and maturation cofactor M2 is a cholesterol-binding protein. Eur. Biophys. J. 34:52–66 10.1007/s00249-004-0424-1 [DOI] [PubMed] [Google Scholar]
- Schuck S., Simons K. 2004. Polarized sorting in epithelial cells: raft clustering and the biogenesis of the apical membrane. J. Cell Sci. 117:5955–5964 10.1242/jcs.01596 [DOI] [PubMed] [Google Scholar]
- Shvartsman D.E., Kotler M., Tall R.D., Roth M.G., Henis Y.I. 2003. Differently anchored influenza hemagglutinin mutants display distinct interaction dynamics with mutual rafts. J. Cell Biol. 163:879–888 10.1083/jcb.200308142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui B., Hakomori S. 1971. A revised structure for the Forssman glycolipid hapten. J. Biol. Chem. 246:5766–5769 [PubMed] [Google Scholar]
- Simons K., van Meer G. 1988. Lipid sorting in epithelial cells. Biochemistry. 27:6197–6202 10.1021/bi00417a001 [DOI] [PubMed] [Google Scholar]
- Simons K., Virta H. 1987. Perforated MDCK cells support intracellular transport. EMBO J. 6:2241–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G., Simons K. 1986. The function of tight junctions in maintaining differences in lipid composition between the apical and the basolateral cell surface domains of MDCK cells. EMBO J. 5:1455–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G., Voelker D.R., Feigenson G.W. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9:112–124 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit M., Thaa B. 2011. Association of influenza virus proteins with membrane rafts. Adv. Virol. 2011:1–14 10.1155/2011/370606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandinger-Ness A., Bennett M.K., Antony C., Simons K. 1990. Distinct transport vesicles mediate the delivery of plasma membrane proteins to the apical and basolateral domains of MDCK cells. J. Cell Biol. 111:987–1000 10.1083/jcb.111.3.987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech T., Ejsing C.S., Gaus K., de Wet B., Shevchenko A., Simons K., Harder T. 2009. Accumulation of raft lipids in T-cell plasma membrane domains engaged in TCR signalling. EMBO J. 28:466–476 10.1038/emboj.2009.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Pekosz A., Lamb R.A. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634–4644 10.1128/JVI.74.10.4634-4644.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]