Abstract
The Ras superfamily is a fascinating example of functional diversification in the context of a preserved structural framework and a prototypic GTP binding site. Thanks to the availability of complete genome sequences of species representing important evolutionary branch points, we have analyzed the composition and organization of this superfamily at a greater level than was previously possible. Phylogenetic analysis of gene families at the organism and sequence level revealed complex relationships between the evolution of this protein superfamily sequence and the acquisition of distinct cellular functions. Together with advances in computational methods and structural studies, the sequence information has helped to identify features important for the recognition of molecular partners and the functional specialization of different members of the Ras superfamily.
Introduction
Both unicellular and multicellular organisms respond to cues expressed by other cells. In metazoans, studies of intercellular signaling during development have revealed the existence of highly conserved signaling pathways. Cellular organization and signaling is heavily influenced by the Ras superfamily of small GTP-binding proteins, which maintain a structurally and mechanistically preserved GTP-binding core despite considerable divergence in sequence and function. These GTP-binding proteins share a common enzymatic activity, producing GDP by the hydrolysis of GTP.
Ras superfamily signaling is dependent on the binding of specific effectors. Thus, minor modifications in sequence, structure, and/or cellular regulation of members of the superfamily will affect binding to regulators and consequently cell signaling. Accordingly, an important goal in studies of Ras superfamily signaling is to identify the determinants of these specific associations. The relationships between Ras superfamily proteins and their effectors have been analyzed using distinct phylogenetic approaches (Li et al., 2004; Jiang and Ramachandran, 2006; Boureux et al., 2007; Langsley et al., 2008; Mackiewicz and Wyroba, 2009; van Dam et al., 2009). To elucidate the influence of sequence evolution on Ras superfamily signaling, we have analyzed complete (or almost complete) genomes representing crucial evolutionary time points, focusing on the phylogenetic inferences gained from both species and protein trees. Using this information, we have generated a representative tree reflecting the evolutionary history of the Ras superfamily, from which we can classify the human Ras sequences. By adopting this approach to study the functional specificity of different superfamily members, we have been able to integrate the mechanistic information derived from these species and protein trees within a structural framework.
To facilitate the reading of this work we have used the following nomenclature: Ras superfamily refers to the highest organizational level that includes different protein families. Ran, Ras, Rab, Rho, and Arf refer to the specific protein families. RAS, RHO, etc. (capitalized) denote specific proteins. The G-domain refers to the structural domain common to proteins of the Ras superfamily.
The Ras superfamily
The Ras superfamily is divided into five major families: Ras, Rho, Arf/Sar, Ran, and Rab. Members of the Ras family function as signaling nodes that are activated by diverse extracellular stimuli and that regulate intracellular signaling. This signaling ultimately controls gene transcription, which in turn influences fundamental processes such as cell growth and differentiation. The human oncogenic members of the Ras family have been reviewed extensively (Karnoub and Weinberg, 2008), and in general they regulate cell proliferation, differentiation, morphology, and apoptosis. The Rho family is involved in signaling networks that regulate actin, cell cycle progression, and gene expression. In addition to cytoskeletal organization (Heasman and Ridley, 2008) and cell polarity (Park and Bi, 2007), members of the Rho family have recently been implicated in hematopoiesis (Mulloy et al., 2010), particularly the RAC protein that is involved in both canonical and noncanonical wnt signaling (Schlessinger et al., 2009). The Rab family participates in vesicular cargo trafficking and it is by far the largest family of the Ras superfamily. Gene duplication has resulted in a large expansion of this protein family, as witnessed by the presence of duplicates in all vertebrate genomes. Rab family proteins regulate intracellular vesicular transport and the trafficking of proteins between different organelles via endocytotic and secretory pathways (Zerial and McBride, 2001). These proteins facilitate budding from the donor compartment, transport to acceptors, vesicle fusion, and cargo release. A key feature of the Rab family is the distinct intracellular distribution of its different members (Stenmark, 2009). By contrast, only one member of the Ran family is found in all eukaryotic lineages, with the exception of plants, which contain several copies. RAN proteins are the most abundant in the cell and they are involved in nuclear transport. Finally, the Arf family of proteins comprises the most divergent proteins, which, like Rab family proteins, are involved in vesicle trafficking (Wennerberg et al., 2005). These proteins signal through a wide range of effectors, including coat complexes (COP, AP-1, and AP-3) and lipid-modifying enzymes (PLD1, phosphatidylinositol 4,5-kinase, and phosphatidylinositol 4-kinase).
Phylogeny of Ras superfamily proteins
The most recent phylogenetic reconstruction of the Ras superfamily was based on sequences obtained from the complete draft of the human genome (Wennerberg et al., 2005). The resulting tree confirmed the general organization of five families (Ras, Rab, Rho, Arf/Sar, and Ran) and pointed to the Ras family as the root of the superfamily. Previous comparisons between human, fly, yeast (Garcia-Ranea and Valencia, 1998), and plant species revealed a similar organizational structure (Li et al., 2004; Jiang and Ramachandran, 2006). However, the recent sequencing of the complete genomes of additional species now enables us to reanalyze the Ras superfamily over a broader phylogenetic range, thereby increasing the likelihood of tracing the origin of the superfamily and correctly classifying sequences that are otherwise difficult to handle. Indeed, analyzing large sets of orthologous sequences is commonly recognized as the best strategy to improve the quality of phylogenetic analysis (Nei and Kumar, 2000).
The second important reason to reassess the organization of the Ras superfamily is the recent availability of novel techniques to build multiple sequence alignments (MSAs) and trees (Kemena and Notredame, 2009), key elements in phylogenetic reconstruction (Phillips et al., 2000).
The procedure.
The generation of accurate MSAs is a key step in the tree-building process, which is based on estimating the similarities between sequences. The quality of multiple alignments is highly dependent upon the degree of similarity of the sequences to be aligned. Although standard methods work reasonably well when the sequence similarity is high (over 40%), very divergent sequences are difficult to align and the MSAs often contain errors (Kemena and Notredame, 2009). Thus, we used a well-established method to construct MSAs and corroborated the results using two additional independent approaches. The main alignment of 919 G-domains (Table S1) was built using the most recent version of HMMER (HMMERV3.0; Eddy, 2009) and it underwent detailed manual curation to detect incorrectly aligned regions. A profile was constructed that contained the statistical features of the amino acids occupying each position in a G-domain seed alignment that was derived from several Ras superfamily proteins obtained from the PFAM database (identifier PF00071; Finn et al., 2010). This profile was used as a template to align the other Ras superfamily sequences, optimizing their correspondence through the probability of each amino acid type being located at each position in the profile. The resulting alignment was compared with alignments obtained using two other methods known to perform reliably in cases of poor sequence similarity (MAFFT: Katoh et al., 2009; and T-COFFEE: Notredame, 2010). These methods confirmed the essential aspects of the alignment (unpublished data).
It should be noted that although various members of the Ras superfamily contain additional protein domains (e.g., BTB domains, N-terminal anchoring regions; see Figs. S2 and S5), our phylogenetic analysis corresponded exclusively to the G-domain, which contains the basic functional and historical core that is common to the superfamily.
The MSA was used as the basis for the phylogenetic analysis. Phylogenetic reconstruction based on protein sequences presents significant challenges that remain to be fully solved (Cavalli-Sforza and Edwards, 1967; Page and Holmes, 1999). Currently, the best approach involves the integration of thousands of trees using an appropriate statistical framework capable of handling the associated probabilities (based on Bayesian statistics and inference; Holder and Lewis, 2003; Ronquist and Huelsenbeck, 2003; Lartillot et al., 2007). The final tree represents the consensus of thousands of carefully chosen independent trees obtained after detailed matching of the probabilities of multiple combinations of branches. This final tree is the most probable representation of the evolutionary history from a statistical viewpoint (Holder and Lewis, 2003).
The statistical properties derived from evaluating thousands of alternative trees are reflected in the values associated with the tree branches. These values represent the statistical confidence in the grouping of the sequences under that branch point, i.e., the probability of that particular grouping of sequences being correct.
The trees presented were generated with MrBAYES3, the most recent implementation of tree-building methods based on Bayesian statistics (Ronquist and Huelsenbeck, 2003) and that which is considered to be the best in the field (Hall, 2005; Gaucher et al., 2010). The downside of the increased accuracy of these new methods is their high computational demands. In the case of the Ras superfamily in particular, building trees from a starting alignment of more than 900 divergent sequences is not feasible, even for large supercomputers (Wang et al., 2011). Thus, we used an alternative procedure that selects key organisms and representative sequences to build independent trees for each of the five distinct families of the Ras superfamily (Ras, Rho, Rab, Ran, and Arf).
Compiling the Ras superfamily.
The current study is based on Ras superfamily proteins directly related to the human Ras proteins, as compiled by Wennerberg et al. (2005). The updated human Ras superfamily contains 167 human proteins: 39 Ras proteins, 30 Arfs, 22 Rhos, 65 Rabs, and 1 Ran family sequence (Table S2). This list includes 10 “unclassified” sequences and for 5 of these sequences, there is only evidence that they exist at the transcriptional level (Table S2, listed as “Unclassified”).
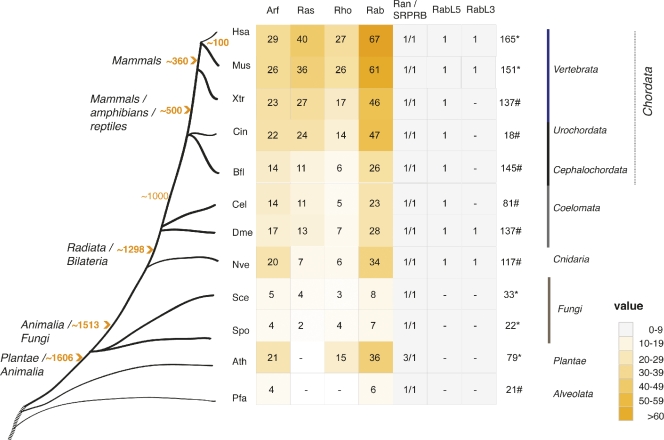
Orthologous sequences correspond to genes separated by species divergence, as opposed to paralogous sequences that are generated by gene duplication. Using the dedicated InParanoid resource (v.4.0; Ostlund et al., 2010), we identified a total of 766 sequences from 11 organisms (excluding human sequences) that correspond to orthologues of the 167 human proteins in the Ras proteins superfamily (orthologues were obtained from various databases, the correspondence between identifiers is given in Tables S3 and S4). These 11 species were selected based on their correspondence to relevant moments in eukaryotic evolution (Fig. 1). Important speciation events were represented by the inclusion of Ras superfamily sequences from the corresponding genomes (Plantae-Animalia and Radiata-Bilateria by A. thaliana and N. vectensis, respectively), or the different Chordata lineages, represented by ascidians (C. intestinalis) and lancelets (B. floridae). Well-annotated genomes are available for some of the species included (e.g., H. sapiens, M. musculus, S. cerevisiae, S. pombe, A. thaliana, D. melanogaster, and C. elegans), whereas for others the genome remains poorly annotated (e.g., P. falciparum) or is only available in a draft format (e.g., N. vectensis, C. intestinalis, B. floridae, and X. tropicalis).
Figure 1.
The orthologues of the human Ras superfamily members in 11 proteomes. Heat map colors indicate the number of the orthologues corresponding to human sequences in a particular species. Numbers inside the boxes indicate the number of orthologous human sequences in the Ras superfamily. Dashes inside the boxes indicate the absence of proteins. Orange numbers in the tree indicate millions of years according to a recently revised scale (Hedges et al., 2004) and the time line is an approximate scale for the purposes of illustration. Arrowheads point to important splits that occurred in the course of evolution. The classical families are represented and for the purpose of clarity, the Ran family and the “unclassified” sequences including SRPRBs, RABL5, and RABL3 proteins are shown independently. The MIRO and RAYL proteins are included in the Rho family and the RABL2 proteins (one per organism) in the Rab family. Numbers on the right of the table indicate G-domain–containing proteins extracted from the PFAM database, therefore some discrepancies are expected due to variability in the synchronization of PFAM and Uniprot databases. Asterisk indicates well-annotated sequences in complete genomes and the associated number reflects the Ras superfamily sequences obtained from PFAM. “#” indicates the presence of the RAS domain in either draft genomes or complete but poorly annotated sequences, and the associated number represents the sequences (fragments have been removed) extracted from PFAM (Finn et al., 2010) in which the G-domain has been identified. Plants: Ath (Arabidopsis thaliana); Alveolata: Pfa (Plasmodium falciparum); Fungi: Spo (Schizosaccharomyces pombe); Yeast: Sce (Saccharomyces cerevisiae); Radiate: Nve (Nematostella vectensis); Worm: Cel (Caenorhabditis elegans); Fly: Dme (Drosophila melanogaster); Lanceolet: Bfl (Brachiostoma floridae, protochordata); Ascidian: Cin (Ciona intestinalis, urochordata); Xtr (Xenopus tropicalis); Human: Hsa (Homo sapiens); and Mouse: Mus (Mus musculus).
The set of orthologous sequences, along with the estimated point in evolution at which they have diverged, is depicted in Fig. 1. The Ras family is entirely absent from plants (A. thaliana), in which the remaining subfamilies are the signaling members of the superfamily (Yang, 2002), while no Rho family orthologues are found in alveolates (P. falciparum). Because our study of the Ras superfamily uses human sequences to retrieve the corresponding orthologues in other species, sequences from these species that are not present in humans may not have been included. Although ascidians would be expected to have a similar number of RAS proteins as humans, based on phylogenetic estimates, there is a noticeable decrease in the number of Ras superfamily orthologues for this organism, probably due to the loss of ancestral genes (Hughes and Friedman, 2005). Similar findings are obtained in coelomates and cnidarians as fewer orthologues are found than in the sea anemone and N. vectensis, although more orthologues are detected for the cnidarian than in the coelomate species. This finding is not unexpected, as gene content and genomic structure has been preserved between N. vectensis and vertebrates (Putnam et al., 2007), whereas extensive gene loss has occurred in the fruit fly and nematodes (Technau et al., 2005).
In some species additional gene-duplication events have produced an accumulation of paralogous sequences, which results in variation between species in terms of the numbers of each Ras superfamily member, as evident in the corresponding family trees (see Figs. S1–S5). For example, three copies of the Ran family sequences were detected in A. thaliana whereas only one was found in the other species analyzed (Figs. 1 and 2; Fig. S1).
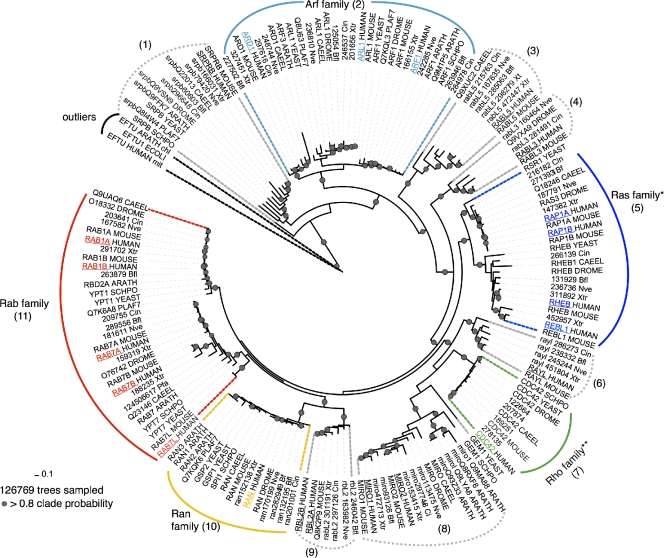
Figure 2.
Consensus phylogenetic tree of selected Ras superfamily members rooted with outliers. The 165 proteins selected after careful individual analyses (see Figs. S1–S5) of species-trees and gene-trees covering the Ras superfamily sequences, plus three sequences (human mitochondrial, plant chloroplast, and bacterial) of representative Elongation factor Tu (a remote homologue of Ras superfamily) used to root the tree, were aligned to the G-domain (see main text). The tree is a consensus of more than 126,769 sampled trees with their associated probabilities. The numbers in brackets indicate stable groups; *, absent in Plants and alveolates; **, absent in alveolates. The founder members are the SRPRB group (1) and the Arf family (2). RABL5 (3; Wu et al., 2002) and RABL3 (4) appear at the basal branches of the Ras family (most likely as founders). This phylogeny proposes SRPRB and the Arf family to be the founder members of the classical Ras superfamily. Underlined names indicate the 22 human sequences. Plants: ARATH (Arabidopsis thaliana); Alveolata: PLAF7 (Plasmodium falciparum); Fungi: SCHPO (Schizosaccharomyces pombe); Yeast: Sce (Saccharomyces cerevisiae); Radiate: Nve (Nematostella vectensis); Worm: CAEEL (Caenorhabditis elegans); Fly: DROME (Drosophila melanogaster); Lanceolet: Bfl (Brachiostoma floridae, protochordata); Ascidian: Cin (Ciona intestinalis, urochordata); Xtr (Xenopus tropicalis); Human (Homo sapiens); and Mouse (Mus musculus).
Rho family proteins expanded extraordinarily in plants (Yang, 2002), and although plant RACs are homologues of RAC, RHO, and CDC42 (Fig. S2), the expansion of RAC in plants after speciation has resulted in the generation of a larger number of RAC proteins (RAC1–RAC11) than in other organisms. The ancestral duplications of RAC in fungi/metazoans led to the appearance of CDC42, which controls cell polarity, and RHO, which is implicated in cytokinesis (Jaffe and Hall, 2005). The CDC42 protein, which promotes the formation of actin microspikes and filopodia, is conserved in all the lineages except plants. Interestingly, the absence of Rho genes in alveolates suggests that other proteins fulfill its role in cell polarity and cytokinesis.
The gene duplication that generated the Ras family proteins in vertebrate genomes (H-Ras, K-Ras; group 7a in Fig. S3) is another example of how variation in the number of Ras superfamily proteins arises (Fig. S3). Indeed, although they are present in Xenopus, the evolutionarily more ancestral genomes only contain one copy (LET60 in C. elegans and RAS1 in D. melanogaster) that is involved in embryogenesis (Ezer et al., 1994). Fungal orthologous sequences (Fig. S3) are only found for RHEB (group 12), RAP (group 4), and RRAS (group 8). Further duplications of these sequences after speciation yielded the MRAS and KRAS groups. Together, these data are consistent with major gene duplication in vertebrates (Kondrashov et al., 2002).
The vertebrate branch of the Rab protein family has expanded considerably (Fig. S4; see also Mackiewicz and Wyroba, 2009) and significantly. We found representatives of each of the different groups of Rab proteins in all lineages, indicating that the appearance of this family was an important evolutionary event. The implication of Rab genes in a complex network of vesicular trafficking events suggests a relationship with multi-cellularity that merits further investigation. The Arf family of proteins (Kahn et al., 2006) is the most divergent member of the superfamily and it is associated with recurrent duplication events (Fig. S5).
Representative phylogenetic tree of the Ras superfamily.
As it was not feasible to generate a full tree with all the sequences orthologous to the proteins of the human Ras superfamily, we generated independent trees for each of the protein families using the sequences from the 12 proteomes (see Figs. S1–S5) and for each of the species. This information was then used to select the sequences that best represented the diversity of species and branches in the tree.
Specific criteria for sequence inclusion were applied to select stable and representative groups from each tree. Thus, information regarding the function of the sequences selected was necessary, as well as that related to any clear orthologues in each of the species analyzed. The selection aimed to respect the variability of the sequences included in each tree (see Figs. S1–S5, which show the groups selected to build the representative tree). For instance, as the Ras family is not represented in plants and alveolates, we selected groups with at least a yeast homologue. Moreover, because the RHEB proteins (Fig. S3, group 12) constitute the most basal branch present in all the organisms of interest, and RAP1 proteins (Fig. S3, group 4) correspond to the most basal group with a yeast orthologue, these two groups were chosen to represent the Ras family in the representative tree.
Given the large number of sequences in the Rab family, we selected two stable groups that covered the phylogeny of the whole family (Fig. S4, stars). RAB7 was chosen on the basis of its well-characterized involvement in Golgi late endosomal transport. RAB7 lineages emerged before the divergence of plants (Mackiewicz and Wyroba, 2009) and this protein is present in amebozoans and ciliates. Analogously, RAB1 (Fig. S4, stars) was selected due to its position in the tree in all species. All the Ran family sequences were included. For the Arf family (Fig. S5, stars) we included the ARD-1 (also known as TRIM23, an Arf-related protein involved in ubiquitination; Mishima et al., 1993) and SRPRB (signal recognition particle receptor subunit β) proteins. These groups cover the phylogenetic range of this family.
Difficult sequences (i.e., those not classified by Wennerberg et al., 2005) were also included for those species in which orthologues corresponding to human sequences could be identified. In addition, we included sequences from the elongation factor Tu (EFTU) family, a distant relative with a G-domain that was used as a guide to situate the root of the Ras superfamily tree. The inclusion of distant sequences (known as outgroups) to define the tree is an accepted procedure to trace the origin of protein families (Nei and Kumar, 2000).
The superfamily tree (Fig. 2) includes 165 sequences, of which 22 are human (Fig. 2, underlined sequences). The remaining sequences belong to 11 species, and 3 additional non-Ras superfamily sequences are included. The first observation is the clear separation of the five main families, which group together with a high degree of confidence. Indeed, of the thousands of independent trees constructed using this procedure, over 80% exhibited the same general tree structure. The clear separation between families is a distinctive feature of the Ras superfamily that is not commonly observed in other large superfamilies, such as protein kinases.
The inclusion of an outgroup (in our case, the EFTU proteins) appears to clarify the overall topology of the superfamily. This new phylogeny places the SRPRBs (Fig. 2, group 1) as the founder members of the Ras superfamily. There is significant support for this structure, even though it differs from that described in previous analyses based on human Ras superfamily sequences alone (Wennerberg et al., 2005). The SRPRB protein is located at membranes and it is an essential component of the signal recognition receptor, which ensures that nascent secretory proteins are targeted to the endoplasmic reticulum (ER) membrane system. This suggests that the original function of the superfamily is related to specific signaling events involving membrane structures.
The human-only Ras superfamily phylogenetic tree.
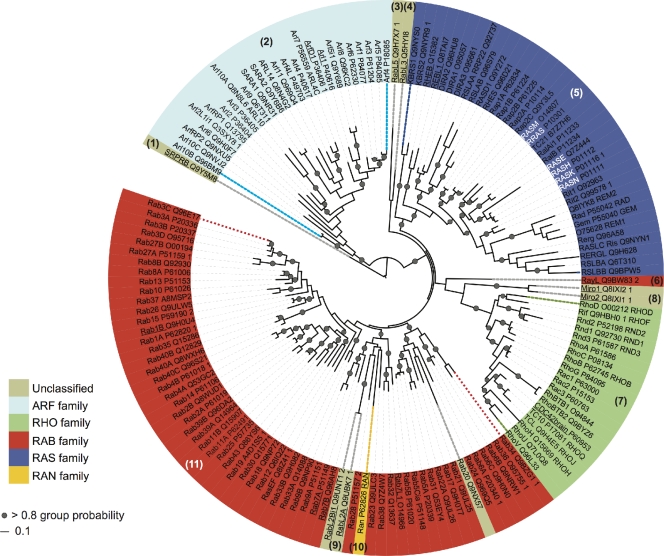
Our most up-to-date representation of the human sequences tree is shown in Fig. 3. As discussed in the previous paragraph, generating an accurate tree of the human sequences requires that their relationship with sequences from other species be taken into account. Thus, we used the topology of the complete species tree represented in Fig. 2 to organize the rest of the human sequences. Additionally, SRPRBs were used to root the human tree based on the information from the representative tree.
Figure 3.
Phylogenetic tree of the human Ras superfamily. Of the 167 sequences defined (Table S2), some proteins are identical in the G-domain while exhibiting differences in other regions of the protein. Thus, identical sequences were removed before performing the alignments. The tree contains 151 sequences that correspond to bona fide unique protein sequences (Table S2) that were aligned with the G-domain profile (PF00071, also RAS) to define the domain boundaries. The alignment was used as the input to generate the probabilistic phylogeny using a Bayesian inference (Ronquist and Huelsenbeck, 2003). Thus, 29,340 trees were sampled and the associated confidence values (group probability values) were obtained for each group. The numbers in brackets indicate the equivalent group numbering, as in Fig. 2. The background colors indicate the original classification (Wennerberg et al., 2005): blue, Ras family; green, Rho; red, Rab; cyan, Arf; and yellow, Ran. Unclassified members are shown in beige. Gray circles indicate group probabilities >80% (confidence value assigned to a group and expressed as a percentage). The white font indicates the archetypal Ras family proteins and the names underlined indicate the human sequences from Fig. 2. The tree was visualized with iTOL (Letunic and Bork, 2007).
Of the 167 defined sequences (Table S2), some genes present alternative splice variants with identical sequences in their G-domains. As we only compared the precise G-domain region, these identical sequences were removed before alignment. The remaining 151 human Ras superfamily sequences were aligned and a phylogenetic analysis was performed following the procedure described in the previous section. The main families were divided into stable groups, as evident by the high confidence values (Fig. 3, gray dots). It is clear that this superfamily has experienced extensive gene duplication, with the Rab family representing the most abundant family in humans (Fig. 3, red background). Previous phylogenetic analyses (Schwartz et al., 2007) divided the Rab family into eight functional groups (Pereira-Leal and Seabra, 2001), whereas a more recent study proposed nine groups (Stenmark, 2009). Incorporating our data with the corresponding subcellular localization described previously (Stenmark, 2009), we can generate a phylogenetic distribution of the Rab repertoire, as shown in Fig. S4. The colored branches represent the functional family to which each particular protein belongs (Stenmark, 2009), and the numbers in brackets represents the previous groupings (Schwartz et al., 2007), in which 14 divisions were established. We found some discrepancies (Fig. S4, group numbers labeled with an asterisk) with the earlier phylogeny proposed (Schwartz et al., 2007), probably due to the inclusion of additional species in our analysis. For instance, group number 10 (Fig. S4, beige branches) contains RAB18, which is traditionally assigned to an independent family (Stenmark, 2009). However, in our analyses RAB18 was grouped within Stenmark’s “RAB3” group (Fig. S4, red branches). A similar situation occurred with “RAB1” (dark brown) and “Rab 28” (dark orange).
A previous phylogenetic analysis of the Arf family described 11 groups: the Arfs, ARL1-6, ARL-8, ARL10/11, ARFRP, and SAR (Li et al., 2004). However, this study included none of the ARD proteins (Arf-like proteins also known as the TRIM23 group; Kahn et al., 2006). We expanded the phylogenetic analysis to include the ARD (TRIM23) proteins, which in our analysis had a high probability of grouping with the ARF proteins (a significant confidence value of the corresponding tree branches). Interestingly, this group contains multidomain proteins with a Ring domain (SMART), a protein interaction domain that shows E3 ubiquitin–protein ligase activity, Zf_boxes that are also protein interaction domains at the N-terminal region (PFAM PF00643), and an ARF domain similarity region in the C-terminal region. The presence of these domains may point to specific functions related to ubiquitination and binding to targets such as DNA, RNA, and proteins.
The analysis of sequences from species not previously included in other studies allowed us to reassess the earlier classifications. For instance, although the NKIRAS (KBRAS) protein (Fig. S3, group 1) was believed to be human specific (Jiang and Ramachandran, 2006), our results indicate that this protein is present in all the eukaryotic lineages except fungi and Plasmodium.
Difficult-to-classify sequences.
The overall classification of the family including information from divergent species may help to elucidate the role of some of the less well-known sequences in the superfamily. For example, the analysis reveals a putative relationship between the MIRO and RAYL sequences within the Rho family, despite the fact that MIRO proteins are considered to be an independent family as they regulate mitochondrial rather than cytoskeletal dynamics (Colicelli, 2004). The position of these proteins within the Rho family (Fig. 2, group 8) suggests that some functional diversification (sub-functionalization) has occurred, although this may also point to a common original mode of action that was later co-opted to perform distinct cellular roles.
The present findings provide some insight into the potential functions of some of the superfamily proteins, for instance Rab-like proteins (named “like” because they are similar to Rab) that lack the conventional C-terminal modification site. Although the role of the RABL5 protein remains a mystery, RAYL protein is thought to be a cell cycle–related protein (Qin et al., 2007), consistent with its phylogenic placement with CDC42 proteins (Fig. 2, group 6). RABL2A/B (Kramer et al., 2010) are Rab-like proteins mapping to the subtelomeric region of chromosome 22q13 (Wong et al., 1999), suggesting that in humans at least, duplicate genes actively express proteins of as yet unknown function in telomeric regions. Curiously, in our phylogeny these proteins grouped with the Ran protein family (Fig. 2, groups 10 and 9). Because the RABL2 proteins are metazoan specific, they may have originated from duplications of Ran proteins after speciation events. The function of RABL3 is also unknown, although it was recently implicated in regulating the proliferation and motility of human cancer cells (Li et al., 2010). Future analysis of additional sequences may clarify certain aspects of this classification.
A particularly interesting case is that of RAG proteins (Sekiguchi et al., 2001) that are believed to be part of the Ras superfamily based on the presence of a GTP domain. Although their structure and overall sequence similarity shows that they contain a G-domain, we found that the lack of conservation and the unusual composition of the GTP-binding sites, otherwise conserved in the superfamily, sheds doubt on their classification as a family of the Ras superfamily. Future studies of this group of proteins using a larger set of sequences from new complete genomes will be required to confirm their classification.
Functional specificity of the proteins in the Ras superfamily
The large functional diversity of the Ras superfamily is perplexing. The conserved 3D structure of the G-domain that is common to the entire superfamily allows these proteins to preserve large structural similarity and common biochemical properties while they recognize their individual binding partners with remarkable affinity and specificity (Colicelli, 2004; Wennerberg et al., 2005). The promiscuity and diversity in this superfamily is illustrated by the multiple upstream (regulators) and downstream proteins to which Ras superfamily proteins bind (Bishop and Hall, 2000; Karnoub and Weinberg, 2008: Table S5). The list of these interacting proteins is continually growing and a plethora of functions have been attributed to both effectors and regulators. Moreover, some GTPases share effectors despite performing distinct functions, leading to another level of regulation within this family (Kiel et al., 2007; Barnekow et al., 2009).
The GTP-binding site is made up of a core of essential residues that also participate in the conformational changes linking GTPase activity to effector binding. The specific distribution of these residues and the differential sequence conservation within families determines the specificity of the association between Ras superfamily proteins and their effectors, interactions that ultimately determine the biological activity of the protein. It is important to note that although structural and sequence-specific features are clearly correlated with function, other factors work together to influence the network of Ras superfamily interactions, like gene expression of the proteins, and the regulation and acquisition of domains that determine subcellular localization (Rodriguez-Viciana et al., 2004; Goldfinger, 2008).
We compared and contrasted the families in the Ras superfamily to identify the residues in the G-domain and to determine any differences that may underlie their specific interactions with effectors. These positions are considered specificity-determining positions (SDPs), as they provide information regarding the branching of the phylogenetic tree (del Sol et al., 2003), also known as tree determinants, and they are associated with ligand-binding sites and protein interaction regions (Rausell et al., 2010). We analyzed the Ras superfamily to identify family-specific residues, using the complete sequence alignment of 919 G-domains, including representatives from each family of the 12 genomes. This analysis was performed using a recently described unsupervised approach (Rausell et al., 2010) that is based on multiple correspondence analysis. This technique represents the sequences of the alignment as vectors (Fig. S6), and their organization is optimized into groups according to their similarities and differences. The groups of sequences resulting from the association of similar vectors with a k-means algorithm are equivalent to the branches of the phylogenetic tree. The advantage of this procedure is that it allows the characteristic residues (SDPs) that dictate the organization of the sequences into groups to be extracted directly. In this way the MSA is transformed into a catalog of groups of sequences with their associated characteristic amino acid and positions.
For example, the sequences corresponding to the Rho, Rab, and Ran families have a characteristic conserved Asp residue within the G1 motif (position 7 in the alignment, corresponding to residues 13, 16, and 18 for the Rho, Rab, and Ran structures indicated in Table 1, respectively). By contrast, the Arf family has a Leu residue in this motif (position 25 for Arf PDB:1HUR), whereas Ras family sequences are not highly conserved (position 11 for RAS PDB:121P). Completely conserved residues (i.e., those binding GTP) cannot be assigned to any particular group of sequences (for more details on this method, see Rausell et al. [2010]).
Table 1.
The position of SDP residues in the structures of the five Ras superfamily members, and their possible biological and functional implications
| Family PDB chain | Arf 1hurA,B | Ras (Fig. 5) 121pA | Rho 1a2bA | Rab 2folA | Ran 1l2MA |
| Residues in the proximity of the GTP/GDP binding pocket | Ala28, Pro47, Gly69 | Ala11, Val14, Pro34, Thr58, Ala59 | Asp13, Cys16, Pro36 | Asp16, Val19, Leu25, Ser39 | Thr21, Asp18, Ala41, Ala67 |
| Residues involved in protein–protein interactions | Pro47, Phe51, Trp66, Val68, Gly69 | Pro34, Glu37, Leu56, Ala59, Arg68 | Asp13, Cys16, Leu22, Pro36, Phe39, Thr58, Thr60, Ala61, Asp67, Arg70, Arg122 | Val43, Trp62, Ala65, Arg71 | Asp18, Val45, Trp64, Ala67, Gly73, Arg76, Asp91 Lys127 |
| Residues coordinating communication between different lobes | Arg75, Val91, Asp93, Pro131 | Ala65, Val81, Ala83, Ala121 | Asp67, Cys83, Ser85, Arg122 | Arg71, Ala81, Val87, Leu89, Ala126 | Gly73, Met89, Asp91, Lys127 |
| Conserved SDP residues with unknown function | Leu25, Leu34, Trp78, Thr85 | Gly75, Thr20 | Thr77 | Thr64, Thr74 | Val27, Thr66, Ala83 |
Numbers correspond to positions in the PDB structures (see Table S5).
The summary of sequence features is presented in Table 1 and Fig. 4. The analysis of SDPs provided independent evidence for the classification of RABL3, RABL5, RAYL, MIRO, and SRPRB proteins (Tables S7 and S8). Significantly, the SDP pattern of these proteins did not correspond well with the characteristic amino acids in the classical families (Table S8). In particular, the conservation of the RABL3, RABL5, and SRPRB sequences reflects important differences at key positions within the groups to which they are assigned, and this is more consistent with their organization as independent groups and it is in agreement with the rooted phylogenetic tree. By contrast, the low intensity SDP signal in the RAYL and MIRO groups (Fig. S7) suggests that they constitute a peculiar group within the Rho family. Additional genomic information will be required to further study this group.
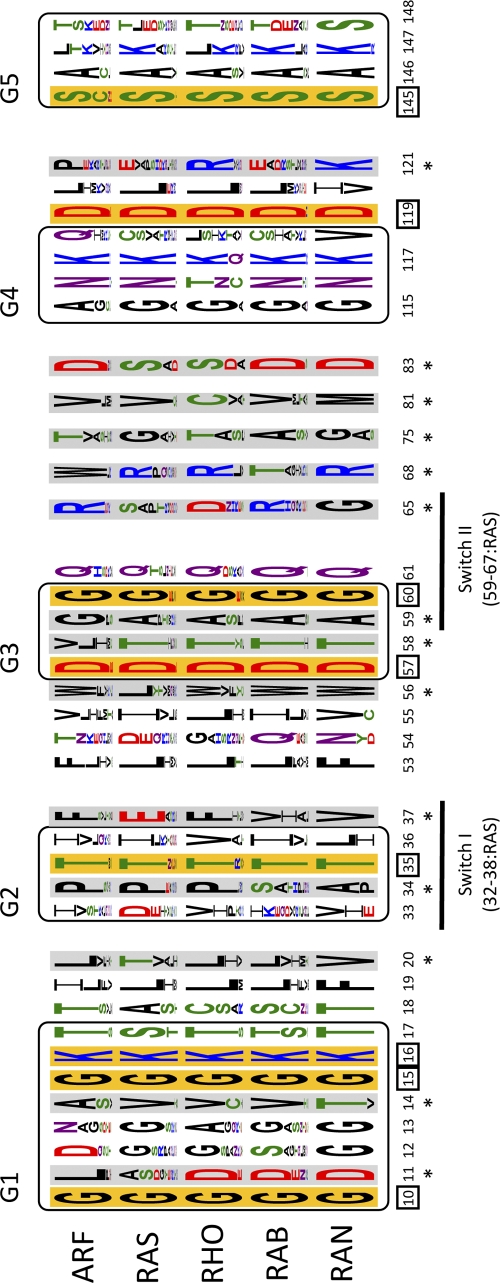
Figure 4.
Specificity-determining positions (SDPs). These residues are displayed in sequence logos corresponding to the different Ras superfamily members and represented in the context of the characteristic G-boxes. The positions are numbered according to the corresponding residue in PDB 121P (H-Ras-1). The SDPs detected according to the classical scenario (the distribution in the classical five subfamilies, Ras, Rab, Ran, Rho, and Arf) are indicated by an asterisk and highlighted against a gray background. The conserved positions are marked with a square and shown on an orange background. The relative size of the amino acid letters in the logos represents the raw frequency for the alignment of the 919 sequences (Table S1), colored as follows: green (polar: S,T,Y,C,Q,N); blue (basic: K,R,H); red (acidic: D,E) and black: G and hydrophobic (A,V,L,I,P,W,F,M). Logos were generated using Weblogo3 (http://weblogo.threeplusone.com; Crooks et al., 2004) and the switch regions are indicated below in the G-boxes.
To further advance our analysis, we focused on the interaction of these SDP residue proteins with the nucleotide-binding pocket, their association with specific binding partners, and their role in communicating between the GTP-binding site and other functional areas of the Ras superfamily (Fig. 5).
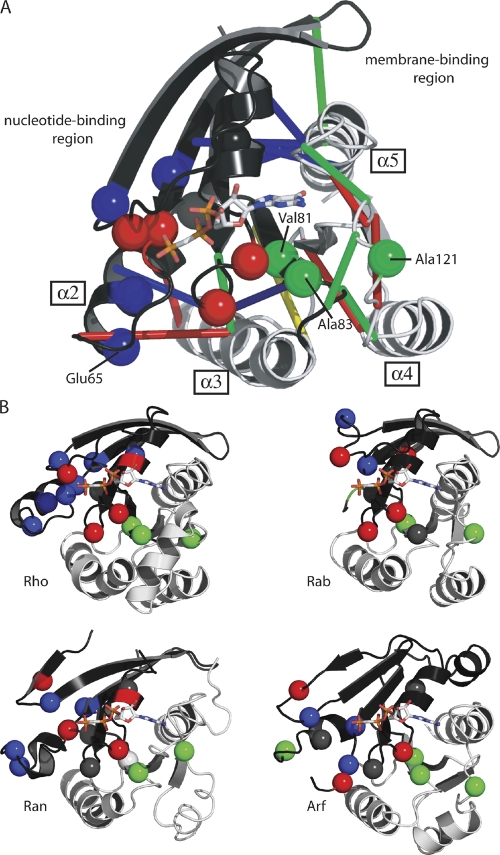
Figure 5.
Location of the SDPs for the different Ras superfamily proteins. (A) SDP residues in the RAS subfamily. The SDPs (Table 1) are shown as spheres. In the RAS protein the three different isoform-specific contacts are indicated by red, blue, and green lines for NRAS, KRAS, and HRAS, respectively. The proposed routes of communication between lobes 1 and 2 linking the nucleotide-binding region with the membrane-binding region are shown in dark gray and pale gray cartoon. (B) SDP residues for the different Ras subfamilies. The SDPs (Table 1) are mapped onto the structure of representative members (Rho, Rab, Ran, and Arf in the four panels). In both panels the colored spheres indicate SDPs mapped into structures. Red: residues in the proximity of the GTP/GDP-binding pocket. Blue: residues involved in protein–protein interaction. Green: SDP residues coordinating the communication between lobes (see explanation in the text). Gray: residues with no identified biological function. Equivalent regions potentially involved in communication between lobes 1 and 2 linking the nucleotide-binding region with the membrane-binding region are shown in dark gray and pale gray cartoon.
SDPs involved in GTP/GDP hydrolysis.
Ras superfamily proteins generally undergo an enzymatic cycle that involves the so-called loaded-spring mechanism, whereby the switch regions relax after release of the hydrolyzed γ-phosphate in the active state, adopting an open-inactive GDP conformation (Vetter and Wittinghofer, 2001). In terms of their 3D structure, the G1–G5 loops form the nucleotide-binding site, with an interface that is responsible for nucleotide specificity and affinity, and that regulates GTP hydrolysis. The SDP residue Val14 reportedly forms hydrogen bonds with the phosphate groups of the nucleotides (Tong et al., 1991). This residue, together with Ala11 and Pro34, is located in regions characterized as hinges that act in the conformational transition between GTP and GDP forms (Díaz et al., 1995, 1997; Futatsugi and Tsuda, 2001), thereby modulating GTP hydrolysis (see Table 1 for the equivalence of these residues in the various families). The residues in switch II (Thr58 and Ala59) may influence nucleotide cycling, in which the movement of neighboring side chains plays a key role, as demonstrated by x-ray diffraction, NMR, spectroscopy, and MD studies (Noé et al., 2005; Gorfe et al., 2008; Lukman et al., 2010). Thus, mutating these residues may alter the structure or dynamics of the system, favoring either active or inactive states, as occurs with oncogenic mutations such as G12V (Gorfe et al., 2008).
SDPs involved in interactions with different binding partners.
Ras superfamily proteins are very specific when transmitting signals to their partners, as illustrated by the extent to which single amino acid changes can alter their individual specificities (Stenmark et al., 1994; Azuma et al., 1999; Bauer et al., 1999; Karnoub et al., 2001). As a general rule, conformational changes induced by nucleotide states are transmitted to the switch I and switch II regions, where they are recognized by the corresponding effectors (Vetter and Wittinghofer, 2001). In addition, the interacting interface is typically comprised of other residues outside the canonical switch regions (Heo and Meyer, 2003; Fuentes and Valencia, 2009), as confirmed by the structures of the Ras–effector complexes (Corbett and Alber, 2001).
To define the SDP residues involved in protein–protein interactions, we inspected the interacting surfaces of a set of complexes within the Ras superfamily using a previously described method (Corbett and Alber, 2001; Biou and Cherfils, 2004; Dvorsky and Ahmadian, 2004; see Table S5 and Fig. S8). It should be noted that the information pertaining to binding surfaces is necessarily incomplete, due to the limited structural characterization of Ras superfamily protein-interacting partner complexes. We considered the static features of the structures, as well as the potential plasticity of neighboring residues (Sprang, 2000). This plasticity of interacting and neighboring residues is important for the binding of Ras superfamily proteins to different partners, and in distinguishing related members within a family (Cherfils et al., 1997).
We analyzed the distribution of SDP residues (Table 1) at the interfaces that interact with both GEF and GAP proteins (Fig. S9). In the case of GAPs, most Ras superfamily proteins interact by means of at least one residue at switch I, which occupies position 85 in the full sequence alignment (Table S6). However, the Rab protein family is an exception as it uses a completely different set of interacting residues, possibly related to other interactions in the context of larger complexes (Goldberg, 1998). Both Rho and Ran families selectively use residues located in their G1 box (residues 13 and 18, respectively), in switch I (residue 39 of RHO), and the β-sheet adjacent to switch II (residue 91 of RAN; Fig. S9).
In the case of binding to GEFs, the binding surface forms an extended patch at the surface and the interactions differ for each family. Residues at position 94 in the full sequence alignment (Table S6) are located at the C terminus of switch I, and they are common to the Arf and Ras families (residues 51 and 37, respectively). Residues at the N terminus of switch II (residue 64 in RAN and 65 in RAB for the PDB entries 1I2M and 2FOL, respectively) form part of the recognition site. The residue at position 85 in the alignment corresponding to residues 47, 34, 36, 39, and 41 for the Arf, Ras, Rho, Rab, and Ran structures, respectively (as indicated in Table 1), and which corresponds to G2 and switch I, is found in the sequences of both Ras and Rho families. The residue at position 219 in the C terminus of the switch II region is shared by proteins of the Ras and Ran families (position 219 in the alignment, corresponding to residues 68 and 76 in Ras and Ran structures, respectively, as indicated in Table 1).
SDPs involved in communication between GTP and membrane-binding regions.
The extension of Arf, Arl, and Sar proteins at the N terminus is required for membrane anchoring (Pasqualato et al., 2002), also serving as a family-specific switch (Goldberg, 1998). Interestingly, recent studies highlighted a more general switch mechanism, involving communication between the membrane-binding domain and the GTP-binding site (Abankwa et al., 2008; Gorfe et al., 2008). Based on sequence variation, two different lobes have been defined in the canonical RAS structure: lobe 1 (comprising the first 86 conserved residues), which lines up the GTP/GDP pocket; and lobe 2 (residues 87–171), which exhibits significant sequence variability and that is associated with Ras anchoring to the membrane (the lobes are shown in dark gray and pale gray in Fig. 5 A). In the 3D structure, different routes of communication between these two regions have been proposed for different isoforms (Gorfe et al., 2008). To characterize residues that mediate the communication between the two lobes, we mapped the proposed GTP–membrane communication routes for the three RAS isoforms onto the canonical G-domain, based on the specific pairwise contacts for each isoform and the location of the SDP residues (Fig. 5 A). Three specific SDP residues may transfer the conformational changes required for the correct transmission of the biological signal. In the 3D structure of RAS, residues Val81 and Ala83 are located in a buried β-sheet, which is sandwiched between the two lobes that coordinate the conformational changes involved in this back-to-front communication mechanism. By contrast, Ala121 is found in a loop that is thought to weave different spatial motifs into the structure, such as the preceding α3 and α5 loops (Fig. 5 A). Although no data are available for the members of other families, the similarity of the SDP patterns suggests that similar routes connect the two regions (see Table 1 for the equivalent residues in the other families).
Based on the location and biological implications of the SDP residues, we conclude that the specificity of the small G protein structural module is characterized by a “canonical” nucleotide switch, multiple specific interactions, and communication with the nucleotide–membrane region. A precise balance between a rigid, high-affinity conformation and conformational flexibility is required to create such an efficient and stringent molecular switch, which may involve residues specific to individual proteins as well as distinct families.
Overview
New genomic information and the improvements to phylogenetic tools have further advanced our understanding of the structure and organization of the Ras protein superfamily. These advances are evident when comparing the current superfamily tree with that initially proposed 20 years ago (Valencia et al., 1991). It is now clear that the separation of the five main families (Ras, Rho, Arf/Sar, Ran, and Rab) was an early evolutionary event that predated the expansion of eukaryotes. Although it was believed that the founding members belonged to the Ras family, our comprehensive phylogenetic analyses of selected, well-defined members of each family in representative species, using EFTU sequences as an outgroup, points to the SRPRB proteins and the Arf family as possible founding members. This implies that the original function of these proteins may have been related to the regulation of membrane trafficking in eukaryotic cells (Munro, 2005), a process potentially linked to the emergence of complex intracellular structures. The presence of representative sequences of each family in the selected genomes indicates that divergence occurred before the emergence of eukaryotes, and strongly suggests that this superfamily expanded very early to generate the functionally distinct families. It is tempting to propose that this ancestral diversification is related to the increasing complexity of intracellular eukaryotic structures. This hypothesis merits further investigation and such studies will require additional information from genomes of more distant species, more precise functional data, and possibly new computational methods.
The general phylogenetic reconstruction proposed here fits well with the known functional properties of the individual families. In some cases, early functional divergence may be related to divisions such as that of the Ran family sequences, which represent a sub-functionalization of Rab to properly achieve nuclear transport capabilities. A number of other observations relating to specific groups in some species, such as the absence of the entire Ras family in A. thaliana, or the loss of genes in coelomates (Technau et al., 2005) and their ascidian vertebrate sisters (Hughes and Friedman, 2005), illustrate the complex gene loss and gain in different lineages (Kondrashov et al., 2002). More functional studies will be necessary to marry the details of functional and evolutionary divergence.
Our review of the phylogenetic classification of the Ras superfamily has provided useful data relating to the classification of a set of more divergent members. According to our analyses, some of these members may be placed in the traditional families, as seen in our reassigning of RAYL and MIRO proteins to the Rho family, and the RAB20 and RABL2A/B proteins to the Rab family. Based on the information available, other sequences such SRPRBs and RABL5/3 can be tentatively classified as independent families.
Phylogenetic approaches such as those described here can further our understanding of the relationships between proteins and protein groups. However, the assignment of function based on orthologous relationships should be approached with caution, and ultimately such facets should be confirmed experimentally.
The protein functions proposed here based on experimental data and phylogenetic inference can be complemented by a combination of structural analyses, and by determining the key residues in functional regions and in the mechanisms adopted by the distinct families (see Pymol Scripts in the Online supplemental material for the mapping of SPDs to structures). Within this new framework, such an approach could identify molecular landmarks that underlie the specific functional differences between families (i.e., the presence of residues such as Val81 and Ala83 in the core of the protein [PDB entry 121P]; or the presence of residues correlated with functional movement, such as Ala121 in the loop regions). These residues may be implicated in signal transmission between GTP-binding sites associated with the GTP state of the protein and the membrane-anchoring regions (N terminus of ARF, ARL, and SAR proteins in the Arf family, and the C terminus in the Ras and Ran families). Similarly, positions 156, 164, and 219 (numbered according to the full sequence alignment and mapping to the switch II region; see Table S6 for correspondence with the representative structures of each family) may be responsible for the recognition of family-specific binding partners (e.g., Arg68 and Arg76 mediate the association of RAS and RAN with GEFs, respectively; Table 1). Positions around the GTP-binding site, such as residues 13 and 18 in Rho and Ran members, respectively, appear to determine the specificity toward GAPs, and to influence the dynamic features of the GTP/GDP-binding pocket. Accordingly, these residues regulate the GTP hydrolysis/exchange mechanisms in each of these families.
Our understanding of the Ras superfamily will be enhanced by future developments in this field, including the incorporation of new complete genomes, elucidation of the structures of Ras superfamily protein–effector complexes, and biophysical studies of signal transmission between GTPase and effector binding sites in individual families. Similarly, a new generation of phylogenetic methods that can accurately organize larger numbers of sequences and refined bioinformatics approaches for the study of structure–sequence relationships will advance our understanding of protein evolution and function.
Online supplemental material
Supplemental figures, tables, and a .zip file that contains pymol scripts are available at http://www.jcb.org/cgi/content/full/jcb.201103008/DC1.
Acknowledgments
We are indebted to Fred Wittinghofer and Anne Spang for their critical reading of the manuscript.
This work was supported by two grants from the Spanish Ministerio de Ciencia e Innovación: BIO2007-66855-E.00408 and FIS PS09/02111.
Footnotes
Abbreviations used in this paper:
- MSA
- multiple sequence alignment
- SDP
- specificity-determining position
- SRPRB
- signal recognition particle receptor subunit β
References
- Abankwa D., Hanzal-Bayer M., Ariotti N., Plowman S.J., Gorfe A.A., Parton R.G., McCammon J.A., Hancock J.F. 2008. A novel switch region regulates H-ras membrane orientation and signal output. EMBO J. 27:727–735 10.1038/emboj.2008.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y., Renault L., García-Ranea J.A., Valencia A., Nishimoto T., Wittinghofer A. 1999. Model of the ran-RCC1 interaction using biochemical and docking experiments. J. Mol. Biol. 289:1119–1130 10.1006/jmbi.1999.2820 [DOI] [PubMed] [Google Scholar]
- Barnekow A., Thyrock A., Kessler D. 2009. Chapter 5: rab proteins and their interaction partners. Int Rev Cell Mol Biol. 274:235–274 10.1016/S1937-6448(08)02005-4 [DOI] [PubMed] [Google Scholar]
- Bauer B., Mirey G., Vetter I.R., García-Ranea J.A., Valencia A., Wittinghofer A., Camonis J.H., Cool R.H. 1999. Effector recognition by the small GTP-binding proteins Ras and Ral. J. Biol. Chem. 274:17763–17770 10.1074/jbc.274.25.17763 [DOI] [PubMed] [Google Scholar]
- Biou V., Cherfils J. 2004. Structural principles for the multispecificity of small GTP-binding proteins. Biochemistry. 43:6833–6840 10.1021/bi049630u [DOI] [PubMed] [Google Scholar]
- Bishop A.L., Hall A. 2000. Rho GTPases and their effector proteins. Biochem. J. 348:241–255 10.1042/0264-6021:3480241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boureux A., Vignal E., Faure S., Fort P. 2007. Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol. Biol. Evol. 24:203–216 10.1093/molbev/msl145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L.L., Edwards A.W. 1967. Phylogenetic analysis. Models and estimation procedures. Am. J. Hum. Genet. 19:233–257 [PMC free article] [PubMed] [Google Scholar]
- Cherfils J., Ménétrey J., Le Bras G., Janoueix-Lerosey I., de Gunzburg J., Garel J.R., Auzat I. 1997. Crystal structures of the small G protein Rap2A in complex with its substrate GTP, with GDP and with GTPgammaS. EMBO J. 16:5582–5591 10.1093/emboj/16.18.5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colicelli J. 2004. Human RAS superfamily proteins and related GTPases. Sci. STKE. 2004:RE13 10.1126/stke.2502004re13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.D., Alber T. 2001. The many faces of Ras: recognition of small GTP-binding proteins. Trends Biochem. Sci. 26:710–716 10.1016/S0968-0004(01)01974-0 [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Sol A., Pazos F., Valencia A. 2003. Automatic methods for predicting functionally important residues. J. Mol. Biol. 326:1289–1302 10.1016/S0022-2836(02)01451-1 [DOI] [PubMed] [Google Scholar]
- Díaz J.F., Wroblowski B., Engelborghs Y. 1995. Molecular dynamics simulation of the solution structures of Ha-ras-p21 GDP and GTP complexes: flexibility, possible hinges, and levers of the conformational transition. Biochemistry. 34:12038–12047 10.1021/bi00037a047 [DOI] [PubMed] [Google Scholar]
- Díaz J.F., Wroblowski B., Schlitter J., Engelborghs Y. 1997. Calculation of pathways for the conformational transition between the GTP- and GDP-bound states of the Ha-ras-p21 protein: calculations with explicit solvent simulations and comparison with calculations in vacuum. Proteins. 28:434–451 [DOI] [PubMed] [Google Scholar]
- Dvorsky R., Ahmadian M.R. 2004. Always look on the bright site of Rho: structural implications for a conserved intermolecular interface. EMBO Rep. 5:1130–1136 10.1038/sj.embor.7400293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. 2009. A new generation of homology search tools based on probabilistic inference. Genome Inform. 23:205–211 10.1142/9781848165632_0019 [DOI] [PubMed] [Google Scholar]
- Ezer S.T., Sahar D., Salzberg A., Lev Z. 1994. Differential expression during embryogenesis of three genes clustered in the Ras1 region of Drosophila melanogaster. Dev. Dyn. 201:179–190 10.1002/aja.1002010208 [DOI] [PubMed] [Google Scholar]
- Finn R.D., Mistry J., Tate J., Coggill P., Heger A., Pollington J.E., Gavin O.L., Gunasekaran P., Ceric G., Forslund K., et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38(Database issue):D211–D222 10.1093/nar/gkp985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes G., Valencia A. 2009. Ras classical effectors: new tales from in silico complexes. Trends Biochem. Sci. 34:533–539 10.1016/j.tibs.2009.07.001 [DOI] [PubMed] [Google Scholar]
- Futatsugi N., Tsuda M. 2001. Molecular dynamics simulations of Gly-12—>Val mutant of p21(ras): dynamic inhibition mechanism. Biophys. J. 81:3483–3488 10.1016/S0006-3495(01)75979-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ranea J.A., Valencia A. 1998. Distribution and functional diversification of the ras superfamily in Saccharomyces cerevisiae. FEBS Lett. 434:219–225 10.1016/S0014-5793(98)00967-3 [DOI] [PubMed] [Google Scholar]
- Gaucher E.A., Kratzer J.T., Randall R.N. 2010. Deep phylogeny—how a tree can help characterize early life on Earth. Cold Spring Harb. Perspect. Biol. 2:a002238 10.1101/cshperspect.a002238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. 1998. Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 95:237–248 10.1016/S0092-8674(00)81754-7 [DOI] [PubMed] [Google Scholar]
- Goldfinger L.E. 2008. Choose your own path: specificity in Ras GTPase signaling. Mol. Biosyst. 4:293–299 10.1039/b716887j [DOI] [PubMed] [Google Scholar]
- Gorfe A.A., Grant B.J., McCammon J.A. 2008. Mapping the nucleotide and isoform-dependent structural and dynamical features of Ras proteins. Structure. 16:885–896 10.1016/j.str.2008.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B.G. 2005. Comparison of the accuracies of several phylogenetic methods using protein and DNA sequences. Mol. Biol. Evol. 22:792–802 10.1093/molbev/msi066 [DOI] [PubMed] [Google Scholar]
- Heasman S.J., Ridley A.J. 2008. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9:690–701 10.1038/nrm2476 [DOI] [PubMed] [Google Scholar]
- Hedges S.B., Blair J.E., Venturi M.L., Shoe J.L. 2004. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol. Biol. 4:2 10.1186/1471-2148-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo W.D., Meyer T. 2003. Switch-of-function mutants based on morphology classification of Ras superfamily small GTPases. Cell. 113:315–328 10.1016/S0092-8674(03)00315-5 [DOI] [PubMed] [Google Scholar]
- Holder M., Lewis P.O. 2003. Phylogeny estimation: traditional and Bayesian approaches. Nat. Rev. Genet. 4:275–284 10.1038/nrg1044 [DOI] [PubMed] [Google Scholar]
- Hughes A.L., Friedman R. 2005. Loss of ancestral genes in the genomic evolution of Ciona intestinalis. Evol. Dev. 7:196–200 10.1111/j.1525-142X.2005.05022.x [DOI] [PubMed] [Google Scholar]
- Jaffe A.B., Hall A. 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21:247–269 10.1146/annurev.cellbio.21.020604.150721 [DOI] [PubMed] [Google Scholar]
- Jiang S.Y., Ramachandran S. 2006. Comparative and evolutionary analysis of genes encoding small GTPases and their activating proteins in eukaryotic genomes. Physiol. Genomics. 24:235–251 [DOI] [PubMed] [Google Scholar]
- Kahn R.A., Cherfils J., Elias M., Lovering R.C., Munro S., Schurmann A. 2006. Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J. Cell Biol. 172:645–650 10.1083/jcb.200512057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnoub A.E., Weinberg R.A. 2008. Ras oncogenes: split personalities. Nat. Rev. Mol. Cell Biol. 9:517–531 10.1038/nrm2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnoub A.E., Worthylake D.K., Rossman K.L., Pruitt W.M., Campbell S.L., Sondek J., Der C.J. 2001. Molecular basis for Rac1 recognition by guanine nucleotide exchange factors. Nat. Struct. Biol. 8:1037–1041 10.1038/nsb719 [DOI] [PubMed] [Google Scholar]
- Katoh K., Asimenos G., Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 537:39–64 10.1007/978-1-59745-251-9_3 [DOI] [PubMed] [Google Scholar]
- Kemena C., Notredame C. 2009. Upcoming challenges for multiple sequence alignment methods in the high-throughput era. Bioinformatics. 25:2455–2465 10.1093/bioinformatics/btp452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel C., Foglierini M., Kuemmerer N., Beltrao P., Serrano L. 2007. A genome-wide Ras-effector interaction network. J. Mol. Biol. 370:1020–1032 10.1016/j.jmb.2007.05.015 [DOI] [PubMed] [Google Scholar]
- Kondrashov F.A., Rogozin I.B., Wolf Y.I., Koonin E.V. 2002. Selection in the evolution of gene duplications. Genome Biol. 3:RESEARCH0008 10.1186/gb-2002-3-2-research0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M., Backhaus O., Rosenstiel P., Horn D., Klopocki E., Birkenmeier G., Schreiber S., Platzer M., Hampe J., Huse K. 2010. Analysis of relative gene dosage and expression differences of the paralogs RABL2A and RABL2B by Pyrosequencing. Gene. 455:1–7 10.1016/j.gene.2010.01.005 [DOI] [PubMed] [Google Scholar]
- Langsley G., van Noort V., Carret C., Meissner M., de Villiers E.P., Bishop R., Pain A. 2008. Comparative genomics of the Rab protein family in Apicomplexan parasites. Microbes Infect. 10:462–470 10.1016/j.micinf.2008.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N., Brinkmann H., Philippe H. 2007. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol. Biol. 7:S4 10.1186/1471-2148-7-S1-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 23:127–128 10.1093/bioinformatics/btl529 [DOI] [PubMed] [Google Scholar]
- Li Y., Kelly W.G., Logsdon J.M., Jr, Schurko A.M., Harfe B.D., Hill-Harfe K.L., Kahn R.A. 2004. Functional genomic analysis of the ADP-ribosylation factor family of GTPases: phylogeny among diverse eukaryotes and function in C. elegans. FASEB J. 18:1834–1850 10.1096/fj.04-2273com [DOI] [PubMed] [Google Scholar]
- Li Q., Wang L., Zeng L., Zhang Y., Li K., Jin P., Su B., Wang L. 2010. Evaluation of the novel gene Rabl3 in the regulation of proliferation and motility in human cancer cells. Oncol. Rep. 24:433–440 [DOI] [PubMed] [Google Scholar]
- Lukman S., Grant B.J., Gorfe A.A., Grant G.H., McCammon J.A. 2010. The distinct conformational dynamics of K-Ras and H-Ras A59G. PLOS Comput. Biol. 6 10.1371/journal.pcbi.1000922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz P., Wyroba E. 2009. Phylogeny and evolution of Rab7 and Rab9 proteins. BMC Evol. Biol. 9:101 10.1186/1471-2148-9-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima K., Tsuchiya M., Nightingale M.S., Moss J., Vaughan M. 1993. ARD 1, a 64-kDa guanine nucleotide-binding protein with a carboxyl-terminal ADP-ribosylation factor domain. J. Biol. Chem. 268:8801–8807 [PubMed] [Google Scholar]
- Mulloy J.C., Cancelas J.A., Filippi M.D., Kalfa T.A., Guo F., Zheng Y. 2010. Rho GTPases in hematopoiesis and hemopathies. Blood. 115:936–947 10.1182/blood-2009-09-198127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. 2005. The Arf-like GTPase Arl1 and its role in membrane traffic. Biochem. Soc. Trans. 33:601–605 10.1042/BST0330601 [DOI] [PubMed] [Google Scholar]
- Nei M., Kumar S. 2000. Molecular Evolution and Phylogenetics. Oxford University Press [Google Scholar]
- Noé F., Ille F., Smith J.C., Fischer S. 2005. Automated computation of low-energy pathways for complex rearrangements in proteins: application to the conformational switch of Ras p21. Proteins. 59:534–544 10.1002/prot.20422 [DOI] [PubMed] [Google Scholar]
- Notredame C.2010. Computing multiple sequence/structure alignments with the T-coffee package. Curr. Protoc. Bioinformatics. Chapter 3:Unit 3.8 1–25.
- Ostlund G., Schmitt T., Forslund K., Köstler T., Messina D.N., Roopra S., Frings O., Sonnhammer E.L. 2010. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 38(Database issue):D196–D203 10.1093/nar/gkp931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R.D.M., Holmes E.C.1999. Molecular Evolution: A Phylogenetic Approach. Blackwell Sciences Ltd., Oxford.
- Park H.O., Bi E. 2007. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 71:48–96 10.1128/MMBR.00028-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualato S., Renault L., Cherfils J. 2002. Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for ‘front-back’ communication. EMBO Rep. 3:1035–1041 10.1093/embo-reports/kvf221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Leal J.B., Seabra M.C. 2001. Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 313:889–901 10.1006/jmbi.2001.5072 [DOI] [PubMed] [Google Scholar]
- Phillips A., Janies D., Wheeler W. 2000. Multiple sequence alignment in phylogenetic analysis. Mol. Phylogenet. Evol. 16:317–330 10.1006/mpev.2000.0785 [DOI] [PubMed] [Google Scholar]
- Putnam N.H., Srivastava M., Hellsten U., Dirks B., Chapman J., Salamov A., Terry A., Shapiro H., Lindquist E., Kapitonov V.V., et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 317:86–94 10.1126/science.1139158 [DOI] [PubMed] [Google Scholar]
- Qin H., Wang Z., Diener D., Rosenbaum J. 2007. Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr. Biol. 17:193–202 10.1016/j.cub.2006.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausell A., Juan D., Pazos F., Valencia A. 2010. Protein interactions and ligand binding: from protein subfamilies to functional specificity. Proc. Natl. Acad. Sci. USA. 107:1995–2000 10.1073/pnas.0908044107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P., Sabatier C., McCormick F. 2004. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol. Cell. Biol. 24:4943–4954 10.1128/MCB.24.11.4943-4954.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Schlessinger K., Hall A., Tolwinski N. 2009. Wnt signaling pathways meet Rho GTPases. Genes Dev. 23:265–277 10.1101/gad.1760809 [DOI] [PubMed] [Google Scholar]
- Schwartz S.L., Cao C., Pylypenko O., Rak A., Wandinger-Ness A. 2007. Rab GTPases at a glance. J. Cell Sci. 120:3905–3910 10.1242/jcs.015909 [DOI] [PubMed] [Google Scholar]
- Sekiguchi T., Hirose E., Nakashima N., Ii M., Nishimoto T. 2001. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J. Biol. Chem. 276:7246–7257 10.1074/jbc.M004389200 [DOI] [PubMed] [Google Scholar]
- Sprang S.R. 2000. Conformational display: a role for switch polymorphism in the superfamily of regulatory GTPases. Sci. STKE. 2000:pe1 10.1126/stke.2000.50.pe1 [DOI] [PubMed] [Google Scholar]
- Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- Stenmark H., Valencia A., Martinez O., Ullrich O., Goud B., Zerial M. 1994. Distinct structural elements of rab5 define its functional specificity. EMBO J. 13:575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau U., Rudd S., Maxwell P., Gordon P.M., Saina M., Grasso L.C., Hayward D.C., Sensen C.W., Saint R., Holstein T.W., et al. 2005. Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet. 21:633–639 10.1016/j.tig.2005.09.007 [DOI] [PubMed] [Google Scholar]
- Tong L.A., de Vos A.M., Milburn M.V., Kim S.H. 1991. Crystal structures at 2.2 A resolution of the catalytic domains of normal ras protein and an oncogenic mutant complexed with GDP. J. Mol. Biol. 217:503–516 10.1016/0022-2836(91)90753-S [DOI] [PubMed] [Google Scholar]
- Valencia A., Kjeldgaard M., Pai E.F., Sander C. 1991. GTPase domains of ras p21 oncogene protein and elongation factor Tu: analysis of three-dimensional structures, sequence families, and functional sites. Proc. Natl. Acad. Sci. USA. 88:5443–5447 10.1073/pnas.88.12.5443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam T.J., Rehmann H., Bos J.L., Snel B. 2009. Phylogeny of the CDC25 homology domain reveals rapid differentiation of Ras pathways between early animals and fungi. Cell. Signal. 21:1579–1585 10.1016/j.cellsig.2009.06.004 [DOI] [PubMed] [Google Scholar]
- Vetter I.R., Wittinghofer A. 2001. The guanine nucleotide-binding switch in three dimensions. Science. 294:1299–1304 10.1126/science.1062023 [DOI] [PubMed] [Google Scholar]
- Wang L.S., Leebens-Mack J., Kerr Wall P., Beckmann K., dePamphilis C.W., Warnow T. 2011. The impact of multiple protein sequence alignment on phylogenetic estimation. IEEE/ACM Trans. Comput. Biol. Bioinformatics. 8:1108–1119 10.1109/TCBB.2009.68 [DOI] [PubMed] [Google Scholar]
- Wennerberg K., Rossman K.L., Der C.J. 2005. The Ras superfamily at a glance. J. Cell Sci. 118:843–846 10.1242/jcs.01660 [DOI] [PubMed] [Google Scholar]
- Wong A.C., Shkolny D., Dorman A., Willingham D., Roe B.A., McDermid H.E. 1999. Two novel human RAB genes with near identical sequence each map to a telomere-associated region: the subtelomeric region of 22q13.3 and the ancestral telomere band 2q13. Genomics. 59:326–334 10.1006/geno.1999.5889 [DOI] [PubMed] [Google Scholar]
- Wu X.S., Rao K., Zhang H., Wang F., Sellers J.R., Matesic L.E., Copeland N.G., Jenkins N.A., Hammer J.A., III 2002. Identification of an organelle receptor for myosin-Va. Nat. Cell Biol. 4:271–278 10.1038/ncb760 [DOI] [PubMed] [Google Scholar]
- Yang Z. 2002. Small GTPases: versatile signaling switches in plants. Plant Cell. 14(Suppl):S375–S388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M., McBride H. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2:107–117 10.1038/35052055 [DOI] [PubMed] [Google Scholar]