Glucose remodels the post-translational modifications of the yeast arrestin-like protein Rod1 to promote glucose-induced transporter endocytosis.
Abstract
Endocytosis regulates the plasma membrane protein landscape in response to environmental cues. In yeast, the endocytosis of transporters depends on their ubiquitylation by the Nedd4-like ubiquitin ligase Rsp5, but how extracellular signals trigger this ubiquitylation is unknown. Various carbon source transporters are known to be ubiquitylated and endocytosed when glucose-starved cells are exposed to glucose. We show that this required the conserved arrestin-related protein Rod1/Art4, which was activated in response to glucose addition. Indeed, Rod1 was a direct target of the glucose signaling pathway composed of the AMPK homologue Snf1 and the PP1 phosphatase Glc7/Reg1. Glucose promoted Rod1 dephosphorylation and its subsequent release from a phospho-dependent interaction with 14-3-3 proteins. Consequently, this allowed Rod1 ubiquitylation by Rsp5, which was a prerequisite for transporter endocytosis. This paper therefore demonstrates that the arrestin-related protein Rod1 relays glucose signaling to transporter endocytosis and provides the first molecular insights into the nutrient-induced activation of an arrestin-related protein through a switch in post-translational modifications.
Introduction
Endocytosis is critical for the ability of cells to adapt to changes in the environment. One of its primary functions is to attenuate intracellular signaling after stimulation, through the down-regulation of plasma membrane receptors. Conversely, intracellular signaling also regulates endocytosis. The endocytosis of receptors often relies on their own signaling activity or that of a close partner, such as for receptor tyrosine kinases or G protein–coupled receptors (Sorkin and von Zastrow, 2009). However, intracellular signaling also influences the endocytosis of transporters (Miranda and Sorkin, 2007; Argenzio et al., 2011; Vina-Vilaseca et al., 2011), but because transporters lack intrinsic signaling activity, this regulation remains poorly understood.
The yeast Saccharomyces cerevisiae is a powerful model system for studying transporter endocytosis in response to nutritional changes (Haguenauer-Tsapis and André, 2004). Numerous examples of nutrient-induced down-regulation of transporters were described since the early studies on yeast genetics and physiology. In particular, amino acid and sugar transporters were shown to undergo “catabolite inactivation” (Holzer, 1976; Grenson, 1983; Haguenauer-Tsapis and André, 2004), in which transporter activity was thought to be inactivated in response to a nutritional change, but which were later revealed as the first examples of signal-induced transporter endocytosis (Hein et al., 1995; Medintz et al., 1996; Horak and Wolf, 1997; Lucero and Lagunas, 1997; Haguenauer-Tsapis and André, 2004).
S. cerevisiae preferentially uses glucose for growth, and glucose-starved yeast cells rapidly adapt upon exposure to glucose by remodeling their enzymatic content. For instance, glucose causes the degradation of enzymes involved in the metabolism of alternate carbon sources. In addition, glucose also induces the endocytosis of various sugar transporters (Horák, 2003) and of Jen1 (Paiva et al., 2002), a monocarboxylate transporter of the SLC16/MCT family (Casal et al., 2008). The endocytosis of transporters requires their ubiquitylation by Rsp5, a ubiquitin ligase of the Nedd4 family that harbors several members in higher eukaryotes, some of which also participate in endocytosis (Rotin and Kumar, 2009). Therefore, the number of possible Rsp5 substrate is tremendous, leading to the question of how these transporters are specifically recognized by Rsp5, and how the timeliness of the ubiquitylation reaction is ensured. Proteins of the Nedd4/Rsp5 family are known to interact, through their WW domains, with proteins harboring a PY motif (usually, a “PPxY” sequence). However, a very limited number of membrane proteins harbor this motif. Instead, it has become clear that the interaction between Rsp5 and the transporters occurs through so-called adaptor proteins that generally display at least one PY motif (Polo and Di Fiore, 2008; Léon and Haguenauer-Tsapis, 2009).
In particular, several yeast proteins with homologies to arrestins (arrestin-related trafficking adaptors, or ARTs, also coined “alpha-arrestins”) were proposed to recruit Rsp5 to transporters in response to changes in the environment (Lin et al., 2008; Polo and Di Fiore, 2008; Nikko and Pelham, 2009). Yeast arrestin-related proteins display human homologues, named ARRDC (arrestin domain-containing), which also act as adaptors of Nedd4-like enzymes (Draheim et al., 2010; Nabhan et al., 2010) and are evolutionary related to β-arrestins from higher eukaryotes, which participate in endocytosis and signaling (DeWire et al., 2007; Alvarez, 2008). Although the discovery of these arrestin-related proteins has provided a molecular basis explaining how Rsp5 interacts with and ubiquitylates transporters, it does not fully explain how transporter ubiquitylation is regulated in a timely manner with respect to the presence of extracellular signals. Indeed, phosphorylation of the metal transporter Smf1 was shown to promote the recruitment of the yeast arrestin-related protein Ecm21/Art2 (Nikko et al., 2008), but although this is required for Smf1 endocytosis it did not appear to be the trigger, suggesting the involvement of an additional regulatory step. Importantly, yeast arrestin-related proteins were described to regulate endocytosis in a signal-specific rather than transporter-specific manner (Lin et al., 2008; Nikko and Pelham, 2009). This raised the possibility that they become activated in response to a specific signal. However, the existence of such an activation mechanism remains unknown.
In this paper, we identified the arrestin-related protein Rod1, also named Art4, as an essential component of the glucose-induced endocytosis of Jen1, the lactate transporter. We show that Rod1 is a direct target of the glucose-signaling pathway composed of the Snf1 kinase, the yeast homologue of 5’-AMP-activated kinase (AMPK), and the protein phosphatase 1 (PP1). As such, Rod1 phosphorylation status is regulated by glucose availability, which consequently modulates its activity. In particular, we show that the glucose-induced dephosphorylation of Rod1 promotes its ubiquitylation, a modification that is essential for Jen1 endocytosis. Moreover, we demonstrate that this cross talk between dephosphorylation and ubiquitylation of Rod1 is coordinated by a phospho-dependent interaction of Rod1 with yeast 14-3-3 proteins, which inhibits Rod1 ubiquitylation. Altogether, our findings establish that Rod1 is a crucial player in glucose-induced transporter endocytosis, and provide the first molecular insights into the mechanism of arrestin-related protein activation in response to intracellular signaling.
Results
The arrestin-like protein Rod1 is required for the glucose-induced endocytosis of the lactate transporter Jen1
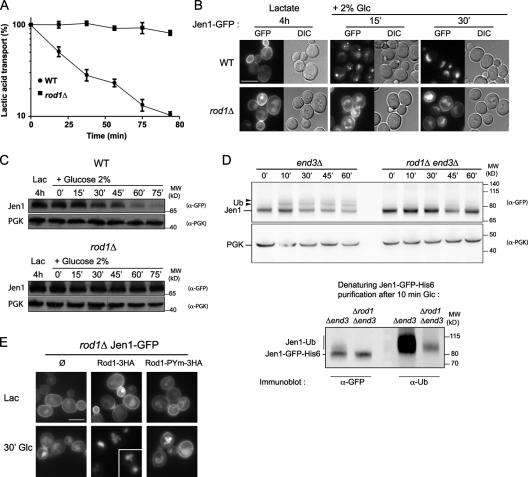
Previous work has shown that the endocytosis of a given transporter can be mediated by various signals through the involvement of different arrestin-related proteins, suggesting that they are involved in signal-specific endocytosis (Lin et al., 2008; Nikko and Pelham, 2009). To gain insight into how an extracellular signal regulates endocytosis, we studied the glucose-induced endocytosis of the lactate transporter, Jen1, as a model, and first aimed to identify whether a yeast arrestin-related protein is involved in this process. We measured the down-regulation of lactate transport activity, i.e., Jen1 internalization away from the plasma membrane, after glucose addition in various mutants in which the genes encoding arrestin-related proteins were disrupted. Specifically, Jen1 was not subjected to glucose-induced inactivation in rod1Δ, whereas mutants for other arrestin-related genes were not affected (Fig. 1 A and Fig. S1 A). Consistent with this observation, the glucose-induced trafficking of Jen1-GFP was abnormal in the rod1Δ mutant, in which Jen1 was stably localized at the plasma membrane (Fig. 1 B), and its degradation in response to glucose was strongly affected (Fig. 1 C). Rod1 was specifically involved in the regulation of glucose-induced endocytosis because neither the glucose-mediated repression of the invertase gene (SUC2) nor the glucose-induced degradation of fructose-1,6-bisphosphatase (Fbp1) were affected in the rod1Δ mutant (Fig. S1, B and C). Likewise, Rod1 was not required for Jen1 endocytosis in response to a glucose-unrelated signal, such as cycloheximide, which stimulates the endocytosis of several transporters (Lin et al., 2008; Nikko and Pelham, 2009) including Jen1 (Fig. S1 D). We conclude that Rod1 is not a general regulator of Jen1 endocytosis but rather participates in its “catabolite inactivation” by glucose.
Figure 1.
The arrestin-related protein Rod1 is involved in the glucose-induced endocytosis of the lactate transporter, Jen1. (A) WT and rod1Δ cells were grown in lactate medium, and 14C-labeled lactate transport activity was followed over time after glucose addition. A representative experiment of more than three repetitions is shown. The values represent the mean ± SEM of triplicate measurements. (B) Cells in which Jen1 is tagged with GFP at the chromosomal locus were grown in lactate medium, and Jen1-GFP subcellular localization was followed after glucose addition for the indicated times in WT and rod1Δ strains. Bar, 5 µm. (C) Total protein extracts from a similar experiment as in B were prepared at the indicated times after glucose addition and immunoblotted with the indicated antibodies. (D, top) A plasmid-encoded, galactose-inducible Jen1-GFP-His6 construct was expressed in end3Δ (control) and end3Δ rod1Δ cells. Total protein extracts were prepared at several time points after glucose addition and immunoblotted with anti-GFP or anti-PGK (loading control) antibodies. Arrowheads indicate ubiquitylated Jen1 species. (Bottom) Jen1-GFP-His6 was purified under denaturing conditions by Ni-based chromatography 10 min after glucose addition. The eluates were loaded twice and incubated with anti-GFP or anti-ubiquitin (P4D1) antibodies. (E) A rod1Δ strain expressing Jen1-GFP tagged at the chromosomal locus was transformed with an empty plasmid (Ø) or plasmid-encoded Rod1-3HA or Rod1-PYm-3HA, and the cells were imaged after 4 h of growth on lactate medium, and 30 min after the addition of glucose. Bar, 5 µm.
The glucose-induced endocytosis of Jen1 involves its ubiquitylation by Rsp5 (Paiva et al., 2009), the sole Nedd4-like ubiquitin ligase of yeast, which was previously shown to interact with Rod1 (Andoh et al., 2002). To establish an eventual contribution of Rod1 in the Rsp5-mediated ubiquitylation of Jen1, we assessed Jen1 ubiquitylation in response to glucose in a strain devoid of Rod1. We used an endocytic mutant, end3Δ, which is known to accumulate transporters in a ubiquitylated state, to improve the detection of ubiquitylated conjugates. As shown in Fig. 1 D, the glucose-induced ubiquitylation of Jen1 occurred several minutes after glucose addition in the end3Δ strain, whereas it was strongly diminished and delayed when the ROD1 gene was further deleted (Fig. 1 D). These results demonstrate that Rod1 is required for the rapid and efficient ubiquitylation of Jen1 in response to glucose, which may explain the endocytosis defects observed in the rod1Δ mutant.
The WW domains of Rsp5 interact with (L/P)PxY sequences (named “PY” motifs; Rotin and Kumar, 2009). Rod1, like most arrestin-related proteins, displays two such PY motifs, and a point mutation in both PPxY sequences resulting in PPxA (Rod1-PYm) abolished the interaction with Rsp5 WW domains in vitro (Fig. S1 E). Although the Rod1-PYm construct was expressed at similar levels to the wild-type (WT) protein (Fig. S1 E), the mutant protein was unable to restore Jen1 endocytosis in a rod1Δ mutant background, suggesting an essential function for the Rod1/Rsp5 interaction in this process (Fig. 1 E). This is consistent with the role of arrestin-related proteins in assisting transporter ubiquitylation by Rsp5 (Lin et al., 2008; Nikko et al., 2008; Hatakeyama et al., 2010). However, because we have not observed a direct interaction between Rod1 and Jen1, we cannot exclude the possibility that Rod1 acts more indirectly on Jen1 ubiquitylation.
Rod1 phosphorylation and ubiquitylation are dynamically regulated by glucose availability
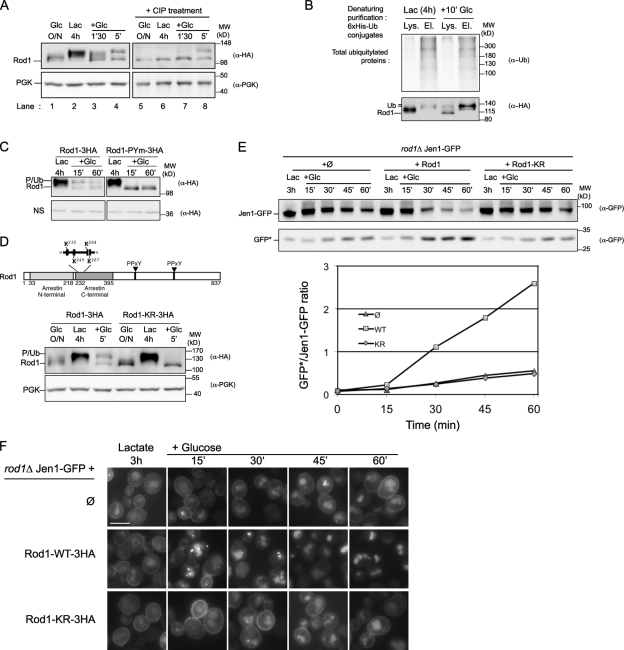
Rod1 is therefore a major actor in the glucose-induced endocytosis of Jen1, consistent with its previous involvement in the glucose-induced endocytosis of the high-affinity glucose transporter Hxt6 (Nikko and Pelham, 2009). We then sought to investigate how Rod1 function in endocytosis is specifically regulated by glucose availability. When cells were cultured overnight in glucose medium, the HA epitope-tagged Rod1 migrated primarily as a diffuse band on SDS-PAGE (Fig. 2 A, lane 1). This was due to Rod1 phosphorylation because phosphatase treatment of protein extracts led to a single band that displayed a faster migration. Rod1 phosphorylation was strongly enhanced when cells were grown in lactate medium (lane 2) but was rapidly lost upon glucose addition (lanes 3 and 4), revealing that Rod1 phosphorylation is controlled by glucose availability in the culture medium.
Figure 2.
Dynamic modifications of Rod1 by phosphorylation and ubiquitylation are regulated by glucose availability. (A) A WT strain expressing a plasmid-encoded Rod1-3HA was grown overnight in glucose medium (exponential phase), then 4 h in lactate medium, before glucose was added for the indicated times. Total protein extracts were either treated or untreated with calf intestinal phosphatase (CIP) and immunoblotted with anti-HA and anti-PGK antibodies. (B) A WT strain expressing plasmid-encoded Rod1-3HA and 6xHis-tagged ubiquitin was grown for 4 h in lactate medium before glucose was added for 10 min. 6xHis-tagged ubiquitin conjugates were purified under denaturing conditions by Ni-based chromatography. The lysates (Lys.) and eluates (El.) were immunoblotted with an anti-ubiquitin antibody, and with an anti-HA antibody to detect Rod1. (C) Strains expressing a plasmid-encoded Rod1-3HA or a variant mutated on both PPxY motifs (Rod1-PYm-3HA) were grown as in B. Total protein extracts were immunoblotted with an anti-HA antibody. A nonspecific, cross-reactive band was used as internal loading control. P/Ub refers to the molecular weight of phosphorylated or ubiquitylated Rod1, respectively. (D, top) Representation of Rod1 primary sequence. The numbers below show the amino acid boundaries of the predicted arrestin-related domains (“Arrestin_N”/PF00339 and “Arrestin_C”/PF02752) as determined by Pfam. The position of the Rod1 lysines that were mutated to raise the Rod1-KR-3HA construct (below) is displayed above the scheme. (Bottom) A strain expressing a plasmid-encoded Rod1-3HA or a mutant form in which the four lysines depicted in the scheme above are mutated to arginine (Rod1-KR-3HA) were grown as in A. Total protein extracts were immunoblotted with anti-HA and anti-PGK antibodies. P/Ub refers to the molecular weight of phosphorylated or ubiquitylated Rod1, respectively. (E) A rod1Δ strain in which Jen1 is tagged with GFP at the chromosomal locus was transformed with an empty plasmid (ø), or plasmids encoding Rod1-3HA or Rod1-KR-3HA, and grown for 3 h in lactate medium before glucose was added for the indicated times. Total protein extracts were immunoblotted with an anti-GFP antibody. Band intensities were quantified using ImageJ and the GFP*/Jen1-GFP ratio is plotted over time. (F) Jen1-GFP subcellular localization was followed in the same strains as in E grown in similar conditions. Bar, 5 µm.
This paper also revealed that exposure to glucose induced an additional post-translational modification on Rod1, which was insensitive to phosphatase treatment (Fig. 2 A, lanes 7 and 8). A common feature of Rsp5 adaptor proteins is their propensity to be ubiquitylated by Rsp5, as described for several yeast arrestin-related proteins (Kee et al., 2006; Lin et al., 2008; Hatakeyama et al., 2010; Herrador et al., 2010) including Rod1 (Andoh et al., 2002). As shown in Fig. 2 B, Rod1 ubiquitylation was indeed most prominent during the lactate–glucose transition, and was dependent on its interaction with Rsp5, as indicated by the inability of the Rod1-PYm point mutant to be ubiquitylated (Fig. 2 C). The use of this mutant also confirmed that Rod1 ubiquitylation accounted for the observed phosphatase-insensitive modification mentioned above. Therefore, Rod1 dephosphorylation in response to glucose is concomitant with its ubiquitylation by Rsp5.
To determine whether this modification is important for Rod1 function, we aimed at identifying the lysine residue targeted for ubiquitylation. Using a lysine-less Rod1 construct, in which 48 lysines are mutated to arginine, and various chimeras (Fig. S2), we mapped the ubiquitylated site(s) to a 33-residue region (aa 235–267) located in the predicted “Arrestin_C” domain and that contains four lysines (Fig. 2 D). This mutant protein (Rod1-KR) was expressed at similar levels to the WT protein, but was not functional as determined by its inability to complement the rod1Δ mutant for the glucose-induced Jen1 degradation and vacuolar targeting (Fig. 2, E and F), indicating an essential role of Rod1 ubiquitylation in its function in endocytosis.
Loss of Rod1 phosphorylation leads to its constitutive ubiquitylation and activation
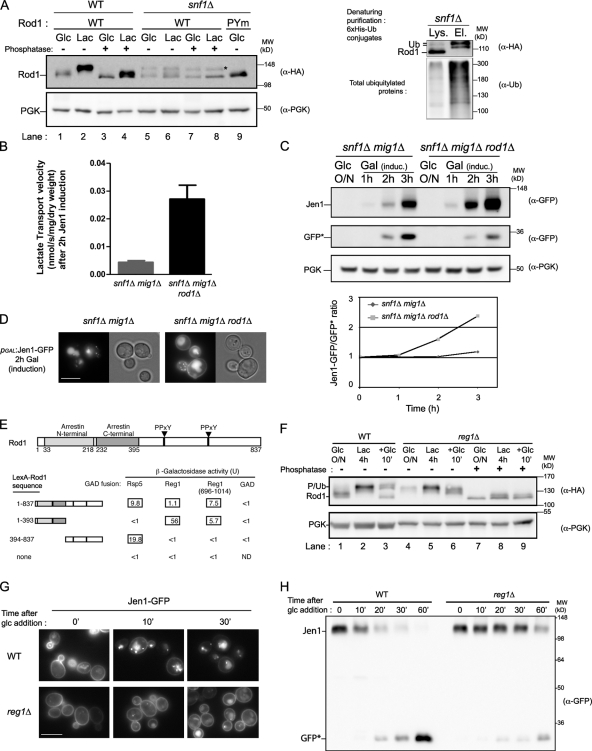
The sudden ubiquitylation of Rod1 in response to glucose and its importance for Jen1 endocytosis prompted us to study how this modification is regulated at the molecular level. We first set out to determine the eventual contribution of Rod1 phosphorylation in this process. Rod1 is a target of the Snf1 kinase (Shinoda and Kikuchi, 2007), the yeast homologue of AMPK, which is a crucial component of nutrient sensing and carbon metabolism regulation in eukaryotes (Hedbacker and Carlson, 2008). We observed that in the snf1Δ mutant, Rod1 was indeed unphosphorylated (Fig. 3 A, lanes 5 and 6), but importantly, it was constitutively ubiquitylated regardless of the presence of glucose, as determined by the appearance of a phosphatase-insensitive band (lanes 7 and 8) whose presence depended on the interaction of Rod1 with Rsp5 (lane 9) and that were identified as Rod1-ubiquitin conjugates (Fig. 3 A, right). This suggested that the lack of Rod1 phosphorylation promotes its ubiquitylation. Because the Rod1 migration profile in the snf1Δ mutant mimics that observed at the lactate–glucose transition during which Rod1 is activated, we expressed Jen1-GFP in a strain devoid of Snf1 to study its endocytosis. However, in this mutant, very little lactate transport activity was measured after induction (Fig. 3 B). Although Jen1-GFP was expressed, it was rapidly degraded in this strain in the course of its synthesis (Fig. 3 C) and localized predominantly to intracellular compartments, including the vacuole, rather than the plasma membrane (Fig. 3 D). Strikingly, the additional deletion of ROD1 in this mutant restored lactate transport activity, as well as Jen1 stabilization and plasma membrane localization (Fig. 3, B–D), showing that the defects observed in the absence of Snf1 were due to a constitutive, Rod1-dependent endocytosis, even in the absence of glucose.
Figure 3.
The Snf1/PP1 glucose-signaling pathway regulates Rod1 activation and Jen1 endocytosis. (A, left) WT and snf1Δ cells expressing plasmid-encoded Rod1-3HA or Rod1-PYm-3HA were grown in glucose overnight and then 4 h in lactate medium. Crude extracts were prepared, treated with phosphatase where indicated, and immunoblotted with the indicated antibodies. The asterisk indicates ubiquitylated Rod1 as determined by the absence of this band in cells expressing Rod1-PYm (lane 9). (Right) snf1Δ cells expressing plasmid-encoded Rod1-3HA and 6xHis-tagged ubiquitin were grown overnight in glucose-containing medium (exponential phase). 6xHis-tagged ubiquitin conjugates were purified under denaturing conditions by Ni-based chromatography. The lysates (Lys.) and eluates (El.) were immunoblotted with an anti-ubiquitin antibody, and with an anti-HA antibody to detect Rod1. (B) The snf1Δ mutant is unable to grow on alternate sugars such as galactose or nonfermentable carbon sources, but the additional deletion of MIG1, encoding a Snf1-regulated transcriptional repressor, allows growth on galactose (Vallier and Carlson, 1994). Transport activity of 14C-labeled lactate was therefore assayed in the snf1Δ mig1Δ and snf1Δ mig1Δ rod1Δ strains transformed with a galactose-inducible Jen1-GFP-His6 construct after 2 h of growth in galactose medium. A representative experiment of three repetitions is shown. The values represent the mean ± SEM of triplicate measurements. (C) Crude extracts were prepared from the strains described in B. Crude extracts were prepared at the indicated times and immunoblotted with anti-GFP and anti-PGK antibodies. GFP* indicates the size of a proteolytic fragment indicative of Jen1-GFP degradation in the vacuole. Band intensities were quantified using ImageJ and the Jen1-GFP/GFP* ratio is plotted over time. (D) Jen1-GFP localization was followed in strains as in B after 2 h of induction. (E) A yeast two-hybrid screen using full-length Rod1 as the bait led to the identification of the C-terminal region of Reg1, a PP1 regulatory subunit. Rsp5 was used as a positive control. Average β-galactosidase activity values (Miller units) for each combination are presented. (F) WT and reg1Δ strains expressing plasmid-encoded Rod1-3HA were grown as indicated and total protein extracts were treated with phosphatase when indicated, and immunoblotted with the indicated antibodies. (G) WT and reg1Δ strains in which Jen1 is tagged with GFP at the chromosomal locus were grown for 4 h in lactate medium, and Jen1-GFP subcellular localization was followed at the indicated time after glucose addition by fluorescence microscopy. Bar, 5 µm. (H) The same strains as in G were grown in similar conditions. Samples were collected at the indicated times after glucose treatments and total protein extracts were immunoblotted with anti-GFP antibodies. GFP* indicates the size of a proteolytic fragment indicative of Jen1-GFP degradation in the vacuole.
Reg1, a regulatory subunit of protein phosphatase 1 (PP1), interacts with Rod1 and is required for Rod1 ubiquitylation and activation
A yeast two-hybrid screen using Rod1 as the bait led to the identification of the C-terminal half of Reg1 (Fig. 3 E), a regulatory subunit of the protein phosphatase PP1 that dephosphorylates and thus inactivates Snf1 in the presence of glucose (Hedbacker and Carlson, 2008). This interaction involves the N-terminal region of Rod1 harboring the arrestin domain. To evaluate a putative role for PP1 in Rod1 regulation, we assessed the Rod1 phosphorylation status in a reg1Δ strain. Indeed, Rod1 was strongly phosphorylated in this strain (Fig. 3 F), even in glucose-grown cells, and the rapid dephosphorylation observed in WT cells during the lactate–glucose transition was strongly diminished (compare lane 3 with lane 6). Although we cannot exclude that PP1 regulates Rod1 phosphorylation through the regulation of Snf1 activity, the physical interaction between Rod1 and Reg1 as well as the rapid kinetics of Rod1 dephosphorylation in response to glucose strongly suggest that Rod1 is a new direct target of PP1. Importantly, Rod1 was not ubiquitylated in response to glucose in the reg1Δ mutant (Fig. 3 F, lane 9), further confirming that Rod1 dephosphorylation is required for its ubiquitylation. Therefore, Rod1 activation requires Reg1. Consistent with this, the glucose-induced endocytosis of Jen1 was impaired in the reg1Δ mutant (Fig. 3, G and H; and Fig. S3 A), as reported previously for other sugar transporters (Jiang et al., 2000b; Horák, 2003; Mayordomo et al., 2003). Altogether, these results show that Rod1 ubiquitylation and activity are modulated by its phosphorylation status through the opposing activities of the Snf1 kinase and the PP1 phosphatase. Noteworthy, we confirmed that the Snf1/PP1 pathway was the only glucose-signaling pathway involved in the glucose-induced endocytosis of Jen1 (Fig. S3, B–G). Indeed, glucose-mediated Jen1 down-regulation was normal in cells deficient for the glucose sensors Snf3 and Rgt2 and the glucose receptor Gpr1 (Fig. S3, H and I).
A phosphorylation-dependent interaction of Rod1 with 14-3-3 proteins regulates Rod1 ubiquitylation
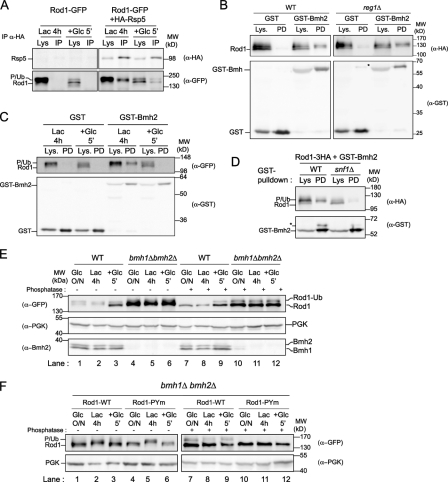
To better understand the molecular basis by which Rod1 dephosphorylation controls its ubiquitylation, we investigated the contribution of Rod1 phosphorylation on its interaction with Rsp5. However, coimmunoprecipitation experiments revealed that Rod1 interacted with Rsp5 under all growth conditions (Fig. 4 A), indicating that Rod1 phosphorylation status does not alter its ability to interact with Rsp5. An alternative explanation to the coupling between Rod1 dephosphorylation and its ubiquitylation came from the identification of Bmh1, one of the two redundant 14-3-3 proteins in yeast (with Bmh2), as a candidate in the yeast two-hybrid screen using Rod1 as a bait. 14-3-3 proteins are a family of conserved eukaryotic proteins involved in many cellular functions, including signaling pathways, and often interact with their partners in a phosphorylation-dependent manner (Bridges and Moorhead, 2005). To confirm this interaction in vivo, we first performed a GST pull-down using a GST-Bmh2 construct expressed in WT cells coexpressing Rod1-3HA. This confirmed that indeed, when cells are grown overnight in glucose medium, Rod1 was able to interact with Bmh2 (Fig. 4 B). Previous reports have shown that 14-3-3 proteins interact with Reg1 (Mayordomo et al., 2003; Dombek et al., 2004); however, the interaction between Rod1 and Bmh2 did not depend on the presence of Reg1 because Rod1 and Bmh2 were still able to interact in a reg1Δ deletion mutant (Fig. 4 B). Given that Rod1 phosphorylation depends on glucose availability, we assessed the dynamics of the interaction between Rod1 and Bmh2 in various conditions. As shown in Fig. 4 C, Rod1 strongly associated with Bmh2 when the cells were grown in lactate medium, but this interaction vanished in response to glucose, when Rod1 is dephosphorylated. Similarly, in the snf1Δ mutant, Rod1 failed to interact with Bmh2 (Fig. 4 D). This confirms that the Snf1-dependent phosphorylation of Rod1 tunes its interaction with the 14-3-3 proteins.
Figure 4.
Rod1 ubiquitylation is regulated by a phospho-dependent binding to 14-3-3 proteins. (A) WT cells expressing a plasmid-encoded Rod1-GFP construct were transformed with an empty plasmid or a plasmid encoding 3HA-Rps5. Immunoprecipitation of 3HA-Rsp5 was performed from cells grown as indicated. The input (Lys) and immunoprecipitated (IP) fractions were immunoblotted with the indicated antibodies. (B) WT and reg1Δ cells expressing Rod1-3HA tagged at the chromosomal locus were transformed with a plasmid encoding GST-Bmh2 or GST alone as a control. GST pull-downs were performed from cells harvested after overnight growth in glucose medium (exponential phase). The input (Lys) and pull-down (PD) fractions were immunoblotted with the indicated antibodies. Asterisk indicates a nonspecific band that sometimes cross-reacted with the anti-GST antibody. (C) WT cells in which Rod1 is tagged with GFP at the chromosomal locus were transformed with a plasmid encoding GST-Bmh2 or GST alone. GST pull-downs were performed from cells grown as indicated, and the input (Lys) and pull-down (PD) fractions were immunoblotted with the indicated antibodies. (D) WT or snf1Δ strains in which Rod1 is tagged with GFP at the chromosomal locus were transformed with a plasmid encoding GST-Bmh2 or GST alone and grown overnight (exponential phase) in glucose medium. GST pull-downs were performed, and input (Lys) and pull-down (PD) fractions were immunoblotted with the indicated antibodies. Asterisk indicates a nonspecific band that sometimes cross-reacted with the anti-GST antibody. (E) WT or bmh1Δ bmh2Δ strains expressing a plasmid-encoded Rod1-GFP were grown as indicated. Total protein extracts were treated with phosphatase when indicated to better visualize ubiquitylation, and immunoblotted with the indicated antibodies. (F) bmh1Δ bmh2Δ strains expressing a plasmid-encoded Rod1-GFP (WT or PYm) were grown as indicated and treated as in E.
To determine the role of the 14-3-3 proteins on Rod1 ubiquitylation, we assessed its post-translational modifications in the bmh1Δ bmh2Δ double mutant (Roberts et al., 1997). In this mutant, Rod1 ubiquitylation was constitutive, regardless of the carbon source provided to the cells and of its phosphorylation status (Fig. 4, E and F), showing that in this mutant, these two modifications are uncoupled. These data indicate that the interaction between Rod1 and 14-3-3 coordinates Rod1 ubiquitylation in response to glucose availability.
Discussion
The endocytosis of yeast transporters has been described for many years to be triggered by nutritional changes, especially in the case of carbon sources transporters (Horák, 2003). Notably, the involvement of signaling pathway components in the regulation of the endocytosis of carbon sources transporters has been observed (Jiang et al., 2000a,b; Horak et al., 2002; Mayordomo et al., 2003), but there were no details as to how this regulation operates at the molecular level. Also, although it is known that the signal-induced ubiquitylation of transporters leads to their endocytosis (Haguenauer-Tsapis and André, 2004), the way by which this ubiquitylation promptly reacts to a change in nutrient availability remained unknown. Our results demonstrate that the yeast arrestin-related protein Rod1 relays intracellular signaling to transporter ubiquitylation and endocytosis.
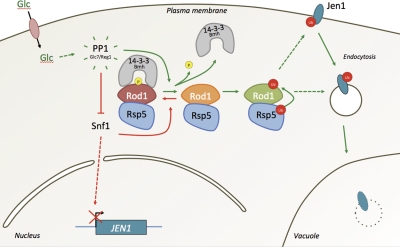
Indeed, we established that Rod1 is a new direct target of both the Snf1 kinase and the PP1 phosphatase, Glc7/Reg1—both of which play an essential role in the regulation of carbon source utilization and metabolism (Gancedo, 2008). Rod1 becomes rapidly activated in response to glucose replenishment, and acts as a downstream effector of the Snf1/PP1 pathway to mediate the catabolite inactivation of transporters for nonglucose carbon sources such as Jen1, providing a novel molecular basis for the regulation of transporter endocytosis by intracellular signaling. So far, the Snf1/PP1 signaling pathway was mostly documented for its role in transcriptional regulation in response to glucose (Gancedo, 2008). Snf1 modulates gene expression upon glucose depletion through the activation of the transcriptional activators Cat8 and Adr1, and inactivation of the Mig1 transcriptional repressor, while PP1 inactivates Snf1 and therefore allows adaptation to glucose medium. Interestingly, JEN1 promoter activity is itself regulated by Snf1, likely through the regulation of Mig1 and Cat8 (Bojunga and Entian, 1999; Lodi et al., 2002). Our results thus illustrate an integrated physiological response to glucose fluctuations where the same actors coordinate transcriptional and post-translational regulations (Fig. 5).
Figure 5.
Model for the regulation of transporter endocytosis by intracellular signaling through arrestin-related protein activation. When yeast cells are grown in lactate medium, Snf1, the yeast AMPK homologue, is active and phosphorylates Rod1 to inactivate it (red arrows). Glucose addition triggers Jen1 endocytosis, which depends on Rod1 activation through its PP1-mediated dephosphorylation and subsequent Rsp5-mediated ubiquitylation, which are coordinated by 14-3-3 proteins (green arrows). The subcellular compartment at which Rod1 acts on Jen1 endocytosis may be the plasma membrane, or internal compartments (dashed lines). See the Discussion for details. Noteworthy, the Snf1/PP1 pathway also controls the transcriptional reprogramming of cells in response to glucose fluctuation, including the expression of the JEN1 gene (Lodi et al., 2002), illustrating a robust physiological regulation in which transcriptional and post-translational events are coordinated.
We also uncovered a new example for the regulation of ubiquitylation by substrate dephosphorylation. Cross talks between various post-translational modifications have been described (Hunter, 2007), notably for substrates of F-box–containing E3 ligases that are ubiquitylated in a phosphorylation-dependent manner. In contrast, PP1-mediated dephosphorylation of Rod1 promotes its ubiquitylation and activation in response to a nutritional change. Rod1 ubiquitylation appears essential for Jen1 endocytosis, although the precise role of this modification on Rod1 function remains to be elucidated. The ubiquitylation of another arrestin-related protein, Art1, is also essential for its function in endocytosis; however, its ubiquitylation appears constitutive in the conditions tested (Lin et al., 2008), whereas Rod1 ubiquitylation is induced by glucose and may therefore provide an additional step for its regulation. The signal-dependent ubiquitylation of arrestin-related proteins has been described in filamentous fungi (Herranz et al., 2005; Hervás-Aguilar et al., 2010) and is reminiscent of β-arrestin ubiquitylation in mammalian cells in response to agonist treatment (DeWire et al., 2007); however, the precise mechanism by which this occurs is still unknown. Our findings are therefore likely to open new avenues toward understanding the regulation of the signal-induced ubiquitylation of arrestin-related proteins.
We further unraveled the involvement of 14-3-3 proteins in the coordination of this molecular switch by protecting phosphorylated Rod1 from ubiquitylation (Fig. 5). 14-3-3 proteins were reported to bind phosphorylated Nedd4-2, which inhibits its interaction with the sodium channel ENaC, and consequently impairs ENaC ubiquitylation and endocytosis (Bhalla et al., 2005; Yang and Kumar, 2010; Chandran et al., 2011). We uncovered a new level of regulation by showing that 14-3-3 proteins can also regulate an intramolecular cross talk between phosphorylation and ubiquitylation on a ubiquitin ligase adaptor protein. Strikingly, a proteomic study identified most of the yeast arrestin-related proteins as phospho-dependent interactants of 14-3-3 proteins, suggestive of their more general involvement in the regulation of arrestin-related proteins (Kakiuchi et al., 2007). Among them, Aly2/Art3 regulates the endocytosis of the basic amino acid transporter Dip5 (Hatakeyama et al., 2010), and is phosphorylated by Npr1 (O’Donnell et al., 2010), a protein kinase controlled by nitrogen limitation known to regulate endocytosis (De Craene et al., 2001; Haguenauer-Tsapis and André, 2004). Aly2 is therefore a likely candidate that could be regulated in a similar manner as Rod1, but in response to nitrogen signaling.
An important question that should now be addressed in more detail is the subcellular localization of yeast arrestin-related proteins, and the compartment at which they perform their function. It was initially proposed that they are recruited and act at the plasma membrane, and at least a partial localization at this compartment was observed for several of them (Lin et al., 2008; Herrador et al., 2010; O’Donnell et al., 2010). The fact that Rod1 is required for the glucose-induced ubiquitylation of Jen1 in an endocytic mutant (end3Δ; Fig. 1 D) suggests that Jen1 ubiquitylation indeed occurs at the plasma membrane. Rod1 is mainly a cytosolic protein, and although we did not observe a significant enrichment of Rod1 at the plasma membrane in response to glucose, this localization is still compatible with a transient function at this compartment (unpublished data). We also observed that a pool of Rod1 localizes to punctate structures (unpublished data). This is reminiscent of the observations concerning Ldb19/Art1, Aly1/Art6, or Aly2/Art3 that also localize to internal structures of an endosomal/trans-Golgi nature (Lin et al., 2008; O’Donnell et al., 2010). This localization does not preclude a direct involvement of yeast arrestin-related proteins in the signal-induced ubiquitylation of transporters at intracellular compartments. Indeed, it has been proposed that Rsp5 ubiquitylates the reductive iron transporter (Fet3/Ftr1 complex) at the level of endosomes (Strochlic et al., 2008). Hence, we cannot exclude that Rod1 acts, at least partially, at intracellular compartments (Fig. 5). Noteworthy, β-arrestins and the arrestin domain-containing protein ARRDC3 were proposed to regulate ubiquitylation of G protein–coupled receptor by Nedd4-like E3 ligases at endosomes (Bhandari et al., 2007; Shenoy et al., 2008; Nabhan et al., 2010), and it may very well be that some yeast arrestin-related proteins act likewise toward certain cargoes.
In mammalian cells, the endocytosis of several solute carriers (SLCs) is known to be influenced by various signaling pathways. Recently, EGF treatment of HeLa cells was reported to lead to an increase in the ubiquitylation of many SLCs, such as members of the monocarboxylate transporter/SLC16 family, which belong to the same family as Jen1 (Argenzio et al., 2011). Interestingly, in this paper SLC3A2 was reported to be modified by K63-linked ubiquitin chains, a type of ubiquitylation performed by several members of the Nedd4/Rsp5 ubiquitin ligase family (Kee et al., 2005; Kim et al., 2011; Maspero et al., 2011), suggesting a possible involvement of Nedd4 in the EGF-induced ubiquitylation of SLCs. Other SLCs, such as the dopamine transporter DAT/SLC6A3 and the cationic amino acid transporter CAT-1/SLC7A1 undergo a signal-induced, Nedd4-dependent ubiquitylation (Sorkina et al., 2006; Vina-Vilaseca et al., 2011). However, how Nedd4 recognizes its target SLCs in response to a specific signal is unknown. Interestingly, mammalian arrestin domain–containing (ARRDC) proteins, like their yeast counterparts, interact with Nedd4-related ubiquitin ligases (Zhang et al., 2010; Rauch and Martin-Serrano, 2011) and localize to the plasma membrane and/or along the endocytic pathway (Oka et al., 2006; Patwari et al., 2009; Rauch and Martin-Serrano, 2011; Vina-Vilaseca et al., 2011). In agreement with this, ARRDC3 was shown to mediate the ubiquitylation and down-regulation of plasma membrane receptors (Draheim et al., 2010; Nabhan et al., 2010). It would be interesting to know whether this family of proteins is also responsible for the signal-induced ubiquitylation of SLCs.
Among these ARRDC proteins, TXNIP/VDUP-1, which regulates insulin sensitivity and energy balance in mice (Chutkow et al., 2010), is particularly relevant to our paper because both TXNIP and Rod1 regulate glucose transport activity (Parikh et al., 2007; Lin et al., 2008; Nikko and Pelham, 2009). Cancer cells are known to display an enhanced glycolytic activity and impaired oxidative phosphorylation even under aerobic conditions (Warburg effect), similarly to S. cerevisiae when grown in the presence of glucose (Diaz-Ruiz et al., 2011). The high-affinity glucose transporters (GLUT1 and GLUT3) are overexpressed in several cancer cell lines, in which they are thought to allow an increase in glycolysis rate and thus promote tumor progression (Macheda et al., 2005). Inhibition of GLUT transporters has been used to impair the growth of tumor cells in vitro and in vivo (Cao et al., 2007; Young et al., 2011), and so was the inhibition of lactate transporters in vivo (Sonveaux et al., 2008). Therefore, addressing the potential involvement of ARRDC proteins in the regulation of carbon sources transporter, and understanding the molecular basis of their activation in this context, might be critical to the strategies of metabolic therapies (Nijsten and van Dam, 2009).
Materials and methods
Yeast strains, transformation, and growth conditions
Strains are listed and detailed in Table S1. They are derivatives of the ∑1278b background for the bmh1bmh2Δ strains, W303 for snf1Δ mig1Δ, snf1Δ mig1Δ rod1Δ, and snf3Δ rgt2Δ gpr1Δ mth1Δ strains, and BY4741/2 background for all other strains. Yeast two-hybrid screening was performed using the CTY10.5d and TAT7 strains. Yeast was transformed by standard lithium acetate/polyethylene glycol procedure. Cells were grown in yeast extract/peptone/glucose (YPD) rich medium, or in synthetic complete (SC) medium containing 2% (wt/vol) Glc, or 0.5% (vol/vol) Na-lactate, pH 5.0 (Sigma-Aldrich).
Cells were grown overnight in SC-Glc, harvested in early exponential phase (A600: 0.3), resuspended in SC-lactate, and grown for 4 h (A600: 0.6) before the addition of glucose (2% wt/vol, final concentration).
For galactose induction, cells were precultured in SC-Glc medium and grown overnight to early exponential phase (A600: 0.3) in SC medium containing 2% raffinose (wt/vol) and 0.02% Glc (wt/vol) to initiate growth. Galactose was then added at a final concentration of 2% (wt/vol) and cells were grown for the indicated times. Chase/endocytosis was started by adding glucose to a final concentration of 2% (wt/vol) and incubating for the indicated times. For galactose induction of Jen1 in Fig. 3 D, cells were directly transferred from an overnight glucose culture to a galactose-containing medium due to the slow growth of the snf1Δ mig1Δ mutant in raffinose.
Plasmid constructs
The plasmids used in this paper are described in Table S2. The pROD1 :ROD1-GFP and the pROD1:ROD1-3HA fusion were amplified from BY4741 genomic DNA, inserted into pCRII-ZeroBlunt (Invitrogen), and subcloned into SacI–XmaI sites in pRS416 (pSL93) and in pRS415 (pSL94), respectively. Site-directed mutageneses were performed by PCR using mutagenic oligonucleotides using pSL93 or pSL94 as templates. Mutagenized plasmids were checked by sequencing, and a region containing the mutation was systematically subcloned into the original pSL93/pSL94 using endogenous restriction sites to prevent mutations that may have occurred elsewhere on the plasmid. Mutation of the PPxY motifs in PPxA led to pSL118 and pSL119, respectively. Synthetic constructs from Eurofins-MWG/Operon (Ebersberg, Germany) were used to construct the lysine mutant plasmids (see Fig. S2). The pROD1:ROD1-K0-Nter-3HA (pSL133) was obtained from a synthetic construct (lysines 10–367 mutated to arginine; #1 in Fig. S2 A) after subcloning into endogenous PacI–BspEI sites in pSL94. The pROD1 :ROD1-K0-Cter-3HA (pSL104) construct was obtained from a synthetic construct (lysines 401–833 mutated to arginine; #2 in Fig. S2 A) after subcloning into endogenous BspEI–XmaI sites in pSL93 and pSL94, respectively. The pROD1:ROD1-K0-3HA (pSL134) was obtained by subcloning a PacI–BspEI fragment of pSL133 (C-terminal KR mutant) into PacI–BspEI sites in pSL104 (N-terminal KR mutant). The pROD1:ROD1-K0-Nter-3HA (pSL133) was digested with NsiI–StuI and subcloned at NsiI–StuI sites in pSL94, giving rise to pROD1:ROD1-K0-ArrN-3HA (pSL137; #3 in Fig. S2 A). The complementary construct was generated by cloning a NsiI–StuI fragment of pSL94 into pSL133 at NsiI–StuI sites, giving pROD1:ROD1-K0-ArrC-3HA (pSL138; #4 in Fig. S2 A). Site-directed mutagenesis was performed on pSL138 to mutate three arginines R361, R365, and R367 back to lysines to obtain the pROD1:ROD1-K16R-3HA (pSL140). The pROD1:ROD1-K7R-3HA (pSL143; #5 in Fig. S2 A) and pROD1:ROD1-K9R-3HA (pSL146; #6 in Fig. S2 A) constructs were obtained from a synthetic gene after subcloning at StuI–BspEI sites in pSL94. The pROD1:ROD1-K(3–9)R-3HA (pSL148; #7 in Fig. S2 A) was obtained by subcloning a PacI–NheI fragment from pSL143 into the PacI–NheI sites of pSL146. The pROD1:ROD1-KR-3HA construct (pSL147; #8 in Fig. S2 A), which contains four lysines mutated to arginine, was constructed by subcloning a PacI–NheI fragment from pSL146 into the PacI–NheI sites of pSL143. For the yeast two-hybrid, plasmids encoding LexA-Rod1 and LexA-Rod1 (394–837) were constructed by cloning the corresponding ROD1 coding sequence in pLexA (1–202)+PL or pBTM116. Plasmids encoding GAD-Rsp5, GAD-Bmh1 and GAD-Reg1 were constructed by cloning the corresponding ORF in the polylinker of pACT2 (Takara Bio Inc.).
Total protein extracts, coimmunoprecipitations, and in vivo GST pull-downs
For total protein extracts, 1 ml of cells were harvested and precipitated on ice for 10 min with trichloroacetic acid (TCA; Sigma-Aldrich) to a final concentration of 10%. Cells were lysed with glass beads in a 100-µl volume for 10 min at 4°C and the pellet was resuspended in TCA-sample buffer (50 mM Tris-HCl, pH 6.8, 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, and 10% glycerol containing 200 mM of unbuffered Tris solution) at a concentration of 50 µl/initial OD unit, before being denatured at 37°C for 10 min.
Coimmunoprecipitations were performed using 20–30 OD units of cells as described below and in Léon et al. (2008). Cells were resuspended in immunoprecipitation (IP) lysis buffer (0.1 M Tris HCl, pH 8, 0.2 M NaCl, 5% glycerol, 1 mM EDTA, 0.1% Triton X-100, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and EDTA-free protease inhibitor cocktail “Complete” (Roche) at a concentration of 1 OD unit/10 µl. Cells were disrupted with glass beads at 4°C for 10 min. Cell debris and unbroken cells were removed by a centrifugation of 5 min at 3,000 g, and the supernatant (“Input”) was incubated with GammaBind-Sepharose beads previously incubated with the required antibody for 1 h at 4°C. The unbound fraction was eliminated by a 10-s centrifugation in a mini-centrifuge; beads were then washed three times for 10 min (at 4°C) with 1 ml of IP lysis buffer before elution of the immunoprecipitate with SDS sample buffer (1 µl/initial OD).
In vivo GST pull-down experiments were performed using the pGST (empty) and pGST-Bmh2 plasmids kindly provided by Dr. Pascual Sanz (Consejo Superior de Investigaciones Científicas [CSIC], Valencia, Spain; Mayordomo et al., 2003) following the same protocol as for the coimmunoprecipitations, but using GSH-Sepharose beads (GE Healthcare).
GST-WW recombinant protein purification and in vitro GST pull-downs
GST-WW protein was expressed and purified from bacteria using a plasmid kindly provided by Dr. Catherine Dargemont (Institut Jacques Monod, Paris, France; Gwizdek et al., 2005) and purified in native conditions as follows. Bacteria were grown overnight, inoculated at 1/100 in fresh medium, and grown for 2 h. Cells were then treated with ethanol (10% final concentration) on ice for 10 min, and transferred back at 37°C for 30 min. After 2 h, protein expression was induced by adding 2.5 mg/ml IPTG and cells were grown overnight at 18°C. Bacteria were centrifuged and lysed by sonication in 10 ml cold PBS 1x/NaCl 250 mM, supplemented with protease inhibitors (Complete, EDTA-free mixture; Roche). After centrifugation (18,000 g for 20 min), the lysate was incubated with GSH-Sepharose beads for 1 h at 4°C. The beads were resuspended with wash buffer (1x PBS, 0.5 M NaCl, 0.1% Triton X-100, and 10% glycerol) and then loaded onto a glass Econo-column chromatography column (Bio-Rad Laboratories). Beads were then washed with an additional 40 ml of wash buffer and recombinant proteins were finally eluted with elution buffer (50 mM Tris, pH 8, and 10 mM glutathione). The eluate was dialyzed overnight at 4°C in a dialysis tubing (MWCO 10,000; Carl Roth Gmbh) in 1,000 volumes of dialysis buffer (1x PBS, 250 mM NaCl, and 10% glycerol) to eliminate the glutathione. Proteins were assayed for concentration and frozen at −80°C.
GST pull-down experiments were performed using 40 µg of recombinant proteins bound to 50 µl GSH-Sepharose beads. Crude extract incubations, washes, and elutions were performed as for the immunoprecipitation experiments.
His6-tagged ubiquitin protein conjugate purification
Cells were transformed with a plasmid expressing 6×Histidine-tagged ubiquitin under the control of the copper-inducible CUP1 promoter, kindly provided by Dr. Catherine Dargemont. These experiments were performed using 100 OD units of cells, grown in exponential phase (A600 = 0.6), without addition of extra copper to avoid His6-tagged ubiquitin overexpression. In this situation, His6-ubiquitin expression level was similar to that of endogenous ubiquitin as determined by immunoblotting of crude extracts using an anti-ubiquitin antibody (clone P4D1; Santa Cruz Biotechnology, Inc.). Purification of His6-tagged ubiquitin protein conjugates was then performed as described below following a protocol adapted from Ziv et al. (2011). Cells were precipitated with TCA to a final concentration of 10% for 30 min on ice, and lysed with glass beads for 20 min at 4°C. After centrifugation, the pellet was resuspended with 1 ml of guanidium buffer (6 M GuHCl, 20 mM Tris-HCl, pH 8, 100 mM K2HPO4, 10 mM imidazole, 100 mM NaCl, and 0.1% Triton X-100) and incubated for 1 h at room temperature on a rotating platform. After centrifugation (16,000 g for 10 min at room temperature), the lysate was incubated for 2.5 h with nickel-nitriloacetic acid beads (Ni-NTA superflow; QIAGEN). The beads were then washed three times with guanidium buffer, three times with wash buffer 1 (20 mM Tris-HCl, pH 8.0, 100 mM K2HPO4, 20 mM imidazole, 100 mM NaCl, and 0.1% Triton X-100) and three times with wash buffer 2 (20 mM Tris-HCl, pH 8.0, 100 mM K2HPO4, 10 mM imidazole, 1 M NaCl, and 0.1% Triton X-100). His6-ubiquitin–conjugated proteins were finally eluted with 40 µl of elution buffer (50 mM Tris-HCl, pH 8.0, and 250 mM imidazole) for 10 min at room temperature.
Purification of His6-tagged Jen1-GFP
The purification of Jen1-GFP-His6 was performed in denaturing conditions from end3Δ or art4Δ end3Δ cells transformed with pRHT373, encoding pGAL:Jen1-GFP-His, using nickel-nitriloacetic acid beads (Ni-NTA superflow; QIAGEN). Cells were first grown overnight in SD medium containing 2% raffinose and 0.02% glucose to initiate growth. Then, galactose was added to a final concentration of 2% for 2 h. Ubiquitylation was induced by addition of glucose (2% final concentration) for 10 min. Cells were harvested and resuspended on ice in 400 µl of cold lysis buffer (100 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 10 mM N-ethylmaleimide, 1 mM phenylmethylsulfonyl fluoride, and EDTA-free protease inhibitor cocktail “Complete” (Roche). Cells were lysed with glass beads and the lysate was centrifuged at 1,500 g for 5 min (at 4°C) to remove cell debris. The supernatant was further centrifuged for 30 min at 13,000 g (at 4°C). The pellet, enriched in plasma membrane proteins, was resuspended in 300 µl of buffer “M” (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% SDS, and 1% Triton X-100), and left on ice for 30 min to allow for membrane proteins solubilization. Then, TCA was added to a final concentration of 10% and the sample was incubated on ice for 30 min. Precipitated proteins were collected by centrifugation (13,000 g for 10 min). The pellet was washed briefly in 500 µl of unbuffered Tris solution (1 M) to neutralize the sample, and was resuspended with 800 µl of buffer “His” (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, and 5 mM imidazole). The sample was then centrifuged for 10 min at 13,000 g to remove unsolubilized material, and the supernatant was incubated with 50 µl of prewashed Ni-NTA beads (Ni-NTA superflow; QIAGEN) for 1 h at 4°C, in rotation. After a brief centrifugation (10 s on a mini-centrifuge), the supernatant (unbound fraction) was collected and the beads were washed three times with buffer “His”. Elution was performed with 100 µl of elution buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.5% SDS, 1% Triton X-100, and 200 mM imidazole) after a 10-min incubation at 4°C. The eluate (8 µl) was loaded on SDS-PAGE gels.
Antibodies and immunoblotting
We used monoclonal antibodies raised against GFP (clones 7.1 and 13.1; Roche), actin (clone C4; MP Biomedicals), HA (clone F7; Santa Cruz Biotechnology, Inc.), anti-ubiquitin antibody coupled to horseradish peroxidase (clone P4D1; Santa Cruz Biotechnology, Inc.), and polyclonal antibodies against GFP (a gift from Naima Belgareh, R.H. Tsapis laboratory), HA (Y11; Santa Cruz Biotechnology, Inc.), 3-phosphoglycerate kinase (PGK; clone 22CS; Invitrogen), GST (Z5; Santa Cruz Biotechnology, Inc.), FBPase (a gift from Dr. Carlos Gancedo, CSIC, Madrid, Spain), and Bmh2 (a gift from Dr. Sandra Lemmon, University of Miami, Coral Gables FL; Gelperin et al., 1995). Images were acquired with the LAS-4000 imaging system (Fujifilm). Quantification was performed using ImageJ (National Institutes of Health) on nonsaturated blots.
Yeast two-hybrid analysis
The two-hybrid screen for LexA-Rod1 (from a pBTM116 derivative; Table S1)–interacting proteins was performed in strain TAT7 (Table S2) co-transformed with a library of S. cerevisiae cDNAs fused to GAD in vector pACT (a gift of S. Elledge, Baylor University, Waco, TX). His+ transformants (2 × 106 clones) were selected in the presence of 5 mM 3-aminotriazole (3-AT) and subsequently screened for β-galactosidase activity using a filter assay. Two-hybrid assays were performed in strain CTY10.5d (Table S2) with pLexA (1–202)+PL and pACT2 plasmid derivatives (Table S1). β-Galactosidase was quantitatively assayed in permeabilized yeast cells grown to mid-log phase and expressed in Miller units. Values are average β-galactosidase activities for at least four transformants and standard errors were <20%.
Fluorescence microscopy
Cells were mounted in medium and observed with a motorized fluorescence microscope (model BX-61; Olympus) equipped with a PlanApochromat 100x oil-immersion objective (1.40 NA; Olympus), a Spot 4.05 charge-coupled device camera, and the MetaVue acquisition software (Molecular Devices). Cells were mounted in SD medium and imaged at room temperature. GFP-tagged proteins were visualized using a GFP II filter (excitation wavelength 440–470 nm; Chroma Technology Corp.). Images were processed in ImageJ and Photoshop (Adobe) for levels.
Transport assays
Cells incubated under induction conditions were harvested by centrifugation and washed twice in ice-cold deionized water to a final concentration of ∼25–40 mg dry weight/ml. Conical centrifuge tubes containing 30 µl of 0.1 M KH2PO4 buffer at pH 5.0 and 10 µl of the yeast suspension were incubated for 2 min at 25°C. The reaction was started by the addition of 10 µl of an aqueous solution of 4,000 d.p.m./nmol of radiolabeled l-[14C]lactic acid (sodium salt; GE Healthcare) at pH 5.0. The reaction was stopped by dilution with 5 ml of ice-cold water. The reaction mixtures were filtered immediately through GF/C membranes (GE Healthcare) and the filters were washed with 10 ml of ice-cold water and transferred to scintillation fluid (Opti-Phase HiSafe II; Pharmacia LKB). Radioactivity was measured in a liquid scintillation spectrophotometer (Tri-Carb 2200 CA; Packard Instrument Co.) equipped with a d.p.m. correction facility. For nonspecific adsorption of 14C, labeled lactic acid was added at time zero after the cold water. All the experiments were repeated at least three times, and the data reported represent the average values. Representative experiments of at least three repetitions are presented. The data obtained are represented as the mean ± SEM of triplicate measurements.
Other methods
Dephosphorylation of yeast protein extracts was performed on protein extracts prepared as described above, and proteins were resuspended in SDS sample buffer. Samples were diluted 1:10 with phosphatase buffer and incubated for 2 h at 37°C with or without calf intestinal alkaline phosphatase, as recommended by the supplier (Roche). Proteins were then precipitated again with 10% trichloroacetic acid, resuspended in sample buffer, and analyzed by immunoblotting.
The invertase assay was performed as in Vallier and Carlson (1994). In brief, glucose-repressed and -derepressed cells were prepared from exponentially growing cultures in high glucose (2%) or after shifting to low glucose (0.05%) for 2.5 h. Secreted invertase was quantitatively assayed by using whole cells washed twice with cold sodium azide at 10 mM. Samples (5–50 µl) were incubated with 200 µl 0.1 M sodium acetate, pH 5.1, for 20 min at 37°C after the addition of 20 µL of 40% sucrose. Glucose standards (2.5–15 µg) were used for calibration. The reaction was stopped by boiling the tubes after addition of 200 µl of potassium phosphate (200 mM, pH 7.0). 1.0 mL glucose oxidase reagent was added, and the tubes were incubated at 37°C for 30 min. The absorbance was measured at 540 nm after addition of 1 mL of HCl 6N. Activity is expressed as micromoles of glucose released per minute per 100 mg (dry weight) of cells.
Online supplemental material
Tables S1 and S2 list yeast strains and plasmids used in this paper, respectively. Fig. S1 describes that Rod1 is the only yeast arrestin-like protein involved in the glucose-induced inactivation of lactate transport activity, but is not a general regulator of glucose signaling. Fig. S2 shows the mapping strategy to identify Rod1 lysines that are targets for ubiquitylation. Fig. S3 demonstrates that the Snf1/PP1 glucose-signaling pathway regulates the glucose-induced endocytosis of Jen1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201109113/DC1.
Acknowledgments
We thank Naima Belgareh, Catherine Dargemont, Gerald Fink, Juana-Maria Gancedo, Carlos Gancedo, Sandra Lemmon, Hans Ronne, Pascual Sanz, Johan Thévelein, and Teresa Zoladek for reagents; Catherine Dargemont, Valérie Doye, and Lionel Pintard (IJM, Paris, France) and members of R.H.T.’s laboratory for helpful discussions and support; and Valérie Doye, Isabelle Jupin and Lionel Pintard (IJM, Paris, France) as well as Bruno André (Université Libre de Bruxelles, Belgium) for critical reading of the manuscript.
Work in R. Haguenauer-Tsapis’s laboratory was supported by the Centre National de la Recherche Scientifique (CNRS) and Université Paris Diderot, as well as by grants from the Ligue Nationale Contre le Cancer (Comité de Paris, grant RS09/75-26) and the EU sixth Framework Program (Role of Ubiquitin & Ubiquitin-like Modifiers in Cellular Regulation: RUBICON NoE, contract LSHG-CT-2005-018683; and Marie Curie RTN: UBIREGULATORS, contract MRTN-CT-2006-034555) to R.H. Tsapis, and by a grant from the Association pour la Recherche sur le Cancer (ARC, grant SFI20101201844) to S. Léon. Work in O. Vincent’s laboratory was supported by grants from the Spanish CICYT (BFU2008-02005), the Comunidad de Madrid and CSIC (CCG08-CSIC/SAL-3611) to O. Vincent. M. Becuwe is the recipient of a PhD fellowship from the French Ministère de l’Enseignement Supérieur et de la Recherche.
Footnotes
Abbreviations used in this paper:
- AMPK
- 5′ adenosine monophosphate-activated protein kinase
- PP1
- protein phosphatase 1
- SLC
- solute carrier protein
- WT
- wild type
References
- Alvarez C.E. 2008. On the origins of arrestin and rhodopsin. BMC Evol. Biol. 8:222 10.1186/1471-2148-8-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andoh T., Hirata Y., Kikuchi A. 2002. PY motifs of Rod1 are required for binding to Rsp5 and for drug resistance. FEBS Lett. 525:131–134 10.1016/S0014-5793(02)03104-6 [DOI] [PubMed] [Google Scholar]
- Argenzio E., Bange T., Oldrini B., Bianchi F., Peesari R., Mari S., Di Fiore P.P., Mann M., Polo S. 2011. Proteomic snapshot of the EGF-induced ubiquitin network. Mol. Syst. Biol. 7:462 10.1038/msb.2010.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla V., Daidié D., Li H., Pao A.C., LaGrange L.P., Wang J., Vandewalle A., Stockand J.D., Staub O., Pearce D. 2005. Serum- and glucocorticoid-regulated kinase 1 regulates ubiquitin ligase neural precursor cell-expressed, developmentally down-regulated protein 4-2 by inducing interaction with 14-3-3. Mol. Endocrinol. 19:3073–3084 10.1210/me.2005-0193 [DOI] [PubMed] [Google Scholar]
- Bhandari D., Trejo J., Benovic J.L., Marchese A. 2007. Arrestin-2 interacts with the ubiquitin-protein isopeptide ligase atrophin-interacting protein 4 and mediates endosomal sorting of the chemokine receptor CXCR4. J. Biol. Chem. 282:36971–36979 10.1074/jbc.M705085200 [DOI] [PubMed] [Google Scholar]
- Bojunga N., Entian K.D. 1999. Cat8p, the activator of gluconeogenic genes in Saccharomyces cerevisiae, regulates carbon source-dependent expression of NADP-dependent cytosolic isocitrate dehydrogenase (Idp2p) and lactate permease (Jen1p). Mol. Gen. Genet. 262:869–875 10.1007/s004380051152 [DOI] [PubMed] [Google Scholar]
- Bridges D., Moorhead G.B. 2005. 14-3-3 proteins: a number of functions for a numbered protein. Sci. STKE. 2005:re10 10.1126/stke.2962005re10 [DOI] [PubMed] [Google Scholar]
- Cao X., Fang L., Gibbs S., Huang Y., Dai Z., Wen P., Zheng X., Sadee W., Sun D. 2007. Glucose uptake inhibitor sensitizes cancer cells to daunorubicin and overcomes drug resistance in hypoxia. Cancer Chemother. Pharmacol. 59:495–505 10.1007/s00280-006-0291-9 [DOI] [PubMed] [Google Scholar]
- Casal M., Paiva S., Queirós O., Soares-Silva I. 2008. Transport of carboxylic acids in yeasts. FEMS Microbiol. Rev. 32:974–994 10.1111/j.1574-6976.2008.00128.x [DOI] [PubMed] [Google Scholar]
- Chandran S., Li H., Dong W., Krasinska K., Adams C., Alexandrova L., Chien A., Hallows K.R., Bhalla V. 2011. Neural precursor cell-expressed developmentally down-regulated protein 4-2 (Nedd4-2) regulation by 14-3-3 protein binding at canonical serum and glucocorticoid kinase 1 (SGK1) phosphorylation sites. J. Biol. Chem. 286:37830–37840 10.1074/jbc.M111.293233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutkow W.A., Birkenfeld A.L., Brown J.D., Lee H.Y., Frederick D.W., Yoshioka J., Patwari P., Kursawe R., Cushman S.W., Plutzky J., et al. 2010. Deletion of the alpha-arrestin protein Txnip in mice promotes adiposity and adipogenesis while preserving insulin sensitivity. Diabetes. 59:1424–1434 10.2337/db09-1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene J.O., Soetens O., Andre B. 2001. The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. J. Biol. Chem. 276:43939–43948 10.1074/jbc.M102944200 [DOI] [PubMed] [Google Scholar]
- DeWire S.M., Ahn S., Lefkowitz R.J., Shenoy S.K. 2007. Beta-arrestins and cell signaling. Annu. Rev. Physiol. 69:483–510 10.1146/annurev.physiol.69.022405.154749 [DOI] [PubMed] [Google Scholar]
- Diaz-Ruiz R., Rigoulet M., Devin A. 2011. The Warburg and Crabtree effects: On the origin of cancer cell energy metabolism and of yeast glucose repression. Biochim. Biophys. Acta. 1807:568–576 10.1016/j.bbabio.2010.08.010 [DOI] [PubMed] [Google Scholar]
- Dombek K.M., Kacherovsky N., Young E.T. 2004. The Reg1-interacting proteins, Bmh1, Bmh2, Ssb1, and Ssb2, have roles in maintaining glucose repression in Saccharomyces cerevisiae. J. Biol. Chem. 279:39165–39174 10.1074/jbc.M400433200 [DOI] [PubMed] [Google Scholar]
- Draheim K.M., Chen H.B., Tao Q., Moore N., Roche M., Lyle S. 2010. ARRDC3 suppresses breast cancer progression by negatively regulating integrin beta4. Oncogene. 29:5032–5047 10.1038/onc.2010.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo J.M. 2008. The early steps of glucose signalling in yeast. FEMS Microbiol. Rev. 32:673–704 10.1111/j.1574-6976.2008.00117.x [DOI] [PubMed] [Google Scholar]
- Gelperin D., Weigle J., Nelson K., Roseboom P., Irie K., Matsumoto K., Lemmon S. 1995. 14-3-3 proteins: potential roles in vesicular transport and Ras signaling in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 92:11539–11543 10.1073/pnas.92.25.11539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenson M. 1983. Inactivation-reactivation process and repression of permease formation regulate several ammonia-sensitive permeases in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 133:135–139 10.1111/j.1432-1033.1983.tb07438.x [DOI] [PubMed] [Google Scholar]
- Gwizdek C., Hobeika M., Kus B., Ossareh-Nazari B., Dargemont C., Rodriguez M.S. 2005. The mRNA nuclear export factor Hpr1 is regulated by Rsp5-mediated ubiquitylation. J. Biol. Chem. 280:13401–13405 10.1074/jbc.C500040200 [DOI] [PubMed] [Google Scholar]
- Haguenauer-Tsapis R., André B.2004. Membrane trafficking of yeast transporters: mechanisms and physiological control of downregulation. In Topics in Current Genetics Control of Transmembrane Transport. Vol 9 R. Boles E and Krämer, editors. Springer Verlag.
- Hatakeyama R., Kamiya M., Takahara T., Maeda T. 2010. Endocytosis of the aspartic acid/glutamic acid transporter Dip5 is triggered by substrate-dependent recruitment of the Rsp5 ubiquitin ligase via the arrestin-like protein Aly2. Mol. Cell. Biol. 30:5598–5607 10.1128/MCB.00464-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedbacker K., Carlson M. 2008. SNF1/AMPK pathways in yeast. Front. Biosci. 13:2408–2420 10.2741/2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein C., Springael J.Y., Volland C., Haguenauer-Tsapis R., André B. 1995. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol. Microbiol. 18:77–87 10.1111/j.1365-2958.1995.mmi_18010077.x [DOI] [PubMed] [Google Scholar]
- Herrador A., Herranz S., Lara D., Vincent O. 2010. Recruitment of the ESCRT machinery to a putative seven-transmembrane-domain receptor is mediated by an arrestin-related protein. Mol. Cell. Biol. 30:897–907 10.1128/MCB.00132-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz S., Rodríguez J.M., Bussink H.J., Sánchez-Ferrero J.C., Arst H.N., Jr, Peñalva M.A., Vincent O. 2005. Arrestin-related proteins mediate pH signaling in fungi. Proc. Natl. Acad. Sci. USA. 102:12141–12146 10.1073/pnas.0504776102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervás-Aguilar A., Galindo A., Peñalva M.A. 2010. Receptor-independent Ambient pH signaling by ubiquitin attachment to fungal arrestin-like PalF. J. Biol. Chem. 285:18095–18102 10.1074/jbc.M110.114371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer H. 1976. Catabolite inactivation. Trends Biochem. Sci. 1:178–181 [Google Scholar]
- Horák J. 2003. The role of ubiquitin in down-regulation and intracellular sorting of membrane proteins: insights from yeast. Biochim. Biophys. Acta. 1614:139–155 10.1016/S0005-2736(03)00195-0 [DOI] [PubMed] [Google Scholar]
- Horak J., Wolf D.H. 1997. Catabolite inactivation of the galactose transporter in the yeast Saccharomyces cerevisiae: ubiquitination, endocytosis, and degradation in the vacuole. J. Bacteriol. 179:1541–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak J., Regelmann J., Wolf D.H. 2002. Two distinct proteolytic systems responsible for glucose-induced degradation of fructose-1,6-bisphosphatase and the Gal2p transporter in the yeast Saccharomyces cerevisiae share the same protein components of the glucose signaling pathway. J. Biol. Chem. 277:8248–8254 10.1074/jbc.M107255200 [DOI] [PubMed] [Google Scholar]
- Hunter T. 2007. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell. 28:730–738 10.1016/j.molcel.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Jiang H., Medintz I., Zhang B., Michels C.A. 2000a. Metabolic signals trigger glucose-induced inactivation of maltose permease in Saccharomyces. J. Bacteriol. 182:647–654 10.1128/JB.182.3.647-654.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Tatchell K., Liu S., Michels C.A. 2000b. Protein phosphatase type-1 regulatory subunits Reg1p and Reg2p act as signal transducers in the glucose-induced inactivation of maltose permease in Saccharomyces cerevisiae. Mol. Gen. Genet. 263:411–422 10.1007/s004380051185 [DOI] [PubMed] [Google Scholar]
- Kakiuchi K., Yamauchi Y., Taoka M., Iwago M., Fujita T., Ito T., Song S.Y., Sakai A., Isobe T., Ichimura T. 2007. Proteomic analysis of in vivo 14-3-3 interactions in the yeast Saccharomyces cerevisiae. Biochemistry. 46:7781–7792 10.1021/bi700501t [DOI] [PubMed] [Google Scholar]
- Kee Y., Lyon N., Huibregtse J.M. 2005. The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J. 24:2414–2424 10.1038/sj.emboj.7600710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y., Muñoz W., Lyon N., Huibregtse J.M. 2006. The deubiquitinating enzyme Ubp2 modulates Rsp5-dependent Lys63-linked polyubiquitin conjugates in Saccharomyces cerevisiae. J. Biol. Chem. 281:36724–36731 10.1074/jbc.M608756200 [DOI] [PubMed] [Google Scholar]
- Kim H.C., Steffen A.M., Oldham M.L., Chen J., Huibregtse J.M. 2011. Structure and function of a HECT domain ubiquitin-binding site. EMBO Rep. 12:334–341 10.1038/embor.2011.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon S., Haguenauer-Tsapis R. 2009. Ubiquitin ligase adaptors: regulators of ubiquitylation and endocytosis of plasma membrane proteins. Exp. Cell Res. 315:1574–1583 10.1016/j.yexcr.2008.11.014 [DOI] [PubMed] [Google Scholar]
- Léon S., Erpapazoglou Z., Haguenauer-Tsapis R. 2008. Ear1p and Ssh4p are new adaptors of the ubiquitin ligase Rsp5p for cargo ubiquitylation and sorting at multivesicular bodies. Mol. Biol. Cell. 19:2379–2388 10.1091/mbc.E08-01-0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.H., MacGurn J.A., Chu T., Stefan C.J., Emr S.D. 2008. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 135:714–725 10.1016/j.cell.2008.09.025 [DOI] [PubMed] [Google Scholar]
- Lodi T., Fontanesi F., Guiard B. 2002. Co-ordinate regulation of lactate metabolism genes in yeast: the role of the lactate permease gene JEN1. Mol. Genet. Genomics. 266:838–847 10.1007/s00438-001-0604-y [DOI] [PubMed] [Google Scholar]
- Lucero P., Lagunas R. 1997. Catabolite inactivation of the yeast maltose transporter requires ubiquitin-ligase npi1/rsp5 and ubiquitin-hydrolase npi2/doa4. FEMS Microbiol. Lett. 147:273–277 10.1111/j.1574-6968.1997.tb10253.x [DOI] [PubMed] [Google Scholar]
- Macheda M.L., Rogers S., Best J.D. 2005. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J. Cell. Physiol. 202:654–662 10.1002/jcp.20166 [DOI] [PubMed] [Google Scholar]
- Maspero E., Mari S., Valentini E., Musacchio A., Fish A., Pasqualato S., Polo S. 2011. Structure of the HECT:ubiquitin complex and its role in ubiquitin chain elongation. EMBO Rep. 12:342–349 10.1038/embor.2011.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayordomo I., Regelmann J., Horak J., Sanz P. 2003. Saccharomyces cerevisiae 14-3-3 proteins Bmh1 and Bmh2 participate in the process of catabolite inactivation of maltose permease. FEBS Lett. 544:160–164 10.1016/S0014-5793(03)00498-8 [DOI] [PubMed] [Google Scholar]
- Medintz I., Jiang H., Han E.K., Cui W., Michels C.A. 1996. Characterization of the glucose-induced inactivation of maltose permease in Saccharomyces cerevisiae. J. Bacteriol. 178:2245–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M., Sorkin A. 2007. Regulation of receptors and transporters by ubiquitination: new insights into surprisingly similar mechanisms. Mol. Interv. 7:157–167 10.1124/mi.7.3.7 [DOI] [PubMed] [Google Scholar]
- Nabhan J.F., Pan H., Lu Q. 2010. Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the beta2-adrenergic receptor. EMBO Rep. 11:605–611 10.1038/embor.2010.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijsten M.W., van Dam G.M. 2009. Hypothesis: using the Warburg effect against cancer by reducing glucose and providing lactate. Med. Hypotheses. 73:48–51 10.1016/j.mehy.2009.01.041 [DOI] [PubMed] [Google Scholar]
- Nikko E., Pelham H.R. 2009. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic. 10:1856–1867 10.1111/j.1600-0854.2009.00990.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E., Sullivan J.A., Pelham H.R. 2008. Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep. 9:1216–1221 10.1038/embor.2008.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell A.F., Apffel A., Gardner R.G., Cyert M.S. 2010. Alpha-arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol. Biol. Cell. 21:3552–3566 10.1091/mbc.E10-07-0636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka S., Masutani H., Liu W., Horita H., Wang D., Kizaka-Kondoh S., Yodoi J. 2006. Thioredoxin-binding protein-2-like inducible membrane protein is a novel vitamin D3 and peroxisome proliferator-activated receptor (PPAR)gamma ligand target protein that regulates PPARgamma signaling. Endocrinology. 147:733–743 10.1210/en.2005-0679 [DOI] [PubMed] [Google Scholar]
- Paiva S., Kruckeberg A.L., Casal M. 2002. Utilization of green fluorescent protein as a marker for studying the expression and turnover of the monocarboxylate permease Jen1p of Saccharomyces cerevisiae. Biochem. J. 363:737–744 10.1042/0264-6021:3630737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva S., Vieira N., Nondier I., Haguenauer-Tsapis R., Casal M., Urban-Grimal D. 2009. Glucose-induced ubiquitylation and endocytosis of the yeast Jen1 transporter: role of lysine 63-linked ubiquitin chains. J. Biol. Chem. 284:19228–19236 10.1074/jbc.M109.008318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh H., Carlsson E., Chutkow W.A., Johansson L.E., Storgaard H., Poulsen P., Saxena R., Ladd C., Schulze P.C., Mazzini M.J., et al. 2007. TXNIP regulates peripheral glucose metabolism in humans. PLoS Med. 4:e158 10.1371/journal.pmed.0040158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwari P., Chutkow W.A., Cummings K., Verstraeten V.L., Lammerding J., Schreiter E.R., Lee R.T. 2009. Thioredoxin-independent regulation of metabolism by the alpha-arrestin proteins. J. Biol. Chem. 284:24996–25003 10.1074/jbc.M109.018093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S., Di Fiore P.P. 2008. Finding the right partner: science or ART? Cell. 135:590–592 10.1016/j.cell.2008.10.032 [DOI] [PubMed] [Google Scholar]
- Rauch S., Martin-Serrano J. 2011. Multiple interactions between the ESCRT machinery and arrestin-related proteins: implications for PPXY-dependent budding. J. Virol. 85:3546–3556 10.1128/JVI.02045-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R.L., Mösch H.U., Fink G.R. 1997. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell. 89:1055–1065 10.1016/S0092-8674(00)80293-7 [DOI] [PubMed] [Google Scholar]
- Rotin D., Kumar S. 2009. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10:398–409 10.1038/nrm2690 [DOI] [PubMed] [Google Scholar]
- Shenoy S.K., Xiao K., Venkataramanan V., Snyder P.M., Freedman N.J., Weissman A.M. 2008. Nedd4 mediates agonist-dependent ubiquitination, lysosomal targeting, and degradation of the beta2-adrenergic receptor. J. Biol. Chem. 283:22166–22176 10.1074/jbc.M709668200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda J., Kikuchi Y. 2007. Rod1, an arrestin-related protein, is phosphorylated by Snf1-kinase in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 364:258–263 10.1016/j.bbrc.2007.09.134 [DOI] [PubMed] [Google Scholar]
- Sonveaux P., Végran F., Schroeder T., Wergin M.C., Verrax J., Rabbani Z.N., De Saedeleer C.J., Kennedy K.M., Diepart C., Jordan B.F., et al. 2008. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J. Clin. Invest. 118:3930–3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A., von Zastrow M. 2009. Endocytosis and signalling: intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 10:609–622 10.1038/nrm2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkina T., Miranda M., Dionne K.R., Hoover B.R., Zahniser N.R., Sorkin A. 2006. RNA interference screen reveals an essential role of Nedd4-2 in dopamine transporter ubiquitination and endocytosis. J. Neurosci. 26:8195–8205 10.1523/JNEUROSCI.1301-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic T.I., Schmiedekamp B.C., Lee J., Katzmann D.J., Burd C.G. 2008. Opposing activities of the Snx3-retromer complex and ESCRT proteins mediate regulated cargo sorting at a common endosome. Mol. Biol. Cell. 19:4694–4706 10.1091/mbc.E08-03-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L.G., Carlson M. 1994. Synergistic release from glucose repression by mig1 and ssn mutations in Saccharomyces cerevisiae. Genetics. 137:49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vina-Vilaseca A., Bender-Sigel J., Sorkina T., Closs E.I., Sorkin A. 2011. Protein kinase C-dependent ubiquitination and clathrin-mediated endocytosis of the cationic amino acid transporter CAT-1. J. Biol. Chem. 286:8697–8706 10.1074/jbc.M110.186858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Kumar S. 2010. Nedd4 and Nedd4-2: closely related ubiquitin-protein ligases with distinct physiological functions. Cell Death Differ. 17:68–77 10.1038/cdd.2009.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young C.D., Lewis A.S., Rudolph M.C., Ruehle M.D., Jackman M.R., Yun U.J., Ilkun O., Pereira R., Abel E.D., Anderson S.M. 2011. Modulation of glucose transporter 1 (GLUT1) expression levels alters mouse mammary tumor cell growth in vitro and in vivo. PLoS ONE. 6:e23205 10.1371/journal.pone.0023205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Wang C., Gao K., Wang D., Mao J., An J., Xu C., Wu D., Yu H., Liu J.O., Yu L. 2010. The ubiquitin ligase itch regulates apoptosis by targeting thioredoxin-interacting protein for ubiquitin-dependent degradation. J. Biol. Chem. 285:8869–8879 10.1074/jbc.M109.063321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv I., Matiuhin Y., Kirkpatrick D.S., Erpapazoglou Z., Leon S., Pantazopoulou M., Kim W., Gygi S.P., Haguenauer-Tsapis R., Reis N., et al. 2011. A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol. Cell Proteomics. 10:M111.009753 10.1074/mcp.M111.009753 [DOI] [PMC free article] [PubMed] [Google Scholar]