Abstract
Addition of short (6 to 16 amino acids) peptide sequences to the N-terminus of p110α induces a gain of function. Such sequences include the common Flag, His and VSV tags as well as random sequences. An N-terminal myristylation signal generally believed to activate p110α by providing a constitutive membrane address is also activating, even if myristylation is mutationally abolished. The gain of function seen with N-terminally tagged (NTT) p110α constructs extends to signaling, oncogenic transformation and stimulation of cell growth. The activating effect of N-terminal tags requires a functional Rasbinding domain in p110α. Mutations in the RBD domain (T208D and K227A) abolish the gains of function in oncogenicity and signaling. The dominant negative mutant of Ras, RasN17, interferes with transformation induced by NTT p110α. In contrast, binding to p85 activity is not required for cellular transformation and enhanced signaling by NTT p110α.
Key words: N-terminal tags, p110a, oncogenicity, Ras binding domain, p85 binding mutations
Introduction
PI3Ks (phosphoinositide-3-kinases of class IA) are dimeric enzymes, consisting of a catalytic subunit and a regulatory subunit; both exist in several isoforms. The interaction between regulatory and catalytic subunit has two principal effects: it stabilizes the catalytic subunit, and it inhibits its enzymatic activity.1,2 Activation of PI3K by upstream signals from RTKs (receptor tyrosine kinases) or G-protein-coupled receptors relieves the inhibitory action of the regulatory subunit.3–5 Cancer-specific mutations have been identified in the p110α isoform of the catalytic subunit and in the p85α isoform of the regulatory subunit.6–13 Most of these mutations induce a gain of function in PI3K which manifests itself in oncogenic activity, enhanced downstream signaling and elevated enzymatic activity.6,7,14–21 The gain of function is achieved by at least two distinct molecular mechanisms; one is characterized by a change in the interaction between p85 and p110α and is exemplified by the helical domain mutations in p110α, the other stems from an altered dependence on Ras and is seen in the kinase domain mutation H1047R of p110α.22–24 The gain of function resulting from a change in the interaction between p110α and p85 reflects essentially a disinhibition of the enzyme, not unlike that resulting from the physiological activation that follows binding of p85 to phosphorylated tyrosines on RTKs.22
Here we have investigated a chance observation on the activating effect of adding N-terminal tags to p110α. This effect was first noticed by Yu and coworkers.1 We have extended the initial observation and analyzed the molecular mechanism of this activation. Our data support the conclusion that N-terminal tails induce a conformational change in p110α, similar to that of helical domain mutations, and disrupt an inhibitory interaction with p85.
Results
N-terminal tags activate the oncogenic potential of p110α.
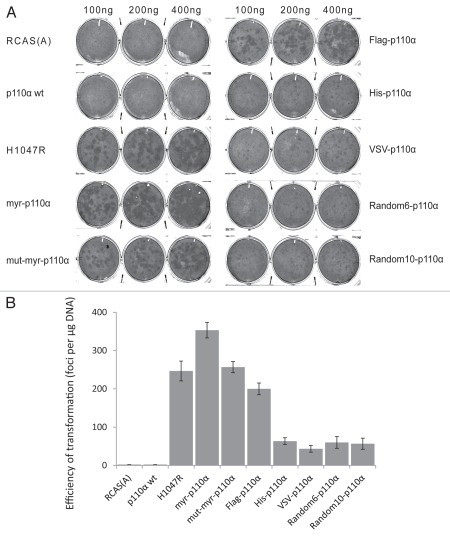
Wild-type (wt) p110α is not oncogenic and does not induce transformation of cultured cells. Unexpectedly, a Flag tag added to the N-terminus of the wt protein activates the oncogenic potential. We explored this observation by generating several N-terminally tagged (NTT) p110α constructs (Fig. 1). These include Flag, His and VSV tags, two random sequences, a myristylation signal and a mutated myristylation signal. All of these constructs show gain of function. When expressed in chicken embryo fibroblasts (CEF) with the RCAS (replication-competent retroviral avian sarcoma-leukosis virus) vector,25,26 they induce oncogenic transformation (Fig. 2A). The myristylation signal is known to activate p110α, and that activation has been ascribed to constitutive membrane localization. However, mutation of the essential glycine in the myristylation signal does not disrupt the ability of this tag to activate the oncogenic potential of p110α. Hence, membrane localization is not an essential feature of activation by the myristylation signal. The generality of the N-terminal tag effect suggests that another mechanism is at work.
Figure 1.
Schematic representation of NTT p110α constructs. In the current study, the following tags were added to the N-terminus of p110α: epitope tags including Flag, His and VSV; a myristylation signal, myr, (N-M(G)QSLTT H-C); a mutated myristylation signal, mut-myr, (N-M(A)QSLTT H-C); and two random sequences, random6: N-QGSGFS-C; random10: N-QVAHGVVLLR-C.
Figure 2.
NTT p110α induces oncogenic transformation in CEF. (A) Representative focus assays of NTT p110α and controls on CEF. (B) The efficiencies of cellular transformation (number of foci per µg DNA) for individual NTT p110α and control constructs.
Although the NTT p110α constructs differ in the efficiencies with which they induce oncogenic transformation (expressed in the number of transformed cell foci normalized to the amount of DNA transfected) (Fig. 2B), the activities fall within the same order of magnitude. The most active construct studied by us is comparable in potency to the H1047R mutation of p110α included in Figures 2 and 3 as positive control. It is likely that any amino acid sequence added to the N-terminus of p110α will have a significant activating effect.
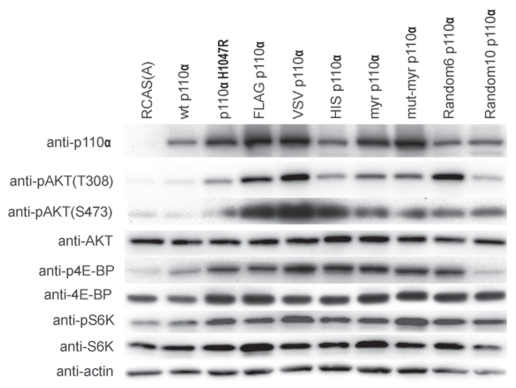
Figure 3.
Increased phosphorylation of Akt, 4EBP and S6K in CEF transfected with NTT p110α. Protein gel blots of NTT p110α-infected and of control CEF. Cells were lysed in NP-40 lysis buffer, and the lysates were clarified by centrifugation. For each lysate, 40 µg of protein were separated by SDS/PAGE and transferred to Immobilon-P membranes. Processing of the membranes is described in the Materials and Methods section. The membranes were blotted with the following antibodies: anti-p110α, anti-pAKT(T308), anti-pAKT(S473), anti-AKT, anti-pS6K, anti-S6K, anti-p4E-BP, anti-4E-BP and anti-β-actin.
N-terminal tags induce elevated downstream signaling of p110α.
PI3K phosphorylates PtdIns(4,5)P2 (phosphoinositide-4,5-bisphosphate) to generate PtdIns(3,4,5)P3 (phosphoinositide-3,4,5-trisphosphate). PtdIns(3,4,5)P3 recruits Akt (cellular homolog of the Akt8 murine leukemia viral oncogene) and its activating kinase PDK1 (phosphoinositide-dependent kinase 1). Activated Akt initiates a cascade of downstream phosphorylations that include TOR (target of rapamycin), S6K (ribosomal protein S6-kinase, 70 kDa) and 4E-BP1 (eukaryotic initiation factor 4E binding protein 1). We have used these activating phosphorylations of Akt, S6K and 4E-BP1 as seen in protein gel blots of transfected CEF to estimate the strength of PI3K signaling (Fig. 3). Transfection with the H1047R mutant of p110α served as positive control, and transfection with empty RCAS vector or wt p110α was used as negative control. The NTT constructs show enhanced downstream signaling as indicated by elevated phosphorylation of Akt, 4E-BP and S6K (Fig. 3). Again, there are differences in potency; these are roughly correlated with the differences in the efficiencies of transformation. Although the NTT constructs signal with increased activity, there is still a possibility that the oncogenic transformation induced by these proteins does not depend on enzymatic function. It is conceivable that these constructs work by re-localizing Ras or by titrating a negative regulator of p110α away from the wild-type enzyme. We therefore examined a kinase-dead NTT mutant (D933A of myristylated p110α). Its expression did not induce oncogenic transformation (data not shown).
Binding to Ras is essential for the gain of function in NTT p110α.
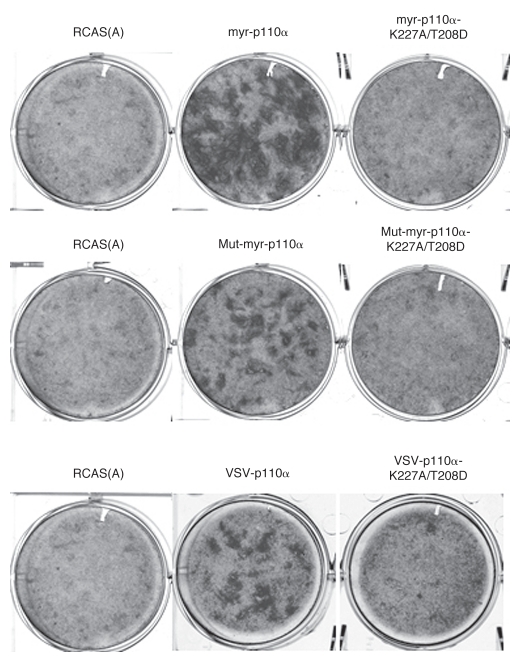
Cancer-derived gain-of-function mutations in p110α are classified according to their dependence on the interactions with p85 and with Ras.23 We applied this principle of classification to the NTT constructs. The p110α mutations K227A/T208D disrupt binding to Ras without affecting the basal activity of the enzyme.27 We introduced these mutations into several NTT constructs and determined their oncogenic activity in focus assays on CEF. All of the constructs in which Ras binding was disabled failed to induce oncogenic transformation (Fig. 4). This result was confirmed with dominant negative Ras (RasN17).28,29 Cells expressing this mutant of Ras became resistant to transformation induced by NTT p110α (Table 1), but remained sensitive to the H1047R mutant of p110α (data not shown). The K227A/T208D mutations also extinguished signaling by NTT p110α (Fig. S1). We conclude that interaction with Ras is essential for the gain of function in NTT p110α.
Figure 4.
Binding to Ras is essential for oncogenic transformation induced by NTT p110α constructs. Representative focus assays of NTT p110α with or without Ras-binding mutations on CEF.
Table 1.
Ras binding activity is critical for NTT p110α induced cell transformation
| Construct | Efficiency of transformation* | ||
| NTT p110α | NTT p110α, Ras binding disabled | NTT p110α plus DN Ras | |
| Myr-p110α | 1.0 | 0.03 ± 0.01 | 0.27 ± 0.08 |
| Mut-myr-p110α | 0.69 ± 0.02 | 0.01 ± 0.01 | 0.36 ± 0.05 |
| VSV-p110α | 0.44 ± 0.02 | 0.03 ± 0.01 | 0.15 ± 0.05 |
Number of transformed cell foci/µg DNA, normalized to the value obtained with Myr-p110α.
Interaction with p85 is not essential for the oncogenic activity of NTT p110α.
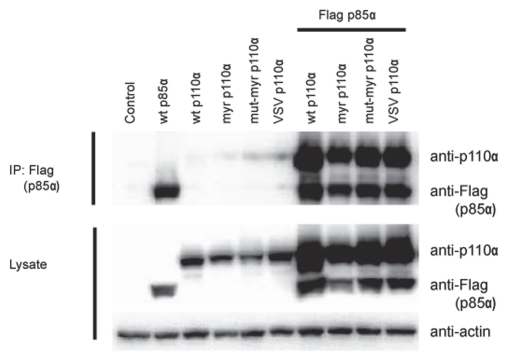
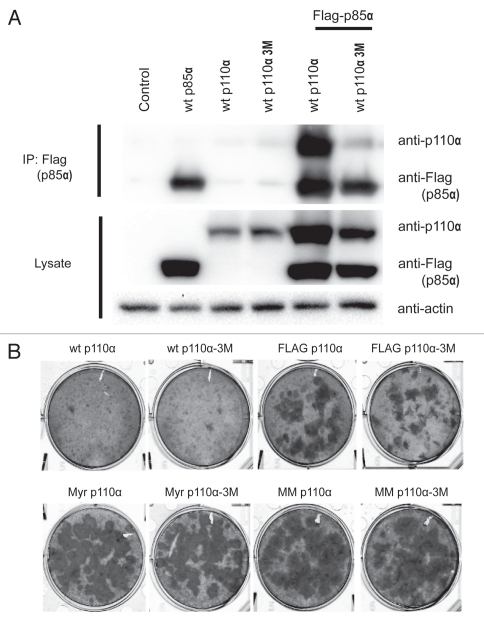
The short sequences added to the N-terminus of p110α abut the p85-binding domain and could affect the interaction of the NTT constructs with p85. In order to examine this possibility, NTT constructs were co-expressed with Flag-tagged p85a in HEK293T cells using the pCAGGS vector.30 Co-immunoprecipitation (IP) and protein gel blots of cell lysates were performed as outlined in the legend to Figure 5. All NTT p110α retained the ability to interact with p85α; there was no significant reduction in binding activity. This observation raised the question whether binding to p85 is essential for the gain of function seen with the NTT constructs. We therefore introduced three point mutations, E23R, P27E and F95A, in critical residues of the p85-binding domain as determined by the published p85-p110α structure.31 The mutations disrupt the contacts between p110α and p85 in the wt protein and the NTT constructs as shown by the greatly reduced co-immunoprecipitation of the two expressed proteins from lysates of HEK293T cells (Fig. 6A). The NTT constructs in which p85-binding was mutationally disabled retained oncogenic activity comparable to that seen with their p85-binding progenitors (Fig. 6B). These observations suggest that interaction with p85 is not required for the oncogenic activity of the NTT constructs. Similar data were obtained by determining signaling activity. Loss of p85-binding did not abolish the enhanced phosphorylation of Akt seen with the NTT constructs (Fig. S2).
Figure 5.
NTT p110α still binds to p85. Co-immunoprecipitation of NTT p110α and wt p85a expressed in HEK293T cells. HEK293T cells were transfected with empty pCAGGS vector (control lane), and in separate transfections with Flag-tagged wt p85a, with wt p110α or NTT p110α constructs as indicated on top of the gel. Lanes 7–10 were co-transfected with the Flag-tagged wt p85 and with the NTT constructs indicated. 48 hrs post transfection, the cells were lysed, and p85a was immunoprecipitated with Flag antibody-conjungated agarose beads. The precipitates were separated by SDS-PAGE and immunoblotted with anti-p110α and anti-Flag antibodies. Separately, protein gel blots of the cell lysates were prepared with anti-p110α and anti-Flag antibodies to reveal the total amounts of p110α and p85a. Actin was used as loading control.
Figure 6.
Binding to p85 is not required for oncogenic transformation induced by NTT p110α. (A) Binding of p85a by p110α is greatly reduced by three point mutations (E23R, P27E and F95A referred to as 3M) in the adapter-binding domain. HEK293T cells were transfected using lipofectamine with empty pCAGGS vector (control lane) and Flag-p85a, wt p110α, p110α-3M alone or in combination as indicated. Forty-eight hours post transfection, the cells were lysed, and p85a was immunoprecipitated with Flag antibody-conjungated agarose beads. The precipitates were separated by SDS-PAGE and immunoblotted with anti-p110α and anti-Flag antibodies. Separately, protein gel blots of the cell lysates were prepared with anti-p110α and anti-Flag antibodies to reveal the total amounts of p110α and p85a. Actin was used as loading control. Substitution of NTT constructs for p110α yielded similar results. (B) Focus assays show that interrupting p85 binding does not affect the oncogenic activity of NTT constructs.
Discussion
The data presented in this study show that diverse short amino acid sequences added to the N-terminus of p110α, the catalytic subunit of PI3K, induce a gain of function. We speculate that the N-terminus of p110α represents a “sweet spot,” where addition of any amino acid sequence triggers an activating conformational change. From the available structure data, it appears that the N-terminus is buried between the p85-binding domain and the catalytic domain. In both the p110α and p110α-H1047R structures, the N-terminal His tag is inserted into another molecule of p110α. While disordered, the path taken must interact with both the iSH2 of p85 and the catalytic domain of p110. Our data confirm and extend a previous report that showed an N-terminal tag attached to bovine p110α induced elevated enzymatic activity.1 The gain of function in the NTT constructs studied here extends to both oncogenicity and signaling. The oncogenic activities recorded with different constructs are not the same, they are characteristic of the tag sequence, but the values lie within the same order of magnitude. The most potent constructs, carrying the Flag, myristylation or mutated myristylation tags, induce oncogenic transformation with efficiencies that approach that of the cancer-derived H1047R mutation of p110α. The fact that a mutated, nonfunctional myristylation signal is effective in activating p110α shows that the myristyl-mediated membrane localization is not a critical feature of this activation mechanism, although it could be an ancillary contributor, as disabling myristylation results in somewhat lower activity. Preliminary studies with p110β reveal a different situation. The wt protein p110β is oncogenic when overexpressed in cell culture. The efficiency of this p110β-mediated transformation can be significantly enhanced by an N-terminal myristylation signal.32 This enhancement is completely abolished if the myristylation site is mutated (Fig. S3).
Further insight into the mechanism of N-terminal activation of p110α has been obtained by studying the interaction of the tagged constructs with Ras and with p85. The activity of p110α is controlled by the interaction with two adapter proteins, the regulatory subunit of PI3K, primarily in its p85 isoform, and Ras. Gain-of-function mutations in PI3K can be characterized by their dependence on these interactions and fall into two classes, those independent of p85-binding and those not requiring Ras-binding.23,24 We have applied this strategy of determining the interaction with adapters to the characterization of the NTT constructs. Mutational disruption of Ras-binding inactivated the oncogenic potential and the enhanced signaling of the constructs. The dependence on an interaction with Ras was confirmed by the effect of dominant negative Ras which strongly reduced cellular transformation by the NTT constructs. Whereas Rasbinding is thus essential to the activity of the NTT constructs, p85-binding is dispensable. The NTT constructs still bound to p85; the tags did not exert a steric interference with this interaction. But this binding was irrelevant to activity. Point mutations in the p85-binding domain of the NTT constructs that greatly reduced the interaction between p110α and p85 had no effect on the oncogenic activity of these proteins nor did they eliminate the gain of function in signaling. The requirement for Ras-binding and the independence from p85 are reminiscent of the cancer-derived helical domain mutations E542K and E545K of p110α. These mutations introduce an electrostatic repulsion that disrupts an inhibitory interaction between the N-terminal SH2 domain of p85 and the helical domain of p110α and thus lead to an activation of the enzyme.33 The activity is dependent on Rasbinding but does not require binding to p85 nor is it affected by phosphopeptides that, in the wt enzyme, lift the p85-mediated inhibition by binding to the N-terminal SH2 domain of p85.22–24 These mutations essentially mimic the activated state of PI3K that is achieved by upstream signaling from RTKs; we would expect that their activity is not further enhanced by an interaction with an RTK-derived phosphopeptide. Our data are in accord with the supposition that the NTT constructs operate by essentially the same mechanism as E542K and E545K, requiring Ras but independent of p85 and p85-mediated recruitment. The precise role of Ras in this mechanism remains to be worked out. A Ras-induced conformational change is the most likely explanation for this dependency. A Ras-mediated recruitment to the plasma membrane is probably not of decisive importance here, because p110α with an active myristylation signal is still inactivated by loss of Ras binding. This is in contrast to p110γ where loss of Ras binding can be compensated by myristylation.34–36 In accordance with this suggestion, we have failed to detect a difference by fluorescence microscopy between the cellular distribution of the NTT constructs with and without Ras binding. However, none of these findings rules out a possible effect of the N-tags on the stability of p110α.
Our data also shed new light and suggest a re-interpretation of other N-terminally modified p110α proteins that have been described in previous publications.37–40 The avian retrovirus ASV16 carries a copy of the cellular p110α-encoding gene PIK3CA as an oncogene. The oncoprotein produced by ASV16 is a fusion of N-terminal viral Gag sequences to the entire coding sequence of p110α. The virus is highly transforming in cultures of CEF. if instead of ASV16, the avian retroviral vector RCAS is used to express wt cellular p110α in CEF, oncogenic transformation can occur, but it is exceedingly rare. Progeny virus from these rare transformants is, however, highly oncogenic. This acquisition of oncogenicity by wt cellular p110α during viral passage is always accompanied by an N-terminal fusion to viral Gag sequences derived from the vector. It is this fusion that activates wt cellular p110α.38 Since Gag sequences can mediate recruitment to cellular membranes, constitutive localization to the plasma membrane was proposed as the mechanism of Gag-mediated activation. However, in the light of our present studies, it is more likely that Gag works like other N-terminal tails and activates the enzyme by inducing a conformational change that leads to disruption of the inhibitory interaction with p85.
A second NTT construct that has been studied in the past is p110*.41 p110* is a cytosolic, constitutively active p110α that is fused, via an N-terminal glycine linker, to the inter-SH2 domain of p85. We posit that the activity of the p110* construct reflects in part the same mechanism that we have outlined here for other NTT p110α. This activity is independent of p85-binding and p85-mediated membrane localization, but in the case of p110*, it can be further enhanced by adding a myristylation signal.37 However, with p110*, additional factors could play a role. Although the inter-SH2 domain that is linked to the N-terminus of p110α does not appear to block the adapter binding domain of the same protein, it could interact with other targets (Gymnopoulos 2009, unpublished). The interpretation of the p110* is also further complicated by possible effects of a Myc tag at the C-terminus of the molecule.
The activation of p110α by a His tag as documented in this communication is also relevant for the interpretation of structural studies on p110α and the oncogenic H1047R mutant of p110α.31,42–45 For crystallization, p110α was purified with the aid of an N-terminal His-TEV tag, and successful crystallization depended on the retention of this tag. The structure derived from these crystals is therefore likely to represent an at least partially disinhibited conformation rather than the basal state of the enzyme.
The studies described in this communication lead us to propose that there exist two basic states of mutational activation of p110α: in one the mutation replaces or mimics the activating function of p85 (helical domain mutations, NTT constructs), and in the other it replaces or mimics the activating function of Ras (p110α H1047R). Additional genetic and structural studies will be needed to confirm or refute this proposal.
Materials and Methods
Plasmid construction.
The construction of the RCAS vector encoding p110α H1047R has been described in references 23 and 32. The NTT RCAS constructs, myr-p110α, mut-myr-p110α, Flag-p110α, His-p110α, VSV-p110α, Random6-p110α and Random10-p110α were constructed by standard PCR and cloning. The Ras-binding mutants (myr-p110α K227A/T208D, mut-myr-p110α K227A/T208D and VSV-p110α K227A/T208D) and p85-binding mutants (p110α-3M, myr-p110α-3M, mut-myr-p110α-3M and FLAG-p110α-3M) were generated by sequential cloning using the QuikChange site-directed mutagenesis kit (Stratagene, 200518). The following primers were used to introduce E23R, P27E and F95A into p110α: E23R (+): 5′-CCC CAA GAA TCC TAG TAA GAT GTT TAC TAC CAA ATG G-3′; E23R (−): 5′-CCA TTT GGT AGT AAA CAT CTT ACT AGG ATT CTT GGG G-3′; P27E (+); 5′-GTA AGA TGT TTA CTA GAA AAT GGA ATG ATA GTG AC-3′; P27E (−): 5′-GTC ACT ATC ATT CCA TTT TCT AGT AAA CAT CTT AC-3′; F95A (+): 5′-GTG ACC TTC GGC TTG CTC AAC CCT TTT TAA AAG-3′; F95A (−): 5′-CTT TTA AAA AGG GTT GAG CAA GCC GAA GGT CAC-3′. The mutated genes were subsequently cloned as SfiI DNA fragments into the avian retrovirus vector RCAS.Sfi.46 All clones were confirmed by sequencing.
Cell culture and transfection.
Fertilized chicken eggs (white Leghorn) were obtained from Charles River Breeding Laboratories. Primary chicken embryo fibroblasts (CEF) were prepared and cultured as described in references 47 and 48. For transfection, cells were plated at 80% confluence in F-10 medium containing 5.8% iron-supplemented fetal calf serum (Omega Scientific, FB-01) and 1% L-glutamine-penicillin-streptomycin solution (Sigma-Aldrich, G1146). On the following day, CEF were transfected with the RCAS vectors using the dimethyl sulfoxide/Polybrene method.49 After two passages in the presence of serum, the cells were harvested for further analysis.
HEK 293T cells were cultured in DMEM (Gibco, Invitrogen, 11965) supplemented with 10% fetal calf serum and 1% L-glutamine-penicillin-streptomycin solution. Transfections were carried using Lipofectamine-PLUS (Invitrogen, 18324-012 and 10964-021) following the manufacturer's protocol. HEK293T cells in MP6 plates at 70% confluency were washed once with Opti-MEM medium and incubated in 0.8 ml of Opti-MEM. A total of 1 µg of plasmid DNA was mixed with 0.1 ml of Opti-MEM and 2 µl of Lipofectamine-PLUS for 15 min at room temperature. In the next step, 0.1 ml Opti-MEM and 12 µl of Lipofectamine were added and the mixture was incubated for another 15 min at room temperature. The mixture was then added to the cells and incubated overnight. The second day, the medium was changed to DMEM containing 10% fetal calf serum and 1% L-glutamine-penicillin-streptomycin solution. Forty hours post transfection, the cells were collected, lysed and analyzed for specific proteins.
Focus assay and interference assay.
Focus assays with infectious retroviral vectors were performed as described in references 47 and 48. CEF were transfected with the appropriate RCAS constructs using the dimethyl sulfoxide/Polybrene method and overlayed with nutrient agar every other day for two to three weeks until focus formation was observed. The plates were stained with crystal violet, and foci of transformed cells were counted. For interference assays, CEF were transfected with RCAS(B) or RCAS(B) RasN17. The transfected cells were transferred three times and were then transfected with NTT p110α using the dimethyl sulfoxide/Polybrene method and overlayed with nutrient agar every other day for one to two weeks until focus formation was observed. The plates were stained with crystal violet, and foci of transformed cells were counted.
Protein gel blots and immunoprecipitation.
Protein gel blotting was performed as described in reference 38, with minor modifications. Cells were lysed in modified NP-40 lysis buffer (20 mM Tris-Cl, 150 mM NaCl, 1 mM MgCl2, 1% NP-40 and 10% glycerol with 1 mM PMSF, 1 mM DTT, 50 mM NaF, 1 mM Na3VO4, 50 mM β-glycerophosphate and a protease inhibitor cocktail from Roche, 11697498001). After centrifugation for 10 min at 18,000x g at 4°C, the protein concentration of the supernatant was determined. For immunoprecipitation, cell lysates containing 40 µg of protein were incubated with anti-FLAG M2 Agarose (Sigma-Aldrich, A2220) overnight at 4°C. Agarose beads were washed 4 times with lysis buffer and heated to 95°C for 5 min before separation on a SDS-PAGE gel. After transfer to Immobilon-P membranes (Millipore, IPVH304F0), the membranes were blocked with 5% BSA in Tris-buffered saline with 0.1% Tween-20 (TBS-T) for 1 h at room temperature and then incubated with a dilution of 1:2,000 of anti-FLAG or 1:1,000 of anti-p110α or anti-p110β primary antibody overnight. Membranes were washed three times in TBS-T and incubated with peroxidase coupled goat-anti-mouse or goat-anti-rabbit (Thermo Scientific) antibody for 1 h in 5% BSA/TBS-T at room temperature. The reactive bands were visualized by SuperSignal West Pico Chemiluminescent substrate (Thermo Scientific, 34078).
For protein gel blotting, cell lysates containing 10 µg of total protein were separated on SDS/PAGE gels and transferred to Immobilon-P membranes (Millipore, IPVH304F0). Membranes were incubated with 1:1,000 dilutions of primary antibodies directed against p85, pAkt (T308), Akt, p4E-BP, 4E-BP and β-actin. Protein gel blots were developed as described above. Anti-FLAG antibody (Flag-M2 #3185) was purchased from Sigma-Aldrich. Anti-p110α antibody (#4255), anti-Akt (#2967), anti-phospho-Akt (Thr-308) (#9275S), anti-4E-BP1 (#9452), and anti-phospho-4E-BP1 (Ser-65) (#9451S) antibodies were obtained from Cell Signaling Technology.
Acknowledgments
The authors are grateful to Lynn Ueno for expert technical assistance. This work was supported by NIH grants CA078045 and CA078230. This is manuscript 21,250 of The Scripps Research Institute.
Abbreviations
- 4E-BP1
eukaryotic initiation factor 4E binding protein 1
- Akt
cellular homolog of the Akt8 murine leukemia viral oncogene
- CEF
chicken embryo fibroblasts
- IP
immunoprecipitation
- NTT
N-terminally tagged
- PDK1
3-phosphoinositide-dependent protein kinase 1
- PI3Ks
phosphoinositide-3-kinases of class IA
- PIP2
phosphoinositide-4,5-bisphosphate
- PIP3
phosphoinositide-3,4,5-trisphosphate
- RasN17
dominant negative Ras
- RCAS
replication-competent retroviral avian sarcoma-leukosis virus
- RTKs
receptor tyrosine kinases
- S6K
ribosomal protein S6-kinase, 70 kDa
- TOR
target of rapamycin
- wt
wild-type
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Regulation of the p85/p110 phosphatidylinositol-3′-kinase: stabilization and inhibition of the p110αlpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol. 1998;18:1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo J, Field SJ, Lee JY, Engelman JA, Cantley LC. The p85 regulatory subunit of phosphoinositide-3-kinase downregulates IRS-1 signaling via the formation of a sequestration complex. J Cell Biol. 2005;170:455–464. doi: 10.1083/jcb.200503088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dbouk HA, Pang H, Fiser A, Backer JM. A biochemical mechanism for the oncogenic potential of the p110beta catalytic subunit of phosphoinositide-3-kinase. Proc Natl Acad Sci USA. 2010;107:19897–19902. doi: 10.1073/pnas.1008739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guillermet-Guibert J, Bjorklof K, Salpekar A, Gonella C, Ramadani F, Bilancio A, et al. The p110beta isoform of phosphoinositide-3-kinase signals downstream of G protein-coupled receptors and is functionally redundant with p110gamma. Proc Natl Acad Sci USA. 2008;105:8292–8297. doi: 10.1073/pnas.0707761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerchner KR, Clay RL, McCleery G, Watson N, McIntire WE, Myung CS, et al. Differential sensitivity of phosphatidylinositol-3-kinase p110gamma to isoforms of G protein betagamma dimers. J Biol Chem. 2004;279:44554–44562. doi: 10.1074/jbc.M406071200. [DOI] [PubMed] [Google Scholar]
- 6.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 7.Samuels Y, Ericson K. Oncogenic PI3K and its role in cancer. Curr Opin Oncol. 2006;18:77–82. doi: 10.1097/01.cco.0000198021.99347.b9. [DOI] [PubMed] [Google Scholar]
- 8.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 9.Shekar SC, Wu H, Fu Z, Yip SC, Nagajyothi, Cahill SM, et al. Mechanism of constitutive phosphoinositide-3-kinase activation by oncogenic mutants of the p85 regulatory subunit. J Biol Chem. 2005;280:27850–27855. doi: 10.1074/jbc.M506005200. [DOI] [PubMed] [Google Scholar]
- 10.Philp AJ, Campbell IG, Leet C, Vincan E, Rockman SP, Whitehead RH, et al. The phosphatidylinositol-3′-kinase p85alpha gene is an oncogene in human ovarian and colon tumors. Cancer Res. 2001;61:7426–7429. [PubMed] [Google Scholar]
- 11.Mizoguchi M, Nutt CL, Mohapatra G, Louis DN. Genetic alterations of phosphoinositide-3-kinase subunit genes in human glioblastomas. Brain Pathol. 2004;14:372–377. doi: 10.1111/j.1750-3639.2004.tb00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jücker M, Sudel K, Horn S, Sickel M, Wegner W, Fiedler W, et al. Expression of a mutated form of the p85alpha regulatory subunit of phosphatidylinositol-3-kinase in a Hodgkin's lymphoma-derived cell line (CO) Leukemia. 2002;16:894–901. doi: 10.1038/sj.leu.2402484. [DOI] [PubMed] [Google Scholar]
- 13.Jimenez C, Jones DR, Rodriguez-Viciana P, Gonzalez-Garcia A, Leonardo E, Wennstrom S, et al. Identification and characterization of a new oncogene derived from the regulatory subunit of phosphoinositide-3-kinase. EMBO J. 1998;17:743–753. doi: 10.1093/emboj/17.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao JJ, Liu Z, Wang L, Shin E, Loda MF, Roberts TM. The oncogenic properties of mutant p110αlpha and p110beta phosphatidylinositol-3-kinases in human mammary epithelial cells. Proc Natl Acad Sci USA. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang S, Bader AG, Vogt PK. Phosphatidylinositol-3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 18.Ikenoue T, Kanai F, Hikiba Y, Obata T, Tanaka Y, Imamura J, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65:4562–4567. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- 19.Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci USA. 2006;103:1475–1479. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun M, Hillmann P, Hofmann BT, Hart JR, Vogt PK. Cancer-derived mutations in the regulatory subunit p85alpha of phosphoinositide-3-kinase function through the catalytic subunit p110αlpha. Proc Natl Acad Sci USA. 2010;107:15547–15552. doi: 10.1073/pnas.1009652107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaiswal BS, Janakiraman V, Kljavin NM, Chaudhuri S, Stern HM, Wang W, et al. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell. 2009;16:463–474. doi: 10.1016/j.ccr.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaussade C, Cho K, Mawson C, Rewcastle GW, Shepherd PR. Functional differences between two classes of oncogenic mutation in the PIK3CA gene. Biochem Biophys Res Commun. 2009;381:577–581. doi: 10.1016/j.bbrc.2009.02.081. [DOI] [PubMed] [Google Scholar]
- 23.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110αlpha of phosphatidylinositol-3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci USA. 2008;105:2652–2657. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao L, Vogt PK. Hot-spot mutations in p110αlpha of phosphatidylinositol-3-kinase (pI3K): differential interactions with the regulatory subunit p85 and with RAS. Cell Cycle. 2010;9:596–600. doi: 10.4161/cc.9.3.10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh J, Julias JG, Ferris AL, Hughes SH. Construction and characterization of a replication-competent retroviral shuttle vector plasmid. J Virol. 2002;76:1762–1768. doi: 10.1128/JVI.76.4.1762-8.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes SH, Vogt PK, Stubblefield E, Bishop JM, Varmus HE. Integration of avian sarcoma virus DNA in chicken cells. Virology. 1981;108:208–221. doi: 10.1016/0042-6822(81)90539-0. [DOI] [PubMed] [Google Scholar]
- 27.Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, et al. Binding of ras to phosphoinositide-3-kinase p110αlpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 28.Stacey DW, Feig LA, Gibbs JB. Dominant inhibitory Ras mutants selectively inhibit the activity of either cellular or oncogenic Ras. Mol Cell Biol. 1991;11:4053–4064. doi: 10.1128/mcb.11.8.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi J, Sun M, Vogt PK. Smooth muscle alpha-actin is a direct target of PLZF: effects on the cytoskeleton and on susceptibility to oncogenic transformation. Oncotarget. 2010;1:9–21. doi: 10.18632/oncotarget.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-D. [DOI] [PubMed] [Google Scholar]
- 31.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, et al. The structure of a human p110αlpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 32.Kang S, Denley A, Vanhaesebroeck B, Vogt PK. Oncogenic transformation induced by the p110beta, -gamma and -delta isoforms of class I phosphoinositide-3-kinase. Proc Natl Acad Sci USA. 2006;103:1289–1294. doi: 10.1073/pnas.0510772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu H, Shekar SC, Flinn RJ, El-Sibai M, Jaiswal BS, Sen KI, et al. Regulation of Class IA PI 3-kinases: C2 domain-iSH2 domain contacts inhibit p85/p110αlpha and are disrupted in oncogenic p85 mutants. Proc Natl Acad Sci USA. 2009;106:20258–20263. doi: 10.1073/pnas.0902369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez-Viciana P, Warne PH, Vanhaesebroeck B, Waterfield MD, Downward J. Activation of phosphoinositide-3-kinase by interaction with Ras and by point mutation. EMBO J. 1996;15:2442–2451. [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, et al. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 36.Chan TO, Rodeck U, Chan AM, Kimmelman AC, Rittenhouse SE, Panayotou G, et al. Small GTPases and tyrosine kinases coregulate a molecular switch in the phosphoinositide-3-kinase regulatory subunit. Cancer Cell. 2002;1:181–191. doi: 10.1016/S1535-6108(02)00033-8. [DOI] [PubMed] [Google Scholar]
- 37.Klippel A, Reinhard C, Kavanaugh WM, Apell G, Escobedo MA, Williams LT. Membrane localization of phosphatidylinositol-3-kinase is sufficient to activate multiple signal-transducing kinase pathways. Mol Cell Biol. 1996;16:4117–4127. doi: 10.1128/mcb.16.8.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aoki M, Schetter C, Himly M, Batista O, Chang HW, Vogt PK. The catalytic subunit of phosphoinositide-3-kinase: requirements for oncogenicity. J Biol Chem. 2000;275:6267–6275. doi: 10.1074/jbc.275.9.6267. [DOI] [PubMed] [Google Scholar]
- 39.Martin SS, Haruta T, Morris AJ, Klippel A, Williams LT, Olefsky JM. Activated phosphatidylinositol-3-kinase is sufficient to mediate actin rearrangement and GLUT4 translocation in 3T3-L1 adipocytes. J Biol Chem. 1996;271:17605–17608. doi: 10.1074/jbc.271.30.17605. [DOI] [PubMed] [Google Scholar]
- 40.Jensen JE, Sorensen HA, Kollerup G, Jensen LB, Sorensen OH. Biological variation of biochemical bone markers. Scand J Clin Lab Invest Suppl. 1994;219:36–39. doi: 10.3109/00365519409088575. [DOI] [PubMed] [Google Scholar]
- 41.Hu Q, Klippel A, Muslin AJ, Fantl WJ, Williams LT. Ras-dependent induction of cellular responses by constitutively active phosphatidylinositol-3-kinase. Science. 1995;268:100–102. doi: 10.1126/science.7701328. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, Vadas O, Perisic O, Anderson KE, Clark J, Hawkins PT, et al. Structure of lipid kinase p110beta/p85beta elucidates an unusual SH2-domain-mediated inhibitory mechanism. Mol Cell. 2011;41:567–578. doi: 10.1016/j.molcel.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gabelli SB, Huang CH, Mandelker D, Schmidt-Kittler O, Vogelstein B, Amzel LM. Structural effects of oncogenic PI3Kalpha mutations. Curr Top Microbiol Immunol. 2010;347:43–53. doi: 10.1007/82_2010_53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mandelker D, Gabelli SB, Schmidt-Kittler O, Zhu J, Cheong I, Huang CH, et al. A frequent kinase domain mutation that changes the interaction between PI3Kalpha and the membrane. Proc Natl Acad Sci USA. 2009;106:16996–17001. doi: 10.1073/pnas.0908444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amzel LM, Huang CH, Mandelker D, Lengauer C, Gabelli SB, Vogelstein B. Structural comparisons of class I phosphoinositide-3-kinases. Nat Rev Cancer. 2008;8:665–669. doi: 10.1038/nrc2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aoki M, Batista O, Bellacosa A, Tsichlis P, Vogt PK. The akt kinase: molecular determinants of oncogenicity. Proc Natl Acad Sci USA. 1998;95:14950–14955. doi: 10.1073/pnas.95.25.14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duff RG, Vogt PK. Characteristics of two new avian tumor virus subgroups. Virology. 1969;39:18–30. doi: 10.1016/0042-6822(69)90344-4. [DOI] [PubMed] [Google Scholar]
- 48.Bos TJ, Monteclaro FS, Mitsunobu F, Ball AR Jr, Chang CH, Nishimura T, et al. Efficient transformation of chicken embryo fibroblasts by c-Jun requires structural modification in coding and noncoding sequences. Genes Dev. 1990;4:1677–1687. doi: 10.1101/gad.4.10.1677. [DOI] [PubMed] [Google Scholar]
- 49.Kawai S, Nishizawa M. New procedure for DNA transfection with polycation and dimethyl sulfoxide. Mol Cell Biol. 1984;4:1172–1174. doi: 10.1128/mcb.4.6.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.