Abstract
The mitotic checkpoint is a specialized signal transduction pathway that monitors kinetochore-microtubule attachment to achieve faithful chromosome segregation. MAD2 is an evolutionarily conserved mitotic checkpoint protein that exists in open (O) and closed (C) conformations. The increase of intracellular C-MAD2 level during mitosis, through O→C-MAD2 conversion as catalyzed by unattached kinetochores, is a critical signaling event for the mitotic checkpoint. However, it remains controversial whether MAD2 is an integral component of the effector of the mitotic checkpoint—the mitotic checkpoint complex (MCC). We show here that endogenous human MCC is assembled by first forming a BUBR1:BUB3:CDC20 complex in G2 and then selectively incorporating C-MAD2 during mitosis. Nevertheless, MCC can be induced to form in G1/S cells by expressing a C-conformation locked MAD2 mutant, indicating intracellular level of C-MAD2 as a major limiting factor for MCC assembly. In addition, a recombinant MCC containing C-MAD2 exhibits effective inhibitory activity toward APC/C isolated from mitotic HeLa cells, while a recombinant BUBR1:BUB3:CDC20 ternary complex is ineffective at comparable concentrations despite association with APC/C. These results help establish a direct connection between a major signal transducer (C-MAD2) and the potent effector (MCC) of the mitotic checkpoint, and provide novel insights into protein-protein interactions during assembly of a functional MCC.
Key words: MAD2, conformer, mitotic checkpoint complex, anaphase promoting complex/cyclosome
Introduction
The mitotic checkpoint (or spindle assembly checkpoint) delays the metaphase-to-anaphase transition in response to defects in microtubule attachment or tension at kinetochores on sister chromatids.1 Proper functioning of the checkpoint guards against aneuploidy and chromosomal instability which are hallmarks of most solid tumor cells.2,3 The mitotic checkpoint can be regarded as a specialized signal transduction pathway that include steps such as signal detection, signal transduction and amplification and formation of effectors.4
A single unattached kinetochore is able to activate the mitotic checkpoint and delay anaphase onset, implicating exquisite and efficient signal transduction and amplification processes in the checkpoint.5 A group of evolutionarily conserved proteins that include MAD1, MAD2, BUBR1(MAD3), BUB1, BUB3 and MPS1 have been identified to mediate signal transduction for the mitotic checkpoint.1,6 Of these proteins, MAD2 has been characterized as the major signal transducer that links signaling events at defective kinetochores with those in the cytoplasm.7–9 MAD2 also plays significant roles in amplifying the checkpoint signals.7 Both functions of MAD2 are dependent on the unique property of the protein to adopt two distinct native conformations, termed open (O) or closed (C) MAD2.8,9 In vitro experiments have demonstrated that O- and C-conformers behave differently in NMR analysis, gel filtration and ion exchange chromatography.8,9 Specifically, O-MAD2 was found to exist as a monomer or form O:C heterodimers. C-MAD2 may also form C:C homodimers, and is the conformation adopted by MAD2 when bound to its binding partners MAD1, CDC20 or BUBR1.8–10 In interphase cells, most MAD2 exists in the O-conformation while a fraction that does adopt the C-conformation localizes at the nuclear envelope in a stable complex with MAD1.11–14 Following nuclear envelope breakdown, the MAD1:C-MAD2 complex, through unknown mechanisms, is targeted to and enriched at unattached kinetochores.12 The kinetochore-localized MAD1:CMAD2 complex is believed to function as the catalyst to convert cytoplasmic O-MAD2 into C-MAD2.15 It has been estimated that intracellular C-MAD2 levels increase from ∼25% of the total MAD2 population in interphase cells to 70% when cells are arrested in mitosis with nocodazole, a microtubule-depolymerizing drug.16 How O→C-MAD2 conversion is accomplished mechanistically is still being investigated, nevertheless it is clear that dimerization between O- and C-MAD2, mediated mainly through the α-C helices present in both conformers, is required.17,18 Converted C-MAD2 molecules are released from unattached kinetochores, as detected by rapid exchange of ∼50% of MAD2 molecules at kinetochores in fluorescence recovery after photobleaching (FRAP) experiments.19,20 C-MAD2 entering the cytosol may form C:C-MAD2 homodimers, however the primary target is thought to be CDC20.1,21 CDC20 is an activator for the anaphase promoting complex/cyclosome (APC/C), a multi-subunit E3 ubiquitin ligase responsible for ubiquitylation of securin and cyclin B, whose subsequent destruction leads to chromosome segregation and mitotic exit.22,23 Increased formation of CDC20:MAD2 complex has been observed in mitotic cells as compared with interphase cells.24,25 Recent results also suggested that the CDC20:C-MAD2 complex may further catalyze O→C-MAD2 conversion, exploiting mechanisms similar to kinetochore-localized MAD1:C-MAD2 complexes.26 Collectively, the role of C-MAD2 in signal transduction and amplification has gained extensive support from multiple lines of evidence.
The effector step of the mitotic checkpoint was previously suggested to be sequestration of CDC20 from APC/C core subunits by C-MAD2 or BUBR1.27–29 However, full sequestration of CDC20 for a complete inhibition of the APC/C activity requires up to hundreds fold more MAD2 and BUBR1 than their respective intracellular concentrations.27–29 In addition, the sequestration model cannot explain the existence of a MAD2:CDC20:APC/C ternary complex.28,30 Currently, the best candidate for the physiologically relevant effector of the mitotic checkpoint is the mitotic checkpoint complex (MCC), an evolutionarily conserved protein complex consisting of CDC20, MAD2 and a cell cycle independent subcomplex BUBR1:BUB3.31–34 The MCC exhibits potent APC/C inhibitory activity (>3,000-fold higher than recombinant MAD2 for human MCC) and most likely functions through stoichiometric binding to APC/C.32,35 However, several discrepancies regarding the MCC as the mitotic checkpoint effector remain to be resolved. One controversy relates to the cell cycle regulation of MCC assembly. Several previous reports have proposed that the MCC is present throughout the cell cycle and/or MCC assembly is independent of unattached kinetochores,32,36–38 raising the question how MCC formation responds to upstream mitotic checkpoint signaling events. The second controversy concerns MCC composition. In several cases, MAD2 was not observed in the MCC either isolated from HeLa cells or reconstituted with recombinant proteins.27,29,39,40 How these results are reconciled with the role of C-MAD2 as a major signal transducer is still unclear.
To resolve the discrepancies and assess the connections between MAD2 as the major signal transducer and the MCC as an effector of the mitotic checkpoint, we investigated how different conformers of MAD2 affected MCC assembly. Our results suggest that MCC selectively incorporates C-MAD2 during mitosis and the intracellular C-MAD2 level is a major limiting factor for MCC assembly.
Results
MCC is sequentially assembled in G2 and mitosis.
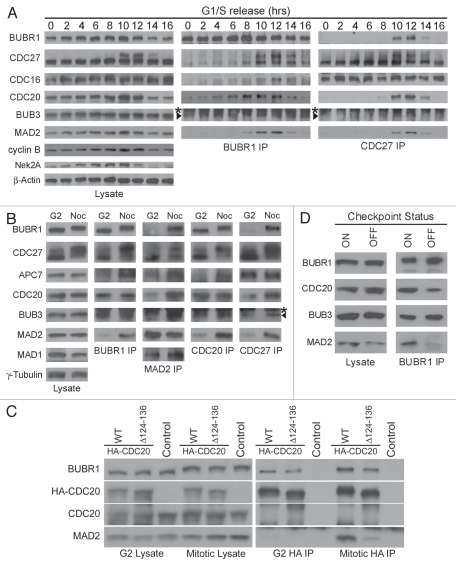
The original characterization of MCC in human cells suggested that the complex forms throughout the cell cycle.32 However, protein levels of individual MCC subunits prepared from interphase and mitotic cells were not compared directly and several reports have since questioned the existence of interphase MCC.1 To address these inconsistencies, MCC formation and MCC:APC/C association were examined by BUBR1 and CDC27 (or APC3, a core APC/C subunit) immunoprecipitations (IP) in synchronized HeLa cells (Fig. 1). BUBR1 associated with BUB3 throughout the cell cycle, confirming previous observations32 (Fig. 1A). In contrast, the levels of MAD2 and CDC20 that co-IPed with BUBR1 clearly peaked between 8–12 h after thymidine release, coinciding with accumulation of G2/M cells as indicated by the rise in cyclin B and Nek2A levels (Fig. 1A, left and middle parts). Further examination found no difference in the levels of BUBR1:CDC20 interaction in G2 or mitotic lysates (Fig. 1B, “CDC20” row in “BUBR1 IP” part). In contrast, the level of MAD2 associated with BUBR1 increased ∼5-fold in mitosis despite equivalent amounts of MAD2 in G2 and mitotic lysates (Fig. 1B, “MAD2” row in “BUBR1 IP” part). Reciprocal IPs using MAD2 or CDC20 antibodies confirmed their distinct patterns of BUBR1 association, but also showed that the MAD2:CDC20 interaction was enhanced during mitosis (Fig. 1B, compare “BUBR1,” “MAD2” and “CDC20” rows in “MAD2 IP” and “CDC20 IP” parts). Although MAD2 was suggested to stimulate the BUBR1:CDC20 interaction,39,41,42 a CDC20 mutant lacking the MAD2-binding region (Δ124–136) still bound to BUBR1 in G2, suggesting that MAD2 is dispensable for BUBR1:CDC20 interaction [Fig. 1C, “G2 HA IP,” notice the presence of BUBR1 in both HA-CDC20 wild-type (WT) and mutant (Δ124–136) IPs]. Taken together, the results indicate that the bulk of MCC assembly is initiated in G2 by recruitment of CDC20 to the BUBR1:BUB3 subcomplex, and is completed by incorporation of MAD2 during mitosis.
Figure 1.
MCC assembly is cell cycle regulated and depends on the mitotic checkpoint. (A) HeLa cells were synchronized at the G1/S boundary by double thymidine block and harvested at the indicated hours (hrs) after release. Lysates were analyzed either directly by immunoblotting or first subjected to BUBR1 or CDC27 immunoprecipitation (IP) then probed. (B) Cells were synchronized as in (A), and then released into medium containing nocodazole. Mitotic cells (Noc) were collected by shake-off 12 h after release and adherent cells (G2) harvested by trypsinization. Lysates were probed to compare levels of protein expression, and protein interactions analyzed by the indicated IP's. Arrowheads in (A and B) denote BUB3 that migrates below the immunoglobulin heavy chain (labeled by asterisks) in IP samples. (C) HeLa cells were transfected with 6x HA-tagged wild-type CDC20 or CDC20 lacking the MAD2-binding region (Δ124–136), and mitotic lysates or G2 lysates were prepared as in (B). Lysates or anti-HA IPs were probed for BUBR1, HA, endogenous CDC20 and MAD2. (D) Mitotic cells were collected as described in (B) and re-plated in medium containing MG132 alone (checkpoint “off”) or both nocodazole and MG132 (checkpoint “on”) for 4 h. Lysates were analyzed directly or by BUBR1 IP. A different percentage of gel was used in this experiment to better resolve BUB3 from the IgG heavy chains.
Once MCC was fully assembled, the complex was found to physically associate with the APC/C (“CDC27 IP” in Fig. 1A, lanes 10 and 12; “CDC27 IP” in Fig. 1B, compare MCC subunits in “G2” and “Noc” lanes). In agreement with recent reports in references 35 and 43, BUBR1 IPs from checkpoint “off” mitotic cells contained ∼3-fold less MAD2 and CDC20 than checkpoint “on” cells (Fig. 1D). The result suggests that MCC disassembles in cells whose spindle checkpoint has been silenced when all kinetochores are properly attached.
C-MAD2 is selectively incorporated into MCC.
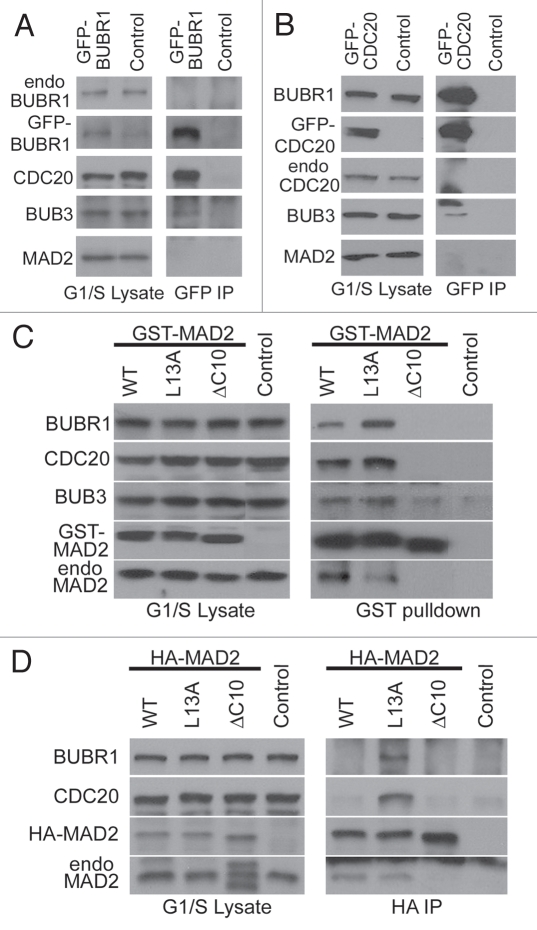
Mild increases in the protein levels of BUBR1 and CDC20 have been previously observed in G2/M cells.44,45 To evaluate whether such increases contribute to cell cycle dependent MCC assembly independently of any G2/M specific posttranslational modifications, GFP-tagged BUBR1 and CDC20 were expressed in G1/S arrested HeLa cells (Fig. 2A and B). Exogenous expression of either protein dramatically enhanced BUBR1:CDC20 interaction in G1/S cells (compare with endogenous BUBR1 IP at time 0 in Fig. 1A), suggesting the BUBR1:CDC20 interaction may be subject to regulation by intracellular protein levels. However, no endogenous MAD2 was found to associate with GFP-BUBR1 or GFP-CDC20 in G1/S cells (Fig. 2A and B).
Figure 2.
Evaluation of MCC assembly following overexpression of MCC subunits in G1/S cells. Cell lysates were prepared from double thymidine arrested G1/S cells transfected with GFP-BUBR1 (A), GFP-CDC20 (B) and GST-tagged (C) or HA-tagged (D) wild-type MAD2 (WT), C-MAD2 (L13A) or O-MAD2 (ΔC10). The lysates were subjected either to IPs using anti-GFP antibody (A and B) or anti-HA antibody (D), or GST-pulldowns (C). MCC assembly was evaluated by probing BUBR1, CDC20, BUB3 and MAD2 in the IPs and pulldowns. Untransfected cells served as controls. In (A) both endogenous BUBR1 (“endo BUBR1”) and GFP-BUBR1 were probed with the same BUBR1 antibody, therefore signals could be compared directly. GFP-CDC20 and endogenous CDC20 (“endo CDC20”) in (B), and GST-tagged and endogenous MAD2 (“GST-MAD2” and “endo MAD2”) in (C) were similarly probed with the antibodies against CDC20 or MAD2 respectively to allow direct comparison of relative expression levels of endogenous and exogenously expressed proteins. Intervening lanes between “control” and “ΔC10” were cropped in (C).
We then focused on how MAD2 is incorporated into the MCC. Given that C-MAD2 conformers are amplified in checkpoint active cells, we tested the effect of increasing intracellular C-MAD2 level on MCC assembly in G1/S cells. A recently characterized MAD2 mutant that adopts the C-conformation (MAD2L13A) 8,9,21 was transfected into cells that were arrested at the G1/S boundary. GST-pulldown of MAD2L13A from these cells contained BUBR1, BUB3 and CDC20; while a MAD2ΔC10 mutant locked in the O-conformation (described in refs. 28 and 46) failed to pull down any other MCC subunits (Fig. 2C). The result suggested selective incorporation of C-MAD2 into the MCC. GST-MAD2WT displayed binding to MCC subunits similarly to GST-MAD2L13A, despite the assumption of MAD2WT being predominantly O-MAD2 (Fig. 2C). As pointed out before, the GST-tag may affect rearrangement of the N terminus of MAD2 that is required for inter-conversion between MAD2WT conformers.8,9 Replacing the GST-tag with a smaller and unstructured HA-tag made MAD2WT behave more like an O-conformer (Fig. 2D). Together, the above results suggest that intracellular C-MAD2 level is a major rate-limiting factor for MCC assembly. The restriction of MCC assembly to checkpoint-active mitotic cells (Fig. 1) therefore may reflect higher C-MAD2 levels in those cells.
C-MAD2 enhances MCC assembly and MCC:APC/C association in mitotic cells and extracts.
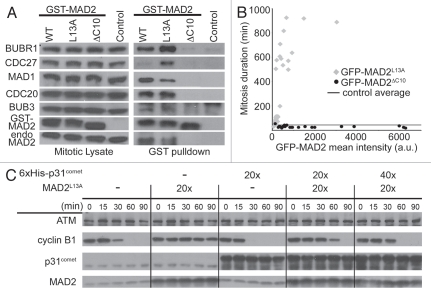
We next examined MCC assembly in mitotic cells expressing GST-tagged MAD2WT, MAD2L13A or MAD2ΔC10 (Fig. 3A). Similar to results obtained from G1/S arrested cells, wild type and C-MAD2, but not the O-MAD2 mutant, pulled down other MCC subunits in nocodazole arrested mitotic cells. The amount of APC/C (represented by CDC27) that associated with MCC was higher in cells expressing MAD2L13A as compared with MAD2WT, and no CDC27 was observed to associate with MAD2ΔC10 (Fig. 3A).
Figure 3.
C-MAD2 enhanced MCC assembly, MCC:APC/C interaction and mitotic checkpoint responses in mitotic cells and mitotic lysates. (A) MCC assembly was evaluated as in Figure 2C except that lysates were from nocodazole arrested mitotic cells. MAD1 and CDC27 were also probed. (B) Comparison of mitosis duration in cells expressing GFP-MAD2L13A or GFP-MAD2ΔC10. The duration of mitosis (in minutes from cell round-up to initiating cleavage furrow) was scatter plotted vs. intracellular GFP mean intensity (a.u., arbitrary units) of individual cells overexpressing GFP-MAD2L13A (diamonds) or GFP-MAD2ΔC10 (circles). The times were compared with average mitosis duration in untransfected control cells (thick line). (C) Opposite effects of C-MAD2 (MAD2L13A) and p31comet on APC/C activation in a mitotic extract. The fold (x) of recombinant proteins added is relative to the endogenous protein levels in the extract. ATM was used as loading control.
Live cell imaging was performed to directly evaluate the effects of GFP-MAD2L13A and GFP-MAD2ΔC10 expression on mitosis progression (Vids. S1–S4). In untransfected control cells, the average time from mitosis entry to anaphase onset was 43 ± 21 min (n = 70, range = 18–150 min). Transfection of GFP vector alone did not significantly affect the duration (51 ± 51 min, n = 34, range = 30–324 min) (Welch's unpaired t-test, p = 0.35 > 0.05) (Vids. S5 and S6). In contrast, cells transfected with GFP-MAD2L13A spent 334 ± 312 min (n = 31, range = 30–924 min) in mitosis before anaphase or cell death (Fig. 3B). The length of mitosis correlated with the C-MAD2 level (as determined by intracellular mean GFP intensity, r = 0.66) and was significantly longer than in untransfected control cells (Welch's unpaired t-test, p < 0.001). Cells expressing GFP-MAD2ΔC10 accelerated through mitosis within 29 ± 8 min (n = 21, range = 18–54 min, p < 0.001 as compared with control) (Fig. 3B).
The role of C-MAD2 as part of the mitotic checkpoint effector was also examined in a mitotic extract prepared from nocodazole arrested HeLa cells.43,47 Incubation of the chromosome-free extract in vitro bypasses upstream signal transduction events (including kinetochore catalyzed O→C-conversion) but recapitulates APC/C activation that is accompanied by disassociation of MCC from APC/C.43 When C-MAD2 was added to the extract at 20-fold over endogenous concentration, a delay of ∼60 min in APC/C activation was observed as reflected in slower degradation of cyclin B (Fig. 3C). Further titration experiments found even addition of an extra 40% of endogenous level of C-MAD2 reproducibly induced a delay of ∼5 min (data not shown). Consistent with selective incorporation of C-MAD2 into the MCC and absence of unattached kinetochores in the extract to catalyze O→C conversion, addition of O-MAD2 had no effect on the degradation of cyclin B.10 The mitotic checkpoint silencing protein p31comet specifically binds to C-MAD2 and silences mitotic checkpoint signaling.48–50 Addition of p31comet itself to the extract indeed accelerated APC/C activation51 (Fig. 3C). Moreover, addition of p31comet alleviated the delay of APC/C activation caused by C-MAD2 (Fig. 3C). These results are consistent with a recent report showing that p31comet may cause MCC disassembly through interaction with MAD2.52
A recombinant MCC containing C-MAD2 inhibits mitotic APC/C in vitro.
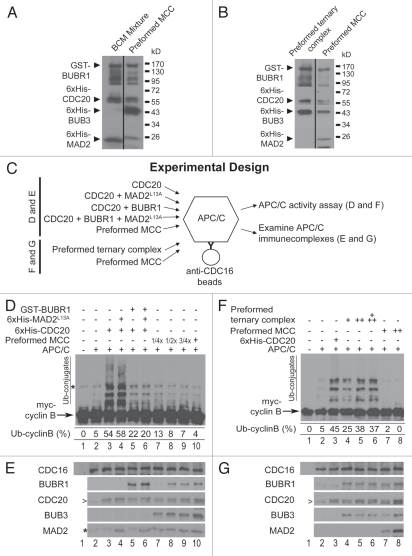
Some earlier observations did not detect or only detected sub-stoichiometric levels of MAD2 in the MCC either isolated from cell lysates or reconstituted from recombinant proteins.27,29,39,40 Based on the above results we reasoned that these observations may be due to the conformational heterogeneity of MAD2WT and/or spontaneous inter-conversion between O- and C-conformers. To test the idea, we co-expressed GST-BUBR1 and 6x His-tagged CDC20, BUB3 and -MAD2L13A by infecting Sf9 insect cells with all four recombinant baculoviruses. The association of C-MAD2 with other components of the MCC was clearly observed in the GSH-agarose affinity-purified MCC (Fig. 4A, right lane).
Figure 4.
Recombinant MCC displays higher inhibitory activity toward mitotic APC/C than individual MCC subunits or a preassembled BUBR1:BUB3:CDC20 ternary complex. (A) A mixture of individual MCC subunits (left lane; BCM: BUBR1, CDC20 and MAD2) at equivalent levels to subunits in a preformed affinity-purified MCC (right lane; BUBR1:BUB3:CDC20:MAD2) were examined by immunoblot. The difference in 6x His-MAD2L13A molecular weight in the two lanes results from the different constructs used for bacterial and insect cell expression. (B) A preformed ternary complex (left lane; BUBR1:BUB3:CDC20) and the preformed MCC (right lane; BUBR1:BUB3:CDC20:MAD2) were affinity-purified and examined by immunoblot. (C) Diagram of experimental design for (D–G). GST-BUBR1 (78 nM), 6x His-CDC20 (198 nM) and 6x His-MAD2L13A (32 nM) were incubated with affinity-purified APC/C in various combinations or as preformed MCC and examined for the ability to inhibit (D) and to interact (E) with mitotic APC/C. Similarly, preformed BUBR1:BUB3:CDC20 ternary complex or preformed MCC were incubated with affinity-purified APC/C and examined for the ability to inhibit (F) and to interact (G) with mitotic APC/C. (D) Comparison of APC/C inhibition by individual MCC components and preformed MCC. APC/C activity was assessed by the levels of myc-cyclin B ubiquitylation (“Ub-conjugates”). Mitotic APC/C was isolated by immunoprecipitation using an anti-CDC16 (a core APC/C subunit) antibody and is present in all lanes except lane 1. In lanes 7–9 the fractions of MCC as used in lane 10 were indicated. The percentage of ubiquitylated cyclin B for each reaction is indicated below corresponding lanes. The asterisk (*) indicates a cross-reacting band present in myc-cyclin B preparation. (E) Recombinant proteins associated with mitotic APC/C were probed. The beads were taken directly from corresponding lanes in (D) and washed before immunoblotting. The lanes are numbered as in (D) and the asterisk (*) indicates a cross-reacting band most obviously seen in lanes 2, 3 and 5. The arrowhead (>) in the CDC20 part may also be a cross-reacting band as APC/C containing this band showed minimal activity as seen in lane 2 of (D). (F) Comparison of APC/C inhibition by preformed MCC and preformed BUBR1:BUB3:CDC20 ternary complex. Increasing concentrations of preformed ternary complex (lanes 4–6) or preformed MCC (lanes 7–8) were tested. Other labels are similar to (D). (G) Recombinant proteins associated with mitotic APC/C were probed. The beads were taken directly from corresponding lanes in (F) and washed before immunoblotting. The lanes are numbered as in (F).
We next compared the affinity-purified MCC with individual MCC subunits for APC/C binding capability and APC/C inhibitory activities (Fig. 4C–E). In contrast to many previous publications, we used mitotic APC/C affinity-isolated from HeLa cells not interphase Xenopus APC/C in these assays (see Materials and Methods for APC/C purification). Using myccyclin B as a substrate, mitotic APC/C essentially devoid of CDC20 and MCC exhibited only minimal ubiquitylation activity (Fig. 4D and E, lane 2). Addition of recombinant CDC20 (∼198 nM final concentration, comparable to the level in recombinant MCC added to the APC/C assay in Fig. 4D and E, lane 10) significantly stimulated the ubiquitylation activity (∼10-fold) (Fig. 4D, lane 3). Pre-incubation of the same amount of CDC20 with C-MAD2 at comparable level to MAD2 in recombinant MCC did not affect CDC20-stimulated APC/C activity, while BUBR1 addition reduced CDC20 stimulation, with ubiquitylated cyclin B percentage dropping from 54 to 22% (Fig. 4D, compare lanes 4 and 5 to lane 3). Simultaneous addition of BUBR1 and C-MAD2, together with CDC20, did not further reduce APC/C activity as compared with BUBR1 alone (Fig. 4D, lane 6). Interestingly, addition of preformed recombinant MCC almost completely inhibited APC/C activity to the background level as shown by APC/C core even though ∼198 nM of CDC20 was contained in the MCC preparation (Fig. 4D, compare lane 10 to lane 2). Use of as low as 4-fold less MCC in the APC/C assays still effectively inhibited APC/C activity (lanes 7–9). Notably, in experiments containing individual MCC subunits or the preformed MCC complexes (Fig. 4D and E, lanes 6–10), both BUBR1 and MAD2 co-IPed with APC/C core subunit CDC16 (Fig. 4E). BUB3 is also present in the CDC16 IP when recombinant MCC was used in the assays (Fig. 4E). Efficient association of checkpoint proteins with mitotic APC/C but no or weak APC/C inhibition (as seen in Fig. 4D and E, lane 6) strongly suggested that certain preformed interactions between MCC subunits are essential for the MCC to acquire potent APC/C inhibitory activity.
To further test the importance of C-MAD2 incorporation into the MCC for the latter's potent APC/C inhibitory activity, we generated a BUBR1:BUB3:CDC20 ternary complex in Sf9 cells (Fig. 4B, left lane). APC/C binding and APC/C inhibition was then compared between affinity-purified MCC and the ternary complex (Fig. 4C, F and G). Strikingly, the ternary complex displayed little inhibition of cyclin B ubiquitylation under several tested concentrations (Fig. 4F, compare lanes 4–6 to lane 3). Incubation of APC/C with higher concentrations of ternary complex seemed even to produce more ubiquitin conjugates; suggesting CDC20 in the ternary complex is still able to enhance APC/C activity. In contrast, addition of recombinant MCC reduced cyclin B ubiquitylation to background level (Fig. 4F, compare lanes 7 and 8 to lane 2), agreeing with results shown in Figure 4D. In all experiments containing the preformed ternary complex or preformed MCC, BUBR1, BUB3 and MAD2, when they are present, associate with APC/C as evidenced by co-immunoprecipitation with CDC16 (Fig. 4G). Despite the ability of the preformed ternary complex to interact with the APC/C, the results suggest that only C-MAD2-containing MCC is sufficient to mediate APC/C inhibition.
Discussion
The results presented in this report are consistent with the idea that in human cells the MCC, as a complex of BUBR1, BUB3, CDC20 and MAD2, is responsible for APC/C inhibition. Furthermore, MCC only incorporates C-MAD2 and the intracellular C-MAD2 level is a major limiting factor of MCC assembly. We suggest that the mitosis-specific and checkpointdependent assembly of MCC is tightly linked with the increase of C-MAD2 level in cells with an activated mitotic checkpoint, primarily due to O→C-MAD2 conversion catalyzed by unattached kinetochores.
The selective incorporation of C-MAD2 into the MCC provides reasonable explanations to several earlier observations. As wild-type MAD2 is primarily O-MAD2, but also contains low levels of C-MAD2 in interphase, a basal level of MCC containing sub-stoichiometric levels of MAD2 may be observed in BUBR1 IPs from interphase cells.32 The functional significance of this basal level of MCC may be reflected in the “timer” roles attributed to BUBR1 and MAD2, but not other mitotic checkpoint proteins.53 Along the same line, if wild-type MAD2 was used in MCC reconstitution experiments, only sub-stoichiometric levels of MAD2 would be expected.40 In principle, the explanation could also be applied to the different levels of MAD2 observed to associate with BUBR1 in mitotic lysates.27,29,32,35 However, as C-MAD2 was regarded as a more stable lower-energy state compared with O-MAD2, spontaneous C→O conformation seems unlikely to occur without external energy input.8 It remains a possibility that variations in experimental protocols may have differentially perturbed the O- and C-MAD2 equilibrium.
Direct BUBR1 interaction with CDC20 has been observed in vitro.29,40 We also showed in cell lysates that BUBR1 interacts with a CDC20 mutant lacking the MAD2-binding domain (Fig. 1D). In addition, exogenous expression of BUBR1 or CDC20 in G1/S cells leads to BUBR1:CDC20 complex formation with no apparent MAD2 incorporation (Fig. 2). Therefore, even though it has been observed using exogenous expression in mouse cells,42 or MAD2 siRNA in human cells39 or recombinant yeast proteins41 that MAD2 stimulates the interaction between BUBR1 (or yeast MAD3) and CDC20, it is likely that MAD2 may promote but is not absolutely required for BUBR1:CDC20 interaction. Our results suggest a two-step model for MCC assembly in human cells, suggesting that CDC20 is recruited to the BUBR1:BUB3 complex in G2 cells and C-MAD2 is then incorporated during mitosis. Malureanu et al. have previously reported that MCC was fully assembled at similar levels in both G2 and mitotic mouse embryonic fibroblast (MEF) cells.36 We cannot completely explain the discrepancy at present except suggesting possible differences between HeLa and MEF cells. The MAD2 level in MEFs is subject to Rb regulation and fluctuates through the cell cycle,54 while MAD2 in HeLa cells is maintained at a constant level irrespective of cell cycle stages. Although G2 MCC may help prevent premature APC/C activation, it should be noticed that another APC/C inhibitor Emi1 also participates in repression of APC/C activity during G2 phase.55
How the MCC is assembled with regard to the stoichiometry and architecture of its individual subunits is still unknown, but we have recently characterized a direct interaction between BUBR1 and C-MAD2 and demonstrated its functional relevance to MCC activity.10 Together with well-characterized BUBR1:CDC20 and C-MAD2:CDC20 interactions,1,7 this indicates all three binary protein-protein interactions are involved in MCC assembly. In addition, the efficiency of MCC formation in checkpoint active cells can certainly be enhanced by other previously described mechanisms even though this work focused on the essential requirement for C-MAD2 to assemble a functional MCC. For example, the increase of BUBR1 and CDC20 protein levels in G2/M cells may promote BUBR1:CDC20 interaction56 (Fig. 1A and B), and phosphorylation of CDC20 in mitosis enhances CDC20 interaction with both BUBR1 and MAD2.25,56–58 The role of BUB3 in the mitotic checkpoint is still controversial,36,59 and BUB3 is absent from the MCC associated with APC/C in S. pombe.60 BUB3, however, was found in CDC27 and CDC16 IPs using either HeLa mitotic cell lysate or recombinant proteins (Figs. 1 and 4). It will be interesting to compare the APC/C inhibitory activity with fully assembled recombinant MCCs or a subcomplex lacking BUB3.
In conclusion, we have presented evidence in this study that C-MAD2 is selectively incorporated into the MCC, establishing a direct connection between a major signal transducer and an effector for mitotic checkpoint signaling. C-MAD2 incorporation into the BUBR1:BUB3:CDC20 ternary complexes, both in vivo and in vitro, is likely to be one crucial step to assemble a fully functional MCC as the potent APC/C inhibitor. Future work will focus on dissecting the interactions between MCC subunits and the mechanisms underlying MCC inhibition of APC/C activity.
Materials and Methods
DNA constructs.
Human full length BUBR1, MAD2, p31comet and CDC20 cDNAs were amplified from a prostate cDNA library (Invitrogen) or freshly prepared reverse transcribed cDNAs provided by Dr. Douglas Leaman (University of Toledo). Full-length cDNAs and fragments were cloned into pENTR-D/TOPO first and then subcloned into various destination vectors using the Gateway recombination reactions (Invitrogen). Point mutations were generated using the QuickChange Site-Directed Mutagenesis Kit (Stratagene). Internal deletion of CDC20 was made through three rounds of PCRs with primers defining N- and C termini and an intermediate primer joining residues 123 and 137. All constructs were confirmed by DNA sequencing. pFastbac-BUB3, pET28-cyclinB-myc and pET28-E1 were generous gifts from Dr. Hongtao Yu (UT Southwestern Medical Center), and a His-tagged UbcH10 construct was from Dr. Michael Rape (UC Berkeley).
Cell culture, synchronization and transfection.
HeLaM,62 a subline of HeLa, was maintained in DMEM with 10% fetal bovine serum at 37°C in 5% CO2. To synchronize at the G1/S boundary, cells were grown in 2.5 mM thymidine for 15 h, released into drug-free medium for 9 h, and then treated with thymidine for another 15 h. To block cells in prometaphase, HeLaM cells were treated with 2.5 mM thymidine for 24 h, and then directly released into medium containing 60 ng/ml nocodazole for 12 h. The proteasome inhibitor MG132 was used at 20 µM. DNA transfection was performed using polyethylenimine (PEI)62 at a DNA:PEI ratio of 1:3 or Fugene6 (Roche) according to the manufacturer's instructions. Sf9 cells were grown at 27°C in SFX medium (Hyclone) supplemented with 10% fetal bovine serum. Cellfectin (Invitrogen) was used to transfect bacmids into Sf9 cells.
Antibodies.
The following antibodies were used at the indicated dilutions. CDC20 (sc-8358, for immunoprecipitation only; Santa Cruz Biotechnology), HA (sc-805, for immunoprecipitation only; Santa Cruz Biotechnology), CDC20 (sc-13162, 1:200; Santa Cruz Biotechnology), MAD2 (A300–301A-2, 1:1,000; Bethyl), Nek2A (1:1,000; Abcam), cyclin B1 (sc-245, 1:1,000; Santa Cruz), GST (26H1, 1:1,000; Cell Signaling), GFP (A11122, 1:500; Invitrogen), γ-Tubulin (T5326, 1:1,000; Sigma), β-actin (ab6276, 1:5,000; Abcam), CDC27 (sc-6392, 1:200; Santa Cruz Biotechnology), CDC16 (sc-6395, 1:200; Santa Cruz Biotechnology), HA (3F10, 1:1,000; Roche) and c-myc (9E10, 1:500; Developmental Studies Hybridoma Bank). Other antibodies included BUBR1 (mouse monoclonal, 1:1,000), BUBR1 (rabbit polyclonal, 1:1,000), MAD2 (mouse monoclonal, 1:500), MAD1 (rabbit polyclonal, 1:2,000), BUB3 (rabbit polyclonal, 1:1,000), CDC27 (rabbit polyclonal, 1:1,000), CDC16 (rabbit polyclonal, 1:1,000), APC7 (rabbit polyclonal, 1:1,000), and have been described previously in references 44 and 63. Alkaline phosphatase conjugated goat-anti-mouse (A3688) and goat-anti-rabbit (A3812) secondary antibodies were purchased from Sigma and used at 1:30,000. Alkaline phosphatase conjugated donkey-anti-goat (sc-2022) secondary antibody was purchased from Santa Cruz Biotechnology and used at 1:10,000.
Cell lysates, immunoblotting, immunoprecipitation and GST-pulldown.
Cells were lysed in Cell Lysis Buffer (1x PBS, 10% glycerol, 0.5% NP-40) supplemented with protease inhibitors (Protease Inhibitor Cocktail set III, EDTA-Free; Calbiochem) and phosphatase inhibitors (100 mM NaF, 1 mM Na3VO4, 60 mM β-glycerophosphate, 100 nM Microcystin LR). The protein concentration of the lysates was measured using the BCA Protein Assay Kit (Pierce). For immunoprecipitation, 200–300 µg of lysates were incubated with appropriate antibodies (0.5–1 µg) at 4°C for 1 h, and then mixed with Protein A-agarose beads (RepliGen) for another 1 h. Immune-complexes were washed four times with Cell Lysis Buffer containing 250 mM NaCl and then subjected to SDS-PAGE separation. For GST-pulldown experiments, cell lysates were directly added to glutathioneagarose (Pierce) and incubated for 1.5 h at 4°C. Immunoblotting was used to probe specific proteins in the cell lysates or immunoprecipitates. In some experiments the blots were scanned and the intensities of bands of interest were quantitated using Kodak Molecular Imaging Software.
Protein expression.
6x His tagged UbcH10, E1, cyclin B (1–102), p31comet and mutant MAD2 were expressed in BL21-CodonPlus (DE3) RIPL (Stratagene) at 25°C. 6x His tagged CDC20 and GST-tagged BUBR1 were expressed in Sf9 cells following the manufacturer's instructions (Invitrogen). Recombinant MCC was generated by simultaneously co-infecting SF9 cells with four recombinant baculoviruses encoding GST-tagged BUBR1, 6x His tagged CDC20, 6x His tagged BUB3 and 6x His tagged MAD2L13A (Invitrogen). Recombinant ternary complex was generated similarly to recombinant MCC, except that the recombinant baculovirus encoding 6x His tagged MAD2L13A was excluded during Sf9 co-infection. Proteins or protein complexes were purified using glutathione-agarose (Pierce), Probond nickel beads (Invitrogen) or cobalt beads (Pierce). Concentrations of individual recombinant proteins were determined by comparing the target band with BSA standards on Coomassie blue stained gels.
APC/C activation assay using concentrated mitotic extracts.
The extracts were prepared and the assay performed as described in reference 10.
In vitro ubiquitylation assay.
Mitotic APC/C was affinity purified by incubating anti-CDC16 bound protein G-agarose beads (2 µg per µl beads) with 20 bead volumes of lysates (at 15–20 mg/ml protein concentration) made from nocodazole arrested HeLaM cells for 2 h at 4°C. Beads were washed three times with 2 bead volumes of TBS-T (20 mM Tris pH 7.5, 150 mM NaCl, 0.01% Tween-20) and four times with 2 bead volumes of TBS-T containing 500 mM NaCl. Washed beads were further treated in 1 ml of Buffer A (50 mM Tris pH 7.5, 0.3 M KCl, 1 mM TCEP, 0.5% NP-40, 1 mg/ml BSA, 10% glycerol) for 10 min at 4°C to remove MCC and CDC20, and then resuspended as 50% slurry in a buffer containing 20 mM Tris pH 7.5, 150 mM NaCl. For ubiquitylation assays, 4 µl bead slurry was used in 10 µl reactions containing 40 mM Tris pH 7.5, 5 mM MgCl2, 5% (vol/vol) glycerol, 1 mM DTT, 0.5 mg/ml BSA, 2 mM ATPγS, 2 mg/ml ubiquitin (Boston Biochem), purified E1 (100 ng) and UbcH10 (200 ng). Recombinant myc-tagged cyclin-B1(1–102) was used at the final concentration of 300 ng/µl as an APC/C substrate. When testing APC/C inhibition (Fig. 4), preformed MCC or preformed BUBR1:BUB3:CDC20 ternary complexes were directly added to the APC/C beads, while mixture of individual recombinant proteins were pre-incubated for 1 h at 37°C before addition. The ubiquitylation reactions were performed in a thermomix shaker (Peqlab, 1,000 rpm at 23°C) for 1 h. The beads were separated from the supernatants, washed and saved for examination of APC/C associated proteins. The supernatants were mixed with SDS-PAGE sample buffer, resolved by SDS-PAGE and analyzed by anti-myc immunoblot to probe cyclin B ubiquitylation.
Live cell imaging and statistical analysis.
Imaging was performed on HeLaM cells within 24–48 h after transfection with GFP vector, GFP-MAD2L13A or GFP-MAD2ΔC10. The medium was replaced with Hepes-buffered DMEM and covered with mineral oil before imaging. Phase contrast and GFP images were collected on an automated Olympus IX-81 microscope using a 20x NA 0.50 UPlanFLN objective lens while cells were maintained at 37°C in a heated weather chamber. Single plane images focusing on mitotic cells in the field were acquired at multiple positions and 6 min intervals by a cooled CCD camera (CoolSNAP HQ2; Photometrics) with 2 × 2 binning. Image acquisition and analysis were performed using Slidebook software (Intelligent Imaging Innovations). Mitosis duration was estimated based on the time from cell round-up till initiation of cleavage furrow ingression or cell death. Intracellular GFP mean intensity was derived by masking transfected cells and subtracting the background. Welch unpaired t-test was used to compare the mitosis durations assuming unequal variances in cells transfected with different MAD2 conformers.
Acknowledgments
We thank Hongtao Yu, Michael Rape and Douglas Leaman for generous gifts. We also thank Avram Hershko for advice on making and using mitotic HeLa cell extracts. T.J.Y. was funded by grants from the National Institutes of Health (GM86877 and GM44762), a core grant (CA06927), an appropriation from the Commonwealth of Pennsylvania, and the Greenberg Fund. S.T.L. was funded by deArce Memorial Fund, Ohio Cancer Research Associates, University of Toledo Interdisciplinary Research Initiation Award and NSF grant MCB-1052413.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 2.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 3.Weaver BA, Cleveland DW. The aneuploidy paradox in cell growth and tumorigenesis. Cancer Cell. 2008;14:431–433. doi: 10.1016/j.ccr.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musacchio A, Hardwick KG. The spindle checkpoint: structural insights into dynamic signalling. Nat Rev Mol Cell Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- 5.Rieder CL, Schultz A, Cole R, Sluder G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J Cell Biol. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang H, Yen TJ. BubR1 is an effector of multiple mitotic kinases that specifies kinetochore: microtubule attachments and checkpoint. Cell Cycle. 2009;8:1164–1167. doi: 10.4161/cc.8.8.8151. [DOI] [PubMed] [Google Scholar]
- 7.Ciliberto A, Shah JV. A quantitative systems view of the spindle assembly checkpoint. EMBO J. 2009;28:2162–2173. doi: 10.1038/emboj.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo X, Yu H. Protein metamorphosis: the two-state behavior of Mad2. Structure. 2008;16:1616–1625. doi: 10.1016/j.str.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mapelli M, Musacchio A. MAD contortions: conformational dimerization boosts spindle checkpoint signaling. Curr Opin Struct Biol. 2007;17:716–725. doi: 10.1016/j.sbi.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Tipton AR, Wang K, Link L, Bellizzi JJ, Huang H, Yen T, et al. BUBR1 and Closed MAD2 (C-MAD2) Interact Directly to Assemble a Functional Mitotic Checkpoint Complex. J Biol Chem. 2011;286:21173–21179. doi: 10.1074/jbc.M111.238543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol Cell. 2002;9:59–71. doi: 10.1016/S1097-2765(01)00435-X. [DOI] [PubMed] [Google Scholar]
- 12.Chen RH, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- 13.Campbell MS, Chan GK, Yen TJ. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J Cell Sci. 2001;114:953–963. doi: 10.1242/jcs.114.5.953. [DOI] [PubMed] [Google Scholar]
- 14.Kitagawa R. The spindle assembly checkpoint in Caenorhabditis elegans: one who lacks Mad1 becomes mad one. Cell Cycle. 2009;8:338–344. doi: 10.4161/cc.8.3.7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 16.Luo X, Tang Z, Xia G, Wassmann K, Matsumoto T, Rizo J, et al. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat Struct Mol Biol. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- 17.Nezi L, Rancati G, De Antoni A, Pasqualato S, Piatti S, Musacchio A. Accumulation of Mad2-Cdc20 complex during spindle checkpoint activation requires binding of open and closed conformers of Mad2 in Saccharomyces cerevisiae. J Cell Biol. 2006;174:39–51. doi: 10.1083/jcb.200602109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mapelli M, Massimiliano L, Santaguida S, Musacchio A. The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell. 2007;131:730–743. doi: 10.1016/j.cell.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 19.Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol. 2000;150:1233–1250. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol. 2004;14:942–952. doi: 10.1016/S0960-9822(04)00381-1. [DOI] [PubMed] [Google Scholar]
- 21.Yang M, Li B, Liu CJ, Tomchick DR, Machius M, Rizo J, et al. Insights into mad2 regulation in the spindle checkpoint revealed by the crystal structure of the symmetric mad2 dimer. PLoS Biol. 2008;6:50. doi: 10.1371/journal.pbio.0060050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu H. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol Cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Lees E. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: model for spindle checkpoint regulation. Mol Cell Biol. 2001;21:5190–5199. doi: 10.1128/MCB.21.15.5190-9.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung E, Chen RH. Phosphorylation of Cdc20 is required for its inhibition by the spindle checkpoint. Nat Cell Biol. 2003;5:748–753. doi: 10.1038/ncb1022. [DOI] [PubMed] [Google Scholar]
- 26.Simonetta M, Manzoni R, Mosca R, Mapelli M, Massimiliano L, Vink M, et al. The influence of catalysis on mad2 activation dynamics. PLoS Biol. 2009;7:10. doi: 10.1371/journal.pbio.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol Biol Cell. 2002;13:755–766. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Z, Bharadwaj R, Li B, Yu H. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell. 2001;1:227–237. doi: 10.1016/S1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Gorbea C, Mahaffey D, Rechsteiner M, Benezra R. MAD2 associates with the cyclosome/anaphase-promoting complex and inhibits its activity. Proc Natl Acad Sci USA. 1997;94:12431–12436. doi: 10.1073/pnas.94.23.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardwick KG, Johnston RC, Smith DL, Murray AW. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p and Mad2p. J Cell Biol. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20 and MAD2. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Millband DN, Hardwick KG. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p- and Mph1p-dependent manner. Mol Cell Biol. 2002;22:2728–2742. doi: 10.1128/MCB.22.8.2728-42.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen RH. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J Cell Biol. 2002;158:487–496. doi: 10.1083/jcb.200204048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herzog F, Primorac I, Dube P, Lenart P, Sander B, Mechtler K, et al. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malureanu LA, Jeganathan KB, Hamada M, Wasilewski L, Davenport J, van Deursen JM. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev Cell. 2009;16:118–131. doi: 10.1016/j.devcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fraschini R, Beretta A, Sironi L, Musacchio A, Lucchini G, Piatti S. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 2001;20:6648–6659. doi: 10.1093/emboj/20.23.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poddar A, Stukenberg PT, Burke DJ. Two complexes of spindle checkpoint proteins containing Cdc20 and Mad2 assemble during mitosis independently of the kinetochore in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:867–878. doi: 10.1128/EC.4.5.867-78.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol. 2008;10:1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell. 2009;16:105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21:655–667. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davenport J, Harris LD, Goorha R. Spindle checkpoint function requires Mad2-dependent Cdc20 binding to the Mad3 homology domain of BubR1. Exp Cell Res. 2006;312:1831–1842. doi: 10.1016/j.yexcr.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 43.Braunstein I, Miniowitz S, Moshe Y, Hershko A. Inhibitory factors associated with anaphase-promoting complex/cylosome in mitotic checkpoint. Proc Natl Acad Sci USA. 2007;104:4870–4875. doi: 10.1073/pnas.0700523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan GK, Jablonski SA, Sudakin V, Hittle JC, Yen TJ. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J Cell Biol. 1999;146:941–954. doi: 10.1083/jcb.146.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinstein J. Cell cycle-regulated expression, phosphorylation and degradation of p55Cdc. A mammalian homolog of CDC20/Fizzy/slp1. J Biol Chem. 1997;272:28501–28511. doi: 10.1074/jbc.272.45.28501. [DOI] [PubMed] [Google Scholar]
- 46.Sironi L, Melixetian M, Faretta M, Prosperini E, Helin K, Musacchio A. Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J. 2001;20:6371–6382. doi: 10.1093/emboj/20.22.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- 48.Xia G, Luo X, Habu T, Rizo J, Matsumoto T, Yu H. Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 2004;23:3133–3143. doi: 10.1038/sj.emboj.7600322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mapelli M, Filipp FV, Rancati G, Massimiliano L, Nezi L, Stier G, et al. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J. 2006;25:1273–1284. doi: 10.1038/sj.emboj.7601033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang M, Li B, Tomchick DR, Machius M, Rizo J, Yu H, et al. p31comet blocks Mad2 activation through structural mimicry. Cell. 2007;131:744–755. doi: 10.1016/j.cell.2007.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Summers MK, Pan B, Mukhyala K, Jackson PK. The unique N terminus of the UbcH10 E2 enzyme controls the threshold for APC activation and enhances checkpoint regulation of the APC. Mol Cell. 2008;31:544–556. doi: 10.1016/j.molcel.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teichner A, Eytan E, Sitry-Shevah D, Miniowitz-Shemtov S, Dumin E, Gromis J, et al. p31comet promotes disassembly of the mitotic checkpoint complex in an ATP-dependent process. Proc Natl Acad Sci USA. 2011;108:3187–3192. doi: 10.1073/pnas.1100023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 54.Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 55.Reimann JD, Freed E, Hsu JY, Kramer ER, Peters JM, Jackson PK. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001;105:645–655. doi: 10.1016/S0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- 56.Wu H, Lan Z, Li W, Wu S, Weinstein J, Sakamoto KM, et al. p55CDC/hCDC20 is associated with BUBR1 and may be a downstream target of the spindle checkpoint kinase. Oncogene. 2000;19:4557–4562. doi: 10.1038/sj.onc.1203803. [DOI] [PubMed] [Google Scholar]
- 57.Kallio M, Weinstein J, Daum JR, Burke DJ, Gorbsky GJ. Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex, and is involved in regulating anaphase onset and late mitotic events. J Cell Biol. 1998;141:1393–1406. doi: 10.1083/jcb.141.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Visconti R, Palazzo L, Grieco D. Requirement for proteolysis in spindle assembly checkpoint silencing. Cell Cycle. 2010;9:564–569. doi: 10.4161/cc.9.3.10581. [DOI] [PubMed] [Google Scholar]
- 59.Elowe S, Dulla K, Uldschmid A, Li X, Dou Z, Nigg EA. Uncoupling of the spindle-checkpoint and chromosome-congression functions of BubR1. J Cell Sci. 2010;123:84–94. doi: 10.1242/jcs.056507. [DOI] [PubMed] [Google Scholar]
- 60.Sczaniecka M, Feoktistova A, May KM, Chen JS, Blyth J, Gould KL, et al. The spindle checkpoint functions of Mad3 and Mad2 depend on a Mad3 KEN box-mediated interaction with Cdc20-anaphase-promoting complex (APC/C) J Biol Chem. 2008;283:23039–23047. doi: 10.1074/jbc.M803594200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tiwari RK, Kusari J, Sen GC. Functional equivalents of interferon-mediated signals needed for induction of an mRNA can be generated by double-stranded RNA and growth factors. EMBO J. 1987;6:3373–3378. doi: 10.1002/j.1460-2075.1987.tb02659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Durocher Y, Perret S, Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu ST, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.