Abstract
The genomics era has yielded great advances in the understanding of cancer biology. At the same time, the immense complexity of the cancer genome has been revealed, as well as a striking heterogeneity at the whole-genome (or omics) level that exists between even histologically similar tumors. The vast accrual and public availability of multi-omics databases with associated clinical annotation including tumor histology, patient response, and outcome are a rich resource that has the potential to lead to rapid translation of high-throughput omics to improved overall survival. We focus on the unique advantages of a multidimensional approach to genomic analysis in this new high-throughput omics age and discuss the implications of the changing cancer demographic to translational omics research.
The remarkable technological breakthroughs of the last 10 yr have reshaped how we view the cancer genome; therefore, so must our approach to the translation of this knowledge. “Cancer genomics” refers to the study of tumor genomes using various profiling strategies including (but not limited to) DNA copy number, DNA methylation, and transcriptome and whole-genome sequencing—technologies that may collectively be defined as omics. The goal of cancer genomics is to survey these omics data to identify genes and pathways deregulated in cancer and reveal those that may be useful for the detection and management of disease. Such discoveries will improve our understanding of the biology of cancer and lead to the discovery of novel diagnostic, prognostic, and therapeutic markers that will ultimately improve patient outcomes. The field of cancer genomics is rapidly evolving and coupled with the ever-increasing efficiency of genomic profiling; this has led to the realization that personalized medicine is likely to soon become a reality. It is hopeful that in the near future, tumors of cancer patients will be profiled in a timely manner and that the tumor omics findings will subsequently be used to inform patient management.
To date, high-resolution and high-throughput technologies have yielded an unprecedented view of cancer omics. This work has led to the identification of biologically important genes and pathways frequently disrupted across many cancer types that has improved our understanding of cancer as a disease and, moreover, has revealed clinically relevant diagnostic, prognostic, and druggable targets. At the same time, these technologies have also unveiled the immense genomic complexity, and striking inter- and intratumor heterogeneity—at the level of mutational load and structural rearrangements—that exists between even histologically similar tumors (Ding et al. 2008; Stephens et al. 2009; Bozic et al. 2010; Pleasance et al. 2010; Swanton et al. 2011). Distinguishing the molecular events that confer oncogenic properties driving cancer biology, for example, a gene mutation that activates a cancer promoting cellular pathway, from those events that are merely passenger events—alterations that do not drive cancer pathway disruptions—is critical for the translation of omics findings (Hudson et al. 2010). Deciphering driver events is key to designing rational therapeutics aimed at specific cancer phenotypes, predicting patient response to traditional modalities, and expanding the pool of patients likely to benefit from existing treatments. Thus, the field of cancer genomics is presently tasked with distinguishing key genes and pathways driving tumorigenesis and drug response from a bewildering background of genomic variability. Currently, two large international research efforts are churning out omics data for several cancer types that will be extremely useful in helping with this task. The goal of both the International Cancer Genome Consortium (ICGC) and The Cancer Genome Atlas (TCGA) is to compile omics data that are openly available to the public in order to rapidly improve our understanding of the molecular mechanisms driving cancer (Cancer Genome Atlas Research Network 2008; Hudson et al. 2010; Verhaak et al. 2010). In an age of targeted therapy and routine tests for prognostic or predictive molecular markers, the availability of these data has profound implications for translating basic research into personalized medicine.
Successes in translating cancer genomics to targeted cancer therapy
Alongside the widespread use of early detection and screening programs, particularly for breast, cervical, prostate, testicular, and colorectal cancers, targeted therapies have been instrumental in extending the lives of millions of cancer patients (Etzioni et al. 2003). A summary of the fortuitous and labored discovery, translation, and rational application of many targeted therapies has been recently described in an excellent essay by Haber et al. (2011). Pioneering work in this field began with the discovery and targeted interference of the oncogenes to which cancer cells are addicted. The first of these, the ERBB2 (also known as HER2) receptor antagonist trastuzumab (Herceptin) for ERBB2-positive breast cancers, was quickly followed by the first kinase inhibitor, imatinib (Gleevec), which targets the BCR–ABL1 fusion gene harbored by 95% of chronic myeloid leukemia (CML) patients. Treatment with these targeted therapies resulted in a dramatic increase in patient response (Druker et al. 2006; Hudis 2007). It was also realized that existing drugs could be used to treat other cancers driven by similar molecular mechanisms (Papaetis and Syrigos 2010). Gleevec was also discovered to inhibit additional tyrosine kinases including KIT, which is constitutively activated by mutation in gastrointestinal stromal tumors (GIST). Treatment of GIST patients with Gleevec also resulted in a significant increase in patient response. The success of Gleevec paved the way for the development and application of other tyrosine kinase inhibitors (TKIs), such as those used to treat non-small-cell lung cancer (NSCLC) and colorectal cancer (CRC) (Nowell and Hungerford 1961; Lynch et al. 2004; Paez et al. 2004; Druker 2008; Keedy et al. 2011). Clearly, the study of cancer genome structure has led to the detection of important molecular alterations, which have been translated into improved patient outcomes.
Encouragingly, the elapsed time between target discovery and clinical utilization of targeted therapies has decreased significantly in the past five years (Gerber and Minna 2010; Chin et al. 2011). The translation of ALK inhibitors, which are used to treat the ∼7% of NSCLCs patients whose tumors harbor EML4–ALK rearrangements, was achieved in a remarkable three years, although the speed of translation was enhanced due to the preexistence of ALK inhibitors (Soda et al. 2007; Gerber and Minna 2010; Kwak et al. 2010). Speed of translation will likely increase further as classic drug development and trial regimes are reshaped to include prospective characterization of patients so that targeted therapies are only applied to patients harboring the specific genetic alteration for which the therapy was designed, and collection and interrogation of tumor and blood specimens throughout clinical trials to assess patient response (Gerber and Minna 2010; Chin et al. 2011; Haber et al. 2011). Had these principles not been applied, the therapeutic potential of gefitinib in EGFR-mutant lung tumors would have likely gone unnoticed (Lynch et al. 2004). Mutations in EGFR frequently occur in NSCLC (10%–30%), resulting in aberrant activation of the tyrosine kinase domain. Thus, targeted therapies directed at mutant EGFR are designed to inhibit its tyrosine kinase activity to which the lung cancer cells are addicted (Pao and Chmielecki 2010).
Fortunately, as target discovery moves genome wide, cancer omics data are rapidly accumulating, providing a valuable resource for identifying novel targets. Promising new drugs, such as PLX-4032 for melanoma patients harboring V600E-BRAF mutations (Flaherty et al. 2010; Poulikakos and Rosen 2011), validate the genomics → target → drug route of translation, whereby a specific molecular target, in this case, BRAF mutation, was identified and an efficacious, selective therapeutic was developed based on the underlying molecular findings. BRAF mutations have since been discovered in other cancers including lung and most recently multiple myeloma, providing a rationale for their future evaluation as therapeutic targets in these cancers (Ding et al. 2008; Chapman et al. 2011).
In addition to the successful translation of individual gene mutations into therapeutic targets, diagnostic, prognostic, and predictive signatures based on gene panels have also emerged from cancer genomics work. For example, expression signatures based on multigene sets are used clinically for breast cancer prognostic prediction (Oakman et al. 2010). Moreover, genomic profiling technologies are being translated into the clinic, allowing clinicians to perform assays to assess several relevant cancer genes in parallel, reducing the time, cost, and materials required for testing (Stricker et al. 2011). The development of a biomarker—a measurable marker that can be used clinically to assess one or more clinical needs, e.g., to diagnose or predict disease behavior or response to therapy—is a complex process that involves assessment of prevalence, sensitivity, specificity and rigorous validation in multiple clinical cohorts. Along with the increasing successful application of cancer biomarkers, several pertinent practical and ethical considerations have been acknowledged, as discussed in detail by Offit related to discovery and appropriate clinical evaluation standards of genomic biomarkers, along with development of new “genomic counselling” models, for example (Offit 2011; Weitzel et al. 2011).
Challenges in translation of cancer omics findings
Many of the successes in translational cancer genomics have occurred in tumors addicted to single genetic alterations. However, for genetically complex tumors characterized by many alterations, prescribing targeted therapies based on the status of a single molecular alteration in a patient's tumor is often not sufficient to predict therapeutic response (Fojo and Parkinson 2010). In these cases, a major challenge in predicting treatment response is the substantial molecular heterogeneity that exists even for histologically similar tumors that may sustain (1) alterations to different cellular pathways, (2) disruption of different components of similar pathways, and/or (3) unique mechanisms of disruption to genes or pathways. These factors contribute substantially to the variable tumor behavior and treatment response observed clinically and therefore complicate the translation of omics findings to the clinic. For example, some lung cancers are characterized by mutations in the oncogene EGFR, while others are affected by KRAS mutations, and although these tumors may have a similar appearance histologically, they exhibit different therapeutic responses to EGFR inhibitors. EGFR and KRAS function at different levels in the same cellular pathway; however, targeting this pathway upstream at the level of EGFR results in shorter progression-free or overall survival in KRAS mutant patients compared with EGFR mutant patients (Karapetis et al. 2008; Allegra et al. 2009). This example illustrates the importance of considering molecular heterogeneity when making decisions regarding therapy. In addition to drug response, the relationship between tumor heterogeneity and variable tumor behavior also applies to other tumor phenotypes including prognosis and outcome.
The use of cancer omics to define molecular subtypes for some genetically heterogeneous tumors has proven clinically useful. For example, five major subtypes of breast cancer have been identified based on molecular profiling, and each is associated with different clinical measures such as treatment response and prognosis (Prat and Perou 2011). Thus, molecular subtyping as well as identification of particular mutations in tumors can be used to identify specific patient cohorts with favorable responses to gene-targeted therapies, thereby improving patient outcomes.
Many of the challenges in translational omics can be summed up by the “cancer biomarker problem,” which refers to the great disparity between the large amount of omics information that has been produced compared with the relatively smaller number of successfully translated diagnostic, prognostic, and especially predictive biomarkers derived from this massive body of work (Sawyers 2008; Poste 2011). The practical challenges hindering translation of omics-generated tumor markers or signatures include the lack of availability of well-defined, clinically characterized cohorts for evaluating the biomarker and lack of standardization regarding how specimens are collected, handled, and stored. Ultimately, these roadblocks can influence whether or not biomarkers validate in well controlled cohorts (Liotta and Petricoin 2011; Poste 2011). Biomarker translation has also been impeded by a lack of mechanistic links to the tumor itself, although some successfully translated biomarkers do not have well-established roles in cancer biology, yet are clinically very useful due to their strong correlations with specific phenotypes (Liotta and Petricoin 2011). These considerations suggest that there will be greater success translating biomarkers to the clinic when (1) validation studies are performed across multiple data sets and (2) the biomarkers have a role in tumorigenesis, so that they are causative rather than correlative.
Access to cohorts with well-annotated clinical information remains a significant barrier and challenge for most omics studies. Nevertheless, the relation of omics data to distinct clinical–pathological phenotypes is paramount to acceleration of translatable findings, particularly for cancers associated with well-known risk factors. For example, lung tumors from smokers and never smokers were found to exhibit distinct molecular and clinical features and different tumorigenic mechanisms, suggesting that they should be managed differently (Lee et al. 2010; Dasgupta et al. 2011; KL Thu, EA Vucic, R Chari, W Zhang, WW Lockwood, JC English, CE MacAulay, AF Gazdar, S Lam, WL Lam, unpubl.). Specifically, distinct mutational profiles for the EGFR–RAS–RAF–MEK–ERK pathway exist in lung adenocarcinoma from smokers and non-smokers. Moreover, patients who responded to TKIs were primarily never smokers (Lynch et al. 2004). These findings were quickly translated to EGFR diagnostic testing to predict which patients are likely to respond to TKI regimens. Another example of the importance of stratifying patients according to risk when pursuing omics studies is the detection of novel susceptibility loci specific to never-smoker lung cancer (Ahn et al. 2011)—findings that would not have been possible without the collection of corresponding detailed clinical information. In summary, these omics studies applied to defined clinical cohorts informed treatment strategies for smoking related cancer and may also lead to improved screening and early detection programs for never smokers. For other cancers, the integration of clinical features, particularly those pertaining to well-known cancer risk factors, into study design will significantly advance our understanding of the biology underlying distinct clinical phenotypes and cohorts, translating to improved disease management.
Deciphering biologically relevant DNA mutations
Whole-genome data allow for the interpretation of sequence variants in the context of a tumor's entire genomic landscape. Interpretation of omics data for any malignancy, in a biologically relevant context, is paramount to the clinical translation of this information (Jones et al. 2010; Lunshof et al. 2010; Link et al. 2011; Pasche and Absher 2011; Welch et al. 2011). The most biologically relevant alterations are “drivers” of tumorigenesis that underlie particular tumor features or behaviors, such as treatment response, and generally represent ideal candidates for therapeutic intervention. Therefore, the goal of many cancer omics studies is to identify these driver molecular events from those changes that may occur by random chance, or are “reactive” to causative perturbations to genes or pathways. Thus, it is important to acknowledge that at the biological level, not all sequence mutations (or other omics alterations) have biological implications. The imminent arrival of $1,000 genome sequences and the increasing availability of sequencing data today allow for the interpretation of DNA level alterations beyond their impact on protein structure (Lunshof et al. 2010). Analytical approaches to interpreting DNA level data vary widely. Some of these approaches, along with various sequencing methodologies, have been recently reviewed (Robison 2010; Hoffmann 2011). Importantly, approaches to distinguish causative events informatically can be confounded by several factors, which are compounded greatly by whole-genome sequencing data. Here we discuss several of the confounding factors that affect such analysis.
In a sequencing experiment, silent mutations are often used to calculate background mutation rates against which all other mutations may be statistically compared to identify genes mutated at frequencies greater than background, because these mutations may be biologically relevant. Non-silent mutations alter amino acid sequences and are therefore more likely to have a functional effect and confer a growth advantage to cancer cells. However, non-silent mutations, which include missense, nonsense, insertions, deletions, and mutations affecting splicing, are not all functionally equivalent, and as several groups have found, not equally selected for (Radivojac et al. 2008; Mort et al. 2010; Fischer et al. 2011; Youn and Simon 2011). For example, mutations introducing stop codons (nonsense), those affecting splicing, or resulting in functional alterations to phosphorylation sites occur at significantly higher rates than other non-silent mutations, which may signify their selection and importance to cancer cells (Greenman et al. 2006; Radivojac et al. 2008; Fischer et al. 2011; Youn and Simon 2011). Moreover, mutations occurring in a tumor sample with very few mutations overall, compared with a tumor sample with thousands, are likely relevant to tumor biology (Youn and Simon 2011). Algorithms that consider such factors will likely yield more biologically informative results and are less biased by overall levels of genomic alteration compared with algorithms that focus on gene mutation frequency alone.
The overall level of genomic complexity differs substantially between tumors. The extent of genomic alteration may indicate underlying biological mechanisms affecting genomic stability that may mask important mutational events. As such, taking into account the overall background in which identified mutations occur may help to identify those events most biologically important to a tumor and related to a particular tumor phenotype. Genomic alterations of pathways responsible for normal maintenance and repair of the genome may also drive sequence-level alterations particularly for tumors with high frequencies of complex genomic rearrangements (Hampson 1997; Bignell et al. 2007; Stephens et al. 2009; Swanton et al. 2011). For example, frequency of tandem duplications was correlated with specific subtypes of breast cancer, suggesting that disruption of crucial DNA maintenance pathways may underlie distinct mechanisms of transformation in different subtypes (Stephens et al. 2009; Swanton et al. 2011). Sequence-level alterations may also be driven by hypomethylation of normally heavily methylated repetitive DNA (Rizwana and Hahn 1999; Eden et al. 2003; Daskalos et al. 2009; Igarashi et al. 2010; De and Michor 2011) or by aberrant promoter hypermethylation (silencing) of DNA repair genes (MLH1, CDKN2A)—as has been observed in several cancer types (Kane et al. 1997; Sengupta et al. 2007; Cancer Genome Atlas Research Network 2008; Vasavi et al. 2010; Shima et al. 2011; Tawfik et al. 2011). In tumors with a high genomic alteration load, DNA alterations may be a consequence of disruption to genes maintaining genomic integrity. In these cases, the driving events, which represent promising therapeutic targets, may be overlooked if all sequence-level events are considered equally between tumor types and the biological role of the affected genes is not considered.
Cancer genomes are, however, not exclusively disrupted at the level of DNA sequence but frequently at multiple levels of genetic regulation and by diverse mechanisms. Therefore, in addition to DNA sequence-level analysis, it is imperative to investigate other genomic dimensions such as DNA methylation and coding and non-coding RNA expression, to better elucidate the full complement of omics alterations underlying cancer biology.
A multidimensional approach to cancer omics
The identification of events driving distinct tumor phenotypes requires interrogation of multiple omics dimensions on appropriate, well-annotated tumor specimens, and the interpretation of these molecular data in a biologically and clinically meaningful way (Hood et al. 2004; Gondek et al. 2007; Yamamoto et al. 2007; Cancer Genome Atlas Research Network 2008; Dunbar et al. 2008; Gandhi et al. 2009; Modrek et al. 2009; Segditsas et al. 2009; Soh et al. 2009; Wang et al. 2010; Vidal et al. 2011). Using a multi-omics systems-based approach to the analysis of patient cancer samples will enhance our understanding of cancer biology and accelerate the translation of omics research. This approach requires the generation of several types of omics data from each individual cancer specimen (i.e., system), including genome sequence and structural information (i.e., DNA copy number), epigenome, transcriptome, proteome, and metabolomics data followed by integration of these dimensions to identify the key genes and pathways driving an associated phenotype, such as tumor progression or drug response, for example.
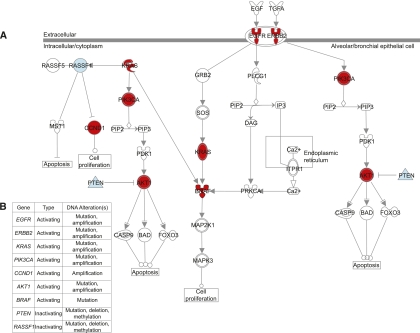
There are several advantages of applying a multi-omics approach to cancer research. For example, multi-omics data allow the identification of genes that are altered at high frequency by multiple mechanisms but at low frequency by any single mechanism (Chari et al. 2010a,b). A single-dimensional approach may overlook frequently disrupted genes, if the frequency of alteration by one mechanism is low. Since aberrations can also occur at multiple points within signaling pathways important to cancer, multi-omics data integration would help to identify signaling pathways frequently disrupted at multiple components, but perhaps at low frequencies at any single component of a pathway (Chari et al. 2010a,b). These concepts are exemplified by the diverse mechanisms of gene disruption at multiple pathway components in the EGFR signaling pathway, which is commonly activated in non-small-cell lung cancer (Fig. 1). Simultaneous interrogation of multiple mechanisms of gene disruption in a single tumor (i.e., mutation, copy number, DNA methylation, miRNA deregulation, etc.) also has the power to identify genes that undergo biallelic disruption, which may be indicative of selection and may therefore signify driver genes. Since it is likely that important cancer genes undergo biallelic disruption, such as inactivation of tumor suppressors by concurrent DNA methylation and copy number loss—disruption of signaling pathways by one or multiple mechanisms may also indicate those most strongly selected for to promote tumor cell growth.
Figure 1.
Activation of EGFR signaling in non-small-cell lung cancer (NSCLC) can occur via disruption of several different components at multiple levels of the pathway. (A) Different proteins in the EGFR pathway can be activated (red) or inactivated (blue) by underlying genetic or epigenetic changes at the DNA level, leading to aberrant pathway activity and oncogenic signaling in NSCLC. Examples of key oncogenes affected include EGFR, RAS, PIK3CA, and AKT. Conversely, examples of tumor suppressors that are inactivated include PTEN and RASSF1. (B) Genetic and epigenetic mechanisms responsible for the disruption of genes in the EGFR signaling pathway in NSCLC include DNA copy number alterations (amplification or deletion), point mutations, and DNA methylation changes. Thus, it is important to consider multiple aspects of the genome and epigenome simultaneously to elucidate the mechanisms driving pathway deregulation. This illustration was generated using Ingenuity Pathway Analysis software.
The aforementioned international genome characterization consortia are generating such data for many tumor types, and already it is clear that even for tumors with thousands of mutations, a limited number of pathways are frequently altered (Boca et al. 2010), echoing the hypothesis put forth by Vogelstein, which suggests that even for tumors with thousands of mutations, a limited number of pathways likely drive cancer biology (Boca et al. 2010). For example, in glioblastoma, one of the first tumor types to be subjected to comprehensive omics profiling, three frequently disrupted pathways (RB, p53, and receptor tyrosine kinase pathways) were found to be independently altered in 78%, 87%, and 88% of tumors, respectively; 74% of all tumors harbored aberrations to all three of these pathways. These observations reveal pathways suitable for targeted therapeutic intervention, including potential inhibition of EGFR and its downstream targets ERBB2, MET, or PI3K
Identifying pathways driving cancer phenotypes
Although some cancers may be driven by alterations to critical cancer genes with unrelated functions, others may be driven by concomitant disruption of a network of genes, which collectively functions to activate or deactivate a specific cellular pathway driving tumor growth. Therefore, in order to better understand the relationships between disrupted genes and the biological mechanisms sustaining cancer growth, it is important to study the complex cellular networks that may underlie distinct clinical phenotypes. Elucidation of deregulated pathways in cancer using pathway analyses are powerful methods that have multifaceted utility: identification of key molecular networks driving cancer phenotypes, development of novel therapeutic targets to inhibit cancer promoting pathways, and guidance of the reappropriation of existing therapies for patients with appropriate predictive pathway profiles. Appreciation of these benefits of pathway analysis has led to an explosion in interest in pathway databases that catalogue various cellular networks (e.g., the Kyoto Encyclopedia of Genes and Genomes, KEGG) (Kanehisa et al. 2010) and analysis tools for identifying pathway perturbations (e.g., Ingenuity Pathways Analysis; http://www.ingenuity.com) that can be used to interpret the genetic disruptions revealed by multi-omics analysis in the context of cancer biology.
The importance of interpreting omics data at the pathway level is highly relevant to translation of new findings. For example, DNA alterations to pathway components downstream from EGFR, as described for glioblastoma (Verhaak et al. 2010), occur in many NSCLC, breast, and colorectal tumors and therefore in patients selected for TKI therapy because of positive predictive EGFR mutations. Frequently, however, these tumors harbor additional mutations in the Ras/Erk pathway including KRAS, PTEN, IGFR1, or BRAF that contribute to TKI non-responsiveness (van Zandwijk et al. 2007; Engelman and Janne 2008; Laurent-Puig et al. 2009; Fojo and Parkinson 2010; Rizzo et al. 2010; Pallis et al. 2011; Park et al. 2011). Many studies and trials continue to emphasize the importance of discerning how additional alterations and pathway disruptions contribute to patient response to targeted therapy (Fojo and Parkinson 2010). These issues can complicate and delay the broad application of targeted therapies. For instance, cetuximab (an EGFR inhibitor) received accelerated FDA approval for treatment of colon cancer. However, it was later shown to result in no improvement in overall survival and/or caused serious harm in the form of high-grade toxicities for ∼40% of patients with tumors harboring KRAS mutations (Karapetis et al. 2008; Fojo and Parkinson 2010). This amounted to an estimated 100,000 patients being treated with a drug from which they likely did not benefit or by which they were harmed. Thus, pathway-level interpretation of omics findings can be critical for determining treatment modalities best suited for individuals whose tumors harbor a complex network of omics aberrations.
Knowledge of how pathways may respond to targeted inhibition is highly relevant to the application and accurate assessment of targeted therapies. Since tumor dependence on amplified receptors may be circumvented by alteration or inactivation of downstream effectors, identifying driver genes and pathways associated with a particular cancer phenotype would undoubtedly accelerate the identification of novel cancer targets aimed at underlying cancer biology—or more likely guide the “repositioning” of already existing drugs (Lussier and Chen 2011).
Exploring genomes of mammalian cancer models
Validation of cancer research findings related to almost all aspects of cancer biology in humans can be performed in animal models. The standard mammalian model is the mouse, which is relatively inexpensive to use, can be genetically modified, and is amendable to pharmacogenomics studies. Several limitations are recognized (Firestone 2010), but perhaps underappreciated considering their importance to drug screening and their function as preclinical models. Most significantly, mouse cancers almost exclusively involve genetic induction in highly inbred strains. Moreover, xenografts do not convey the genetic variation of a complex human disease like cancer nor the interaction between the tumor and its microenvironment and immune system cells. To account for some of these limitations, genetically complex and clinically relevant genetically engineered and inducible mouse models have been developed and are commonly used, as recently described (Politi et al. 2006; DuPage et al. 2009; Zhou et al. 2010; Politi and Pao 2011). Moreover, recombinant inbred strains are instrumental for identifying susceptibility genes and modifiers (Perez-Losada et al. 2011; Philip et al. 2011; Politi and Pao 2011). It is noteworthy that the emerging ability to rapidly and inexpensively characterize genomes of new organisms (e.g., whole genome and transcriptome sequencing) may propel the development of new mammalian models for studying human cancer. For example, the domesticated dog; there are some 80 million dogs in the United States, many of which receive a good level of health care, age five to eight times faster than humans, and share their owners' environmental exposure; in addition, different breeds have dramatically varied risks for diverse types of cancers (Chen et al. 2009; Boyko et al. 2010; Parker et al. 2010; Rowell et al. 2011).
Anticipating shifts in patient demographics
With the advances and widespread implementation of cancer screening and early detection programs, early-stage tumors are becoming more readily detectable, and survival rates for many cancers are improving (Aberle et al. 2011). These changes will also bring new opportunities for omics analyses of pre-malignant lesions. This shift will also put more emphasis on the genomics of early disease and should lead to the development of new omics-based strategies for cancer detection and early intervention. Ultimately, this will lead to improvements in cancer prevention and will further our understanding of the molecular mechanisms driving cancer initiation and development. For example, omics profiling of early-stage disease may reveal gene signatures capable of predicting which lesions progress to prostate cancer or whether a CT-detected lung nodule would become invasive.
Another example of shifting patient demographics involves tobacco-related cancers. With anticipated success in smoking prevention and cessation campaigns, in the coming decades, lung cancer in North America will increasingly become a disease of former and never smokers (Halpern et al. 1993; Tong et al. 1996; Peto et al. 2000). The use of cancer omics to discover novel early-detection biomarkers and to assemble gene signatures capable of stratifying risk for these patients will be immensely important to serve these emerging cohorts.
Translating omics to improved healthcare
The goals of cancer research are also advanced by high-throughput profiling efforts of non-disease cells and tissues, such as the NIH Roadmap Epigenomics Mapping Consortium, a public resource of epigenomics maps for normal tissues and stem cells (Bernstein et al. 2010), and the Personal Genome Project, which aim to create highly comprehensive and integrated human genome maps integrated with phenome data (Church 2005; Lunshof et al. 2010). The generation of 100,000 individual genome sequences with associated medical records will contribute immensely to our understanding of how our genomes contribute to normal phenotypic variation, interact with environments to contribute to disease susceptibility, and function directly in disease initiation and propagation. The knowledge and tools developed by these studies have great potential to provide similar insight into the causes of cancer or responses to targeted chemotherapies, for example, when applied to the study of individual cancer genomes.
The central tenet of omics investigation is that it allows for open (not only hypothesis driven) discovery. Integrating omics data with epidemiological data from well-defined cohorts improves our ability to associate genetic alterations with environmental exposures and specific clinical phenotypes, which has the potential to improve our current understanding of cancer biology and ultimately patient management. Now that technological developments have enabled such multidimensional studies, much of the focus will shift to study design, interpretation, and clinical applicability. Crucial to furthering the goals of this field and to the continued support and funding of this multidisciplinary work is the communication of findings to the public by the scientific community. While translational success of cancer research is judged by improved survival for cancer patients, its effective implementation will require educating the medical establishment and the public at large about the power of omics to transform medicine and improve patient outcomes.
Acknowledgments
This work was supported by funds from the Canadian Institutes for Health Research (CIHR; MOP 86731, MOP 94867, MOP-110949), Canadian Cancer Society (CCS20485), U.S. Department of Defense (CDMRP W81XWH-10-1-0634), NIH (R01 DE015965), NCI Early Detection Research Network, and the Canary Foundation. C.E.A. and L.A.R. were supported by the U.S. National Institutes of Health (HG004663). E.A.V. was supported by Frederick Banting and Charles Best Canada Graduate Scholarship from CIHR, K.L.T. by a Vanier Canada Graduate Scholarship, and R.C. by a Banting Postdoctoral Fellowship.
Footnotes
Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.124354.111.
References
- Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM, Sicks JD 2011. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 365: 395–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn MJ, Won HH, Lee J, Lee ST, Sun JM, Park YH, Ahn JS, Kwon OJ, Kim H, Shim YM, et al. 2011. The 18p11.22 locus is associated with never smoker non-small cell lung cancer susceptibility in Korean populations. Hum Genet doi: 10.1007/s00439-011-1080-z [DOI] [PubMed] [Google Scholar]
- Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL 2009. American Society of Clinical Oncology provisional clinical opinion: Testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 27: 2091–2096 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. 2010. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 28: 1045–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Santarius T, Pole JC, Butler AP, Perry J, Pleasance E, Greenman C, Menzies A, Taylor S, Edkins S, et al. 2007. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res 17: 1296–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boca SM, Kinzler KW, Velculescu VE, Vogelstein B, Parmigiani G 2010. Patient-oriented gene set analysis for cancer mutation data. Genome Biol 11: R112 doi: 10.1186/gb-2010-11-11-r112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, Quignon P, Li L, Schoenebeck JJ, Degenhardt JD, Lohmueller KE, Zhao K, Brisbin A, Parker HG, vonHoldt BM, et al. 2010. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol 8: e1000451 doi: 10.1371/journal.pbio.1000451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozic I, Antal T, Ohtsuki H, Carter H, Kim D, Chen S, Karchin R, Kinzler KW, Vogelstein B, Nowak MA 2010. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci 107: 18545–18550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. 2011. Initial genome sequencing and analysis of multiple myeloma. Nature 471: 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari R, Coe BP, Vucic EA, Lockwood WW, Lam WL 2010a. An integrative multi-dimensional genetic and epigenetic strategy to identify aberrant genes and pathways in cancer. BMC Syst Biol 4: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari R, Thu KL, Wilson IM, Lockwood WW, Lonergan KM, Coe BP, Malloff CA, Gazdar AF, Lam S, Garnis C, et al. 2010b. Integrating the multiple dimensions of genomic and epigenomic landscapes of cancer. Cancer Metastasis Rev 29: 73–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WK, Swartz JD, Rush LJ, Alvarez CE 2009. Mapping DNA structural variation in dogs. Genome Res 19: 500–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Andersen JN, Futreal PA 2011. Cancer genomics: From discovery science to personalized medicine. Nat Med 17: 297–303 [DOI] [PubMed] [Google Scholar]
- Church GM 2005. The personal genome project. Mol Syst Biol 1: 2005.0030 doi: 10.1038/msb4100040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Soudry E, Mukhopadhyay N, Shao C, Yee J, Lam S, Lam W, Zhang W, Gazdar AF, Fisher PB, et al. 2011. Mitochondrial DNA mutations in respiratory complex-I in never-smoker lung cancer patients contribute to lung cancer progression and associated with EGFR gene mutation. J Cell Physiol doi: 10.1002/jcp.22980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalos A, Nikolaidis G, Xinarianos G, Savvari P, Cassidy A, Zakopoulou R, Kotsinas A, Gorgoulis V, Field JK, Liloglou T 2009. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int J Cancer 124: 81–87 [DOI] [PubMed] [Google Scholar]
- De S, Michor F 2011. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat Struct Mol Biol 18: 950–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. 2008. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker BJ 2008. Translation of the Philadelphia chromosome into therapy for CML. Blood 112: 4808–4817 [DOI] [PubMed] [Google Scholar]
- Druker BJ, Guilhot F, O'Brien SG, Gathmann I, Kantarjian H, Gattermann N, Deininger MW, Silver RT, Goldman JM, Stone RM, et al. 2006. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med 355: 2408–2417 [DOI] [PubMed] [Google Scholar]
- Dunbar AJ, Gondek LP, O'Keefe CL, Makishima H, Rataul MS, Szpurka H, Sekeres MA, Wang XF, McDevitt MA, Maciejewski JP 2008. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res 68: 10349–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M, Dooley AL, Jacks T 2009. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc 4: 1064–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden A, Gaudet F, Waghmare A, Jaenisch R 2003. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300: 455. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Janne PA 2008. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res 14: 2895–2899 [DOI] [PubMed] [Google Scholar]
- Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L 2003. The case for early detection. Nat Rev Cancer 3: 243–252 [DOI] [PubMed] [Google Scholar]
- Firestone B 2010. The challenge of selecting the ‘right' in vivo oncology pharmacology model. Curr Opin Pharmacol 10: 391–396 [DOI] [PubMed] [Google Scholar]
- Fischer A, Greenman C, Mustonen V 2011. Germline fitness based scoring of cancer mutations. Genetics 188: 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, et al. 2010. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 363: 809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fojo T, Parkinson DR 2010. Biologically targeted cancer therapy and marginal benefits: Are we making too much of too little or are we achieving too little by giving too much? Clin Cancer Res 16: 5972–5980 [DOI] [PubMed] [Google Scholar]
- Gandhi J, Zhang J, Xie Y, Soh J, Shigematsu H, Zhang W, Yamamoto H, Peyton M, Girard L, Lockwood WW, et al. 2009. Alterations in genes of the EGFR signaling pathway and their relationship to EGFR tyrosine kinase inhibitor sensitivity in lung cancer cell lines. PLoS ONE 4: e4576 doi: 10.1371/journal.pone.0004576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber DE, Minna JD 2010. ALK inhibition for non-small cell lung cancer: From discovery to therapy in record time. Cancer Cell 18: 548–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondek LP, Dunbar AJ, Szpurka H, McDevitt MA, Maciejewski JP 2007. SNP array karyotyping allows for the detection of uniparental disomy and cryptic chromosomal abnormalities in MDS/MPD-U and MPD. PLoS ONE 2: e1225 doi: 10.1371/journal.pone.0001225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Wooster R, Futreal PA, Stratton MR, Easton DF 2006. Statistical analysis of pathogenicity of somatic mutations in cancer. Genetics 173: 2187–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber DA, Gray NS, Baselga J 2011. The evolving war on cancer. Cell 145: 19–24 [DOI] [PubMed] [Google Scholar]
- Halpern MT, Gillespie BW, Warner KE 1993. Patterns of absolute risk of lung cancer mortality in former smokers. J Natl Cancer Inst 85: 457–464 [DOI] [PubMed] [Google Scholar]
- Hampson R 1997. Selection for genome instability by DNA damage in human cells: Unstable microsatellites and their consequences for tumourigenesis. Radiat Oncol Investig 5: 111–114 [DOI] [PubMed] [Google Scholar]
- Hoffmann S 2011. Computational analysis of high throughput sequencing data. Methods Mol Biol 719: 199–217 [DOI] [PubMed] [Google Scholar]
- Hood L, Heath JR, Phelps ME, Lin B 2004. Systems biology and new technologies enable predictive and preventative medicine. Science 306: 640–643 [DOI] [PubMed] [Google Scholar]
- Hudis CA 2007. Trastuzumab—mechanism of action and use in clinical practice. N Engl J Med 357: 39–51 [DOI] [PubMed] [Google Scholar]
- Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabe RR, Bhan MK, Calvo F, Eerola I, Gerhard DS, et al. 2010. International network of cancer genome projects. Nature 464: 993–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi S, Suzuki H, Niinuma T, Shimizu H, Nojima M, Iwaki H, Nobuoka T, Nishida T, Miyazaki Y, Takamaru H, et al. 2010. A novel correlation between LINE-1 hypomethylation and the malignancy of gastrointestinal stromal tumors. Clin Cancer Res 16: 5114–5123 [DOI] [PubMed] [Google Scholar]
- Jones SJ, Laskin J, Li YY, Griffith OL, An J, Bilenky M, Butterfield YS, Cezard T, Chuah E, Corbett R, et al. 2010. Evolution of an adenocarcinoma in response to selection by targeted kinase inhibitors. Genome Biol 11: R82 doi: 10.1186/gb-2010-11-8-r82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R 1997. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res 57: 808–811 [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M 2010. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 38: D355–D360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, et al. 2008. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 359: 1757–1765 [DOI] [PubMed] [Google Scholar]
- Keedy VL, Temin S, Somerfield MR, Beasley MB, Johnson DH, McShane LM, Milton DT, Strawn JR, Wakelee HA, Giaccone G 2011. American Society of Clinical Oncology Provisional Clinical Opinion: Epidermal growth factor receptor (EGFR) mutation testing for patients with advanced non-small-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol 29: 2121–2127 [DOI] [PubMed] [Google Scholar]
- Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, et al. 2010. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363: 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, Rougier P, Lievre A, Landi B, Boige V, et al. 2009. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol 27: 5924–5930 [DOI] [PubMed] [Google Scholar]
- Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, Yue P, Zhang Y, Pant KP, Bhatt D, et al. 2010. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature 465: 473–477 [DOI] [PubMed] [Google Scholar]
- Link DC, Schuettpelz LG, Shen D, Wang J, Walter MJ, Kulkarni S, Payton JE, Ivanovich J, Goodfellow PJ, Le Beau M, et al. 2011. Identification of a novel TP53 cancer susceptibility mutation through whole-genome sequencing of a patient with therapy-related AML. JAMA 305: 1568–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liotta LA, Petricoin E 2011. Cancer biomarkers: Closer to delivering on their promise. Cancer Cell 20: 279–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunshof JE, Bobe J, Aach J, Angrist M, Thakuria JV, Vorhaus DB, Hoehe MR, Church GM 2010. Personal genomes in progress: From the human genome project to the personal genome project. Dialogues Clin Neurosci 12: 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier YA, Chen JL 2011. The emergence of genome-based drug repositioning. Sci Transl Med 3: 96ps35 doi: 10.1126/scitranslmed.3001512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. 2004. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139 [DOI] [PubMed] [Google Scholar]
- Modrek B, Ge L, Pandita A, Lin E, Mohan S, Yue P, Guerrero S, Lin WM, Pham T, Modrusan Z, et al. 2009. Oncogenic activating mutations are associated with local copy gain. Mol Cancer Res 7: 1244–1252 [DOI] [PubMed] [Google Scholar]
- Mort M, Evani US, Krishnan VG, Kamati KK, Baenziger PH, Bagchi A, Peters BJ, Sathyesh R, Li B, Sun Y, et al. 2010. In silico functional profiling of human disease-associated and polymorphic amino acid substitutions. Hum Mutat 31: 335–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC, Hungerford DA 1961. Chromosome studies in human leukemia. II. Chronic granulocytic leukemia. J Natl Cancer Inst 27: 1013–1035 [PubMed] [Google Scholar]
- Oakman C, Santarpia L, Di Leo A 2010. Breast cancer assessment tools and optimizing adjuvant therapy. Nat Rev Clin Oncol 7: 725–732 [DOI] [PubMed] [Google Scholar]
- Offit K 2011. Personalized medicine: New genomics, old lessons. Hum Genet 130: 3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. 2004. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 304: 1497–1500 [DOI] [PubMed] [Google Scholar]
- Pallis A, Briasoulis E, Linardou H, Papadimitriou C, Bafaloukos D, Kosmidis P, Murray S 2011. Mechanisms of resistance to epidermal growth factor receptor tyrosine kinase inhibitors in patients with advanced non-small-cell lung cancer: clinical and molecular considerations. Curr Med Chem 18: 1613–1628 [DOI] [PubMed] [Google Scholar]
- Pao W, Chmielecki J 2010. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer 10: 760–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaetis GS, Syrigos KN 2010. Targeted therapy for gastrointestinal stromal tumors: Current status and future perspectives. Cancer Metastasis Rev 29: 151–170 [DOI] [PubMed] [Google Scholar]
- Park JH, Han SW, Oh DY, Im SA, Jeong SY, Park KJ, Kim TY, Bang YJ, Park JG 2011. Analysis of KRAS, BRAF, PTEN, IGF1R, EGFR intron 1 CA status in both primary tumors and paired metastases in determining benefit from cetuximab therapy in colon cancer. Cancer Chemother Pharmacol 68: 1045–1055 [DOI] [PubMed] [Google Scholar]
- Parker HG, Shearin AL, Ostrander EA 2010. Man's best friend becomes biology's best in show: Genome analyses in the domestic dog. Annu Rev Genet 44: 309–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasche B, Absher D 2011. Whole-genome sequencing: A step closer to personalized medicine. JAMA 305: 1596–1597 [DOI] [PubMed] [Google Scholar]
- Perez-Losada J, Castellanos-Martin A, Mao JH 2011. Cancer evolution and individual susceptibility. Integr Biol (Camb) 3: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peto R, Darby S, Deo H, Silcocks P, Whitley E, Doll R 2000. Smoking, smoking cessation, and lung cancer in the UK since 1950: Combination of national statistics with two case-control studies. BMJ 321: 323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip VM, Sokoloff G, Ackert-Bicknell CL, Striz M, Branstetter L, Beckmann MA, Spence JS, Jackson BL, Galloway LD, Barker P, et al. 2011. Genetic analysis in the Collaborative Cross breeding population. Genome Res 21: 1223–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. 2010. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politi K, Pao W 2011. How genetically engineered mouse tumor models provide insights into human cancers. J Clin Oncol 29: 2273–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politi K, Zakowski MF, Fan PD, Schonfeld EA, Pao W, Varmus HE 2006. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev 20: 1496–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poste G 2011. Bring on the biomarkers. Nature 469: 156–157 [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, Rosen N 2011. Mutant BRAF melanomas—dependence and resistance. Cancer Cell 19: 11–15 [DOI] [PubMed] [Google Scholar]
- Prat A, Perou CM 2011. Deconstructing the molecular portraits of breast cancer. Mol Oncol 5: 5–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, Baenziger PH, Kann MG, Mort ME, Hahn MW, Mooney SD 2008. Gain and loss of phosphorylation sites in human cancer. Bioinformatics 24: i241–i247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizwana R, Hahn PJ 1999. CpG methylation reduces genomic instability. J Cell Sci 112: 4513–4519 [DOI] [PubMed] [Google Scholar]
- Rizzo S, Bronte G, Fanale D, Corsini L, Silvestris N, Santini D, Gulotta G, Bazan V, Gebbia N, Fulfaro F, et al. 2010. Prognostic vs predictive molecular biomarkers in colorectal cancer: Is KRAS and BRAF wild type status required for anti-EGFR therapy? Cancer Treat Rev (Suppl 3) 36: S56–S61 [DOI] [PubMed] [Google Scholar]
- Robison K 2010. Application of second-generation sequencing to cancer genomics. Brief Bioinform 11: 524–534 [DOI] [PubMed] [Google Scholar]
- Rowell JL, McCarthy DO, Alvarez CE 2011. Dog models of naturally occurring cancer. Trends Mol Med 17: 380–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyers CL 2008. The cancer biomarker problem. Nature 452: 548–552 [DOI] [PubMed] [Google Scholar]
- Segditsas S, Rowan AJ, Howarth K, Jones A, Leedham S, Wright NA, Gorman P, Chambers W, Domingo E, Roylance RR, et al. 2009. APC and the three-hit hypothesis. Oncogene 28: 146–155 [DOI] [PubMed] [Google Scholar]
- Sengupta S, Chakrabarti S, Roy A, Panda CK, Roychoudhury S 2007. Inactivation of human mutL homolog 1 and mutS homolog 2 genes in head and neck squamous cell carcinoma tumors and leukoplakia samples by promoter hypermethylation and its relation with microsatellite instability phenotype. Cancer 109: 703–712 [DOI] [PubMed] [Google Scholar]
- Shima K, Nosho K, Baba Y, Cantor M, Meyerhardt JA, Giovannucci EL, Fuchs CS, Ogino S 2011. Prognostic significance of CDKN2A (p16) promoter methylation and loss of expression in 902 colorectal cancers: Cohort study and literature review. Int J Cancer 128: 1080–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. 2007. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature 448: 561–566 [DOI] [PubMed] [Google Scholar]
- Soh J, Okumura N, Lockwood WW, Yamamoto H, Shigematsu H, Zhang W, Chari R, Shames DS, Tang X, MacAulay C, et al. 2009. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS ONE 4: e7464 doi: 10.1371/journal.pone.0007464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. 2009. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker T, Catenacci DV, Seiwert TY 2011. Molecular profiling of cancer—the future of personalized cancer medicine: A primer on cancer biology and the tools necessary to bring molecular testing to the clinic. Semin Oncol 38: 173–185 [DOI] [PubMed] [Google Scholar]
- Swanton C, Burrell RA, Futreal PA 2011. Breast cancer genome heterogeneity: A challenge to personalised medicine? Breast Cancer Res 13: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawfik HM, El-Maqsoud NM, Hak BH, El-Sherbiny YM 2011. Head and neck squamous cell carcinoma: Mismatch repair immunohistochemistry and promoter hypermethylation of hMLH1 gene. Am J Otolaryngol 32: 528–536 [DOI] [PubMed] [Google Scholar]
- Tong L, Spitz MR, Fueger JJ, Amos CA 1996. Lung carcinoma in former smokers. Cancer 78: 1004–1010 [DOI] [PubMed] [Google Scholar]
- van Zandwijk N, Mathy A, Boerrigter L, Ruijter H, Tielen I, de Jong D, Baas P, Burgers S, Nederlof P 2007. EGFR and KRAS mutations as criteria for treatment with tyrosine kinase inhibitors: Retro- and prospective observations in non-small-cell lung cancer. Ann Oncol 18: 99–103 [DOI] [PubMed] [Google Scholar]
- Vasavi M, Kiran V, Ravishankar B, Prabhakar B, Ahuja YR, Hasan Q 2010. Microsatellite instability analysis and its correlation with hMLH1 repair gene hypermethylation status in esophageal pathologies including cancers. Cancer Biomark 7: 1–10 [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. 2010. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Cusick ME, Barabasi AL 2011. Interactome networks and human disease. Cell 144: 986–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Lee I, Carlson G, Hood L, Galas D 2010. Systems biology and the discovery of diagnostic biomarkers. Dis Markers 28: 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzel JN, Blazer KR, Macdonald DJ, Culver JO, Offit K 2011. Genetics, genomics, and cancer risk assessment: State of the art and future directions in the era of personalized medicine. CA Cancer J Clin doi: 10.3322/caac.20128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JS, Westervelt P, Ding L, Larson DE, Klco JM, Kulkarni S, Wallis J, Chen K, Payton JE, Fulton RS, et al. 2011. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA 305: 1577–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto G, Nannya Y, Kato M, Sanada M, Levine RL, Kawamata N, Hangaishi A, Kurokawa M, Chiba S, Gilliland DG, et al. 2007. Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of affymetrix single-nucleotide-polymorphism genotyping microarrays. Am J Hum Genet 81: 114–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn A, Simon R 2011. Identifying cancer driver genes in tumor genome sequencing studies. Bioinformatics 27: 175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Rideout WM III, Zi T, Bressel A, Reddypalli S, Rancourt R, Woo JK, Horner JW, Chin L, Chiu MI, et al. 2010. Chimeric mouse tumor models reveal differences in pathway activation between ERBB family- and KRAS-dependent lung adenocarcinomas. Nat Biotechnol 28: 71–78 [DOI] [PubMed] [Google Scholar]