Abstract
Whole-exome sequencing (Exome-seq) has been successfully applied in several recent studies. We here sequenced the exomes of 15 pancreatic tumor cell lines and their matched normal samples. We captured 162,073 exons of 16,954 genes and sequenced the targeted regions to a mean coverage of 56-fold. This study identified a total of 1517 somatic mutations and validated 934 mutations by transcriptome sequencing. We detected recurrent mutations in 56 genes. Among them, 41 have not been described. The mutation rates varied widely among cell lines. The diversity of the mutation rates was significantly correlated with the distinct MLH1 copy-number status. Exome-seq revealed intensive genomic instability in a cell line with MLH1 homozygous deletion, indicated by a dramatically elevated rate of somatic substitutions, small insertions/deletions (indels), as well as indels in microsatellites. Notably, we found that MLH1 expression was decreased by nearly half in cell lines with an allelic loss of MLH1. While these cell lines were negative in conventional microsatellite instability assay, they showed a 10.5-fold increase in the rate of somatic indels, e.g., truncating indels in TP53 and TGFBR2, indicating MLH1 haploinsufficiency in the correction of DNA indel errors. We further analyzed the exomes of 15 renal cell carcinomas and confirmed MLH1 haploinsufficiency. We observed a much higher rate of indel mutations in the affected cases and identified recurrent truncating indels in several cancer genes such as VHL, PBRM1, and JARID1C. Together, our data suggest that MLH1 hemizygous deletion, through increasing the rate of indel mutations, could drive the development and progression of sporadic cancers.
The current understanding of cancer is that it arises as a result of the accumulation of genetic and epigenetic mutations that confer a selective advantage to the cells in which they occur (Vogelstein and Kinzler 2004; Greenman et al. 2007; Stratton et al. 2009). Over the past quarter of a century, many efforts have been made to learn about the causative mutations that drive various types of cancer, including pancreatic cancer, one of the most lethal forms of human cancer. By using the Sanger sequencing method, i.e., PCR amplification followed by plasmid subcloning and DNA sequencing, previous studies have identified thousands of genetic alterations in the cancer genome and provided important insights into the pancreatic cancer biology (Jones et al. 2008; Maitra and Hruban 2008). However, because Sanger sequencing is performed on single amplicons, its throughput is limited, and large-scale sequencing projects are expensive and laborious (Schuster 2008; Metzker 2010). Moreover, it has been reported that it has a limited sensitivity to recognize the mutant DNA allele if it is present in a minor population of cancer cells (Nakahori et al. 1995; Thomas et al. 2006; Qiu et al. 2008). In addition, the bacterial cloning workflows tend to be complex and time-consuming, and bias can be introduced into this step (Thomas et al. 2006).
The advent of next-generation sequencing (NGS) technologies has brought a high level of efficiency to genome sequencing (Schuster 2008; Metzker 2010). The enriched DNA is sequenced directly, avoiding the cloning step (Ng et al. 2009). While whole-genome sequencing is the most complete, it remains sufficiently expensive that cost-effective alternatives are important. Target-enrichment strategies allow the selective capture of the genomic regions of interest. Whole-exome sequencing (Exome-seq) through integrating two systems has enabled us to concentrate our sequencing efforts on the protein-coding exons in the human genome. This approach is substantially cost- and labor-efficient (Schuster 2008; Metzker 2010; Biesecker et al. 2011). Moreover, by taking advantage of deep coverage of target regions, it shows an excellent sensitivity for the detection of variants with a minor allele frequency down to 2% (Li et al. 2010). Recent studies have successfully applied Exome-seq to identify genetic changes involved in Mendelian diseases (Choi et al. 2009; Ng et al. 2010). In addition to Exome-seq, full-length transcriptome sequencing (mRNA-seq) offers a fast and inexpensive alternative. It is an easier method to identify coding sequences and capture variants in genes that are expressed, as well as to generate additional information, such as gene expression level and splicing patterns (Sugarbaker et al. 2008; Cirulli et al. 2010).
Genomic instability is a characteristic feature of almost all human cancers (Lengauer et al. 1998; Negrini et al. 2010). Its molecular basis is well understood in hereditary cancers, in which it has been linked to mutations in DNA mismatch repair (MMR) genes. One of the best-documented examples is the hereditary nonpolyposis colon cancer (HNPCC). In general, MMR defects are the result of a germline mutation in one of the MMR genes followed by a hit on the second allele of that gene, or methylation of the promoter of a MMR gene, usually MLH1, resulting in the loss of protein function (Fishel et al. 1993; Hemminki et al. 1994). In contrast, the molecular basis of genomic instability in sporadic cancers remains unclear (Negrini et al. 2010).
In the past few years, by use of Sanger sequencing, several consortia have scanned the coding sequences of 18,191–20,661 genes in carcinomas of the colon, breast, and pancreas and in glioblastomas (Sjoblom et al. 2006; Wood et al. 2007; Jones et al. 2008; Parsons et al. 2008). These genome-wide studies reported that mutations targeting caretaker genes (DNA repair genes and mitotic checkpoint genes) were infrequent. To date, no statistical correlation has been described in sporadic cancers between the allelic loss of a caretaker gene and the increased rate of genomic instability. It has been thought that a single copy of the wild-type allele of a caretaker gene is sufficient to perform its normal function, and both alleles of the gene would have to be inactivated before the genome becomes unstable (Bodmer et al. 2008; Negrini et al. 2010). Since the occurrence of two independent somatic mutations at both alleles of the same gene is likely to represent a very rare event (Bodmer et al. 2008), these studies argued that mutations in caretaker genes probably do not account for the presence of genomic instability in many sporadic cancers (Negrini et al. 2010).
We here performed Exome-seq on 15 pancreatic ductal adenocarcinoma (PDAC)–derived cell lines. This study identified 1517 somatic mutations and validated 934 of them by mRNA-seq. We notably found a significant correlation between MLH1 allelic loss and the increased rate of somatic indel mutations, and we further confirmed this finding in primary renal cell carcinomas (RCCs). In the affected cases, we detected recurrent truncating indels that inactivate tumor suppressor genes, such as TP53, TGFBR2, and VHL. We also observed a higher prevalence of indels in the coding microsatellite sequences. Our data, therefore, indicate that deletion of one copy of the MLH1 gene results in haploinsufficiency in the correction of DNA indel errors and could be a driving force in pancreatic and renal carcinogenesis.
Results
The performance of Exome-seq
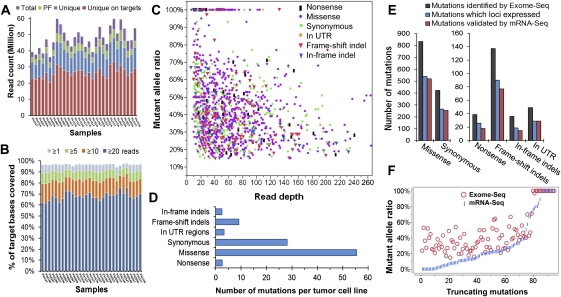
We sequenced the exomes of 15 PDAC-derived cell lines and their matched normal samples (Table 1). On average, 6.6 Gb of high-quality sequence data (about 44.2 million paired 75-base reads) were generated per sample. More than 88% of the sequence reads were uniquely aligned to the human reference genome with the expected insert size and correct orientations, and 68.4% of them fell within the targeted regions (Fig. 1A; Supplemental Fig. S1). The average fold-coverage of each exome was 56× (Supplemental Fig. S2). On average per exome, 96.9% of targeted bases were covered by at least one read, and 83.4% of targeted bases were covered by at least 10 reads (Fig. 1B; Supplemental Fig. S3).
Table 1.
Characteristics of pancreatic tumor cell lines
Figure 1.
The performance of Exome-seq and a summary of somatic mutations. (A) The summary of Exome-seq data. For each sample, the number of raw sequence reads (total), passing filter reads (PF), unique reads that mapped in consistent read pairs (unique), and the unique reads that fall within the targeted regions (unique on target) are shown. (B) The sequence coverage of targeted bases. The fraction of the targeted bases that were covered by unique reads at the sequence depth of 1×, 5×, 10×, and 20× is shown. (C) An overview of the somatic mutations identified by Exome-seq. Different markers and colors were used to show different mutation types. (D) The average number of somatic mutations identified per tumor cell line. (E) The performance of mRNA-seq in verification of somatic mutations identified by Exome-seq. The mutations that loci expressed represent those mutations that loci covered by five or more cDNA sequence reads. (F) Validation of the truncating mutations that introduced premature termination codons. The abundance of the mutant alleles in genomic DNA was compared with that of their corresponding cDNA.
An overview of somatic mutations
By using Exome-seq, we identified a total of 1517 somatic mutations, including 39 nonsense, 833 missense, 423 synonymous substitutions, and 49 substitutions in untranslated regions (UTRs), 137 frame-shift indels and 36 in-frame indels (Fig. 1C). The complete list of 1517 somatic mutations is shown in Supplemental Table S1. On average, each cell line contains 101 somatic mutations, 89% of which are base substitutions (Fig. 1D). The frequencies of mutant alleles ranged from 15%–100%, with a median of 41%. The depth of coverage at the mutation loci ranged from 10× to 637×, with a median of 42× (Fig. 1C; Supplemental Table S1). The lengths of somatic small indels varied from 1–29 bp. Seventy-eight percent of the indels were 1–3 bp in length (Supplemental Fig. S4). By using genome-wide SNP array, we identified more than 50 focal homozygous deletions (Supplemental Table S2). The CDKN2A locus at 9p21.3 and the SMAD4 locus at 18q21.2 were frequently deleted in the tumor cell lines analyzed (Supplemental Fig. S5). The somatic mutations mainly clustered in nine signaling pathways, as shown in Supplemental Figure S6A. The background mutation rate estimated for targeted exonic regions was 2.7 mutations per megabase of DNA sequences.
Validation of somatic mutations using mRNA-seq
In total, 61.6% (934 out of 1517) of the mutations identified by Exome-seq were validated by mRNA-seq. If we focus on the expressed genes, 94.3% (914 out of 969) of the mutations at those loci covered by five or more cDNA sequence reads were successfully validated by mRNA-seq (Fig. 1E). Additionally, 20 mutations at the loci with a lower coverage (less than five reads, but three reads or more) were also confirmed by mRNA-seq. The percentages of mutations validated by mRNA-seq varied across mutation types. Generally, the validation ratio of truncating mutations is lower than that of nontruncating mutations.
For truncating mutations (Fig. 1F), the abundance of the mutant allele in the cDNA appears to be relatively lower than that of their corresponding genomic DNA (gDNA). Despite the lower abundance, mRNA-seq was still able to confirm 81 of those 94 (86.2%) truncating mutations at loci covered by five or more cDNA sequence reads. The remaining 13 truncating mutations were all heterozygous. Their loci were covered moderately well, but no mutant alleles were observed in the cDNA sequences. We performed Sanger sequencing to confirm if they resulted from the false-positive events of Exome-seq. As shown in Supplemental Figure S7, 12 of the 13 truncating mutations were successfully validated by Sanger sequencing. The mutant alleles were only detected in the gDNA of the tumor cell lines rather than in their cDNA, suggesting the transcripts carrying the mutant alleles were probably degraded through the nonsense-mediated mRNA decay (NMD) pathway (Holbrook et al. 2004). One mutation was found to be false-positive, possibly caused by mapping errors.
The recurrently mutated genes
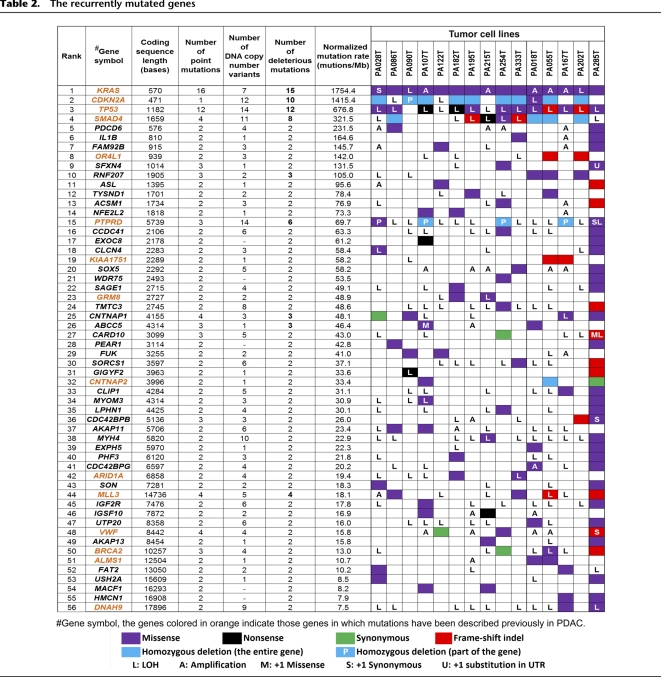
In this study, 1359 genes were identified with somatic mutations. Among them, 56 genes were recurrently mutated in two or more cell lines (Table 2). The mutation rate of these genes was much higher than the background level. The most frequently mutated gene was KRAS, followed by CDKN2A, TP53, and SMAD4. Mutation of these four genes and 11 other genes has been reported either in the COSMIC database (http://www.sanger.ac.uk/genetics/CGP/cosmic/) or in a previous study (Jones et al. 2008), as shown in Supplemental Figure S8, while mutation of the remaining 41 genes, to our knowledge, has not been described in PDAC. Totally, 150 point mutations were identified in the 56 recurrently mutated genes. Among them, 109 mutations in 40 genes were confirmed by mRNA-seq (Supplemental Table S1). For the remaining 41 mutations that were not confirmed by mRNA-seq, seven loci were poorly expressed (covered by two or fewer cDNA sequence reads) and 34 loci were not expressed at all.
Table 2.
The recurrently mutated genes
The widely varied mutation rates
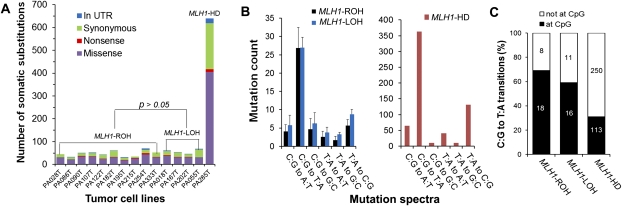
Exome-seq revealed that the mutation rates varied significantly among cell lines (Figs. 2C, 3A). The number of somatic substitutions identified from each cell line ranged from 31–640, and the number of somatic indels varied from zero to 100. Accordingly, we classified the cell lines into three subgroups. Cell lines in group 1 (n = 10) showed a modest level of somatic mutations, while cell lines in group 2 (n = 4) showed a significantly elevated rate of small indels (P = 0.005) (Fig. 2C); a cell line in group 3 (n = 1) showed dramatically increased rates of both indels and substitutions (Figs. 2C, 3A). In the group-3 cell line, we observed a much higher prevalence of mutations involved in all nine core signaling pathways (P = 0.0007) (Supplemental Fig. S6B). In group-2 cell lines, the normalized mutation rate was slightly but significantly increased (P = 0.037) in seven of the nine pathways.
Figure 2.
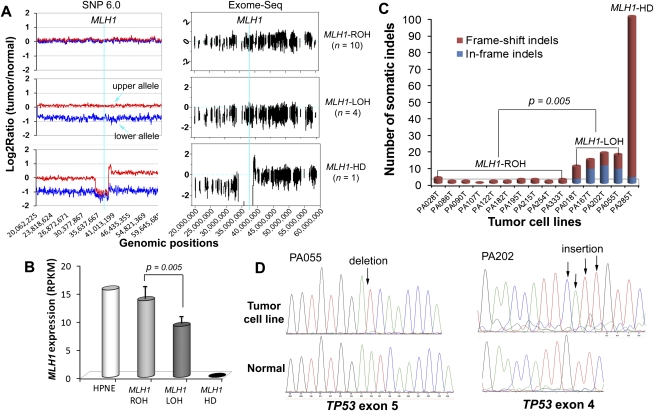
Allelic loss of MLH1 and the increased rate of somatic indel mutations. (A) The distinct DNA copy-number status of MLH1. The left and right panels show the DNA copy-number status inferred from SNP array and Exome-seq data, respectively. The line in light blue indicates the approximate genomic location of MLH1. For graphs in the left panel, the y-axis indicates the adjusted log2 ratios of signal intensities between the tumor cell line and its matched normal sample for perfect match probes. The red line represents the allele with a higher copy number, and the blue line represents the allele with a lower copy number. The log2 ratio of −1 and 0 theoretically corresponds to 0 and 1 copy, respectively. For graphs in the right panel, the y-axis indicates the log2 ratios of the sequence coverage between the tumor cell line and its matched normal sample for targeted exonic regions. (B) The differential expression of MLH1. The gene expression level was examined by mRNA-seq. (RPKM) Reads per kilobase per million mapped reads. (Bars) Mean ± SD. (C) The somatic indels. The number of somatic small indels identified in the targeted exonic regions is shown for each tumor cell line. (D) Validation of the truncating indels identified in TP53 in two MLH1-LOH cell lines. (Left) 1-bp deletion; (right) 4-bp insertion. The positions of indels are indicated by arrows in the sequence electropherograms.
Allelic loss of MLH1 and the increased mutation rate
To find out the genetic factors that accounted for the increased mutation rate in the group-2 and group-3 cell lines, we first screened the MMR genes for somatic alterations. We found that the gene MLH1 was differentially expressed among the subgroups, and the expression levels appeared to be reversely correlated with the mutation rates. As shown in Figure 2B, the expression of MLH1 decreased by nearly half in group-2 cell lines (P = 0.005) and was almost lost in the group-3 cell line. We did not observe any significant differences in the expression of other DNA MMR genes among the subgroups (Supplemental Fig. S9), nor did we detect somatic point mutations of other MMR genes in any of the cell lines. We then quantitatively measured the methylation status of the MLH1 promoter using MassARRAY, but none of the cell lines showed promoter hypermethylation of this gene (Supplemental Fig. S10). We further examined DNA copy-number changes of MLH1 and found a clue to its differential expression. As shown in the left panel of Figure 2A, cell lines in group 1 retained both alleles of MLH1 (MLH1-ROH [retention of heterozygosity]), while cell lines in group 2 lost one of the two alleles of this gene (MLH1-LOH [loss of heterozygosity]); the cell line in group 3 lost both alleles (MLH1-HD [homozygous deletion]). The distinct DNA copy-number status of MLH1 was also well demonstrated by the read-depth–based Exome-seq data (Fig. 2A, right panel).
Characterization of somatic indels in the MLH1-LOH and MLH1-HD cell lines
We identified an average of 1.4 ± 0.8 indels per MLH1-ROH cell line, 14.8 ± 3.5 indels per MLH1-LOH cell line, and 100 indels in the MLH1-HD cell line. The mutation rate of the somatic indels was 10.5- and 72.1-fold higher in MLH1-LOH and MLH1-HD cell lines, respectively, compared with that of the MLH1-ROH cell lines (P = 0.005) (Fig. 2C). Among the total of 173 somatic indels, 94 were detected in the coding microsatellites (Supplemental Table S1). Prevalence of the indels in the microsatellites was increased sixfold and 154-fold, respectively, in the MLH1-LOH and MLH1-HD cell lines. Nearly half of the indels that were detected in MLH1-LOH cell lines and the majority of indels that were detected in the MLH1-HD cell line were frame-shift mutations. Some of the frame-shift indels were present in cancer-related genes such as TP53, BRCA2, TGFBR2, and MLL3 and were predicted to be protein truncating. We identified a 1-bp insertion in the poly(A)10 tract of TGFBR2 in one of the MLH1-LOH cell lines and validated it by mRNA-seq. We detected two truncating indels in TP53 in two other MLH1-LOH cell lines and validated them by both Sanger sequencing and mRNA-seq (Fig. 2D; Supplemental Table S1). Both indels were accompanied by LOH and introduced premature termination codons (PTCs), resulting in a dramatic reduction of TP53 expression (Supplemental Fig. S11).
The mutation spectra
The pattern of mutation spectra was quite similar among the subgroups. As shown in Figure 3B, the predominant type of base substitution was the C:G to T:A transition, followed by the T:A to C:G transition. Many cancer genes such as KRAS, TP53, SMAD4, and APC were mutated by a C:G to T:A transition. In the MLH1-HD cell line, the mutation rate of the C:G to T:A transitions was markedly increased, especially at non-CpG sites (Fig. 3C). The frequency of other classes of base substitution was also dramatically higher except for the C:G to G:C and T:A to G:C transversions.
Figure 3.
Characterization of the somatic base substitutions. (A) The number of somatic base substitutions. The MLH1-HD cell line showed a dramatically elevated mutation rate of somatic substitutions. (B) The pattern of mutation spectra. (C) The distribution of the C:G to T:A transitions at and not at the CpG dinucleotides. For B, the data are shown as mean ± SD. As for C, the mean values are marked on corresponding columns.
Evaluation of genomic instability using Exome-seq
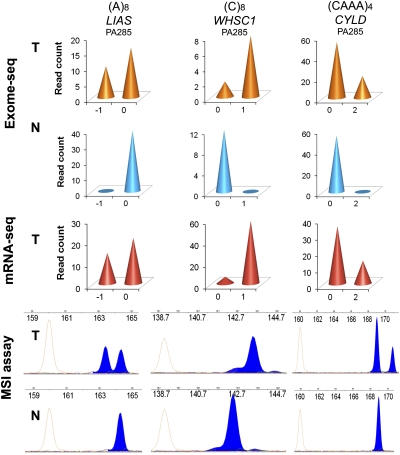
Based on the Exome-seq data, we determined the microsatellite instability (MSI) status of MLH1-ROH, MLH1-LOH, and MLH1-HD cell lines as “stable,” “intermediately unstable,” and “highly unstable,” respectively (Supplemental Table S1). We then performed the conventional MSI assay for the same sample set (Supplemental Fig. S12). The assay revealed that all seven markers were stable in the MLH1-ROH cell lines, and two of the markers, D17S250 and D2S123, were unstable in the MLH1-HD cell line. However, none of the markers showed instability in any of the MLH1-LOH cell lines. Using the conventional MSI assay, MLH1-LOH cell lines were indistinguishable from MLH1-ROH cell lines. To further evaluate the performance of Exome-seq, we selected three representative coding microsatellites, within which somatic indels have been identified by Exome-seq and validated by mRNA-seq. We designed fluorescence-labeled primers and performed the MSI assay. The conventional assay confirmed instability for all three microsatellites (Fig. 4).
Figure 4.
MSI analysis using Exome-seq. The data for three representative microsatellites are shown. (Top) Read-depth based Exome-seq data; (middle) mRNA-seq data; (bottom) electropherograms of the conventional MSI assay. For the top and middle panels, the x-axis indicates the lengths of indels. The negative value indicates base deletion, and the positive value indicates base insertion, while 0 indicates no indel. The numbers marked at the y-axis indicate the number of sequence reads that carry the mutant allele or the wild-type allele. (Bottom) x-axis is the size in bases; y-axis is the fluorescence intensity. The red peaks are internal size standards.
Discussion
In this study, we analyzed 15 PDAC-derived cell lines and their matching normal tissues using Exome-seq. We detected more than 1500 point mutations and showed that 1359 genes were somatically altered in at least one of the cell lines. KRAS, TP53, CDKN2A, and SMAD4, known as the “master” genes for PDAC, were the top four most frequently mutated genes identified in this study. These results are consistent with an early study performed by Jones and colleagues (2008) using the Sanger sequencing method, indicating a good performance of Exome-seq, as well as our mutation detection pipeline.
Mutation of the four key players, although being of paramount importance, may not be sufficient to drive the development and progression of PDAC, since variability can occur among tumors arising in the same organ and among cell populations within the same tumor. Recent studies have reported the intertumoral heterogeneity among PDACs and the intratumoral heterogeneity in a hepatocellular carcinoma (Kim et al. 2011; Totoki et al. 2011). The number of mutated genes that drive development of cancer was found to be far greater than previously thought (Greenman et al. 2007). By using Exome-seq, we identified additional 52 genes that recurrently mutated in PDAC. Among them, the mutation of 41 genes has not been described in this cancer type. More than half of these genes have been suggested to play a role in carcinogenesis. For example, a recent study showed NFE2L2 is frequently mutated in lung cancers (Shibata et al. 2010). The overexpression of SOX5 is associated with prostate tumor progression and early development of distant metastasis (Ma et al. 2009). EXOC8 has been shown to foster oncogenic Ras-mediated tumorigenesis (Issaq et al. 2010). Mutation screening of these genes in a large sample size would help us gain a further understanding of their biological contribution to PDAC.
The application of NGS technologies to cancer genomics has dramatically increased the efficiency of mutation discovery. Since a variety of factors, such as sequencing platforms, data mapping, and variant calling algorithms can affect the final output of identified mutation candidates, validation of the numerous proposed mutations has consequently become a common issue to be considered. We here evaluated the performance of mRNA-seq in verification of mutations identified in coding regions. If we simply consider all somatic mutations identified by Exome-seq, 61.6% of them were validated by mRNA-seq. If we focus, however, on those mutations in expressed genes, 94.3% of them can be successfully confirmed by mRNA-seq. For truncating mutations, despite a lower abundance of the mutant allele in cDNA, mRNA-seq was still able to confirm 86.2% of the mutations. This suggests that although it may miss mutations in poorly expressed regions, mRNA-seq may be a workable alternative to Sanger sequencing for the validation of mutations identified in expressed genes. In addition to learn about gene expression and splicing variants, groups who run NGS on both gDNA and cDNA for the same sample set may get an extra benefit from such an application.
Allelic loss at the short arm of chromosome 3 is one of the most common genetic alterations observed in human cancers. It has been reported in over 30% of PDAC and nearly 90% of RCC cases (Yamano et al. 2000; Harada et al. 2008; Toma et al. 2008). Many potential cancer genes have been identified on chromosome 3p. The DNA MMR gene MLH1 is located at chromosome 3p22.2. In mammals, the MLH1 protein is an essential component of the MMR complex. MLH1 protein binds to either PMS1 or PMS2, and both heterodimers bind either to the MSH2/MSH6 heterodimers to correct mismatches or to the MSH2/MSH3 heterodimers to correct indel errors (Jiricny 1998; Kolodner and Marsischky 1999; Raschle et al. 1999). Among the MMR proteins, the loss of MLH1 is by far the most common cause of MSI. To date, a variety of genetic and epigenetic alterations in MLH1 has been discovered in many different types of cancers (Bronner et al. 1994; Cunningham et al. 1998; Kuismanen et al. 2000; Suter et al. 2004; Arnold et al. 2009). In pancreatic cancers, the mutation of MLH1 and MSI has been reported in a histologically distinct subset of poorly differentiated adenocarcinomas, called medullary carcinomas, which usually have a wild-type KRAS. The sporadic PDAC, however, seldom, if ever, has MSI (Wilentz et al. 2000). To our knowledge, the profile of MSI has yet to be fully demonstrated in a genome-wide manner in pancreatic cancers.
Homozygous deletion of MLH1 is a rare case and has not been documented previously. In one of the cell lines analyzed in this study, we incidentally detected a focal homozygous deletion spanning the entire MLH1 locus. Exome-seq revealed intensive genomic instability in this cell line, indicated by a dramatically elevated mutation rate of somatic substitutions, small indels, as well as the indels presented in coding microsatellites. The number of C:G to T:A transitions was markedly increased, especially at non-CpG sites, suggesting an impaired recognition/repair of G:T mismatches (Marra and Schar 1999; Kumar et al. 2009). The mutation spectrum of the cell line was quite similar to that of other types of MMR-deficient tumors previously reported (Greenman et al. 2007).
Although allelic loss of MLH1 has been reported in over 30% of PDACs (Yamano et al. 2000; Harada et al. 2008), no statistical correlation has been described between MLH1 allelic loss and an increased mutation rate. It was previously thought that mutations in MLH1 and other DNA MMR genes are recessive; i.e., a single copy of the wild-type MLH1 allele is sufficient to perform its normal function (Bodmer et al. 2008; Negrini et al. 2010). In this study, we notably found that MLH1 expression was decreased by nearly half in cell lines with an allelic loss of MLH1. While these cell lines were negative in a conventional MSI assay, they showed a 10.5-fold increase in the rate of somatic indels. We also observed a higher prevalence of indels in the coding microsatellites. Moreover, we identified truncating indels that inactivate tumor suppressor genes, such as TP53 and TGFBR2. These results indicate that deletion of one copy of MLH1 gene results in haploinsufficiency in the correction of DNA indel errors.
An earlier study performed in vitro could support our argument that hemizygous deletion of MLH1 may lead to an impaired DNA repair and genomic instability. Edelmann and colleagues (1996) generated mice with a null mutation of the MLH1 gene and measured the MMR activity in vitro using the cell-free extracts from the mouse embryo-derived fibroblast (MEF). They found that the embedded errors in the reporter gene were repaired 2.3-fold less efficiently in MEF extracts of mlh1+/− mice compared with that of mlh1+/+ mice.
To further address the significance of MLH1 hemizygous deletion in in vivo tumors, we examined the primary RCC samples, which usually exhibit LOH on chromosome 3p. All patients provided informed consent for the research use of their samples, and the study was approved by the institutional review board of the National Cancer Center Research Institute. We enriched the exonic sequences of 15 primary RCCs and their matched normal samples using the Agilent Human All Exon 50 Mb Kit and sequenced the exomes using the HiSeq 2000 sequencing system. Among the 15 RCCs analyzed, 13 cases showed LOH at the MLH1 locus on chromosome 3p, and two cases showed ROH. The data are shown in Supplemental Figure S13 and Supplemental Table S3. On average, we identified 1.5 somatic indels in the MLH1-ROH cases, which is consistent with a previous report (Varela et al. 2011). However, in the MLH1-LOH tumors, we observed a 4.6-fold increased rate of somatic indel mutations (P = 0.0008). A total of 90 somatic indels were identified in 13 MLH1-LOH cases. Among them, 85 were frame-shift indels and 68 were truncating indels. Moreover, we detected recurrent truncating indels in several well-characterized cancer genes, such as VHL (four cases), PBRM1 (four cases), and JARID1C (four cases). These data suggest that the correlation we observed between MLH1 allelic loss and the increased mutation rate of somatic indels is more likely to be the true rather than a simple coincidence. Our data also indicate that MLH1 allelic deletion, through increasing the frequency of somatic indel mutations in cancer genes, could drive the development and progression of cancer. It is potentially significant that the correlation we observed was only with somatic indels, and not base substitutions. Presumably, MLH1 protein may play a pivotal role in correction of DNA indel errors, while its function for MMR can be partially compensated by other MMR proteins or mechanisms. Nevertheless, we could not exclude the possibility that factors that predispose to DNA copy-number losses might also associate with indel frequency.
In human cancers, LOH at chromosome 3p is frequently observed (Yamano et al. 2000; Harada et al. 2008; Toma et al. 2008). However, the association between MLH1 allelic loss and the increased rate of somatic indel mutations has not been notified. There are several possible reasons. First, depending on the platform, sequencing indels can be difficult. Second, reads arising from indel sequence are generally more difficult to be aligned to the reference genome. Without a good coverage, indels are more difficult to be detected. Third, the MSI assay is conventionally used to evaluate the occurrence of indels at microsatellites as genome-wide mutation analysis was not available until recently (Boland et al. 1998). The MSI assay is insufficient since only several microsatellites are selected. In addition, technical limits exist in the conventional assay (Hatch et al. 2005; Fujii et al. 2009). For example, the assay system employs capillary electrophoresis and autoradiography, making it sometimes difficult to recognize small changes in the microsatellite sequences. Some artificial fragment peaks were usually introduced after 32 cycles of PCR amplification. The choice of markers may also affect the sensitivity of the assay (Hatch et al. 2005; Fujii et al. 2009). In contrast, our data suggest that Exome-seq may be an acceptable alternative for microsatellite analysis.
Methods
The samples
PDAC-derived cell lines
We analyzed a total of 15 PDAC-derived cell lines and their matched normal samples. Primary pancreatic tumor tissue contains a high admixture of contaminating non-neoplastic inflammatory and stromal cells. To remove the non-neoplastic cells and facilitate the detection of somatic mutations, microdissected primary tumors were passaged in vitro as cell lines prior to extracting DNA and RNA for sequence analysis. The characteristics of the PDAC-derived cell lines are listed in Table 1. All cell lines were established by researchers at the Cancer Institutes, Japanese Foundation of Cancer Research (JFCR). The matching normal tissues were surgically resected from tumor-negative pancreas. All normal samples were histologically reviewed by two pathologists and were confirmed to be free of tumor tissues. All patients provided informed consent for the research use of their samples, and the study was approved by the institutional review board of the JFCR and the University of Tokyo. The DNA and RNA were extracted by standard protocols. The pair matching of each tumor cell line and the normal sample was confirmed by genome-wide SNP array (Affymetrix).
HPNE cell line
The human telomerase reverse transcriptase (hTERT)-immortalized pancreas duct epithelial cell line (hTERT-HPNE, CRL-4023) was purchased from The American Type Culture Collection (ATCC). The cells were cultured in low-glucose DMEM media (Invitrogen) supplemented with 25% Medium M3 Base (Incell), 5% fetal bovine serum, and 10 ng/mL human recombinant epithelial growth factor (Sigma Aldrich) at 37°C and with 5% carbon dioxide. HPNE serves as the normal control for gene expression analysis.
Exome-seq and data analysis
Exome-seq
Targeted enrichment was performed with Agilent SureSelect Human All Exon Kit V1.0 (Agilent Technologies). This kit is designed to enrich 162,073 exons of 16,954 protein-coding genes, more than 700 microRNAs and 300 noncoding RNAs, covering ∼37.6 Mb of the human genome (Supplemental Fig. S14). SureSelect Biotinlyated RNA baits were designed to be 120-mer long and end-to-end tiled (1× tilling). The gDNA libraries were prepared using an Illumina paired-end DNA sample prep kit (Illumina) following the manufacturer's protocols with slight modifications. In brief, 3 μg gDNA was fragmented using Covaris Acoustic Solubilizer (Covaris) with 20% duty cycle, 4 intensity, and 200 cycles per burst for 160 sec, at 16°C to get DNA fragments with a mean size of 200 bp. Fragmented DNA was then purified using Agencourt AMPure XP magnetic beads (Beckman Coulter). The concentration of the library was measured using a Bioanalyzer (Agilent Technologies). The adapter-ligated libraries were amplified with six PCR cycles, and 500 ng of each amplified library was hybridized with Biotinlyated RNA baits in solution for 24 h for target enrichment. Subsequently, hybridized libraries were cleaned up and further amplified with 12 cycles of PCR; 5–6 pM/lane DNA was applied to the flow cell, and paired-end 76-nucleotide (nt)-long reads were generated using the Illumina Genome Analyzer IIx Platform (GAIIx). Each sample was run on a single lane of Illumina flow cell except for samples PA028N and PA167T, which were each run on two lanes.
Data alignment and variant calling
The detail workflow for data alignment and mutation detection was described in Supplemental Figure S15. For each cell line and matched normal sample, the sequence reads were mapped to the human NCBI Build 36 reference sequence (hg18, downloaded from http://genome.ucsc.edu) initially with the Illumina sequencing pipeline (version 1.6) for quality recalibration. The passing filter (PF) reads were then mapped again using BWA (version 0.5.8) (Li and Durbin 2009). Any potential PCR duplicates, ambiguous reads, inconsistent read pairs, and singletons were excluded. Only the unique reads that mapped in consistent read pairs (with proper insert size and orientations) were selected for further analysis. The bases substitutions were called using SAMtools (version 0.1.7) (Li et al. 2009), and the indels were called using both SAMtools and Pindel algorithms (Ye et al. 2009).
Variant filtering and somatic variant identification
To pick out the high-confident somatic variants, we applied the following rigorous filters and rules to the data set (Supplemental Fig. S15). The first filter applied is the “quality filter.” Variants with a mapping quality of 20 or more, a phred-like consensus quality of 20 or more, a base call quality of more than 10, and a sequence coverage of 10× or more for both the cell line and matched normal sample were considered as high-quality variants. The setting for the filter conditions were optimized by comparing common SNPs detected by BWA (Li and Durbin. 2009) with those genotyped using Affymetrix Human SNP Array 6.0 (Affymetrix), ensuring a high concordance (99.84%) across two analyses (Supplemental Fig. S16).
The second filter applied, referred to as the “somatic filter,” seeks to pick out the somatically acquired variants. All the high-quality variants produced from the above steps were passed through the “somatic filter,” and only those meeting the threshold were considered as the somatic variants. The mutant allele (nonreference allele) ratio was calculated as follows:
The setting for the “somatic filter” is as described in Supplemental Figure S15; for the cell line sample, it is required that four or more reads supporting the mutant allele and the mutant allele ratio should be 15% or more. Moreover, the mutant allele should be supported by reads that aligned in both the forward and reverse directions. For the matched normal sample, given the potential sequencing errors and mapping errors, the mismatch should not be detected in more than 3% of the aligned reads and should not be detected in more than two reads. The indel, however, should not be detected in any of the aligned reads.
The third filter, referred to as the “false-positive filter,” was then applied. This filter is used to remove the potential false-positive events that result from the homologous sequences within the human genome, mapping errors, and so on. For each of the somatic mutations produced in the above steps, we extracted 200–300 bases of DNA sequences flanking its mutation locus and mapped the sequences to hg18 using the BLAT algorithm. Subsequently, the mutations identified within the regions rich for homologous sequences were removed from the list. The somatic mutations were further examined using the integrated genome viewer (IGV), and any mutations found in a “noisy” background (multiple mismatches or indels in flanking sequences) were removed from the list.
As for detection of indels, one more step, called “rescue,” was applied since the sequence read carrying a long indel toward its end is usually difficult to be aligned properly. We use the Pindel algorithm to rescue those possibly missed indels.
Variant annotation
Functional effects of filtered somatic variants were predicted using the SIFT algorithm (Kumar et al. 2009; http://sift.jcvi.org). The SIFT algorithm predicts whether an amino acid substitution affects protein function based on sequence homology and the physical properties of amino acids.
Mutation rate calculation and normalization
The background mutation rate (mutations/per Mb coding sequences) was calculated as follows:
The mutation rate of each gene was normalized by the frequency of mutations and the length of its coding sequences. Only somatic deleterious mutations, including missense substitutions, nonsense substitutions, frame-shift indels, and focal homozygous deletions were counted. The normalized mutation rate for each gene was calculated as follows, and a priority list was made accordingly:
 |
Pathway analysis
The genes with somatic mutations were classified into different functional pathways using the Gene Ontology (GO) database (http://www.geneontology.org/). Only somatic deleterious mutations were counted. The normalized mutation rate for each pathway was calculated as below:
 |
mRNA-seq and data analysis
Library preparation and mRNA-seq
Total RNA was extracted from PDAC-derived cell lines and the HPNE cells using the protocol outlined in the RNeasy Kit (Qiagen). Total RNA integrity was measured using a 2100 Bioanalyzer (Agilent Technologies), and all samples were confirmed to have an RNA Integrity Number (RIN) greater than 8.0 prior to further analysis. The mRNA-seq libraries were prepared using a paired-end mRNA Sequencing Sample Prep Kit (Illumina) following the manufacturer's protocols with slight modifications. Briefly, 2 μg of total RNA was used as the starting material, and the polyadenylated RNAs were selected using Sera-Mag Magnetic Oligo(dT) Beads (Illumina). The Poly(A)+ RNA was then fragmented by heating for 90 sec at 94°C in the supplied fragmentation buffer. Fragmented RNA was mixed with random primers, incubated for 5 min at 65°C, and placed on ice briefly before starting cDNA synthesis. First-strand cDNA synthesis was performed using SuperScript II, and second-strand cDNA synthesis was performed using DNA Pol I in the supplied GEX second-strand reaction buffer. Subsequently, cDNA ends were repaired, and adenine was added to the 3′ end of the cDNA fragments to allow adaptor ligation. Paired-end adaptors were ligated to the cDNA fragments. The ligated product was run on a 2% agarose gel, and a 300 ± 20 bp fragment was cut out and extracted. PCR (eight cycles) was performed with Phusion High-Fidelity DNA Polymerase (Finnzymes Oy) following the manufacturer's protocols. The PCR products were cleaned up with Agencourt AMPure XP magnetic beads (Beckman Coulter); 6.0–6.7 pM/lane cDNA was applied to the flow cell and paired-end 76-nt-long reads were generated using Illumina GAIIx. Each sample was run on two lanes of Illumina flow cell.
Data alignment
All PF reads were aligned to hg18 using TopHat spliced aligner (Trapnell et al. 2009). Meanwhile, all PF reads were aligned to NCBI Reference Sequence (RefSeq) mRNA sequences using BWA. A merged file was generated for each sample by integrating the output of TopHat with that of BWA for an optimal alignment for each sequence read. The ambiguously mapped reads and the duplicates were excluded. The level of gene expression was calculated in reads per kilobase of exonic sequence per million aligned reads (RPKM).
Mutation validation
For each of the somatic mutations identified by Exome-seq, we extracted the aligned mRNA-seq reads at its corresponding locus and examined if the mutant allele was also present in the cDNA sequences. The substitutions were called using SAMtools (Li et al. 2009). The small indels were called by both the SAMtools and Pindel algorithms (Ye et al. 2009). We focus on those loci covered by at least five reads, since it is rather difficult to call the variant accurately for poorly expressed genes. The mutation is supposed to be verified by mRNA-seq if at least two reads carried the mutant allele, and the mutant allele was detected in no less than 5% of the total reads aligned. For those loci covered by less than five reads but two or more reads, the mutation was also supposed to be verified if at least two reads carried the mutant allele.
Genome-wide SNP genotyping and DNA copy-number analysis
Genome-wide SNP genotyping was performed using the Affymetrix Genome-wide Human SNP Array 6.0 (Affymetrix) according to the manufacturer's instructions. SNPs were genotyped using the Birdseed version 2 module of the Affymetrix Genotyping Console software GTC 4.0.1, together with data from 45 HapMap-JPT samples (CEL files obtained from Affymetrix). DNA copy-number changes were analyzed using the Genome Imbalance Map (GIM) algorithm, as we previously described (Ishikawa et al. 2005).
The conventional MSI assay
The conventional MSI assay was performed using the proposed “Bethesda” panel of fluorescence-labeled markers, including BAT25, BAT26, D2S123, D5S346, and D17S250 and an additional two markers, NR21 and NR27. The primer sequences and PCR conditions have been previously described (Murayama-Hosokawa et al. 2010). In this study, we selected an additional three coding microsatellites and designed 6-carboxyfluorescein–labeled primers. Sequences of oligonucleotide primers for these three microsatellites are listed in Supplemental Table S4. PCR reactions were performed using the previously described reagents (Murayama-Hosokawa et al. 2010) under the following thermal cycle conditions: initial denature for 2 min at 94°C, followed by 32 cycles of denature for 15 sec at 94°C, annealing for 30 sec at 58°C, and primer extension for 30 sec at 68°C; the final extension step was carried out for 2 min at 68°C. After PCR, 1 μL of the properly diluted PCR product was mixed with 10 μL of Hi-Di Formamide and GeneScan 500 LIZ Size Standard (Applied Biosystems) mixture (37:1). This product was then denatured for 5 min at 95°C and put on ice immediately for 5 min before loading onto ABI 3130xl Genetic Analyzer (Applied Biosystems). The output data files were analyzed by GeneMapper Software Version 4.0 (Applied Biosystems). Determination of MSI status was made according to the presence of mutant alleles in tumor DNA compared with matched normal DNA.
MSI analysis by Exome-seq
We established a data analysis pipeline to identify small indels in the microsatellites. For each of the somatic indels identified in this study, we extracted the 50 bases of DNA sequences flanking its locus and examined if the indel was present in microsatellite sequences. Only those indels detected in the protein-coding microsatellites with at most 6 nt and repeated at least five times for mono- and dinucleotide microsatellites and at least three times for multiple-nucleotide microsatellites were counted. As shown in Figure 4, a graph was plotted for the indels in coding microsatellites according to the lengths of the indels and the number of sequence reads that supported the mutant alleles or the wild-type alleles. The microsatellite was suggested to be instable if a shorter allele (deletion) or a longer allele (insertion) was detected only in the tumor DNA. The sequence homology of each supporting read was further examined by the BLAT algorithm, and the reads rich of homologous sequences were discarded. The mutant allele ratio was then calculated using a formula as mentioned above.
MLH1 promoter methylation analysis
The methylation status of MLH1 promoter was quantitatively measured using MassARRAY (Sequenom), as previously described (Yagi et al. 2010). Briefly, 500 ng gDNA was bisulfite converted using an EZ DNA Methylation Kit (Zymo Research) according to the manufacturer's instruction manual. Bisulfite-treated DNA was PCR amplified, and the PCR product was transcribed by in vitro transcription (IVT) prior to cleavage using RNase A. Unmethylated cytosine was converted to uracil by bisulfite treatment, while the methylated cytosine was not converted. Methylation status was then determined by the mass difference between A and G in the cleaved RNA product. Quantitative methylation scores were obtained at each analytic unit of a cleaved product, referred to as “CpG unit.” The amplified DNA that was not methylated at all in any CpG sites was used as an unmethylated (0%) control. The amplified DNA, methylated by SssI methylase, was used as a fully methylated (100%) control.
Sanger sequencing
Oligo primers were designed to amplify the genome fragments containing the candidate nucleotide mutations from tumor cell line DNA and the matched normal DNA. PCR was performed using the high-fidelity DNA polymerase KOD-plus (TOYOBO) under optimized thermal conditions. PCR products were evaluated on a 2% agarose gel, purified and sequenced in both directions using Big Dye Terminator reactions, and subsequently loaded on an ABI 3130xl capillary sequencer (Applied Biosystems).
Statistical analysis
The P-value was calculated by Student's t-test when the data were normally distributed or by the nonparametric Wilcoxon signed-rank test when the data were not normally distributed. P-values less than 0.05 were considered to be statistically significant.
Data access
The Exome-seq data, mRNA-seq data, and SNP array data have been submitted to the European Genome-Phenome Archive (EGA; http://www.ebi.ac.uk/ega/), which is hosted at the European Bioinformatics Institute (EBI), under accession no. EGAS00001000149.
Acknowledgments
We thank Dr. Teruhiko Yoshida for having organized collaboration on the Exome-seq of the RCC project. We thank Ms. Kaori Shiina, Ms. Hiroko Meguro, Ms. Kaoru Nakano, and Ms. Saori Kawanabe for their excellent technical assistance. We acknowledge Dr. Michael Jones for the critical reading of the manuscript. This study was supported by Grants-in-Aid for Scientific Research (H.A.) and Scientific Research on Priority Areas (H.A., T.N.); a grant for Translational Systems Biology and Medicine Initiative (TSBMI; H.A.) from the Ministry of Education, Culture, Sports, Science and Technology; the NFAT project from the New Energy and Industrial Technology Development Organization (NEDO; H.A., T.N.), Japan; and the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NIBIO; T.S., H.S.,Y.K.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.123109.111.
References
- Arnold S, Buchanan DD, Barker M, Jaskowski L, Walsh MD, Birney G, Woods MO, Hopper JL, Jenkins MA, Brown MA, et al. 2009. Classifying MLH1 and MSH2 variants using bioinformatic prediction, splicing assays, segregation, and tumor characteristics. Hum Mutat 30: 757–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker LG, Shianna KV, Mullikin JC 2011. Exome sequencing: the expert view. Genome Biol 12: 128 doi: 10.1186/gb-2011-12-9-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer W, Bielas JH, Beckman RA 2008. Genetic instability is not a requirement for tumor development. Cancer Res 68: 3558–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, et al. 1998. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58: 5248–5257 [PubMed] [Google Scholar]
- Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A, et al. 1994. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature 368: 258–261 [DOI] [PubMed] [Google Scholar]
- Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloglu A, Ozen S, Sanjad S, et al. 2009. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci 106: 19096–19101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Singh A, Shianna KV, Ge D, Smith JP, Maia JM, Heinzen EL, Goedert JJ, Goldstein DB 2010. Screening the human exome: a comparison of whole genome and whole transcriptome sequencing. Genome Biol 11: R57 doi: 10.1186/gb-2010-11-5-r57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, Thibodeau SN 1998. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res 58: 3455–3460 [PubMed] [Google Scholar]
- Edelmann W, Cohen PE, Kane M, Lau K, Morrow B, Bennett S, Umar A, Kunkel T, Cattoretti G, Chaganti R, et al. 1996. Meiotic pachytene arrest in MLH1-deficient mice. Cell 85: 1125–1134 [DOI] [PubMed] [Google Scholar]
- Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R 1993. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 75: 1027–1038 [DOI] [PubMed] [Google Scholar]
- Fujii K, Miyashita K, Yamada Y, Eguchi T, Taguchi K, Oda Y, Oda S, Yoshida MA, Tanaka M, Tsuneyoshi M 2009. Simulation-based analyses reveal stable microsatellite sequences in human pancreatic cancer. Cancer Genet Cytogenet 189: 5–14 [DOI] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. 2007. Patterns of somatic mutation in human cancer genomes. Nature 446: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Chelala C, Bhakta V, Chaplin T, Caulee K, Baril P, Young BD, Lemoine NR 2008. Genome-wide DNA copy number analysis in pancreatic cancer using high-density single nucleotide polymorphism arrays. Oncogene 27: 1951–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch SB, Lightfoot HM Jr, Garwacki CP, Moore DT, Calvo BF, Woosley JT, Sciarrotta J, Funkhouser WK, Farber RA 2005. Microsatellite instability testing in colorectal carcinoma: choice of markers affects sensitivity of detection of mismatch repair-deficient tumors. Clin Cancer Res 11: 2180–2187 [DOI] [PubMed] [Google Scholar]
- Hemminki A, Peltomaki P, Mecklin JP, Jarvinen H, Salovaara R, Nystrom-Lahti M, de la Chapelle A, Aaltonen LA 1994. Loss of the wild type MLH1 gene is a feature of hereditary nonpolyposis colorectal cancer. Nat Genet 8: 405–410 [DOI] [PubMed] [Google Scholar]
- Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE 2004. Nonsense-mediated decay approaches the clinic. Nat Genet 36: 801–808 [DOI] [PubMed] [Google Scholar]
- Ishikawa S, Komura D, Tsuji S, Nishimura K, Yamamoto S, Panda B, Huang J, Fukayama M, Jones KW, Aburatani H 2005. Allelic dosage analysis with genotyping microarrays. Biochem Biophys Res Commun 333: 1309–1314 [DOI] [PubMed] [Google Scholar]
- Issaq SH, Lim KH, Counter CM 2010. Sec5 and Exo84 foster oncogenic ras-mediated tumorigenesis. Mol Cancer Res 8: 223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J 1998. Eukaryotic mismatch repair: an update. Mutat Res 409: 107–121 [DOI] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. 2008. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321: 1801–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MP, Fleming JB, Wang H, Abbruzzese JL, Choi W, Kopetz S, McConkey DJ, Evans DB, Gallick GE 2011. ALDH activity selectively defines an enhanced tumor-initiating cell population relative to CD133 expression in human pancreatic adenocarcinoma. PLoS ONE 6: e20636 doi: 10.1371/journal.pone.0020636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner RD, Marsischky GT 1999. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev 9: 89–96 [DOI] [PubMed] [Google Scholar]
- Kuismanen SA, Holmberg MT, Salovaara R, de la Chapelle A, Peltomaki P 2000. Genetic and epigenetic modification of MLH1 accounts for a major share of microsatellite-unstable colorectal cancers. Am J Pathol 156: 1773–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081 [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B 1998. Genetic instabilities in human cancers. Nature 396: 643–649 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Vinckenbosch N, Tian G, Huerta-Sanchez E, Jiang T, Jiang H, Albrechtsen A, Andersen G, Cao H, Korneliussen T, et al. 2010. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat Genet 42: 969–972 [DOI] [PubMed] [Google Scholar]
- Ma S, Chan YP, Woolcock B, Hu L, Wong KY, Ling MT, Bainbridge T, Webber D, Chan TH, Guan XY, et al. 2009. DNA fingerprinting tags novel altered chromosomal regions and identifies the involvement of SOX5 in the progression of prostate cancer. Int J Cancer 124: 2323–2332 [DOI] [PubMed] [Google Scholar]
- Maitra A, Hruban RH 2008. Pancreatic cancer. Annu Rev Pathol 3: 157–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra G, Schar P 1999. Recognition of DNA alterations by the mismatch repair system. Biochem J 338: 1–13 [PMC free article] [PubMed] [Google Scholar]
- Metzker ML 2010. Sequencing technologies—the next generation. Nat Rev Genet 11: 31–46 [DOI] [PubMed] [Google Scholar]
- Murayama-Hosokawa S, Oda K, Nakagawa S, Ishikawa S, Yamamoto S, Shoji K, Ikeda Y, Uehara Y, Fukayama M, McCormick F, et al. 2010. Genome-wide single-nucleotide polymorphism arrays in endometrial carcinomas associate extensive chromosomal instability with poor prognosis and unveil frequent chromosomal imbalances involved in the PI3-kinase pathway. Oncogene 29: 1897–1908 [DOI] [PubMed] [Google Scholar]
- Nakahori S, Yokosuka O, Ehata T, Chuang WL, Imazeki F, Ito Y, Ohto M 1995. Detection of hepatitis B virus precore stop codon mutants by selective amplification method: frequent detection of precore mutants in hepatitis B e antigen positive healthy carriers. J Gastroenterol Hepatol 10: 419–425 [DOI] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD 2010. Genomic instability: an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11: 220–228 [DOI] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, et al. 2009. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461: 272–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, et al. 2010. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet 42: 30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Tong GX, Manolidis S, Close LG, Assaad AM, Su GH 2008. Novel mutant-enriched sequencing identified high frequency of PIK3CA mutations in pharyngeal cancer. Int J Cancer 122: 1189–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle M, Marra G, Nystrom-Lahti M, Schar P, Jiricny J 1999. Identification of hMutLβ, a heterodimer of hMLH1 and hPMS1. J Biol Chem 274: 32368–32375 [DOI] [PubMed] [Google Scholar]
- Schuster SC 2008. Next-generation sequencing transforms today's biology. Nat Methods 5: 16–18 [DOI] [PubMed] [Google Scholar]
- Shibata T, Saito S, Kokubu A, Suzuki T, Yamamoto M, Hirohashi S 2010. Global downstream pathway analysis reveals a dependence of oncogenic NF-E2–related factor 2 mutation on the mTOR growth signaling pathway. Cancer Res 70: 9095–9105 [DOI] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274 [DOI] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA 2009. The cancer genome. Nature 458: 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugarbaker DJ, Richards WG, Gordon GJ, Dong L, De Rienzo A, Maulik G, Glickman JN, Chirieac LR, Hartman ML, Taillon BE, et al. 2008. Transcriptome sequencing of malignant pleural mesothelioma tumors. Proc Natl Acad Sci 105: 3521–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter CM, Martin DI, Ward RL 2004. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet 36: 497–501 [DOI] [PubMed] [Google Scholar]
- Thomas RK, Nickerson E, Simons JF, Janne PA, Tengs T, Yuza Y, Garraway LA, LaFramboise T, Lee JC, Shah K, et al. 2006. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med 12: 852–855 [DOI] [PubMed] [Google Scholar]
- Toma MI, Grosser M, Herr A, Aust DE, Meye A, Hoefling C, Fuessel S, Wuttig D, Wirth MP, Baretton GB 2008. Loss of heterozygosity and copy number abnormality in clear cell renal cell carcinoma discovered by high-density affymetrix 10K single nucleotide polymorphism mapping array. Neoplasia 10: 634–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totoki Y, Tatsuno K, Yamamoto S, Arai Y, Hosoda F, Ishikawa S, Tsutsumi S, Sonoda K, Totsuka H, Shirakihara T, et al. 2011. High-resolution characterization of a hepatocellular carcinoma genome. Nat Genet 43: 464–469 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al. 2011. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469: 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW 2004. Cancer genes and the pathways they control. Nat Med 10: 789–799 [DOI] [PubMed] [Google Scholar]
- Wilentz RE, Goggins M, Redston M, Marcus VA, Adsay NV, Sohn TA, Kadkol SS, Yeo CJ, Choti M, Zahurak M, et al. 2000. Genetic, immunohistochemical, and clinical features of medullary carcinoma of the pancreas: A newly described and characterized entity. Am J Pathol 156: 1641–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. 2007. The genomic landscapes of human breast and colorectal cancers. Science 318: 1108–1113 [DOI] [PubMed] [Google Scholar]
- Yagi K, Akagi K, Hayashi H, Nagae G, Tsuji S, Isagawa T, Midorikawa Y, Nishimura Y, Sakamoto H, Seto Y et al. 2010. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res 16: 21–33 [DOI] [PubMed] [Google Scholar]
- Yamano M, Fujii H, Takagaki T, Kadowaki N, Watanabe H, Shirai T 2000. Genetic progression and divergence in pancreatic carcinoma. Am J Pathol 156: 2123–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]