Abstract
Biomarkers in breast cancer to monitor minimal residual disease have remained elusive. We hypothesized that genomic analysis of circulating free DNA (cfDNA) isolated from plasma may form the basis for a means of detecting and monitoring breast cancer. We profiled 251 genomes using Affymetrix SNP 6.0 arrays to determine copy number variations (CNVs) and loss of heterozygosity (LOH), comparing 138 cfDNA samples with matched primary tumor and normal leukocyte DNA in 65 breast cancer patients and eight healthy female controls. Concordance of SNP genotype calls in paired cfDNA and leukocyte DNA samples distinguished between breast cancer patients and healthy female controls (P < 0.0001) and between preoperative patients and patients on follow-up who had surgery and treatment (P = 0.0016). Principal component analyses of cfDNA SNP/copy number results also separated presurgical breast cancer patients from the healthy controls, suggesting specific CNVs in cfDNA have clinical significance. We identified focal high-level DNA amplification in paired tumor and cfDNA clustered in a number of chromosome arms, some of which harbor genes with oncogenic potential, including USP17L2 (DUB3), BRF1, MTA1, and JAG2. Remarkably, in 50 patients on follow-up, specific CNVs were detected in cfDNA, mirroring the primary tumor, up to 12 yr after diagnosis despite no other evidence of disease. These data demonstrate the potential of SNP/CNV analysis of cfDNA to distinguish between patients with breast cancer and healthy controls during routine follow-up. The genomic profiles of cfDNA infer dormancy/minimal residual disease in the majority of patients on follow-up.
Breast cancer is one of the most common forms of cancer in women in Western industrial countries. Although advances in diagnosis and treatment have improved survival (Early Breast Cancer Trialists' Collaborative Group 2005), it is not possible to reliably identify breast cancer patients who will relapse with metastatic disease, and relapse can occur up to 20 yr after primary treatment (Karrison et al. 1999). This potentially long period between resection and relapse is not likely to be explained by growth of secondary tumors (Meltzer 1990; Demicheli et al. 1998; Chambers and Goss 2008) but more likely suggests a period of dormancy, where there is growth restriction of unseen micrometastases (Murray 1995). Although this long latency between resection and relapse is common in breast cancer, the associated biological mechanisms are poorly understood. However, it is well established that treatment is more effective when given before overt metastatic disease develops, underscoring the need for markers of minimal disease, preferably one that also identifies a molecular target, as disclosed by gene amplification, for example.
A number of classical factors (e.g., type, grade, node status, and hormone receptor status) and prognostic and predictive markers (e.g., HER2, Ki-67) are used to determine individual risk, but these are assessed in the primary tumor removed by surgery and are not useful in monitoring minimal disease. Moreover, genetic changes can occur between metastases and the primary tumor. Therefore, the development of tests with a clinical relevance for risk estimation and monitoring is of great interest (Levenson 2007). Stroun et al. (1987) first reported that circulating DNA in cancer patients could be distinguished from other patients with non-neoplastic disease. Measurement of levels of circulating free DNA (cfDNA) were subsequently suggested for the diagnosis of breast cancers (Huang et al. 2006), but elevated levels are sometimes seen in benign disease (Zanetti-Dällenbach et al. 2008). In breast cancer, gene expression analysis has disclosed that multiple changes can occur in micrometastases in the bone marrow, compared with metastatic disease in draining lymph nodes (Gangnus et al. 2004). Thus, it would be hugely advantageous to be able to detect specific changes indicative of progression in cfDNA.
Copy number (CN) variations (CNVs) are amplified or deleted regions of the genome, of variable size, which are recognized as a major source of normal human genome variability (Iafrate et al. 2004; Sebat et al. 2004) and contribute significantly to phenotypic variation (Redon et al. 2006). Hence, specific CNVs may be characteristic of different tumor types. Loss of heterozygosity (LOH) is also common in many tumors and can reveal recessive alleles (Wang et al. 2004). The Affymetrix SNP 6.0 array contains 906,600 probes for SNPs and 946,000 probes for CNVs and represents more genetic variation on a single array than any other array platform. Analysis of SNP 6.0 array results can generate SNP genotypes, CNVs, and LOH data in a single hybridization experiment. Due to the problems inherent in obtaining sequential samples as the cancer progresses to metastatic disease, little is known of the nature of dynamic changes of the cancer genome over time. We hypothesized that in patients on follow-up who are otherwise disease free, evidence of tumor DNA detected in cfDNA would suggest that this is derived from or related to micrometastases in the bone marrow. Therefore, the aim of this study was to compare SNP 6.0 whole-genome profiles of the primary tumor with paired plasma cfDNA samples of breast cancer patients on follow-up and related findings to plasma cfDNA profiles of primary breast cancer patients for whom we collected presurgical blood samples and healthy female controls. This aim was achieved by the successful profiling of 251 genomes to determine CNVs and LOH in paired tumors and cfDNA and by comparison with matched normal leukocyte DNA samples from the same patients.
Results
Low levels of cfDNA were detected in all plasma samples from patients and healthy female controls, consistent with our previous studies (Page et al. 2011; Shaw et al. 2011). There was no significant difference in mean cfDNA concentration between the healthy controls and either presurgical patients or patients on follow-up as assessed by absolute quantitation of a 96-bp amplicon (Shaw et al. 2011) and by ROC curve analysis.
We surveyed 251 DNA samples, isolated from normal leukocytes, plasma, and tumor from 65 breast cancer patients and eight healthy female controls, using the Affymetrix Genome-Wide Human SNP Array 6.0. We analyzed plasma prior to any surgery or treatment in 15 breast cancer patients. The other 50 patients were on follow-up after surgical removal of their primary tumor (Table 1). We compared cfDNA in two separate plasma samples (P1 and P2) for each of these taken a mean of 6.1 and 9 yr after surgery. None of these 50 patients had any evidence of metastases or recurrent disease using standard radiologic or other clinical parameters. The 251 DNA samples were hybridized in two batches only to reduce interassay variability. We validated the approach by repeating 13 samples for the entire procedure from DNA isolation through array hybridization. The results showed excellent correlation between the replicated samples by three independent measures: quality-control (QC) call-rates (P = 0.0001), median of the absolute values of all pairwise differences (MAPD) (P = 0.0005; two tailed, paired t-tests), and mean Spearman correlation (0.783; range, 0.600–0.984), confirming the reproducibility of our approach. There was also high agreement for both the range and frequency of detected CNVs (Supplemental Fig. 1).
Table 1.
Clinicopathologic details of 50 breast cancer patients on follow-up
Plasma SNP profiles distinguish between patients with breast cancer and healthy female controls
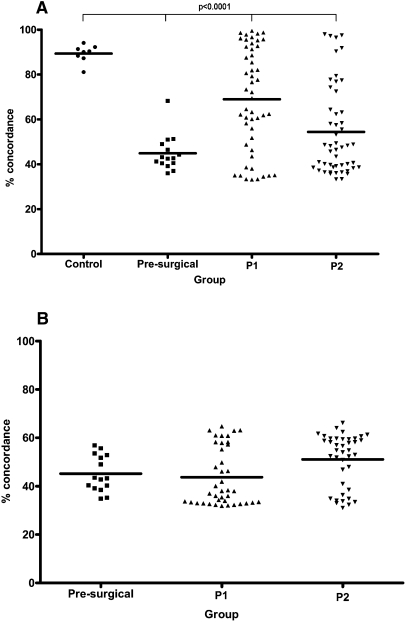
We first reviewed SNP call-rates for all samples as an indicator of successful array hybridization. The highest call-rates were for the normal leukocyte DNA samples (mean, 96.89%), with similar high call-rates in cfDNA from blood plasma and formalin-fixed paraffin-embedded (FFPE) tumor DNA (Supplemental Table 1). We next compared the concordance in SNP genotype calls. The normal leukocyte and plasma DNA samples from the healthy female controls showed an average of 64.23% and 63.50% concordance, respectively, with 15 female Caucasian HapMap samples (range, 62.75%–66.13% and 60.87%–65.38%; http://hapmap.ncbi.nlm.nih.gov/), underscoring the validity of our normal controls (Oldridge et al. 2010). Next, we compared SNP concordance between paired leukocyte and plasma cfDNA in all patients. The healthy controls had the highest mean concordance of SNP genotype calls (89.35%; range, 81.10%–94.08%; 95% confidence interval [CI], 0.09–2.74), and this was significantly lower for the presurgical breast cancer patients and patients on follow-up (P < 0.0001, one-way ANOVA), due to constitutional heterozygosity at multiple SNPs being converted to a hemizygous state in patients' plasma DNA (Fig. 1A). In the patients on follow-up, a total of 25 plasma samples (18 P1 and seven P2) showed high concordance (>80%) with their paired leukocytes, within the range observed for plasmas of the healthy controls, suggesting these plasma samples were derived largely from normal cells (Fig. 1A; Supplemental Table 2). Concordance of SNP genotype calls was low for all paired plasma and primary tumor samples (mean, 46.89%; range, 31.04%–66.20%; 95% CI, 0.12–3.78) (Fig. 1B), indicating significant differences between these.
Figure 1.
Plasma of breast cancer patients shows low SNP concordance with paired normal DNA. (A) Percentage of concordant SNP genotype calls for paired plasma and normal leukocyte DNA samples of patients and healthy controls. Percentage of concordance was significantly lower than controls in breast cancer patients (P < 0·0001, one-way ANOVA). (B) Percentage of concordant SNP genotype calls for paired plasma and microdissected tumor (available for all presurgical patients and 40 patients on follow-up; mean 47.00%; range, 31.04%–66.20%; 95% CI, 0.07–2.28). In A, concordance was lowest for the 15 preoperative primary breast cancer patients (mean, 44.88%; range, 36.00%–68.27%; 95% CI, 0.13–4.02) but remained low for the 50 patients on follow-up using both P1 (mean, 69.10%; range, 33.17%–99.44%; 95% CI, 0.21–6.51) and P2 plasma samples (mean, 54.22%; range, 33.31%–97.96%; 95% CI, 0.18–5.65). Control indicates healthy female controls; presurgical, plasma of presurgical breast cancer patients; and P1 and P2, first and second plasma samples of patients on follow-up.
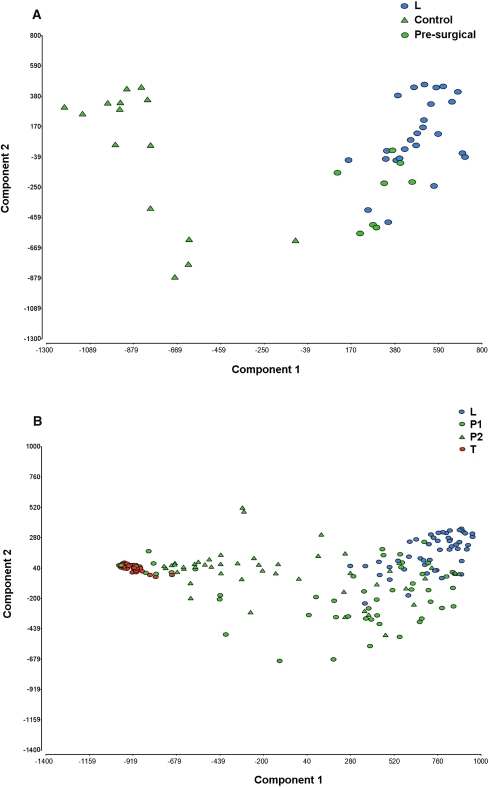
A significant difference was also seen between the concordance of SNP genotype calls between the paired leukocyte and plasma DNA of the presurgical patients and the patients on follow-up (P = 0.0016, one-way ANOVA). Hence by concordance of SNP genotype calls, plasma of the presurgical breast cancer patients differs from healthy controls, and preoperative patients differ from those who have had surgery and treatment. Principal component analysis (PCA), which takes both CN and SNP markers into account, also showed clear separation between the plasma of the healthy controls and presurgical breast cancer patients (Fig. 2A). In the patients on follow-up, the plasma PCA profiles were scattered between the matched normal leukocyte and tumor DNA samples, which grouped separately (Fig. 2B). The 25 plasma samples that showed high SNP concordance with their paired leukocytes also clustered with these by PCA, suggesting a more “normal” genome profile in these samples.
Figure 2.
Principal component analysis (PCA) of SNP/CN markers separates plasma DNA of presurgical breast cancer patients from healthy female controls. (A) PCA profiles of 15 presurgical breast cancer patients and eight healthy controls showing clear separation of the plasma DNA profiles. The plasmas of healthy female controls clustered with normal leukocytes (blue circles). (B) PCA profiles of 50 patients on follow-up, showing separation of normal leukocytes and tumor DNA, with P1 and P2 samples scattered between these. Control indicates healthy female controls; presurgical, plasma of presurgical breast cancer patients; L, normal leukocyte DNA; P1 and P2, first and second plasma samples of patients on follow-up; and T, FFPE tumor DNA.
We also compared the PCA profiles for P1 and P2 in the 50 patients on follow-up, based on the following sample groupings: (1) ER-positive versus ER-negative primary tumor status, (2) PR-positive versus PR-negative primary tumor status, (3) HER2-positive versus HER2-negative primary tumor status, (4) triple-negative (10 patients) versus any receptor-positive primary tumor status, (5) type of surgery (mastectomy versus wide local excision), and (6) endocrine therapy (tamoxifen/arimidex) prior to blood sampling versus none. There were no obvious trends observed in the cfDNA profiles of either the P1 or the P2 samples by PCA, for any of these variables, with samples again scattered between the matched normal leukocyte and tumor DNA samples (data not shown).
Plasma and tumor DNA show heterogeneous CNVs
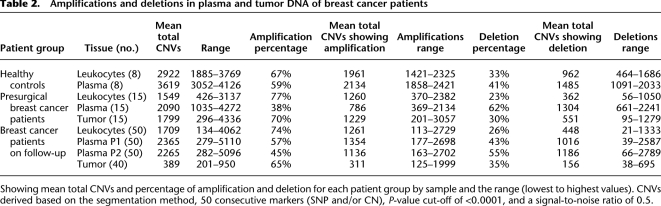
We identified 7131 copy number (CN) segments in the plasma of the 15 presurgical patients and 38,560 CN segments in the plasma of the 50 patients on follow-up. Of these 55.20% completely or partially overlap with known CNVs listed in the Toronto Database of Genomic Variants (DGV) (Iafrate et al. 2004) and 44.80% were novel. The majority of CNVs detected were amplifications, with a mean of 67.25% and 58.75% in tumor and plasma, respectively (Table 2). Both the presurgical patients and patients on follow-up showed significant differences in the frequency and range of amplification and deletions detected between cfDNA and matched leukocytes, again providing evidence of genomic change in patients' cfDNA, whereas CNV results were more similar for paired cfDNA and normal leukocytes of the healthy female controls. We examined the CNV data by applying a Gaussian smoothed signal threshold of >6.0 to filter out lower-level changes, which revealed 634 CNVs common to more than one patient. Filtering these by amplification in >10% of patients identified 23 chromosomal intervals, showing amplification in plasma and tumor DNA with little or no amplification in the plasma of healthy controls (Fig. 3; Table 3). The results were reproducible across three software platforms (Affymetrix Genotyping Console, Partek Genomics Suite, and Nexus Copy Number Discovery Edition). The majority of the 23 CNVs were >50 kb in size with more than 50 markers (Supplemental Table 3): 18 have known overlapping genes, and five have none as defined by the HUGO Gene Nomenclature Committee (HGNC) gene database (http://www.genenames.org/). By applying a lower smoothed signal threshold of >4.0, seven of these intervals showed amplification in >90% of tumor and >25% of plasma samples of patients on follow-up (Supplemental Table 4). These seven CNV intervals were more frequently detected in the plasma of node-positive patients than T1N0 patients.
Table 2.
Amplifications and deletions in plasma and tumor DNA of breast cancer patients
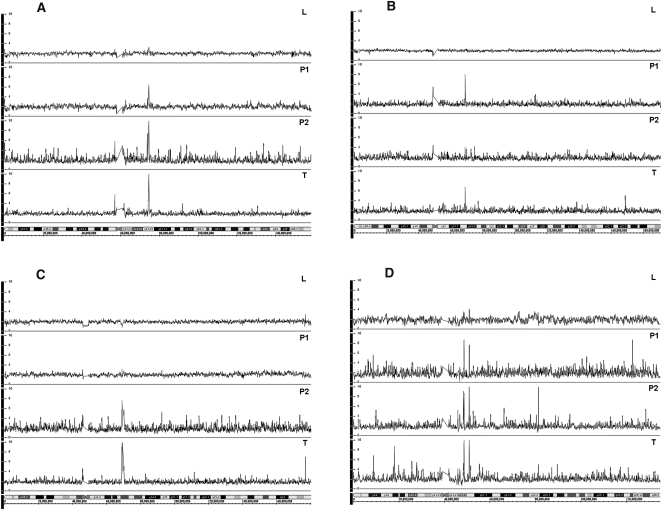
Figure 3.
High-level amplification in plasma and primary tumor DNA of breast cancer patients on follow-up. (A) Patient 44, amplification at 7q11.23 in tumor P1 and P2; (B) patient 27, amplification at 4q13.2 in tumor and P1; (C) patient 35, amplification at 5q13.2 in tumor and P2; and (D) patient 47, amplification at 10q11, showing two clear peaks (10q11.22 and 10q11.23) in tumor, P1 and P2. Top to bottom: L indicates normal leukocyte DNA; P1 and P2, paired plasma DNA samples; and T, FFPE tumor DNA.
Table 3.
Chromosomal intervals with CNVs showing common amplification in plasma and tumor
We also used linear regression analysis to compare the relationship between the presence (or not) of each of the 23 CNVs (from Table 3) in both the cfDNA and tumor DNA samples with tumor phenotype, type of surgery, and therapy. We classed each DNA sample as positive or negative at each CNV interval based on the presence or absence of a peak with a CN > 6.0 by Gaussian smooth signal. The majority of CNVs detected in cfDNA were significantly associated with breast cancer (for both the presurgical patients and 50 patients on follow-up). Of note, a number of CNVs, including 1p36.33, 1q21.1, 9p11.2, 9q12, and 19p13.3, were significantly associated with relapse. In cfDNA, 4q13.2 was associated with ER-positive cancer, and 9q12 was associated with triple-negative cancer. However, there were no significant associations with HER2 and PR (Table 4).
Table 4.
Linear regression analysis for 23 CNVs
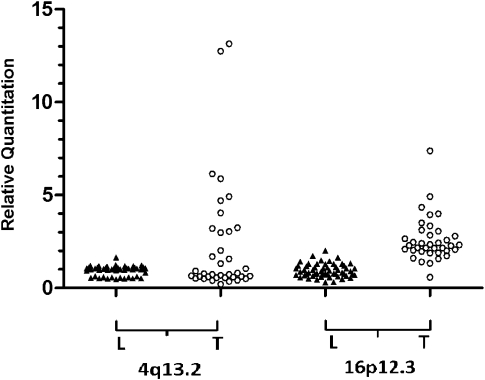
To validate CNVs, we developed locus-specific assays to 4q13.2 and 16p12.3 and used real-time quantitative PCR (qPCR) to analyze the unamplified tumor DNA from 37 primary breast cancers (from an independent series) and compared results with 56 normal leukocyte DNA samples. Ten of 37 tumor DNA samples (27%) showed amplification at 4q13.2, and 14 tumor DNA samples (38%) showed amplification at 16p12.3. In contrast, there was no amplification seen in any of the 56 normal leukocyte DNA samples, confirming the importance of the selected CNVs (Fig. 4). As the HER2 status of the primary tumor was known for many patients, we reviewed the results for the HER2 gene interval. The normal leukocyte DNA samples showed mostly diploid CN (mean CN state = 2.0), whereas the tumor and plasma samples of HER2 3+ patients showed a mean CN state of 2.5–3.0 by Gaussian smooth signal, indicating a low level of amplification (Page et al. 2011).
Figure 4.
Detection of amplification at two CNV intervals in tumor DNA. Real-time qPCR was used to analyze locus-specific assays that map within the CNVs at 4q13.2 and 16p12.3 using unamplified template DNA. Each amplicon was measured relative to the mean of four reference loci, by relative quantitation. Unamplified tumor DNA from 37 primary breast cancers (from an independent series) was compared with 56 normal leukocyte DNA samples. Amplification (RQ > 2.5) was detected in tumor DNA only.
Plasma SNP/CNV changes with time
There was a significant difference in SNP concordance between the first and second paired plasma samples (P = 0·0002; paired t-test) of the 50 patients on follow-up, and all patients showed changes in CNVs between the first and second plasma samples. Thirty patients showed a decrease and 20 patients an increase in the total number of CNVs detected. Some CNVs were common between paired plasma samples (common amplification is shown in Fig. 3), but there were also many sample-specific CNVs detected (Supplemental Fig. 2). Eight patients relapsed 2–9 yr after diagnosis. For these patients, the second plasma sample surveyed was the last blood sample taken prior to relapse. These patients showed the most CNV changes with time in plasma DNA. Figure 5 illustrates the CNV gains and losses in one patient who relapsed. There was an increase in the number of CNVs between the first (1386) and second (2482) plasma sample and a change from gain to loss at multiple CNVs. Two of the eight patients who relapsed were triple-negative; the rest were ER-positive. However, there was no obvious correlation between CNVs and relapse other than for the intervals noted previously (Table 4).
Figure 5.
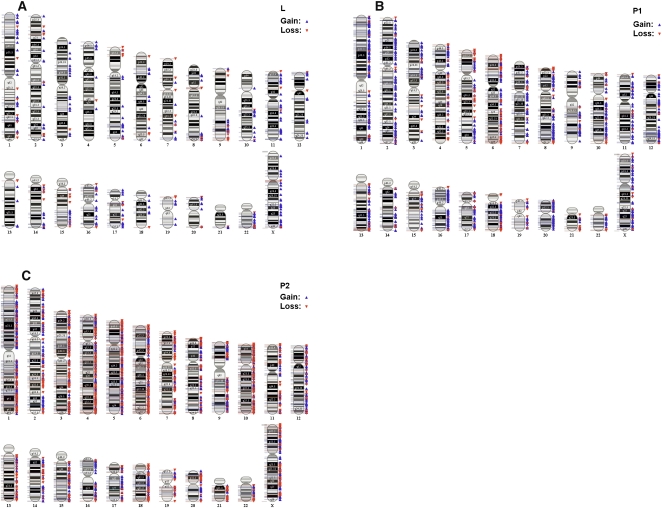
Chromosomal abnormalities in plasma preceding relapse. CNVs based on 50 consecutive markers (SNP and/or CN) and a minimum segment size of 50,000 bp. Example of array karyotypes of cfDNA for one patient preceding relapse: (A) normal leukocyte DNA sample, (B) P1 cfDNA sample taken 6 yr after diagnosis, and (C) P2 cfDNA taken 1 mo before the patient was diagnosed with metastatic disease. There was a significant increase in CNVs detected between P1 and P2: P1, 387 (79.08%) amplifications and 96 (20.92%) deletions; P2, 1332 (53.67%) amplifications and 1150 (46.33%) deletions.
Detection of LOH
There was wide heterogeneity in LOH detected both between patients and samples. The extent of the LOH overlap between paired plasma and tumor DNA also varied widely between patients, ranging from 10%–35% overlap. When we looked at LOH within exons, there were 36 LOH regions found overlapping with genes in two or more of the 15 presurgical patients' plasma samples, and 34 LOH regions found overlapping with genes in two or more plasmas of the 50 patients on follow-up (Supplemental Table 5). There was generally more LOH detected in the node-positive patients than T1N0 patients and an overall increase in LOH detected between P1 and P2 samples. Combining CN and LOH data showed that a small percentage of CN segments called (1.47%) exhibited copy-neutral LOH.
Discussion
We demonstrate for the first time that over a decade since diagnosis there is evidence of specific tumorigenic CNVs within cfDNA in plasma during routine follow-up of breast cancer patients.
At the present time, there are no accepted methods, using body fluids, that can reliably distinguish between patients with primary breast cancer and healthy controls, nor is there a method for monitoring patients after the completion of surgery, radiation therapy, and chemotherapy. Several groups, including ourselves, have reported that measuring circulating tumor cells (CTCs), bone marrow, or total circulating DNA can help in this regard (Meng et al. 2004; Braun et al. 2005; Slade et al. 2005; Schwarzenbach et al. 2009), but we and others only find one to two cells in 7.5 mL blood intermittently present, and other tests aimed at either increasing the number of cells detected or quantifying DNA size or other more straightforward characteristics thus far have not proved sufficiently reliable for clinical use. The results of this study suggest plasma cfDNA analysis is potentially more informative.
First, results from patients on follow-up are striking, since up to 12 yr after diagnosis many patients clearly have cfDNA in plasma with specific CNVs that mirror those in their primary cancer (Fig. 3), despite the fact that they have no clinically evident recurrent disease. Second, concordance of SNP genotype calls from whole-genome array analysis distinguished between patients with primary breast cancer and healthy controls (P < 0·0001) (Figs. 1, 2) and between preoperative cancer patients and patients on follow-up who have had surgery and treatment (P = 0·0016). Third, the paired plasma and leukocytes from the healthy female controls showed the highest concordance of SNP genotype calls (Fig. 1), as would be expected when the cfDNA in plasma DNA is derived from normal cells. This confirms that a representative genome sample can be obtained from plasma, even when the DNA isolated is in limiting amounts. Although whole-genome amplification (WGA) was necessary due to limiting template DNA, we pooled triplicate WGA samples to reduce the imbalance in allele ratios and differential amplification of different parts of the genome (Rook et al. 2004). In addition, we confirmed the reproducibility of the SNP array approach by QC call-rate (P = 0.0001), MAPD (P = 0.0005), and mean Spearman correlation for 13 repeated samples; hence, the results show that it is possible to reliably interrogate the entire circulating genome in a single experiment.
One important feature emerging from previous studies is the observation that tumor-specific DNA as evidenced by LOH and methylation (Levenson 2007) can persist in plasma following treatment. This finding provided the impetus for us to attempt to characterize the entire circulating genome from plasma. Compelling research, including recent parallel sequencing data, also indicates that the cancer genome can change with the evolution of metastatic disease (Gangnus et al. 2004; Ding et al. 2010), thus providing us with another reason to suppose that changes in plasma DNA might provide us with an important indicator of impending onset of life-threatening overt metastatic relapse. When we compared paired plasmas from 50 patient on follow-up, some 25 samples had an essentially normal profile, confirmed by PCA, although the remainder did not. A “normal” SNP profile would be expected if these patients are cured. Conversely, dominant oncogenes, persisting in plasma, could potentially transform stem cells in target organs and initiate metastases, as suggested by animal and in vitro cell models, the “genometastasis hypothesis” (García-Olmo et al. 1999, 2010). In support of this, we saw the most striking changes in CNVs between the P1 and P2 samples of the eight patients who had relapsed (Fig. 5), although this is too small a group to reliably identify the specific markers predictive of relapse. As with other studies concerning cfDNA, we did not try to separate DNA derived from normal cells from tumor or micrometastases prior to analysis. The CN data were supported by LOH data, which also showed an overall increase in LOH detected between P1 and P2 samples of the patients on follow-up with evidence of infrequent copy-neutral LOH. The complex CNV and LOH profiles identified from plasma suggests a mixed origin of this circulating DNA.
One other critical finding that we have made is that plasma DNA characterization may provide important information for clinicians in choosing subsequent therapies; we are able to demonstrate amplified areas of the genome, thus potentially indicating which gene products to target. There were more amplifications than deletions in most plasmas and tumors, as was found in a recent SNP 6.0 analysis of 17 different human embryonic stem cell lines (Närvä et al. 2010). In our data, by applying a Gaussian smoothed signal threshold of >6.0, we identified 23 chromosomal intervals (Table 3) showing common amplification in plasma and tumor of both the presurgical breast cancer patients and patients on follow-up. Some of these appear to discriminate between node-positive and node-negative patients, ER-positive cancer, triple-negative cancer and presence of relapse (Table 4) and may therefore be extremely helpful in deciding on chemotherapy. Applying a lower threshold >4.0 revealed many more CNVs and more frequent amplification in seven of the 23 chromosomal intervals (Supplemental Table 4). We also saw amplification in 69.23% of patients' plasma samples in four markers from the 10-kb interval that spans ZNF703, although amplification was also seen in 50.00% of plasmas from the healthy controls and 32.30% of patients' normal leukocytes. This gene has recently been shown to be a novel oncogene in Luminal B breast cancer (Holland et al. 2011). Overall, the pattern of genomic alteration seen, with focal high-level DNA amplification clustered at several chromosome arms, resembles the “amplifier” or “firestorm” type of DNA CN alterations, detected in previous genomic profiling of breast tumors (Kwei et al. 2010). The CNV of repetitive elements may be important for the five intervals identified that have no known associated gene targets, supporting the only other related study that we are aware of, which focused on repetitive elements in serum of breast cancer patients using next-generation sequencing (Beck et al. 2010). Of note, both studies have shown that there are specific breast cancer–related CNV markers, which could lead to the development of a blood-based test for breast cancer screening and monitoring.
There are many potential gene targets revealed by this genomic profiling of cfDNA (Table 3). A number are of potential interest. Expression of UGT2B15 (at 4q13.2) has been shown to be up-regulated by 17β-estradiol in MCF-7 breast cancer cells. This gene may normally maintain steroid hormone homeostasis and prevent excessive estrogen signaling (Hu and Mackenzie 2009). Hence deregulation of UGT2B15 by amplification might have the opposite effect. Neuronal apoptosis inhibitory protein (NAIP) at 5q13.2 increases in vitro and in vivo in response to androgen deprivation therapy and may be associated with enhanced survival of prostate cancers (Chiu et al. 2010). The DUB3 gene at 8p23.1 has recently been shown to be a major regulator of CDC25A (Pereg et al. 2010), which is overexpressed in many human cancers. DUB3 knockdown significantly reduced growth of breast tumor xenografts in nude mice. Hence, amplification of DUB3 might lead to CDC25A overexpression and increased oncogenesis.
The CNV detected at 14q32.33 contains a number of gene targets of potential interest. Amplification at this interval was found in 67% of the presurgical breast cancer patients' plasma samples but was absent from the healthy controls, which suggests this is a suitable interval for a more targeted study. The BRF1 gene encodes a transcription factor of the RNA polymerase III complex, which, when overexpressed, can transform cells in vitro and cause tumor formation in vivo (Berns 2008). Metastasis-associated tumor antigen 1 (MTA1), is known to be up-regulated in several cancers and has been shown to lead to the transcriptional repression of BRCA1, with resulting abnormalities in centrosome number and chromosomal instability (Molli et al. 2008). Finally, the expression of the Notch ligand JAG2 has been correlated recently with vascular development and angiogenesis (Pietras et al. 2011). Our future studies will focus on validation of these key gene targets and intervals (Table 3) in plasma cfDNA.
The finding that tumor-specific DNA persists in plasma up to 12 yr after diagnosis, although the patient remains disease free, raises important questions regarding the issue of dormancy in breast cancer. Our own studies, as well as those of other groups, have also shown that rare disseminated tumor cells (DTCs) and CTCs can persist for many years after the end of breast cancer treatment (Slade et al. 2009; Criscitiello et al. 2010). Further, the presence of these few cells represents a balance between replication and cell death, since the half-life of these cells in the plasma is 1–2 h (Meng et al. 2004). Our findings in breast cancer may also apply to other cancers where dormancy is a feature, such as melanoma, non-Hodgkin's lymphoma, and renal cancer; all of these are characterized by the development of late recurrences, and the analysis of plasma could help in the management of these conditions. In as much as plasma DNA in part reflects the nature of dying dormant cells, the information from patient samples could help elucidate the molecular determinants of survival.
These findings now require prospective valuation, preferably as part of ongoing adjuvant studies during the follow-up of a larger group of patients. In conclusion, we have demonstrated that SNP 6.0 array analysis of plasma DNA distinguishes between patients with primary breast cancer and healthy controls and between preoperative cancer patients and those who have had surgery and treatment. We have identified focal high-level DNA amplification in paired tumor and plasma, targeting specific CNVs clustered at several chromosome arms, and have shown that these are detectable in plasma up to 12 yr after diagnosis in patients on follow-up. This finding implies dormancy/minimal residual disease in the majority of patients on follow-up. Our future studies will focus on developing high-throughput approaches to target common CNVs for screening and monitoring.
Methods
Patients and samples
The protocols were approved by the Riverside Regional Ethics Committee and conducted in accordance with the Declaration of Helsinki. All patients gave written informed consent prior to participation.
The samples were blinded for analysis, and the patients understood that the results would not be made available to them. We collected blood samples from 15 women attending a clinic who had just been diagnosed with primary breast cancer and from eight age-matched healthy female volunteers. We also retrospectively analyzed stored plasma samples from 50 breast cancer patients who had been operated on for breast cancer at least 3 yr previously (Table 1). Eight of these patients developed recurrent disease between 2 and 9 yr after diagnosis.
Following plasma separation by centrifugation at 850g for 10 min (×2), plasma and cell pellets were separated and stored at −80°C. For the analysis of tumor samples, hematoxylin and eosin–stained FFPE tissue sections were reviewed, and the foci of tumor cells were isolated by manual microdissection.
DNA extraction, amplification, and SNP 6.0 arrays
DNA was extracted from blood cell pellets, 1 mL plasma, and foci of tumor cells, as described previously (Page et al. 2006; Shaw et al. 2011). WGA was performed in triplicate with the Illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare Life Sciences) and pooled (Rook et al. 2004). WGA DNA samples were hybridized on Affymetrix GeneChip Human Mapping SNP 6.0 arrays, using the Human mapping SNP 6.0 assay kit following the Genome-Wide Human SNP Nsp/Sty 6.0 protocol. Samples were hybridized in two batches only to reduce interassay variability.
Data processing and analysis
The analysis of raw data microarray CEL files was performed using Partek Genomics Suite 6.5, build 6.10.1129 (Partek Inc., http://www.partek.com/) with SNP and QC call-rates used as indicators of sample quality. Genotyping analysis and SNP/CN marker calls were performed using the Birdseed v2 algorithm (Broad Institute, Harvard–Massachusetts Institute of Technology, http://www.broadinstitute.org/mpg/birdsuite/index.html), incorporating regional GC correction. The International HapMap (build 270 na30 r1 a5, International HapMap Project, http://hapmap.ncbi.nlm.nih.gov/) was used as the initial reference model file.
Genomic segmentation was performed using a minimum of 50 markers per segment, P-value cut-off of <0.0001, and a signal-to-noise ratio of 0.5. Minimum segment sizes of 1000 bp, 50,000 bp, 100,000 bp, and 1,000,000 bp were used for viewing different-sized amplifications and deletions across different samples.
PCA was performed using Partek Genomics Suite 6.5, build 6.10.1129 (Partek). Principal components were determined using a covariance matrix method with normalized eigenvector scaling. An ANOVA P-value < 0.0001 (followed in some cases by a Bonferroni-corrected P-value < 0.0001 for multiple comparisons) was used to filter out probes of insignificance. In addition, a fold-change larger than |4| was applied to further filter data. LOH using a hidden Markov model (HMM) was also analyzed using this software on a paired basis (matched to lymphocyte) using the following parameters—genomic decay of 1 Mbp, maximum probability of 0.98, genotype error of 0.02—and was filtered using a Hardy-Weinberg Equilibrium P-value < 0.001 or <0.0001. The frequency analysis for CNVs was performed using Nexus Copy Number 5.1 Discovery Edition (BioDiscovery Inc., http://www.biodiscovery.com/).
Statistics
Data were analyzed using GraphPad Prism 5.0. Paired, two-tailed t-tests were used as appropriate. Nonparametric tests were used for further analysis; unpaired t-tests and one-way ANOVAs were followed by Mann-Whitney and Kruskal-Wallis tests, respectively. For all statistical analyses, the α value was set at 0.05.
Real-time qPCR
To confirm amplification at 4q13.2 and 16p12.3 identified by SNP 6.0 array, DNA samples were analyzed in triplicate by real-time qPCR using locus-specific assays designed in house in a 10 μL reaction volume. Reactions were run on an Applied Biosystems thermal cycler (Step One Plus) and analyzed with Step One v2.1 software and Microsoft Excel. The ΔCt was determined (average Ct value of the target locus minus the mean Ct value of four independent reference loci) and used to calculate the ΔΔCt for each DNA sample, using the mean relative quantitation (RQ) value derived from normal human genomic DNA (Roche) as the experimental calibrator. RQ values were calculated as 2−ΔΔCt as described previously (Page et al. 2011).
Data access
All microarray raw and processed data files have been deposited at ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under accession no. E-MTAB-624.
Acknowledgments
The study was supported by grants from Cancer Research UK (BIDD), the Cancer Research UK/Department of Health Experimental Cancer Medicine Centres (at Leicester University and Imperial College), and the BRC (Imperial College). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Professor Sir Alec Jeffreys, Dr. J. Howard Pringle, and Dr. Tim Gant (Leicester) for helpful discussion. Probe preparation and array hybridizations were carried out as a service by Almac Diagnostics following appropriate quality-control checks.
Authors' contributions: J.A.S. jointly conceived of and supervised the study and prepared the manuscript with R.C.C.; K.P., D.G., J.B., and N.H. carried out the study with contributions from C.R., R.P., C.P., J.S., and S.C.; K.B. and K.P. performed all bioinformatics; and R.A.W. and C.R. reviewed FFPE tumor sections. All authors contributed to the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.123497.111.
References
- Beck J, Urnovitz HB, Mitchell WM, Schütz E 2010. Next generation sequencing of serum circulating nucleic acids from patients with invasive ductal breast cancer reveals differences to healthy and nonmalignant controls. Mol Cancer Res 3: 335–342 [DOI] [PubMed] [Google Scholar]
- Berns A 2008. A tRNA with oncogenic capacity. Cell 133: 29–30 [DOI] [PubMed] [Google Scholar]
- Braun S, Vogl FD, Naume B, Janni W, Osborne MP, Coombes RC, Schlimok G, Diel IJ, Gerber B, Gebauer G, et al. 2005. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med 353: 793–802 [DOI] [PubMed] [Google Scholar]
- Chambers AF, Goss PE 2008. Putative growth characteristics of micrometastatic breast cancer. Breast Cancer Res 10: 114 doi: 10.1186/bcr2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu HH, Yong TM, Wang J, Wang Y, Vessella RL, Ueda T, Wang YZ, Sadar MD 2010. Induction of neuronal apoptosis inhibitory protein expression in response to androgen deprivation in prostate cancer. Cancer Lett 292: 176–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscitiello C, Sotiriou C, Ignatiadis M 2010. Circulating tumor cells and emerging blood biomarkers in breast cancer. Curr Opin Oncol 22: 552–558 [DOI] [PubMed] [Google Scholar]
- Demicheli R, Terenziani M, Bonadonna G 1998. Estimate of tumor growth time for breast cancer local recurrences: rapid growth after wake-up? Breast Cancer Res Treat 51: 133–137 [DOI] [PubMed] [Google Scholar]
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, et al. 2010. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464: 999–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early Breast Cancer Trialists' Collaborative Group 2005. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365: 1687–1717 [DOI] [PubMed] [Google Scholar]
- Gangnus R, Langer S, Breit E, Pantel K, Speicher MR 2004. Genomic profiling of viable and proliferative micrometastatic cells from early-stage breast cancer patients. Clin Cancer Res 10: 3457–3464 [DOI] [PubMed] [Google Scholar]
- García-Olmo D, García-Olmo DC, Ontañón J, Martinez E, Vallejo M 1999. Tumor DNA circulating in the plasma might play a role in metastasis. The hypothesis of the genometastasis. Histol Histopathol 4: 1159–1164 [DOI] [PubMed] [Google Scholar]
- García-Olmo DC, Domínguez C, García-Arranz M, Anker P, Stroun M, García-Verdugo JM, García-Olmo D 2010. Cell-free nucleic acids circulating in the plasma of colorectal cancer patients induce the oncogenic transformation of susceptible cultured cells. Cancer Res 70: 560–567 [DOI] [PubMed] [Google Scholar]
- Holland DG, Burleigh A, Git A, Goldgraben MA, Perez-Mancera PA, Chin SF, Hurtado A, Bruna A, Ali HR, Greenwood W, et al. 2011. ZNF703 is a common Luminal B breast cancer oncogene that differentially regulates luminal and basal progenitors in human mammary epithelium. EMBO Mol Med 3: 167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu DG, Mackenzie PI 2009. Estrogen receptor α, Fos-related antigen-2, and c-Jun coordinately regulate human UDP glucuronosyltransferase 2B15 and 2B17 expression in response to 17β-estradiol in MCF-7 cells. Mol Pharmacol 76: 425–439 [DOI] [PubMed] [Google Scholar]
- Huang ZH, Li LH, Hua D 2006. Quantitative analysis of plasma circulating DNA at diagnosis and during follow-up of breast cancer patients. Cancer Lett 243: 64–70 [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C 2004. Detection of large-scale variation in the human genome. Nat Genet 36: 949–951 [DOI] [PubMed] [Google Scholar]
- Karrison TG, Ferguson DJ, Meier P 1999. Dormancy of mammary carcinoma after mastectomy. J Natl Cancer Inst 91: 80–85 [DOI] [PubMed] [Google Scholar]
- Kwei KA, Kung Y, Salari K, Holcomb IN, Pollack JR 2010. Genomic instability in breast cancer: pathogenesis and clinical implications. Mol Oncol 3: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson VV 2007. Biomarkers for early detection of breast cancer: What, when, and where? Biochim Biophys Acta 1770: 847–856 [DOI] [PubMed] [Google Scholar]
- Meltzer A 1990. Dormancy and breast cancer. J Surg Oncol 43: 181–188 [DOI] [PubMed] [Google Scholar]
- Meng S, Tripathy D, Frenkel EP, Shete S, Naftalis EZ, Huth JF, Beitsch PD, Leitch M, Hoover S, Euhus D, et al. 2004. Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res 10: 8152–8162 [DOI] [PubMed] [Google Scholar]
- Molli PR, Singh RR, Lee SW, Kumar R 2008. MTA1-mediated transcriptional repression of BRCA1 tumor suppressor gene. Oncogene 27: 1971–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C 1995. Tumor dormancy: not so sleepy after all. Nat Med 1: 117–118 [DOI] [PubMed] [Google Scholar]
- Närvä E, Autio R, Rahkonen N, Kong L, Harrison N, Kitsberg D, Borghese L, Itskovitz-Eldor J, Rasool O, Dvorak P, et al. 2010. High-resolution DNA analysis of human embryonic stem cell lines reveals culture-induced copy number changes and loss of heterozygosity. Nat Biotechnol 4: 371–377 [DOI] [PubMed] [Google Scholar]
- Oldridge DA, Banerjee S, Setlur SR, Sboner A, Demichelis F 2010. Optimizing copy number variation analysis using genome-wide short sequence oligonucleotide arrays. Nucleic Acids Res 38: 3275–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page K, Powles T, Slade MJ, Tamburo de Bella M, Walker RA, Coombes RC, Shaw JA 2006. The importance of careful blood processing in isolation of cell-free DNA. Ann N Y Acad Sci 1075: 313–317 [DOI] [PubMed] [Google Scholar]
- Page K, Hava N, Ward B, Brown J, Guttery DS, Ruangpratheep C, Blighe K, Sharma A, Walker RA, Coombes RC, et al. 2011. Detection of HER2 amplification in circulating free DNA in patients with breast cancer. Br J Cancer 104: 1342–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereg Y, Liu BY, O'Rourke KM, Sagolla M, Dey A, Komuves L, French DM, Dixit VM 2010. Ubiquitin hydrolase Dub3 promotes oncogenic transformation by stabilizing Cdc25A. Nat Cell Biol 12: 400–406 [DOI] [PubMed] [Google Scholar]
- Pietras A, von Stedingk K, Lindgren D, Påhlman S, Axelson H 2011. JAG2 induction in hypoxic tumor cells alters Notch signaling and enhances endothelial cell tube formation. Mol Cancer Res 9: 626–636 [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. 2006. Global variation in copy number in the human genome. Nature 444: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook MS, Delach SM, Deyneko G, Worlock A, Wolfe JL 2004. Whole genome amplification of DNA from laser capture-microdissected tissue for high-throughput single nucleotide polymorphism and short tandem repeat genotyping. Am J Pathol 164: 23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach H, Pantel K, Kemper B, Beeger C, Otterbach F, Kimmig R, Kasimir-Bauer S 2009. Comparative evaluation of cell-free tumor DNA in blood and disseminated tumor cells in bone marrow of patients with primary breast cancer. Breast Cancer Res 11: R71 doi: 10.1186/bcr2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Månér S, Massa H, Walker M, Chi M, et al. 2004. Large-scale copy number polymorphism in the human genome. Science 305: 525–528 [DOI] [PubMed] [Google Scholar]
- Shaw JA, Brown J, Coombes RC, Jacob J, Payne R, Lee B, Page K, Hava N, Stebbing J 2011. Circulating tumor cells and plasma DNA analysis in patients with indeterminate early or metastatic breast cancer. Biomarkers Med 5: 87–91 [DOI] [PubMed] [Google Scholar]
- Slade MJ, Singh A, Smith BM, Tripuraneni G, Hall E, Peckitt C, Fox S, Graham H, Lüchtenborg M, Sinnett HD, et al. 2005. Persistence of bone marrow micrometastases in patients receiving adjuvant therapy for breast cancer: Results at 4 years. Int J Cancer 114: 94–100 [DOI] [PubMed] [Google Scholar]
- Slade MJ, Payne R, Riethdorf S, Ward B, Zaidi SA, Stebbing J, Palmieri C, Sinnett HD, Kulinskaya E, Pitfield T, et al. 2009. Comparison of bone marrow, disseminated tumour cells and blood-circulating tumour cells in breast cancer patients after primary treatment. Br J Cancer 100: 160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroun M, Anker P, Lyautey J 1987. Isolation and characterization of DNA from the plasma of cancer patients. Eur J Cancer Clin Oncol 23: 707–712 [DOI] [PubMed] [Google Scholar]
- Wang ZC, Lin M, Wei LJ, Li C, Miron A, Lodeiro G, Harris L, Ramaswamy S, Tanenbaum DM, Meyerson M, et al. 2004. Loss of heterozygosity and its correlation with expression profiles in subclasses of invasive breast cancers. Cancer Res 64: 64–71 [DOI] [PubMed] [Google Scholar]
- Zanetti-Dällenbach R, Wight E, Fan AX, Lapaire O, Hahn S, Holzgreve W, Zhong XY 2008. Positive correlation of cell-free DNA in plasma/serum in patients with malignant and benign breast disease. Anticancer Res 28: 921–925 [PubMed] [Google Scholar]