Abstract
DNA amplification, particularly of chromosomes 8 and 11, occurs frequently in breast cancer and is a key factor in tumorigenesis, often associated with poor prognosis. The mechanisms involved in the amplification of these regions are not fully understood. Studies from model systems have demonstrated that palindrome formation can be an early step in DNA amplification, most notably seen in the breakage–fusion–bridge (BFB) cycle. Therefore, palindromes might be associated with gene amplicons in breast cancer. To address this possibility, we coupled high-resolution palindrome profiling by the Genome-wide Analysis of Palindrome Formation (GAPF) assay with genome-wide copy-number analyses on a set of breast cancer cell lines and primary tumors to spatially associate palindromes and copy-number gains. We identified GAPF-positive regions distributed nonrandomly throughout cell line and tumor genomes, often in clusters, and associated with copy-number gains. Commonly amplified regions in breast cancer, chromosomes 8q and 11q, had GAPF-positive regions flanking and throughout the copy-number gains. We also identified amplification-associated GAPF-positive regions at similar locations in subsets of breast cancers with similar characteristics (e.g., ERBB2 amplification). These shared positive regions offer the potential to evaluate the utility of palindromes as prognostic markers, particularly in premalignant breast lesions. Our results implicate palindrome formation in the amplification of regions with key roles in breast tumorigenesis, particularly in subsets of breast cancers.
DNA copy-number gain and amplification are essential drivers of tumorigenesis, particularly in epithelial cancers such as breast cancer. In breast tumors, about half of highly amplified genes are also overexpressed (Hyman et al. 2002; Pollack et al. 2002). High-throughput, genome-wide profiling of copy-number alterations has led to the discovery of regions recurrently amplified in breast cancers including 1q, 8q, 11q, 12q, 16p, 17q, and 20q, (Kallioniemi et al. 1994; Isola et al. 1995; Courjal and Theillet 1997; Knuutila et al. 1998; Forozan et al. 2000; Rennstam et al. 2003; Loo et al. 2004; Nessling et al. 2005; Chin et al. 2006). These regions house key oncogenes involved in breast cancer progression including, but not limited to, MYC in 8q24, CCND1 in 11q13, and ERBB2 in 17q12 (Slamon et al. 1989; DePinho et al. 1991; Dickson et al. 1995; Deming et al. 2000; Futreal et al. 2004). Focal amplification of these regions and increased frequency of amplification genome-wide are associated with poor disease prognosis (Chin et al. 2006; Hicks et al. 2006). For this reason, many studies have examined amplification as a potential early marker indicating likelihood of invasion; however, detection of amplification of key regions in premalignant breast lesions is inconsistent (Lu et al. 1998; Werner et al. 1999; Robanus-Maandag et al. 2003; Corzo et al. 2006; Yao et al. 2006; Burkhardt et al. 2009). Additionally, the primary mechanisms by which copy-number gain and amplification occur in breast cancer remain to be elucidated. Ultimately, the initiators and regulators of amplification could provide candidate prognostic markers or present novel therapeutic targets.
Studies from model systems have demonstrated that palindrome formation can be an early and potentially rate-limiting step in DNA amplification. Large, de novo palindromes, resulting in gene duplication, can form via multiple mechanisms. First, DNA double-strand breaks (DSB) can promote inter- or intramolecular recombination between normally occurring inverted repeats (IR) or regions with short sequence homology, leading to hairpin-capped chromosome fragments and subsequent palindrome formation following DNA replication (Yasuda and Yao 1991; Butler et al. 1996, 2002; Lobachev et al. 2002; Tanaka et al. 2002, 2005; Narayanan et al. 2006; VanHulle et al. 2007). In addition, de novo palindromes can form by template switching at an IR when a replication fork is blocked, as has been demonstrated recently in yeast (Mizuno et al. 2009; Paek et al. 2009). Last, dicentric, palindromic chromosomes can also be generated by sister-chromatid fusion subsequent to a DSB or telomere erosion (Smith et al. 1992; Ma et al. 1993; Coquelle et al. 1997). Palindromes in the form of dicentric chromosomes can be substrates for additional rearrangements, including amplification by breakage–fusion–bridge (BFB) cycles (McClintock 1941). Repeated BFB cycles, driven by selection for the amplification of a gene(s) providing a growth advantage to the cell, propagate de novo palindrome formation at the sites of novel DSBs. Palindromic rearrangements marking the breakpoints of focal, high-level amplifications, a typical byproduct of BFB cycles, have been observed in cancer cells (Gisselsson et al. 2000; Saunders et al. 2000; Shuster et al. 2000; Ciullo et al. 2002; Hellman et al. 2002; Lo et al. 2002; Murnane and Sabatier 2004; Prentice et al. 2005; Shimizu et al. 2005; Reshmi et al. 2007). However, it is not known whether palindrome-associated mechanisms are dominant pathways of copy-number gain and amplification in cancers such as breast cancer.
The overall objective of the current study was to identify de novo palindromes in breast cancers and assess their localization relative to regions of copy-number gain and amplification. We assessed palindromes by an assay previously developed and optimized by our group called Genome-wide Analysis of Palindrome Formation (GAPF) (Tanaka et al. 2005; Diede et al. 2010a,b). In light of our previous studies, we first re-evaluated genome-wide distributions of palindromes in human cancers, specifically the cancer cell lines MCF7 and Colo320DM, using GAPF on Affymetrix tiling arrays. We demonstrated the utility of integrating GAPF profiling with high-resolution profiling of copy-number alterations to spatially associate palindromes with copy-number gains. We next profiled palindromes and copy-number gains in a set of breast cancer cell lines and primary tumors, focusing on chromosomes 8, 11, and 12, which contain genes that are recurrently amplified in breast tumors and strongly implicated in breast tumorigenesis. From these analyses, we confirmed that palindrome formation is a nonrandom event in cancers. Further, we demonstrated that palindromes are frequently associated with regions of copy-number gain and amplifications, particularly at their breakpoints, in a subset of breast cancers, suggesting an active role for palindrome-associated amplification in breast tumorigenesis.
Results
GAPF-positive regions distributed nonrandomly in two cancer cell lines, Colo320DM and MCF7
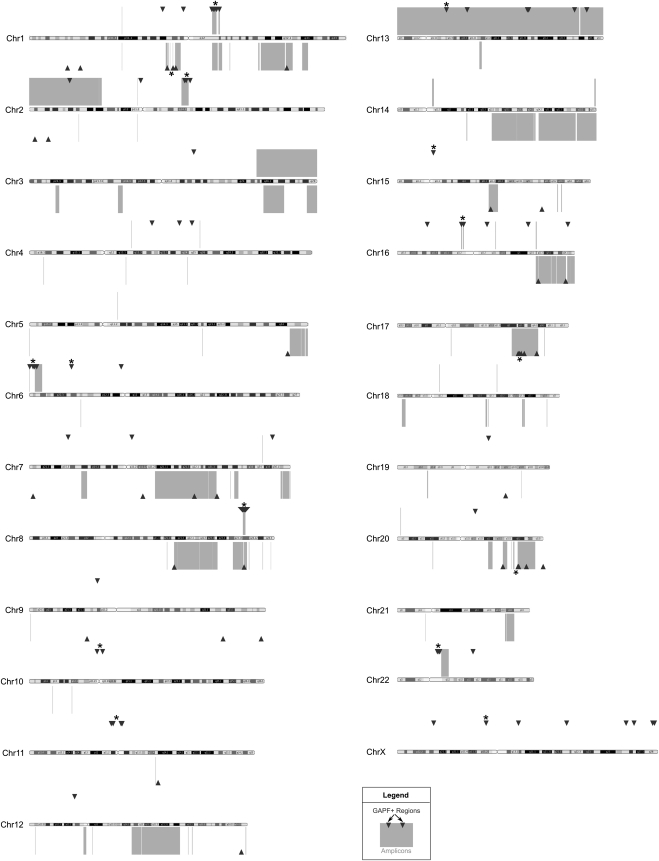
Based on our previous studies optimizing GAPF to detect de novo palindromes (Diede et al. 2010a,b), we re-examined genome-wide palindrome profiles in Colo320DM and MCF7 by performing GAPF with the addition of 50% formamide. To map palindromes at high resolution across the genomes, GAPF-enriched DNA was hybridized to Affymetrix human tiling arrays. These arrays are comprised of 25-mer probes spaced 10-bp apart (35-bp resolution) generated from both inter- and intragenic RepeatMasked sequence, thereby affording extensive, high-resolution detection of palindromic centers, while minimizing the potential artifacts introduced by simple repeat sequences in the genome. In parallel, we performed GAPF on cultured, normal human fibroblasts (HFs) and compared the cancer cell line profiles with the HF profile to detect “GAPF-positive regions.” We defined GAPF-positive regions as those regions enriched in the cancer cell lines relative to the normal HFs at P-value <0.001 and log2 signal ratio >1.5. GAPF-positive regions within 10 kb of other regions were grouped together to be designated as a single GAPF-positive region. By these metrics, we also detected regions that were enriched more in the HFs than the cancer cell lines that mapped predominantly to segments of the genome that are repetitive in nature (e.g., simple tandem repeats, segmental duplications). Therefore, it is likely that GAPF is detecting sample-to-sample genomic variation in addition to tumor-specific palindromes, though for this study we focused on the positive regions representative of de novo palindrome formation in cancer. Overall, we identified 139 GAPF-positive regions in the Colo320DM genome (Supplemental Table 1), 25 of which were located in the double minute containing ∼1.6 Mb around the MYC gene locus of chromosome 8 (Lin et al. 1985; Bianchi et al. 1991), which has previously been shown to have palindromic structures (Ford and Fried 1986; Tanaka et al. 2005). In MCF7, we detected 52 GAPF-positive regions throughout the genome. GAPF-positive regions were distributed nonrandomly throughout both genomes, with some chromosomes having more GAPF-positive regions than others (Supplemental Table 3). Chromosomes that had many GAPF-positive regions in both cell lines included chromosomes 1, 2, 7, and 15.
Closer examination of the distribution of GAPF-positive regions in the MCF7 and Colo320DM genomes revealed evidence of clustering of those regions in discrete genomic locations (Fig. 1). For example, we detected 28 GAPF-positive regions within a 10-Mb window on 1q21 in Colo320DM, which included the previously identified and validated de novo palindromes at the CTSK and ECM1 loci (Tanaka et al. 2005, 2007). The probability that these 28 regions would have all randomly occurred within this 10-Mb window approached zero (P = 1 × 10−16). We identified additional clusters with more than three GAPF-positive regions throughout the Colo320DM and MCF7 genomes using sliding 10-Mb windows (Supplemental Table 5). Clusters that had a <5% probability of occurring randomly in a 10-Mb region are denoted by the asterisks in Figure 1. In addition to 1q21, we observed statistically significant clusters in Colo320DM in chromosomal regions 1p21, 2q14-q21, 6p25, 6p22, 8q24, 10p11-q11, 11p11, 15q11, 16p11, Xp11, and Xq22-q23; and in MCF7 in chromosomal regions 1p13, 17q23, and 20q13. Overall, these findings indicate that in cancer genomes, palindromes occur nonrandomly and often in clusters.
Figure 1.
GAPF-positive regions cluster in copy-number gains in cell lines Colo320DM and MCF7. Genome graph depicting locations of GAPF-positive regions (P < 0.001, log2 signal ratio > 1.5; triangles) and copy-number gains (log2 signal ratio > 0.3; boxes) across the genomes of colon-cancer cell line Colo320DM and breast-cancer cell line MCF7 as compared with cultured HFs. GAPF-positive regions and copy-number gains detected in Colo320DM are shown above and regions identified in MCF7 are shown below each chromosome ideogram. Clusters of at least three GAPF-positive regions that had a <5% probability of randomly occurring within a 10-Mb window are marked by asterisks (*). Chromosomes are drawn approximately to scale.
Associations between GAPF-positive regions and copy-number gains in Colo320DM and MCF7
To determine the spatial association between GAPF-positive regions and copy-number gains, we integrated GAPF profiles and high-resolution, copy-number profiles generated on the Affymetrix SNP arrays. We performed a wavelet-based, nonparametric analysis (Yu et al. 2010) on the normalized SNP-array data to detect copy-number segments and breakpoints. Copy-number segments with an average log2 signal ratio >0.3 were designated as gains, while amplifications were defined as segments <5 MB in size and having an average log2 signal ratio >1.0. We noted genome-wide colocalization of GAPF-positive regions with copy-number gains and amplifications in Colo320DM and MCF7 (Fig. 1). In Colo320DM, 70 out of 124 (56%) GAPF-positive regions on autosomal chromosomes were associated with segments with increased copy number, and in MCF7, 35 out of 52 (67%) GAPF-positive regions were located in copy-number gains, P-value < 0.001 and = 0.0076 based on 10,000 simulations (see Methods), respectively. In MCF7, we identified clusters of GAPF-positive regions in amplifications on 1p, 17q, and 20q, in agreement with paired-end sequencing studies of the MCF7 genome (Raphael et al. 2003; Volik et al. 2006) and consistent with the model of BFB cycles. On the autosomal chromosomes of these cell lines, we also observed significant overlap between GAPF-positive regions and copy-number gain breakpoints identified by wavelet-based analyses; 24 out of the 124 (19%) GAPF-positive regions in Colo320DM and 11 out of the 52 (21%) GAPF-positive regions in MCF7 colocated with breakpoints, P-value < 0.001 and = 0.005, respectively (see Methods). These results suggest that palindrome formation could have been an initiating event in the generation of regional copy-number gains and amplifications in these cancer genomes.
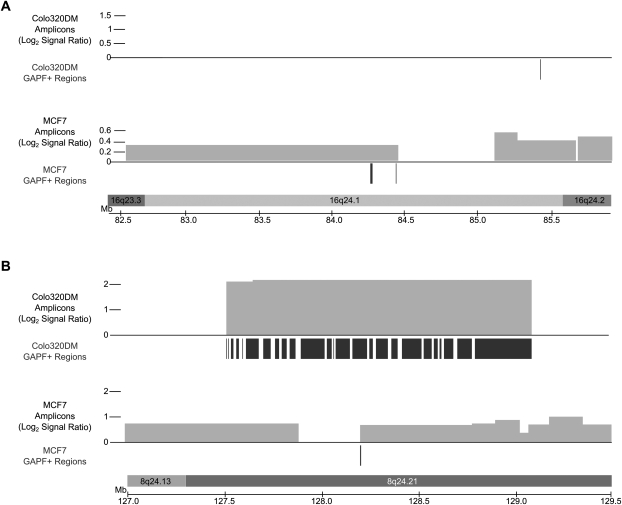
Next, we compared GAPF profiles between MCF7 and Colo320DM to determine whether the distribution of GAPF regions was similar between two cell lines representing different types of cancer. We grouped GAPF-positive regions into cytogenetic bands and detected 49 and 31 GAPF-positive cytogenetic bands in Colo320DM and MCF7, respectively (Supplemental Table 4). Two cytogenetic bands were commonly positive between MCF7 and Colo320DM: 16q24.1 (Fig. 2A) and 8q24.21 (Fig. 2B). The GAPF-positive region in 16q24.1 was associated with copy-number gain in MCF7, but not in Colo320DM. The GAPF-positive region located in 8q24.21 was shared at the same genomic location and was associated with segments of increased copy number in both cell lines. As mentioned previously, in Colo320DM this region is in the form of a high-copy double minute in Colo320DM (Lin et al. 1985; Bianchi et al. 1991) and contains palindromic rearrangements (Ford and Fried 1986; Tanaka et al. 2005). In MCF7, the amplified region from 8q21 to 8qter is neither a double minute nor a homogenously staining region, but is present on multiple, normal, and derivative chromosomes often translocated with segments from other chromosomes (Rummukainen et al. 2001). To our knowledge, palindromic rearrangements have not been observed in the MYC amplicon of MCF7. From these findings, we conclude that the majority of palindromes form at different genomic locations in cancers from different tissues; however, a subset of regions commonly amplified in cancers, such as 8q24, may undergo recurrent palindrome formation in tumorigenesis.
Figure 2.
GAPF-positive regions located in copy-number gains in 16q24.1 (A) and 8q24.21 (B) in Colo320DM and MCF7. Locations of GAPF-positive regions (P < 0.001, log2 signal ratio > 1.5) are denoted by the dark bars under the axes and the copy-number gains (log2 signal ratio > 0.3) by the lighter boxes above the axes, with the height of the box corresponding to the log2 signal ratio of the segment. GAPF-positive regions were determined by tiling array analysis of Colo320DM and MCF7 compared with cultured human fibroblasts. The copy-number segments were identified with SNP arrays coupled with wavelet-based statistical analysis. Regions in Colo320DM are displayed above the corresponding regions for MCF7 (as labeled). (A) The GAPF-positive regions detected in 16q24.1 in MCF7 were located in a copy-number gain, one of which was located near the boundary of the segment. In contrast, there was no amplification of 16q24.1 in Colo320DM, but one GAPF-positive region was identified. (B) The amplification in 8q24 in Colo320DM contains many GAPF-positive regions, while MCF7 has one GAPF-positive region located at a copy-number breakpoint.
Palindrome-associated copy-number gain of a genomic region containing the MYC oncogene in MCF7
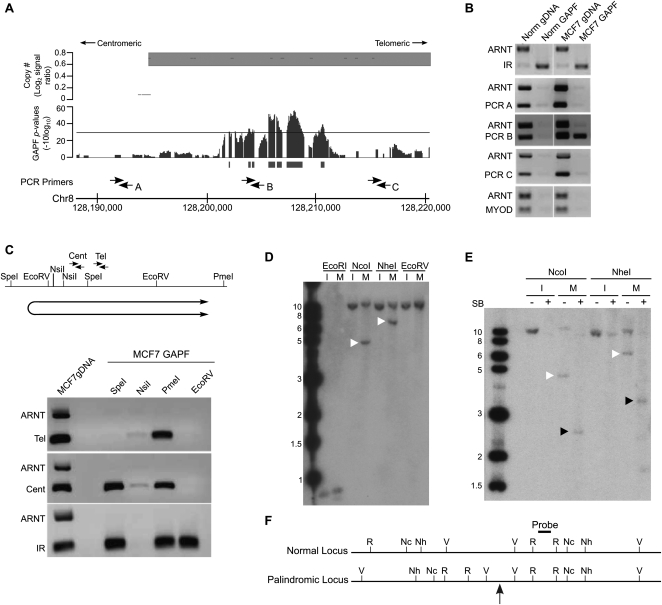
In the MCF7 cell line, we detected a GAPF-positive region at the centromeric breakpoint of a complex copy-number gain in 8q24.21 that contains the MYC oncogene (Fig. 3A). We next confirmed that the GAPF signal represented a cancer-specific palindrome. A regional PCR-based enrichment analysis following the S1 digestion step of GAPF demonstrated that the positive region (PCR B) (see Fig. 3A) was enriched by GAPF over a nonpalindromic region (ARNT) in MCF7 preparations (Fig. 3B, middle), similar to the pattern of enrichment seen with a normally occurring IR (Fig. 3B, top). Locations centromeric to the GAPF signal (PCR A) and telomeric, but within the same copy-number segment (PCR C), were not enriched over the ARNT region in MCF7 GAPF samples. Furthermore, the GAPF-positive region in 8q24.21 in MCF7 was eliminated following repeated rounds of GAPF, or cycled GAPF (Diede et al. 2010b; Supplemental Fig. 1), in a pattern consistent with that seen with a normally occurring IR in cycled GAPF. Using restriction-enzyme mapping coupled with PCR-enrichment analysis, we further determined the orientation of the novel palindrome by locating the palindromic junction centromeric to the GAPF-positive region (Fig. 3C). Additionally, Southern analysis confirmed that the GAPF-positive region at 8q24.21 was part of a rearrangement consistent with the presence of a palindrome. The abnormally sized fragments we obtained from digesting MCF7 genomic DNA with NcoI or NheI (Fig. 3D), migrated as half-sized fragments under GAPF conditions (Fig. 3E). As depicted in the inferred map of this locus (Fig. 3F), we determined that the palindromic junction was located just upstream of the EcoRV digest site, consistent with the previous restriction-enzyme mapping (Fig. 3C). Finally, DNA-sequence analysis of the center of the palindrome revealed a novel, intrachromosomal rearrangement (Supplemental Fig. 2), likely generated by a homologous recombination event between simple repeats present on opposite strands in the normal genomic sequence. This rearrangement has resulted in proximal, inverted Alu elements that have the capacity to induce genomic instability, such as hairpin formation and subsequent large palindrome generation. Overall, we concluded that MCF7 contains a palindromic rearrangement of the region in 8q24.21. This palindrome appears to mark the boundary of a complex copy-number gain in MCF7 that houses the MYC locus, implicating palindrome-associated mechanisms of amplification in this region.
Figure 3.
Validation of a de novo DNA palindrome in chromosome 8q24.21 of breast cancer cell line MCF7. (A) GAPF analysis on tiling arrays comparing MCF7 with cultured HFs. The lower graph displays GAPF P-values (−10log10). The solid bars under the graphs represent GAPF-positive regions (P-value [−10log10] > 30, run > 50 bp, gap < 100 bp). The upper graph displays wavelet-derived copy-number segments (average log2 signal ratio). Segments with a log2 signal ratio >0.3 are designated as gained. The locations of PCR primers used in the PCR-enrichment analysis are shown. (B) PCR-based analyses to detect enrichment of genomic loci in 8q24.21 over the nonpalindromic region (ARNT) following GAPF preparations of MCF7 (MCF7 GAPF) and pooled PBL cells (Norm GAPF). Analysis of a normally occurring inverted repeat (IR) and nonpalindromic region (MYOD) are included to confirm efficient GAPF preparations. Note that the only loci that are enriched by GAPF are IR in MCF7 and pooled PBLs, and PCRB in MCF7, located in the GAPF-positive region. (C) PCR-enrichment analysis following targeted restriction-enzyme digestion preceding GAPF. Genomic DNA from MCF7 cells was predigested with SpeI, NsiI, PmeI, or EcoRV, shown on the map, and processed by GAPF. Enrichment of the GAPF-positive region in 8q24.21 over the nonpalindromic ARNT region was examined using two primer pairs, Cent and Tel. Digestion in the nonpalindromic spacer of a palindrome or IR will eliminate enrichment of the sequence by GAPF, as shown by the lack of PCR product when DNA was first digested with NsiI and enrichment of the IR was assessed (bottom gel). Note that enrichment of the 8q24.21 region was seen with the Cent primers when MCF7 DNA was digested with SpeI, but not observed when the Tel primers were used. Based on these analyses, the inferred location and orientation of palindrome places the palindromic center between SpeI and EcoRV restriction sites, shown below the map. (D–F) Southern analysis to detect rearrangements of the GAPF-positive region in 8q24.21. (D) Genomic DNA from MCF7 (M) and normal human fibroblast cell line IMR90 (I) was digested with EcoRI (R), NcoI (Nc), NheI (Nh), or EcoRV (V) shown on the map (F). Digesting genomic DNA from MCF7 with NcoI and NheI and hybridizing with a probe in the GAPF-positive region in 8q24.21 yielded abnormally sized fragments of 5 and 7 kb, respectively (white arrowheads), which differed from the expected 12-kb fragments based on the normal genome sequence. Note that we also detected normal-sized fragments, indicating the presence of rearranged and normally arranged alleles. (E) Snapback Southern to confirm palindromic nature of GAPF-positive region in 8q24.21. Genomic DNA from MCF7 (M) and IMR90 (I) were first digested with either NcoI or NheI. The digested samples were evenly split and half of the sample was processed through GAPF to create snapback DNA, shown in the + lanes. Note that the rearranged fragments in MCF7 (white arrowheads) were converted to half-sized fragments (black arrowheads) following GAPF. (F) Inferred restriction enzyme map of normal and palindromic loci is shown with the location of the palindromic center marked by the gray arrow. Probe location is denoted on inferred map.
GAPF-positive regions are highly associated with amplifications in a set of primary breast tumors
Amplification of regions housing oncogenes on chromosomes 8, 11, and 12 occurs frequently in breast cancers and is associated with poor prognosis (Courjal and Theillet 1997; Rennstam et al. 2003; Letessier et al. 2006). However, the mechanisms that initiate and generate amplification of these regions in breast cancer remain to be elucidated, and understanding the initiators of amplification could provide early prognostic markers. To implicate a role for palindrome formation in the amplification of regions of chromosomes 8, 11, and 12 in breast cancers, we performed high-resolution GAPF and copy-number analysis on a set of primary breast tumors. In our analysis, GAPF-enriched samples were hybridized to a tiling array with oligo probes covering only chromosomes 8, 11, and 12 to obtain high-resolution coverage of these chromosomes.
We examined six primary invasive ductal carcinomas (IDCs), three estrogen receptor-positive (ERP) and three estrogen receptor-negative (ERN). We compared the GAPF profiles of the IDCs with the GAPF profile of normal peripheral blood lymphocytes (PBLs) pooled from multiple female individuals. We chose pooled PBLs as the normal reference for this analysis to avoid palindromic rearrangements specific to cell culture and to dilute any polymorphic palindromes present in individuals in the pool. GAPF-positive regions were detected and defined as previously described in the genome-wide GAPF analysis in Colo320DM and MCF7. Using these metrics, we did not detect any regions that were GAPF-positive on chromosomes 8, 11, and 12 when comparing PBL samples prepared in parallel to each other. Finally, we compared GAPF and copy-number profiles, generated on Affymetrix SNP Arrays, to identify regions with evidence of palindrome-associated amplification. We analyzed the copy-number data for all samples using a wavelet-based analytical tool that detects breakpoints and copy-number segments (see Methods).
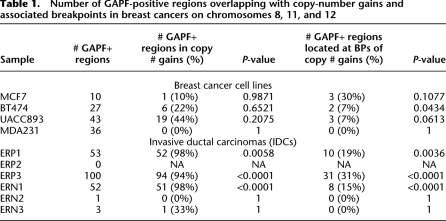
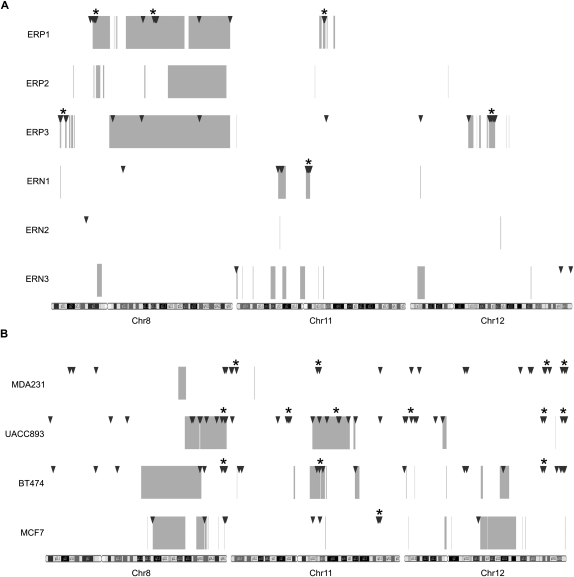
We observed that GAPF-positive regions were distributed nonrandomly and were overwhelmingly associated with copy-number gains along chromosomes 8, 11, and 12 in the primary breast tumors. In three of the IDCs (ERP1, ERP3, and ERN1), we detected 50–100 GAPF-positive regions per sample on these chromosomes (Table 1). We observed clustering of at least three GAPF-positive regions in multiple chromosomal locations in these tumors, particularly in 8p12, 8q21, and 11q13 in ERP1, 8p23 and 12q14-q15 in ERP3, and 11q12 in ERN1 (Fig. 4A; Supplemental Table 5). The GAPF-positive regions in these cytogenetic bands clustered within 5-Mb windows; for all of these clusters the probability that the number of observed GAPF-positive regions would randomly occur in the same 5-Mb window approached zero. These clusters were all located in amplicons and, overall, >90% of the GAPF-positive regions in ERP1, ERP3, and ERN1 colocated with copy-number gains (Table 1), the majority of which are amplifications. Simulation-based analyses (see Methods) were performed to assess the significance of the overlaps between GAPF-positive regions and copy-number gains in these IDCs, and in all three samples the associations were highly significant (ERP1, P = 0.0058; ERP3, P < 0.0001; ERN1, P < 0.0001). Furthermore, the spatial associations between GAPF-positive regions and copy-number gain breakpoints were also highly significant (ERP1, P = 0.0036; ERP3, P < 0.0001; and ERN1, P < 0.0001). In the three ER-positive tumors, we observed segments of 8q with increased copy number, and we also identified GAPF-positive regions located in these segments in ERP1 and ERP3 (Fig. 4A). However, we detected no GAPF-positive regions in ERP2 in the 8q copy-number gains nor on any of the three interrogated chromosomes, suggesting that palindrome-formation is not a prerequisite for amplification. Finally, we identified two GAPF-positive regions in 8q13 and 8q24.12 that were shared between two of the IDCs, ERP1 and ERP3, and were also located in copy-number gains. Other than these two GAPF-positive regions, there were no additional regions shared between more than one IDC.
Table 1.
Number of GAPF-positive regions overlapping with copy-number gains and associated breakpoints in breast cancers on chromosomes 8, 11, and 12
Figure 4.
Distribution of GAPF-positive regions and copy-number gains on chromosomes 8, 11, and 12 in breast cancers. Genome graphs of GAPF-positive regions (P < 0.001, log2 signal ratio > 1.5; triangles) and copy-number gains (average log2 signal ratio > 0.3; boxes) in primary invasive ductal carcinomas (A) and breast cancer cell lines (B). Locations of GAPF-positive regions were determined from tiling array analysis comparing breast cancer samples with pooled PBLs. Copy-number gains were detected by SNP arrays coupled with wavelet-based statistical analyses. Clusters of at least three GAPF-positive regions that had a <5% probability of randomly occurring within a 5-Mb window are marked by asterisks (*). Note the colocation of clusters and copy-number gains, especially in the primary tumors.
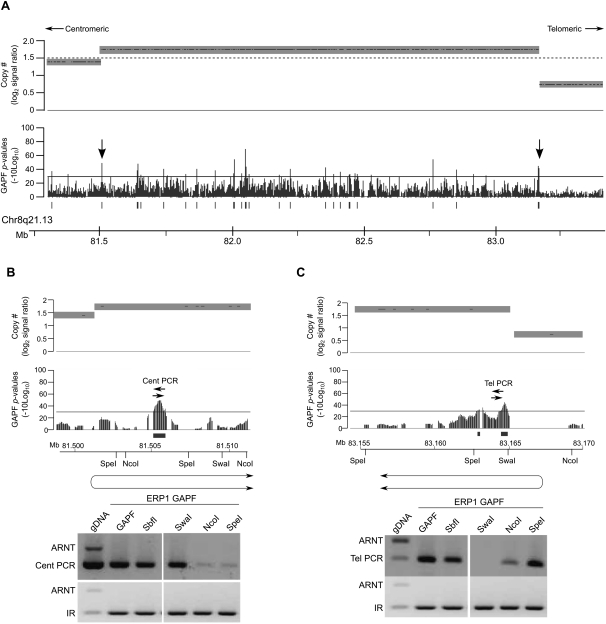
We further examined two clusters of GAPF-positive regions located in high-level amplicons (log2 signal ratio > 1.5) in 8q21.13 (Fig. 5A) and 8p12 (Supplemental Fig. 3) in the IDC ERP1 to confirm evidence of palindrome-associated amplification. Based on restriction-enzyme mapping, we both validated the palindromic nature of the interrogated loci and determined the orientation of the de novo palindromes. In 8q21.13, we confirmed that the GAPF-positive region marking the centromeric boundary of the amplicon was oriented with the novel junction located on the centromeric side of the GAPF-positive region, upstream of the SpeI restriction site, and the palindromic arms extended in the direction of the telomere (Fig. 5B). Examining the GAPF-positive region located at the telomeric breakpoint of the same amplicon revealed that the novel palindrome was in the opposite orientation with the junction of this palindrome located between the SwaI and NcoI sites telomeric to the GAPF-positive region (Fig. 5C). The junctions of both of these palindromes colocated with copy-number breakpoints identified by the wavelet-based analysis. We also validated and located the centers of three palindromes detected by GAPF at copy-number breakpoints of the complex amplicon in 8p12 in ERP1 (Supplemental Fig. 3A). Two of the GAPF-positive regions were oriented with the palindromic centers located telomeric of the regions (Supplemental Fig. 3B,C). The third GAPF-positive region, at the centromeric end of the high-level amplicon, was oriented in the opposite direction with the palindromic center located centromeric of the region (Supplemental Fig. 3D). Overall, these findings are consistent with BFB cycles creating these highly amplified regions in a primary breast tumor.
Figure 5.
Highly amplified regions in an invasive ductal carcinoma have palindromes at amplicon breakpoints. GAPF analysis on tiling arrays of the invasive ductal carcinoma (IDC) ERP1 compared with the normal, pooled PBL reference. Graphs display GAPF P-values (−10log10) and wavelet-derived copy-number segments (average log2 signal ratio). The solid bars under the graphs represent GAPF-positive regions (P-value [−10log10] > 30, run > 50 bp, gap < 100 bp). The dashed line marks where log2 signal ratio = 1.5. (A) A highly amplified region (log2 signal ratio > 1.5) in 8q21.13 in the IDC ERP1 has GAPF-positive regions located throughout the amplicon and at the breakpoints (arrows). (B,C) PCR-based enrichment analyses of GAPF-positive regions following targeted restriction-enzyme digestion prior to GAPF. Genomic DNA from ERP1 was digested with SbfI, SwaI, NcoI, or SpeI and processed by GAPF. The GAPF-positive regions at the centromeric (B) and telomeric (C) boundaries of the amplicon were assessed for enrichment over a nonpalindromic region (ARNT) using primer pairs Cent PCR and Tel PCR, respectively. A known inverted repeat (IR) was assessed to confirm enrichment of palindromic sequences by GAPF. Also assessed was ERP1 DNA not processed through GAPF (gDNA) and processed through the standard GAPF protocol (GAPF). The inferred orientations of the de novo palindromes are shown below the maps of the restriction enzymes, each placing the palindromic junction at the wavelet-derived breakpoint (P < 0.1).
Association between GAPF-positive regions and copy-number gain in breast cancer cell lines
We also examined and compared GAPF and copy-number profiles of chromosomes 8, 11, and 12 of four breast-cancer cell lines, each with different clinicopathological characteristics, specifically estrogen receptor (ER) expression and amplification of ERBB2. The aforementioned MCF7 cell line is ER positive, and ERBB2 negative. The BT474 and UACC893 cell lines are both ERBB2 positive, but the former is ER positive, while the latter is ER negative. Finally, the MDA231 cell line is both ER negative and ERBB2 negative. GAPF profiles of chromosomes 8, 11, and 12 were generated and compared with the profile of the aforementioned PBLs. We identified GAPF-positive regions unique to the cell lines with the same metrics used previously in this report. To locate copy-number gains and associated breakpoints in the cell lines, we obtained copy-number data, generated on Affymetrix SNP arrays from the Wellcome Trust Sanger Institute Cancer Genome Project website (http://www.sanger.ac.uk/genetics/CGP), and performed wavelet-based analyses (see Methods).
In these cell lines, we observed GAPF-positive regions distributed throughout chromosomes 8, 11, and 12, a subset of which colocated with copy-number gains. GAPF-positive regions were distributed both in clusters of at least three regions and independently along these chromosomes (Fig. 4B; Supplemental Table 5). The percentage of GAPF-positive regions that were located in copy-number gains varied appreciably between the cell lines (Table 1). For example, in UACC893 44% or 19 of the 43 GAPF-positive regions were associated with copy-number gains, whereas in MDA231, none of the 36 GAPF-positive regions colocated with gains. Overall, none of the cell lines had significant associations between GAPF-positive regions and copy-number gains and breakpoints at a P-value <0.05 as determined by simulation-based analyses (see Methods; Table 1). Most of the GAPF-positive regions associated with copy-number gains were located in recurrently amplified regions of 8q and 11q centered on the oncogenes MYC (8q24) and CCND1 (11q13), respectively (Supplemental Fig. 4). For example, we detected GAPF-positive regions in 8q copy-number gains in three of the four cell lines (MCF7, BT474, and UACC893), which included the novel palindrome in 8q24.21 in MCF7. In addition, BT474 and UACC893 had GAPF-positive regions colocated with copy-number gains in 11q13 both at the breakpoints and interspersed throughout (Supplemental Fig. 4B). On the whole, these results indicate that palindrome formation might have an important role in the development of copy-number gain of these key regions in breast cancer.
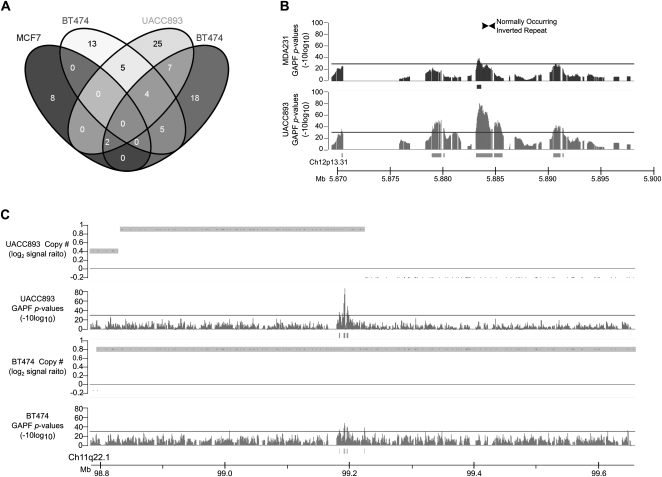
To identify palindromes that might represent precursors of amplification, we compared GAPF profiles between the four cell lines to detect GAPF-positive regions that were both present at the same genomic locations in multiple samples and consistently colocated with copy-number gains. We did not identify any GAPF-positive regions that were shared across all lines, but several regions were common to at least two or three cell lines (Fig. 6A). One of the GAPF-positive regions shared between ER-negative cell lines UACC893 and MDA231 contained an ∼700-bp, normally occurring IR (Fig. 6B). Short IRs can be an originating sequence for large palindrome formation (Yao et al. 1990; Butler et al. 1996; Tanaka et al. 2002; Narayanan et al. 2006; Mizuno et al. 2009; Paek et al. 2009); therefore, this GAPF-positive region could be a palindrome whose formation was initiated at this IR. This shared GAPF-positive region was the only one that overlapped with a known IR. In addition, we detected four GAPF-positive regions that were shared between the ERBB2-positive cell lines UACC893 and BT474. All four of these regions were associated with copy-number gain. One shared GAPF-positive region, located in the CNTN5 gene on chromosome 11, marked the telomeric boundary of a segment with increased copy-number in UACC893 and was located ∼200 kb from a centromeric breakpoint of a copy-number gain in BT474 (Fig. 6C). This region was eliminated after cycled GAPF (Supplemental Fig. 1); however, we were unable to locate the palindromic center based on restriction mapping or confirm that the detected rearrangement was palindromic, suggesting that the rearrangements of this region in UACC893 and BT474 are complex. Overall, we have identified several shared, copy-number-gain-associated GAPF-positive regions in cell lines with similar characteristics (e.g., ERBB2 amplification). These shared GAPF-positive regions offer the potential to evaluate palindromes as recurrent precursors of amplification of chromosomes 8, 11, and 12 in breast cancer.
Figure 6.
Shared GAPF-positive regions in breast cancer cell lines. (A) Venn diagram depicting the number of overlapping GAPF-positive regions (P <0.001, log2 signal ratio > 1.5) between four breast cancer cell lines MCF7, BT474, UACC893, and MDA231. (B) GAPF-positive region in 12p13 shared between UACC893 (lower graph) and MDA231 (upper graph) containing a normally occurring inverted repeat (Warburton et al. 2004). Graphs display P-values (−10log10) of GAPF analyses comparing cell lines with normal PBL reference. The solid bars under the graphs mark GAPF-positive regions (P-value [−10log10] > 30, run > 50 bp, gap < 100 bp). (C) A GAPF-positive region was present in UACC893 (upper graphs) and BT474 (lower graphs) at the same location and associated with copy-number gain. Graphs display GAPF P-values (−10log10) and wavelet-derived copy-number segments (average log2 signal ratio). Copy-number gains are defined by the shaded boxes (log2 signal ratio > 0.3).
Discussion
In this study, we assessed palindrome formation in cancer genomes, focusing on breast cancers, using our microarray-based assay, GAPF, and spatially associated GAPF-positive regions with copy-number gains and amplifications. The data presented here support a role for palindrome-associated mechanisms of amplification in the development of cancer, particularly breast cancer. We observed GAPF-positive regions (i.e., putative palindromes) distributed nonrandomly, often in clusters that colocated with amplicon breakpoints, suggestive of BFB cycles. For example, we indentified GAPF-positive regions clustered in amplicons and at amplicon boundaries on chromosomes 1, 17, and 20 in MCF7 cells, consistent with published paired-end sequencing analyses of the MCF7 genome (Raphael et al. 2003; Volik et al. 2006), which reflect iterative BFB cycles occurring across the genome. Furthermore, we identified genomic regions with GAPF-positive regions located throughout segments of copy-number gain, though not localized to the breakpoint regions. Given the propensity for palindromic sequences to induce genomic instability, these novel palindromes could be initiating the generation of low copy-number increases by mechanisms such as erroneous homologous recombination or DNA replication. On the whole, these data suggest that palindrome formation is a frequent occurrence and is often associated with the development of copy-number gain and amplification in a subset of breast cancers.
In addition, a small subset of GAPF-positive regions were located at the same genomic location in separate samples. Notably, the samples with shared positive regions typically had similar phenotypes (e.g., ERBB2 positive). For example, we identified four positive regions associated with copy-number gain that were common to the two ERBB2-positive cell lines, UACC893 and BT474, including one region located near the boundaries of copy-number gains in 11q. Given that palindrome formation can be a rate-limiting step in amplification (Tanaka and Yao 2009), recurrent palindromes could represent precursors of amplification in breast tumorigenesis. To our knowledge this study is one of the first presentations of a conserved mechanism that could be creating recurrent copy-number gain of these critical regions in breast cancers.
In our initial analysis of palindromes in breast cancers, we selected a small set of tumors and cell lines from phenotypically distinct subtypes based on ER expression and ERBB2 amplification. To obtain a more comprehensive assessment of palindrome formation, there is a need to profile palindromes in much larger sets of primary tumors and to determine whether those profiles cluster into groups that overlap with known subtypes or define novel subtypes. In our study, one commonality between the breast-cancer samples with many amplification-associated GAPF-positive regions was that they all had focal, high-level amplification of multiple portions of their genomes. Related studies have examined global levels of copy-number instability as a means to differentiate novel subtypes of breast cancer (Hicks et al. 2006; Russnes et al. 2010). The investigators proposed that different patterns of amplification are reflective of distinct mechanisms in the evolution of cancer genomes. From these analyses, they defined a novel subtype, termed “firestorm,” which was distinguished by having at least one region in the genome with narrow, closely spaced, high-level amplicons. Similarly, in a recent study, Jonsson et al. (2010) identified a group of breast cancers that was characterized by frequent, high-level amplifications, particularly in 8p12, which they termed the “amplifier” subtype. Both of these groups of investigators compared their amplification-based subtypes with gene-expression subtypes, originally described by Perou et al. (2000). They discovered that their firestorm and amplifier groups often included tumors from multiple gene-expression subtypes, revealing a subset of breast tumors that were better distinguished by the presence of high-level amplifications than gene-expression profiles. Based on our findings, we propose that tumors with frequent high-level amplifications (i.e., in the amplifier subtype) might also have a high frequency of palindrome formation and, furthermore, palindrome-associated amplification might have a key role in the development of these tumors.
An unanticipated observation in this study was the differences in the patterns of GAPF-positive regions and their associations with copy-number gains between the breast cancer cell lines as compared with the primary tumors. In the primary IDCs, we observed high associations between positive regions and copy-number gains and amplifications with very few GAPF-positive regions located outside of amplicons. In contrast, the cell lines had less overlap between positive regions and copy-number gain than the IDCs. It is possible that the nonamplified GAPF-positive regions represent palindromes involved in other types of rearrangements such as deletions (Henthorn et al. 1986) and translocations (Kurahashi et al. 2006; Gotter et al. 2007). However, given that the nonamplified, GAPF-positive regions were almost exclusively in the cell lines and not in the primary tumors, they could also represent rearrangements that accumulate with cell line immortalization and/or tissue culturing as opposed to tumorigenesis (Macieira-Coelho 1996). It is not clear at this time whether palindrome formation in cell lines represents processes involved in in vivo tumorigenesis.
In this study we have achieved a high-resolution evaluation of palindromes and copy-number gain; however, we acknowledge that, in addition to the GAPF signals generated by palindrome formation, there are also some GAPF signals in the data that we currently cannot attribute to palindromes. First, as we discussed earlier, regions of the genome with repeat structure, particularly simple repeats such as Alus, LINEs, or short tandem repeats, can obfuscate the identification of palindromes. In our analysis, these repeats have proven to be a source of false positives, but given that repeat sequences can be the site of novel palindrome formation, eliminating them from the analysis might lead to missed palindromes. An additional source of false negatives could be explained by heterogeneity in the samples. Palindromes or copy-number alterations that exist in a subpopulation of cells might not be detected by the methods utilized in this study due to limitations in the sensitivity. To achieve a more comprehensive assessment of palindromes, we propose adapting GAPF to utilize high-throughput sequencing modalities as opposed to microarray-based platforms.
In summary, we have demonstrated from our integrative analysis of GAPF profiles with copy-number profiles that putative palindromes predominantly cluster in copy-number gains and often colocate with amplicon breakpoints, implicating palindrome-associated mechanisms of amplification. Furthermore, we have identified regions that are susceptible to palindrome formation, including both larger chromosomal regions that are commonly amplified in breast cancer and specific genomic locations. This study has expanded our understanding of the mechanisms creating copy-number gain and amplification in breast cancer, particularly of regions involved in breast tumorigenesis, such as 8q and 11q. It has also opened the door for future comprehensive assessments of the potential of palindromes as early markers in tumorigenesis. In addition, these findings have highlighted the molecular complexity of breast cancer, demonstrating that palindrome formation is another genomic rearrangement that varies in frequency and location in different subtypes of tumors and palindromic profiles could be useful in further refining and defining this disease.
Methods
Cell lines and cancer tissues
MCF7, BT474, UACC893, and MDA231 cells were obtained from American Type Culture Collection. Pre-existing, de-identified invasive ductal carcinomas were obtained from the Breast Specimen Repository and Registry (University of Washington/Fred Hutchinson Cancer Research Center) in accordance with IRB protocol. All cell lines and tumors in this study were from female individuals. The genomic DNA from peripheral blood lymphocytes (PBLs) was extracted by and obtained from Promega Corporation and represents DNA from six to seven female individuals.
GAPF assay
The GAPF assay was performed with 50% formamide as described previously (Diede et al. 2010a) with the following modifications. For the genome-wide GAPF analyses, 2 μg of genomic DNA from Colo320DM cells were split and digested with 10 U of SbfI or KpnI. Two micrograms of genomic DNA from MCF7 cells were split evenly three ways and digested with 10 U of SbfI, KpnI, or PmeI. For the GAPF analyses of breast-cancer cell lines and primary tumors of chromosomes 8, 11, and 12, 2 μg of genomic DNA were split evenly and digested with 10 U of SbfI or PmeI. Two different reference genomes were used in this study. Cultured human fibroblasts were used for genome-wide comparisons with Colo320DM and MCF7. For the GAPF analysis of breast cancer cell lines and primary tumors, genomic DNA from PBLs was used as the normal reference. All arrays in this study were run as singletons. Cancer and normal-reference arrays were matched and run on the same day to avoid batch effects. DNA sequences of the primers used in this study are available upon request.
GAPF statistical analysis
Affymetrix Human Tiling 2.0R Arrays were analyzed using Tiling Array Software (Affymetrix). Probe locations were mapped using the NCBI36/hg18 genome build from March 2006. Raw-intensity data were scaled to a target intensity of 100 and normalized by quantile normalization. Normalized probe intensities were analyzed by Wilcoxon rank sum one-sided test, detecting probes with intensities significantly different between the cancer and normal reference samples. The probe analysis for determining signal ratios and P-values was performed using a bandwidth of 500 bp. Regions were assigned as GAPF positive if the P-value was <0.001 and log2 signal ratio >1.5 with a minimum contiguous run of significant probes of 50 bp and with a <100-bp gap between runs. Also, GAPF-positive regions that mapped to simple tandem repeats (STRs), identified by Benson (1999), were removed. GAPF-positive regions within 10 kb of other regions were grouped together. These data were viewed using the Integrated Genome Browser (Affymetrix, version 1.0).
Clustering analysis
The probabilities that GAPF-positive regions occur in clusters for window sizes of 5 and 10 Mb were calculated assuming that GAPF-positive regions are randomly distributed. A cluster here was defined as at least three GAPF-positive regions. For each sample, we defined the average size of the GAPF-positive regions as a unit. We then divided the length of the chromosome by the average size to obtain the number of units on the chromosome. The probability that a unit could be hit by a GAPF-positive region, denoted by p, was calculated by dividing the actual number of GAPF-positive regions detected by the total number of the units. For a fixed window size, we calculated the number of units, denoted by n, in that window size. We assumed that all units had an equal chance of being hit by a GAPF-positive region (i.e., the number of GAPF-positive regions was assumed to be distributed according to a binomial distribution with n and p.) The probability of observing at least three GAPF-positive regions was calculated by 1 − Prob(observed no hit) − Prob(observed 1 hit) − Prob(observed 2 hits). Specifically, Prob(observed no hit) = (1 − p)n, Prob(observed 1 hit) = np × (1 − p)(n − 1), and Prob(observed two hits) = n × (n − 1)/2 × p2 × (1 − p)(n − 2).
Copy number analysis
Copy-number data were generated on Affymetrix Genome-Wide Human SNP Arrays 6.0. The raw data for MCF7, BT474, UACC893, and MDA231 were obtained from the Wellcome Trust Sanger Institute Cancer Genome Project website (http://www.sanger.ac.uk/genetics/CGP). Genomic DNA from Colo320DM and the six IDCs were processed and hybridized onto the Affymetrix Genome-wide Human SNP Array 6.0 in the Gene Expression and Genotyping Facility at Case Comprehensive Cancer Center in accordance with the manufacturer's protocols (http://www.affymetrix.com). Raw-intensity data were normalized with the Genotyping Console (Affymetrix, v4.0) and compared with a reference file generated from pooled HapMap individuals to generate log2 signal ratios. Copy-number breakpoints and segments were detected using multiscale wavelet products at P < 0.1 (Yu et al. 2010). Segments with average log2 signal ratios >0.3 were designated as copy-number gains and segments <5 Mb in size with an average log2 signal ratio ≥1.0 were designated as amplifications.
Statistical analysis of associations between GAPF-positive regions and amplicons or amplicon breakpoints
The statistical significance of the associations between GAPF-positive regions and copy-number gains and breakpoints was assessed for each sample. The significance was determined by comparing the observed number of overlapping regions against a null distribution that was obtained by simulation. First, the locations of GAPF-positive regions were randomly assigned using the number and the mean size of GAPF-positive regions observed. Next, the locations of the simulated GAPF-positive regions were compared with the actual locations of copy-number gains identified from the copy-number data, and the number of overlapping regions was counted. These steps were repeated 10,000 times, generating the null distribution of the number of overlapping regions assuming no association between GAPF-positive regions and copy-number gains. Finally, the observed number of overlapping GAPF-positive regions and copy-number gains detected was compared with this null distribution, and the P-value was calculated by the frequency among the 10,000 simulated runs that the number of overlaps was greater than or equal to the actual overlap. This simulation-based approach was also used to assess the statistical significance of the associations between GAPF-positive regions and copy-number gain breakpoints. The breakpoints were expanded to 100-kb regions centered on the midpoint of the breakpoint to account for the resolution of the probe locations on the SNP arrays.
PCR analysis of GAPF enrichment
The enrichment of specific genomic loci over a nonpalindromic region (ARNT) following GAPF was accomplished by PCR-based analysis. This analysis was also used for quality-control testing of GAPF preparations prior to processing for hybridization to tiling arrays. Fifty-microliter PCR reactions contained 5 μL of 10× PCR buffer (FastStart Taq polymerase; Roche), 1 μL of dNTP mix (10 mM; Roche), 10 μL of 5× GC-rich solution (Roche), 0.5 μL each of forward and reverse primers (50 μM), 0.5 μL of FastStart Taq Polymerase (Roche) and ddH2O. Primers to the ARNT region were included in each reaction, which should produce little to no product. PCR conditions were as follows: 6 min at 96°C; 30 to 32 cycles of 30 sec at 96°C, 30 sec at 55°C, 30 sec at 72°C, and 7 min at 72°C. PCR products were run on 1.5% agarose gels for 1 h at 90V and the relative amounts of PCR products were assessed qualitatively. DNA sequences of the primers used in this study are available upon request.
Southern Blotting
Southern blotting and Snapback Southerns were performed as previously described (Tanaka et al. 2005).
Data access
The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov) under accession no. GSE29876.
Acknowledgments
This study was supported by the National Institutes of Health grant NCI RO1 CA098415 and by the FHCRC/UW Cancer Consortium Breast Cancer Pilot Project Fund made possible through a donation from Safeway. J.G. was supported as a FHCRC predoctoral, interdisciplinary fellow (NIH T32 CA80416). S.J.D. was supported as an American Society for Clinical Oncology (ASCO) Young Investigator, as a University of Washington Child Health Research Center Scholar (NIH grant K12 HD43376), and by the National Institutes of Health under Ruth L. Kirschstein National Research Service Award T32CA009351. H.T. was supported by NCI ROICA1493835. We thank Barbara Stein for tissue procurement support, Kelly Wirtala and Alyssa Dawson for technical contributions, and the Shared Genomic Resource (FHCRC) and the Gene Expression and Genotyping Facility (Case Comprehensive Cancer Center supported by P30 CA43703).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.117226.110.
References
- Benson G 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27: 573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi NO, Bianchi MS, Kere J 1991. DNA discontinuities in the domain of amplified human MYC oncogenes. Genes Chromosomes Cancer 3: 136–141 [DOI] [PubMed] [Google Scholar]
- Burkhardt L, Grob TJ, Hermann I, Burandt E, Choschzick M, Janicke F, Muller V, Bokemeyer C, Simon R, Sauter G, et al. 2009. Gene amplification in ductal carcinoma in situ of the breast. Breast Cancer Res Treat 123: 757–765 [DOI] [PubMed] [Google Scholar]
- Butler DK, Yasuda LE, Yao MC 1996. Induction of large DNA palindrome formation in yeast: implications for gene amplification and genome stability in eukaryotes. Cell 87: 1115–1122 [DOI] [PubMed] [Google Scholar]
- Butler DK, Gillespie D, Steele B 2002. Formation of large palindromic DNA by homologous recombination of short inverted repeat sequences in Saccharomyces cerevisiae. Genetics 161: 1065–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, et al. 2006. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 10: 529–541 [DOI] [PubMed] [Google Scholar]
- Ciullo M, Debily MA, Rozier L, Autiero M, Billault A, Mayau V, El Marhomy S, Guardiola J, Bernheim A, Coullin P, et al. 2002. Initiation of the breakage-fusion-bridge mechanism through common fragile site activation in human breast cancer cells: the model of PIP gene duplication from a break at FRA7I. Hum Mol Genet 11: 2887–2894 [DOI] [PubMed] [Google Scholar]
- Coquelle A, Pipiras E, Toledo F, Buttin G, Debatisse M 1997. Expression of fragile sites triggers intrachromosomal mammalian gene amplification and sets boundaries to early amplicons. Cell 89: 215–225 [DOI] [PubMed] [Google Scholar]
- Corzo C, Corominas JM, Tusquets I, Salido M, Bellet M, Fabregat X, Serrano S, Sole F 2006. The MYC oncogene in breast cancer progression: from benign epithelium to invasive carcinoma. Cancer Genet Cytogenet 165: 151–156 [DOI] [PubMed] [Google Scholar]
- Courjal F, Theillet C 1997. Comparative genomic hybridization analysis of breast tumors with predetermined profiles of DNA amplification. Cancer Res 57: 4368–4377 [PubMed] [Google Scholar]
- Deming SL, Nass SJ, Dickson RB, Trock BJ 2000. C-myc amplification in breast cancer: a meta-analysis of its occurrence and prognostic relevance. Br J Cancer 83: 1688–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePinho RA, Schreiber-Agus N, Alt FW 1991. myc family oncogenes in the development of normal and neoplastic cells. Adv Cancer Res 57: 1–46 [DOI] [PubMed] [Google Scholar]
- Dickson C, Fantl V, Gillett C, Brookes S, Bartek J, Smith R, Fisher C, Barnes D, Peters G 1995. Amplification of chromosome band 11q13 and a role for cyclin D1 in human breast cancer. Cancer Lett 90: 43–50 [DOI] [PubMed] [Google Scholar]
- Diede SJ, Guenthoer J, Geng LN, Mahoney SE, Marotta M, Olson JM, Tanaka H, Tapscott SJ 2010a. DNA methylation of developmental genes in pediatric medulloblastomas identified by denaturation analysis of methylation differences. Proc Natl Acad Sci 107: 234–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede SJ, Tanaka H, Bergstrom DA, Yao MC, Tapscott SJ 2010b. Genome-wide analysis of palindrome formation. Nat Genet 42: 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford M, Fried M 1986. Large inverted duplications are associated with gene amplification. Cell 45: 425–430 [DOI] [PubMed] [Google Scholar]
- Forozan F, Mahlamaki EH, Monni O, Chen Y, Veldman R, Jiang Y, Gooden GC, Ethier SP, Kallioniemi A, Kallioniemi OP 2000. Comparative genomic hybridization analysis of 38 breast cancer cell lines: a basis for interpreting complementary DNA microarray data. Cancer Res 60: 4519–4525 [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR 2004. A census of human cancer genes. Nat Rev Cancer 4: 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson D, Pettersson L, Hoglund M, Heidenblad M, Gorunova L, Wiegant J, Mertens F, Dal Cin P, Mitelman F, Mandahl N 2000. Chromosomal breakage-fusion-bridge events cause genetic intratumor heterogeneity. Proc Natl Acad Sci 97: 5357–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter AL, Nimmakayalu MA, Jalali GR, Hacker AM, Vorstman J, Conforto Duffy D, Medne L, Emanuel BS 2007. A palindrome-driven complex rearrangement of 22q11.2 and 8q24.1 elucidated using novel technologies. Genome Res 17: 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman A, Zlotorynski E, Scherer SW, Cheung J, Vincent JB, Smith DI, Trakhtenbrot L, Kerem B 2002. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell 1: 89–97 [DOI] [PubMed] [Google Scholar]
- Henthorn PS, Mager DL, Huisman TH, Smithies O 1986. A gene deletion ending within a complex array of repeated sequences 3′ to the human beta-globin gene cluster. Proc Natl Acad Sci 83: 5194–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, Leibu E, Esposito D, Alexander J, Troge J, Grubor V, et al. 2006. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res 16: 1465–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, Ringner M, Sauter G, Monni O, Elkahloun A, et al. 2002. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res 62: 6240–6245 [PubMed] [Google Scholar]
- Isola JJ, Kallioniemi OP, Chu LW, Fuqua SA, Hilsenbeck SG, Osborne CK, Waldman FM 1995. Genetic aberrations detected by comparative genomic hybridization predict outcome in node-negative breast cancer. Am J Pathol 147: 905–911 [PMC free article] [PubMed] [Google Scholar]
- Jonsson G, Staaf J, Vallon-Christersson J, Ringner M, Holm K, Hegardt C, Gunnarsson H, Fagerholm R, Strand C, Agnarsson BA, et al. 2010. Genomic subtypes of breast cancer identified by array comparative genomic hybridization display distinct molecular and clinical characteristics. Breast Cancer Res 12: R42 doi: 10.1186/bcr2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Piper J, Tanner M, Stokke T, Chen L, Smith HS, Pinkel D, Gray JW, Waldman FM 1994. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci 91: 2156–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuutila S, Bjorkqvist AM, Autio K, Tarkkanen M, Wolf M, Monni O, Szymanska J, Larramendy ML, Tapper J, Pere H, et al. 1998. DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol 152: 1107–1123 [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H, Inagaki H, Ohye T, Kogo H, Kato T, Emanuel BS 2006. Palindrome-mediated chromosomal translocations in humans. DNA Repair 5: 1136–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letessier A, Sircoulomb F, Ginestier C, Cervera N, Monville F, Gelsi-Boyer V, Esterni B, Geneix J, Finetti P, Zemmour C, et al. 2006. Frequency, prognostic impact, and subtype association of 8p12, 8q24, 11q13, 12p13, 17q12, and 20q13 amplifications in breast cancers. BMC Cancer 6: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Alitalo K, Schwab M, George D, Varmus HE, Bishop JM 1985. Evolution of karyotypic abnormalities and C-MYC oncogene amplification in human colonic carcinoma cell lines. Chromosoma 92: 11–15 [DOI] [PubMed] [Google Scholar]
- Lo AW, Sabatier L, Fouladi B, Pottier G, Ricoul M, Murnane JP 2002. DNA amplification by breakage/fusion/bridge cycles initiated by spontaneous telomere loss in a human cancer cell line. Neoplasia 4: 531–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193 [DOI] [PubMed] [Google Scholar]
- Loo LW, Grove DI, Williams EM, Neal CL, Cousens LA, Schubert EL, Holcomb IN, Massa HF, Glogovac J, Li CI, et al. 2004. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res 64: 8541–8549 [DOI] [PubMed] [Google Scholar]
- Lu YJ, Osin P, Lakhani SR, Di Palma S, Gusterson BA, Shipley JM 1998. Comparative genomic hybridization analysis of lobular carcinoma in situ and atypical lobular hyperplasia and potential roles for gains and losses of genetic material in breast neoplasia. Cancer Res 58: 4721–4727 [PubMed] [Google Scholar]
- Ma C, Martin S, Trask B, Hamlin JL 1993. Sister chromatid fusion initiates amplification of the dihydrofolate reductase gene in Chinese hamster cells. Genes Dev 7: 605–620 [DOI] [PubMed] [Google Scholar]
- Macieira-Coelho A 1996. Proliferative cell senescence, transformation, and the recombination potential of the genome. Exp Gerontol 31: 227–234 [DOI] [PubMed] [Google Scholar]
- McClintock B 1941. The stability of broken ends of chromosomes in Zea Mays. Genetics 26: 234–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Lambert S, Baldacci G, Murray JM, Carr AM 2009. Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes Dev 23: 2876–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane JP, Sabatier L 2004. Chromosome rearrangements resulting from telomere dysfunction and their role in cancer. Bioessays 26: 1164–1174 [DOI] [PubMed] [Google Scholar]
- Narayanan V, Mieczkowski PA, Kim HM, Petes TD, Lobachev KS 2006. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell 125: 1283–1296 [DOI] [PubMed] [Google Scholar]
- Nessling M, Richter K, Schwaenen C, Roerig P, Wrobel G, Wessendorf S, Fritz B, Bentz M, Sinn HP, Radlwimmer B, et al. 2005. Candidate genes in breast cancer revealed by microarray-based comparative genomic hybridization of archived tissue. Cancer Res 65: 439–447 [PubMed] [Google Scholar]
- Paek AL, Kaochar S, Jones H, Elezaby A, Shanks L, Weinert T 2009. Fusion of nearby inverted repeats by a replication-based mechanism leads to formation of dicentric and acentric chromosomes that cause genome instability in budding yeast. Genes Dev 23: 2861–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. 2000. Molecular portraits of human breast tumours. Nature 406: 747–752 [DOI] [PubMed] [Google Scholar]
- Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO 2002. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci 99: 12963–12968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice LM, Shadeo A, Lestou VS, Miller MA, deLeeuw RJ, Makretsov N, Turbin D, Brown LA, Macpherson N, Yorida E, et al. 2005. NRG1 gene rearrangements in clinical breast cancer: identification of an adjacent novel amplicon associated with poor prognosis. Oncogene 24: 7281–7289 [DOI] [PubMed] [Google Scholar]
- Raphael BJ, Volik S, Collins C, Pevzner PA 2003. Reconstructing tumor genome architectures. Bioinformatics 19: ii162–ii171 doi: 10.1093/bioinformatics/btg1074 [DOI] [PubMed] [Google Scholar]
- Rennstam K, Ahlstedt-Soini M, Baldetorp B, Bendahl PO, Borg A, Karhu R, Tanner M, Tirkkonen M, Isola J 2003. Patterns of chromosomal imbalances defines subgroups of breast cancer with distinct clinical features and prognosis. A study of 305 tumors by comparative genomic hybridization. Cancer Res 63: 8861–8868 [PubMed] [Google Scholar]
- Reshmi SC, Huang X, Schoppy DW, Black RC, Saunders WS, Smith DI, Gollin SM 2007. Relationship between FRA11F and 11q13 gene amplification in oral cancer. Genes Chromosomes Cancer 46: 143–154 [DOI] [PubMed] [Google Scholar]
- Robanus-Maandag EC, Bosch CA, Kristel PM, Hart AA, Faneyte IF, Nederlof PM, Peterse JL, van de Vijver MJ 2003. Association of C-MYC amplification with progression from the in situ to the invasive stage in C-MYC-amplified breast carcinomas. J Pathol 201: 75–82 [DOI] [PubMed] [Google Scholar]
- Rummukainen J, Kytola S, Karhu R, Farnebo F, Larsson C, Isola JJ 2001. Aberrations of chromosome 8 in 16 breast cancer cell lines by comparative genomic hybridization, fluorescence in situ hybridization, and spectral karyotyping. Cancer Genet Cytogenet 126: 1–7 [DOI] [PubMed] [Google Scholar]
- Russnes HG, Vollan HK, Lingjaerde OC, Krasnitz A, Lundin P, Naume B, Sorlie T, Borgen E, Rye IH, Langerod A et al. 2010. Genomic architecture characterizes tumor progression paths and fate in breast cancer patients. Sci Transl Med 2: 38ra47 doi: 10.1126/scitranslmed.3000611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Shuster M, Huang X, Gharaibeh B, Enyenihi AH, Petersen I, Gollin SM 2000. Chromosomal instability and cytoskeletal defects in oral cancer cells. Proc Natl Acad Sci 97: 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N, Shingaki K, Kaneko-Sasaguri Y, Hashizume T, Kanda T 2005. When, where and how the bridge breaks: anaphase bridge breakage plays a crucial role in gene amplification and HSR generation. Exp Cell Res 302: 233–243 [DOI] [PubMed] [Google Scholar]
- Shuster MI, Han L, Le Beau MM, Davis E, Sawicki M, Lese CM, Park NH, Colicelli J, Gollin SM 2000. A consistent pattern of RIN1 rearrangements in oral squamous cell carcinoma cell lines supports a breakage-fusion-bridge cycle model for 11q13 amplification. Genes Chromosomes Cancer 28: 153–163 [PubMed] [Google Scholar]
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al. 1989. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 244: 707–712 [DOI] [PubMed] [Google Scholar]
- Smith KA, Stark MB, Gorman PA, Stark GR 1992. Fusions near telomeres occur very early in the amplification of CAD genes in Syrian hamster cells. Proc Natl Acad Sci 89: 5427–5431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Yao MC 2009. Palindromic gene amplification–an evolutionarily conserved role for DNA inverted repeats in the genome. Nat Rev Cancer 9: 216–224 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Tapscott SJ, Trask BJ, Yao MC 2002. Short inverted repeats initiate gene amplification through the formation of a large DNA palindrome in mammalian cells. Proc Natl Acad Sci 99: 8772–8777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Bergstrom DA, Yao MC, Tapscott SJ 2005. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nat Genet 37: 320–327 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Cao Y, Bergstrom DA, Kooperberg C, Tapscott SJ, Yao MC 2007. Intrastrand annealing leads to the formation of a large DNA palindrome and determines the boundaries of genomic amplification in human cancer. Mol Cell Biol 27: 1993–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanHulle K, Lemoine FJ, Narayanan V, Downing B, Hull K, McCullough C, Bellinger M, Lobachev K, Petes TD, Malkova A 2007. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol Cell Biol 27: 2601–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volik S, Raphael BJ, Huang G, Stratton MR, Bignel G, Murnane J, Brebner JH, Bajsarowicz K, Paris PL, Tao Q, et al. 2006. Decoding the fine-scale structure of a breast cancer genome and transcriptome. Genome Res 16: 394–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Giordano J, Cheung F, Gelfand Y, Benson G 2004. Inverted repeat structure of the human genome: the X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res 14: 1861–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M, Mattis A, Aubele M, Cummings M, Zitzelsberger H, Hutzler P, Hofler H 1999. 20q13.2 amplification in intraductal hyperplasia adjacent to in situ and invasive ductal carcinoma of the breast. Virchows Arch 435: 469–472 [DOI] [PubMed] [Google Scholar]
- Yao MC, Yao CH, Monks B 1990. The controlling sequence for site-specific chromosome breakage in Tetrahymena. Cell 63: 763–772 [DOI] [PubMed] [Google Scholar]
- Yao J, Weremowicz S, Feng B, Gentleman RC, Marks JR, Gelman R, Brennan C, Polyak K 2006. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res 66: 4065–4078 [DOI] [PubMed] [Google Scholar]
- Yasuda LF, Yao MC 1991. Short inverted repeats at a free end signal large palindromic DNA formation in Tetrahymena. Cell 67: 505–516 [DOI] [PubMed] [Google Scholar]
- Yu X, Randolph TW, Tang H, Hsu L 2010. Detecting breakpoints using multi-scale wavelet products. Biometrics 66: 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]