Abstract
While genetic mutation is a hallmark of cancer, many cancers also acquire epigenetic alterations during tumorigenesis including aberrant DNA hypermethylation of tumor suppressors, as well as changes in chromatin modifications as caused by genetic mutations of the chromatin-modifying machinery. However, the extent of epigenetic alterations in cancer cells has not been fully characterized. Here, we describe complete methylome maps at single nucleotide resolution of a low-passage breast cancer cell line and primary human mammary epithelial cells. We find widespread DNA hypomethylation in the cancer cell, primarily at partially methylated domains (PMDs) in normal breast cells. Unexpectedly, genes within these regions are largely silenced in cancer cells. The loss of DNA methylation in these regions is accompanied by formation of repressive chromatin, with a significant fraction displaying allelic DNA methylation where one allele is DNA methylated while the other allele is occupied by histone modifications H3K9me3 or H3K27me3. Our results show a mutually exclusive relationship between DNA methylation and H3K9me3 or H3K27me3. These results suggest that global DNA hypomethylation in breast cancer is tightly linked to the formation of repressive chromatin domains and gene silencing, thus identifying a potential epigenetic pathway for gene regulation in cancer cells.
Breast cancer is characterized by both genetic and epigenetic alterations (Sjoblom et al. 2006; Esteller 2007, 2008; Wood et al. 2007; Stephens et al. 2009). While a large number of genetic mutations are linked to breast cancer, there is clear evidence that epigenetic alterations, such as hypo- or hypermethylation of DNA, occur early in the initiation or development of the tumors (Kohonen-Corish et al. 2007). Some genes commonly hypermethylated in breast cancers are involved in evasion of apoptosis (RASSF1, HOXA5, TWIST1) and cellular senescence (CCND2, CDKN2A), while others regulate DNA repair (BRCA1), cell growth (ESR1, PGR), and tissue invasion (CDH1) (Dworkin et al. 2009; Jovanovic et al. 2010). Further underscoring the role of the epigenetic mechanisms in tumorigenesis, such epigenetic events have been exploited for early diagnosis or treatment. For example, a therapeutic strategy blocking DNA methylation with 5-azacytidine (Jones et al. 1983) has been approved for treatment of preleukemic myelodysplastic syndrome (Kaminskas et al. 2005) and is in clinical trials for several forms of cancer (Kelly et al. 2010).
DNA methylation (Holliday 1979; Feinberg and Vogelstein 1983; Laird 2003, 2010) is the most studied epigenetic event in cancer. Bisulfite sequencing (Frommer et al. 1992) of targeted loci such as the breast cancer susceptibility gene BRCA1 (Tapia et al. 2008) supports the notion that tumor suppressors are frequently inactivated by DNA methylation at CpG islands and promoters. Genome-scale methods including MeDIP-seq (Ruike et al. 2010) and CHARM (Irizarry et al. 2009) have confirmed global hypomethylation and focal hypermethylation as hallmarks of breast and colon cancer. More recently, whole genome shotgun bisulfite sequencing offers single-nucleotide resolution of DNA methylation in human cells (Lister et al. 2009). This unprecedented resolution has revealed that cytosines methylated in the CG context (mCG) are nearly completely methylated in pluripotent cells but are frequently in a partially methylated state in somatic cells. These partially methylated cytosines are clustered to form partially methylated domains (PMDs), which can span nearly 40% the genome (Lister et al. 2009). Interestingly, genes within PMDs are found to be generally repressed, though the mechanism is unclear (Lister et al. 2011). This method has also recently been used to observe increased epigenetic variation at hypomethylated regions in cancer cells (Hansen et al. 2011). Finally, further underscoring the role of DNA methylation in cancer, a recent genetic study showed that mutations in the DNA methyltransferase DNMT3A are frequently found in acute myeloid leukemia (Ley et al. 2010).
Chromatin state can also be altered in cancer cells (Parsons et al. 2010; Jiao et al. 2011; Varela et al. 2011). For example, heterochromatin-associated H3K9me3 and Polycomb-associated H3K27me3 mark large repressed domains in somatic cells (Hawkins et al. 2010) that are misregulated in cancer. H3K9me3 is deposited by a family of histone methyltransferases including SUV39H1, inhibition of which in acute myeloid leukemia cells is sufficient for re-expression of the transcriptionally silenced tumor suppressor genes CDKN2B and CDH1 marked by H3K9me3 (Lakshmikuttyamma et al. 2009). EZH2, the enzyme responsible for depositing H3K27me3, is often overexpressed in aggressive breast cancers (Kleer et al. 2003; Chang et al. 2011), and mutations in the H3K27me3 demethylase KDM6A are common in clear cell renal cell carcinoma (Dalgliesh et al. 2010). Thus, misregulation of repressive chromatin modifications may also play a role in tumorigenesis.
While both DNA methylation and chromatin modifications have been associated with tumorigenesis, few studies have integrated both aspects on a global scale to investigate their coordinated role in cancer progression. Here, we report use of high-throughput sequencing technology to map DNA methylation at base resolution and two repressive chromatin modifications in a breast cancer cell line and primary mammary epithelial cells. Comparative analysis of the two epigenomes reveals widespread DNA hypomethylation that is tightly coupled to the formation of repressive chromatin domains and gene silencing in the cancer cells. We propose that such large scale alterations of the epigenetic landscape may play an important role in tumorigenesis by inhibiting expression of tumor suppressor genes. We also suggest that global hypomethylation may occur through a passive mechanism. Further, we show that, while hypomethylation of repetitive elements is common, it is not the only explanation for increased transcription from such repetitive sequences.
Results
Global DNA hypomethylation in the breast cancer cell line HCC1954
The HCC1954 cell line is derived from the primary tissue of a ductal breast carcinoma of an East Indian female (Neve et al. 2006). It belongs to the subtype of estrogen receptor (ER)/progesterone receptor (PR) negative and ERBB2 (HER2) positive breast cancers characterized by poor prognosis. We performed whole genome shotgun sequencing of bisulfite-treated DNA [MethylC-seq (Lister et al. 2008, 2009)] in HCC1954, generating 889,012,059 uniquely mapped monoclonal reads, with an average of 27-fold genome coverage. As a control, we also performed MethylC-seq on human mammary epithelial cells (HMECs) at 20-fold coverage.
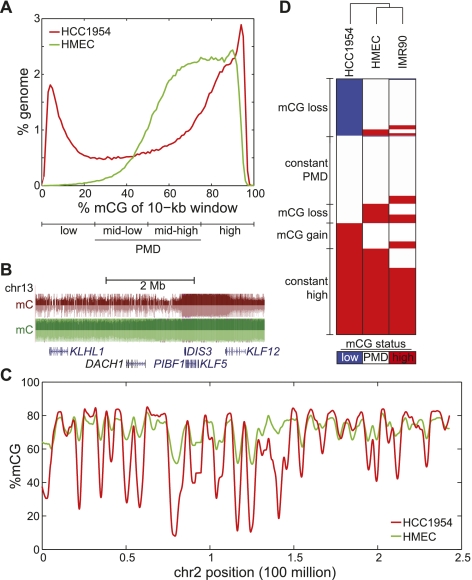
DNA methylation in both HCC1954 and HMEC exists almost exclusively (>99.8%) in the CG context. To compare the global profiles of DNA methylation between HCC1954 and HMEC, we quantified mCG by dividing the genome into 10-kb windows and calculating the percentage of cytosine bases methylated in sequenced reads (%mCG). We found that a large fraction of the HCC1954 genome is differentially methylated as compared to normal cells (Fig. 1A; Supplemental Fig. S1). In agreement with previous studies (Esteller 2008; Ruike et al. 2010), we observed global hypomethylation and local hypermethylation in HCC1954 compared to HMEC. Denoting the four quartiles of %mCG from 0% to 100% as low, mid-low, mid-high, and high (Fig. 1A), we observed that a striking 22.3% of these 10-kb windows have low mCG in HCC1954, compared to 0.64% in HMEC. On the other hand, hypermethylation is more limited, with 3.1% of HCC1954 windows exhibiting %mCG >95%, compared to 0.3% in HMEC.
Figure 1.
Global hypomethylation in breast cancer. (A) Distribution of %mCG for all 10-kb windows in the human genome. Quartiles of increasing %mCG are labeled as low, mid-low, mid-high, and high %mCG, with mid-low and mid-high representing partially methylated domains (PMDs). (B) A large domain of hypomethylation near the DACH1 tumor suppressor. (Red) HCC1954, (green) HMEC. (C) Distribution of %mCG on chromosome 2 for the breast cancer cell line HCC1954 and the normal breast line HMEC. (D) A heatmap of low, PMD, and high %mCG for all 10-kb windows in the human genome. Each of the 282,109 rows represents the mCG status for a 10-kb window in HCC1954, HMEC, and IMR90 fibroblast cells. The dendrogram represents the similarity (Pearson correlation) of the profiles across different cells.
These domains of hypomethylation span known tumor suppressors such as DACH1 (Fig. 1B). On a chromosome-wide view, it is clear that hypomethylated regions form large domains, which are punctuated by regions where DNA methylation is high (Fig. 1C).
Chromosome-wide views also suggest that loci with the most pronounced hypomethylation in HCC1954 coincide with regions that are not fully methylated in HMEC (Fig. 1C). Genome-wide, 88.2% of all low mCG windows in HCC1954 are in a PMD state (defined as mid-low or mid-high %mCG) in HMEC (Fig. 1D). Also, of all hypermethylated HCC1954 regions that are not highly methylated in HMEC, 99.6% belong to the PMD state in HMEC. These results suggest that PMDs in HMEC are the most likely regions to either gain or lose DNA methylation during tumorigenesis.
To test if epigenetically unstable regions in breast cancer (BC) might coincide with those in other cancers, we compared our results to those obtained by a recent study which cataloged epigenetically unstable regions in colon cancer (CC)(Irizarry et al. 2009). Regions showing hypomethylation in CC (N = 994) are also generally hypomethylated in BC: Of 730 hypomethyated CC loci showing methylation bias in BC (Fisher's P-value ≤ 0.01), 537 (73.6%) are hypomethylated compared to 193 that are hypermethylated (Supplemental Fig. S2) (P < 10−100, binomial). In contrast, hypermethylated CC regions (N = 704) tend to be hypermethylated in BC: of 555 hypermethylated CC loci with methylation bias in BC, 485 (87.4%) are hypermethylated and 70 are hypomethylated (Supplemental Fig. S2) (P < 10−100, binomial). This suggests that certain regions of the genome exhibit inherent epigenetic instability and consistently gain or lose mCG in multiple types of cancers. In agreement with Irizarry et al. (2009), overlapping hypo- and hypermethylated genes are enriched for embryonic and developmental proteins, including the reprogramming factor SOX2, neural developmental genes NEGR1 and NRG1, and members of the HOXA cluster (HOXA1 through HOXA7, and HOXA9).
DNA hypomethylation is associated with decreased gene expression in breast cancers
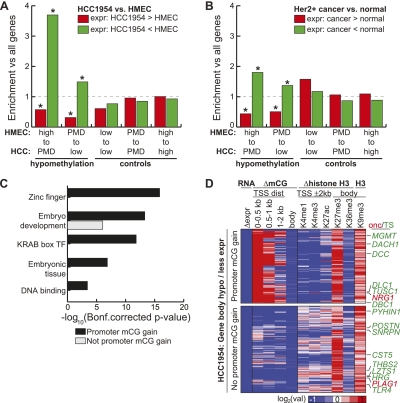
DNA hypermethylation at promoters is generally correlated with gene silencing. Recently, DNA methylation at gene bodies has been shown to be prevalent in mammalian cells, and some evidence has suggested it is correlated with gene activity (Hellman and Chess 2007; Ball et al. 2009), but the mechanisms have not been clearly understood (Jones 1999; Maunakea et al. 2010; Wu et al. 2010). To explore the effect of DNA hypomethylation on gene expression in breast cancer cells, we performed RNA-seq experiments to determine genome-wide steady-state transcript abundance in both HCC1954 and HMEC cells. We then examined the transcript abundance for genes overlapping domains with DNA hypomethylation. Unexpectedly, these genes have a greater tendency to be repressed in HCC1954: While only 3.8% of all genes transition from highly methylated to PMD states, this represents 14.1% of all genes losing expression (phigh→PMD < 10−16, hypergeometric) (Fig. 2A). Likewise, the 220 genes undergoing PMD to low %mCG transition represent 12.1% of all down-regulated genes, compared to 8.1% expected by chance (pPMD→low = 1.08 × 10−9, hypergeometric). In contrast, hypomethylated genes are less likely to gain expression HCC1954 (Fig. 2A) (phigh→PMD = 5.7 × 10−6, pPMD→low = 1.03 × 10−30, hypergeometric). There is also significant overlap between down-regulated, hypomethylated genes in HCC1954 with hypomethylated genes in colon cancer (P = 1.03 × 10−7, hypergeometric).
Figure 2.
Gene body hypomethylation is associated with gene repression. (A) Genes were overlapped with 10-kb domains undergoing hypomethylation (high to PMD, PMD to low) or control (low to low, PMD to PMD, high to high) transitions. Shown is the enrichment of each transition state with genes having HCC1954 expression at least eightfold more (red) or less (green) than HMEC, when compared to the global enrichment of all transition states. (*) P-value ≤ 0.01 (hypergeometric). (B) Using the same transition states in A, but comparing the expression of genes in a panel of 50 ERBB2+ (HER2+) breast cancers compared to a panel of 23 normal breast samples (Weigelt et al. 2005; Oh et al. 2006; Perreard et al. 2006; Herschkowitz et al. 2007, 2008; Hoadley et al. 2007; Mullins et al. 2007; Hennessy et al. 2009; Hu et al. 2009; Parker et al. 2009; Prat et al. 2010). Significantly more (red) or less (green) expressed genes are defined by a Wilcoxon rank sum test (P-value ≤ 0.01) between the expression values of the two different panels. (*) Enrichment P-value ≤ 0.01 (hypergeometric). (C) Functional enrichment of hypomethylated genes having loss of expression by the DAVID analysis tool (Dennis et al. 2003). (D) Epigenetic status of genes undergoing hypomethylation in gene bodies with loss of expression. With the exception of H3K9me3, all values are of HCC1954 relative to HMEC. RNA, log2 (HCC1954 RPKM/HMEC RPKM); ΔmCG, log2 (HCC1954%mCG/HMEC %mCG); Δhistone, log2 (HCC1954 ChIP/input) − log2 (HMEC ChIP/input); H3K9me3, log2 (HCC1954 ChIP/input). (Onc) oncogene, (TS) tumor suppressor.
To test if this pattern of gene expression is representative of a broader set of breast cancers, we examined the gene expression profiles of a panel of 50 ERBB2 positive breast tumors and 23 normal breast samples (Hennessy et al. 2009; Parker et al. 2009; Prat et al. 2010). In this data set, 13,262 genes could be unambiguously assigned to transcripts from our RNA-seq experiments. At 935 of these genes undergoing hypomethylation from HMEC to HCC1954, we similarly observe an enrichment of cancer-specifically repressed genes (phigh→PMD = 8.86 × 10−7, pPMD→low = 1.38 × 10−3, hypergeometric) and a depletion of cancer-specifically expressed genes (Fig. 2B) (phigh→PMD = 2.79 × 10−4, pPMD→low = 4.43 × 10−5, hypergeometric). Interestingly, similar results are also observed when comparing non-ERBB2 positive breast tumors with normal breast samples (data not shown), suggesting that genes undergoing hypomethylation in HCC1954 are generally repressed in breast cancers.
To understand whether specific cellular pathways are particularly affected by the abnormal hypomethylation in HCC1954, we performed gene ontology (GO) analysis for the genes found within the hypomethylated domains. Consistent with previous analysis of colon cancer-specifically methylated regions (Irizarry et al. 2009), these genes are significantly enriched in embryonic development, including the highly divergent homeobox gene HDX along with several neuronal growth factors NEGR1, NETO1, and NTM. In addition, we observe significant enrichment for zinc finger genes, Kruppel-associated box (KRAB) transcription factors, and DNA binding proteins (Dennis et al. 2003; Fig. 2C). This enrichment for transcription factors indicates a drastic alteration of the HCC1954 regulatory program from HMEC concordant with hypomethylation.
Formation of repressive chromatin domains at hypomethylated genomic regions
To explore the molecular processes potentially responsible for lower transcript abundance of genes in the hypomethylated domains, we examined the status of DNA methylation at promoters of the genes. Among the 627 gene-body hypomethylated genes with lower transcript abundance in HCC1954, promoters of 289 (46%) genes are hypermethylated, consistent with a role for promoter DNA hypermethylation in transcriptional repression. These genes include many well-known tumor suppressors such as MGMT (Esteller et al. 1999; Everhard et al. 2009; Hibi et al. 2009a, b), DCC (deleted in colorectal carcinoma) (Fearon et al. 1990), and DLC1 (deleted in liver cancer 1) (Yuan et al. 1998).
Surprisingly, the remaining 54% repressed genes (N = 338) exhibit no change (N = 70) or loss (N = 268) of DNA methylation at promoters, yet still display lower transcript abundance in cancer cells. These genes also include well-known tumor suppressor genes, including the interferon-inducible gene PYHIN1 (Ding et al. 2006), the leucine zipper gene LZTS1 (Ishii et al. 1999), the antiangiogenesis factors THBS2 (Streit et al. 1999) and HRG (Rolny et al. 2011), and the cysteine protease inhibitor CST5 (Alvarez-Diaz et al. 2009). Importantly, genes with intragenic hypomethylation (HMEChigh → HCC1954PMD) but lacking promoter hypermethylation are also significantly enriched in down-regulated genes (P = 2.0 × 10−7, hypergeometric) and significantly depleted in up-regulated genes (P = 5.0 × 10−3, hypergeometric), therefore excluding the possibility that promoter hypermethylation explains these observations. We hypothesized that these genes may be aberrantly repressed by other epigenetic mechanisms, such as repressive chromatin modifications. To address this possibility, we performed chromatin immunoprecipitation followed by sequencing (ChIP-seq) for several histone modifications including the repressive H3K9me3 and H3K27me3 marks in HCC1954 and the active chromatin marks H3K4me1, H3K4me3, H3K27ac, and H3K36me3. Comparing these modifications to those of HMEC produced by the ENCODE Consortium (Birney et al. 2007; Ernst et al. 2011), we observed increased H3K27me3 in HCC1954 at hypomethylated gene bodies, independent of promoter methylation status (Fig. 2D). Although H3K9me3 was not mapped in HMECs, we also observed enrichment of this modification at both sets of genes in HCC1954.
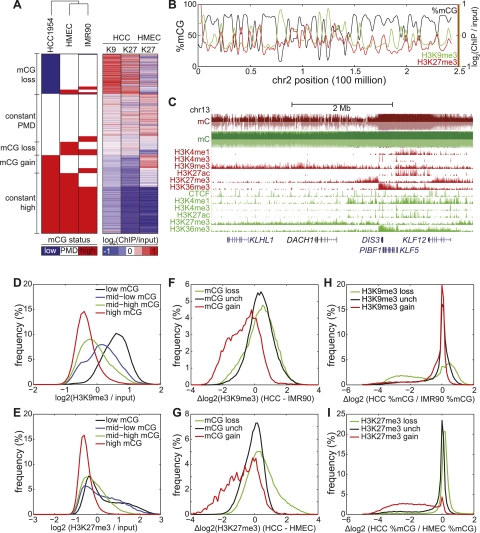
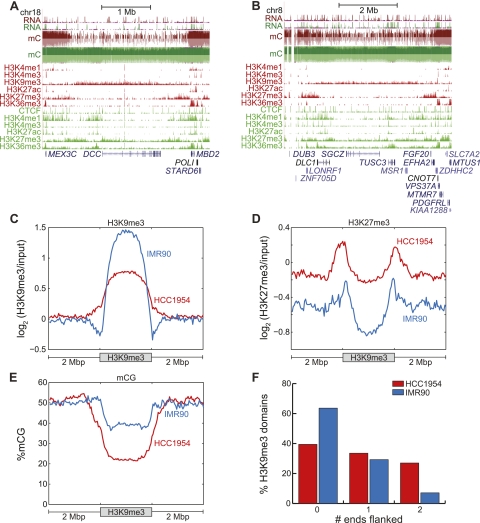
The results reveal that large-scale hypomethylation is correlated with an increase in repressive chromatin formation. To further verify this observation on a global scale, we plotted the enrichment of H3K9me3 and H3K27me3 relative to input [log2(ChIP/input)]. The genomic loci with the strongest H3K9me3 and H3K27me3 enrichment also correspond to low mCG. Conversely, the greatest depletion of these repressive modifications occurs at loci with high mCG for both HCC1954 and HMEC (Fig. 3A). As seen on a chromosome-wide view (Fig. 3B), regions showing hypomethylated DNA exhibit the greatest enrichment of H3K9me3 or H3K27me3 in HCC1954. Inspection of the DACH1 tumor suppressor gene illustrates strong enrichment of these two repressive marks in a large hypomethylated domain, which abruptly transitions to a fully methylated domain coincident with loss of the repressive modifications and gain of the mCG-associated histone modification H3K36me3 (Ball et al. 2009; Lister et al. 2009) (Fig. 3C).
Figure 3.
Repressive chromatin is depleted of mCG. (A) (Left) A reproduction of Figure 1D; (right) each of the 282,109 rows represents the H3K9me3 (denoted K9) and H3K27me3 (denoted K27) status in HCC1954 and HMEC, in the same order as the left panel. (B) Distribution of %mCG, H3K9me3, and H3K27me3 on chromosome 2 for HCC1954. (C) A large domain of DNA hypomethylation near the DACH1 tumor suppressor coincides with H3K9me3 and H3K27me3 in HCC1954. (Red) HCC1954, (green) HMEC. (D) Distribution of H3K9me3 enrichment for four quantiles of DNA methylation status in HCC1954. (E) As in D, but for H3K27me3. (F) Distribution of change in H3K9me3 in HCC1954 compared with IMR90 for 10-kb windows that lose mCG, gain mCG, or are unchanged for mCG. Δlog2 (H3K9me3) = log2 (HCC1954 H3K9me3/input) − log2 (IMR90 H3K9me3/input). (G) As in F, but for H3K27me3 comparing HCC1954 with HMEC. (H) Distribution of change in mCG in HCC1954 compared with IMR90 for 10-kb windows that lose H3K9me3, gain H3K9me3, or are unchanged for H3K9me3. (I) As in H, but for H3K27me3 comparing HCC1954 with HMEC.
It has previously been observed that regions of hypomethylation are biased toward gene poor regions (Aran et al. 2011). We also observe a small but positive correlation between %mCG with promoters (R = 0.03), exons (R = 0.17), and gene bodies (R = 0.26) (Supplemental Fig. S3). However, the magnitude of correlation between %mCG with H3K9me3 (R = −0.66) and H3K27me3 (R = −0.53) is noticeably greater, suggesting a stronger link between hypomethylation with chromatin than with genic features.
Allelic basis of repressive chromatin and DNA methylation
The above observations led us to hypothesize that formation of H3K9me3 and H3K27me3 domains are closely coupled to the loss of DNA methylation in HCC1954 cells. This hypothesis is supported by several global analyses: (1) For HCC1954, H3K9me3/H3K27me3 enrichment increases as mCG decreases (Fig. 3D,E; Supplemental Fig. S4); (2) 10-kb windows gaining the most mCG also lose the most H3K9me3 and H3K27me3, and vice versa (Fig. 3F,G); and (3) 10-kb windows gaining repressive chromatin also tend to lose mCG, and vice versa (Fig. 3H,I).
However, it has been observed that loci exhibiting partial methylation overlap with H3K27me3, as previously reported in IMR90 fetal lung fibroblasts (Lister et al. 2009). To resolve this apparent contradiction, we tested the possibility that PMDs in HCC1954 coinciding with repressive chromatin may have one allele in a fully methylated state and another allele marked with repressive chromatin.
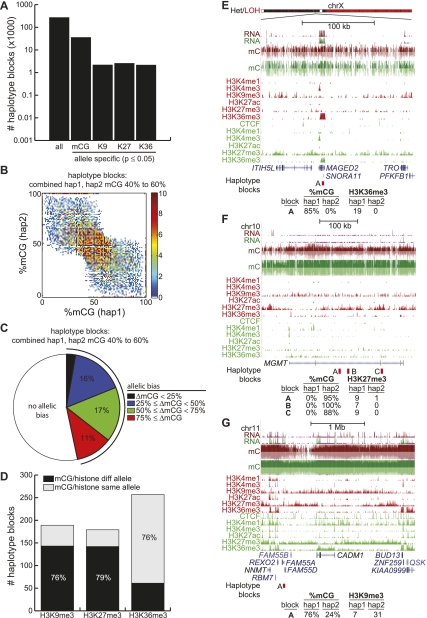
We first distinguished the different alleles in HCC1954 by identifying the haplotype blocks in the genome of these cancer cells. We obtained 1,000,880,493 paired-end, nonclonal, uniquely mapped reads corresponding to a 27.6-fold genome coverage (Supplemental Fig. S5). Using the bam2mpg genotyping program (Teer et al. 2010), we found 1,211,258 high-confidence single nucleotide polymorphisms (SNPs) in the genome. Together with the sequenced reads to link SNPs together, we employed the previously developed error-correcting algorithms HASH and HapCUT to identify 269,392 phased haplotype blocks (Bansal and Bafna 2008; Bansal et al. 2008) with an N50 of 290 bp (Fig. 4A).
Figure 4.
Allelic distribution of epigenetic modifications. (A) Number of haplotype blocks found by SNP phasing (denoted all), and the number of these blocks showing significant allelic bias for mCG, H3K9me3 (denoted K9), H3K27me3 (denoted K27), and H3K36me3 (denoted K36) (Fisher's exact test P-value ≤ 0.05). (B) For haplotype blocks where the combined frequency of methylation of both haplotypes is between 40% and 60%, shown is the density plot of %mCG on haplotype 1 versus %mCG on haplotype 2. (C) For haplotype blocks in B, shown is the fraction with allelic bias (Fisher's exact test P-value ≤ 0.05), for different possible values of allelic bias. ΔmCG = |%mCG(hap1) − %mCG(hap2)|. (D) Of haplotype blocks simultaneously showing allelic bias in mCG (Fisher's exact test P-value ≤ 0.05, ΔmCG ≥ 50%) and a histone modification (Fisher's exact test P-value ≤ 0.05), the number of haplotype blocks where mCG is on the same or different allele as the histone modification. (E) Allelic H3K36me3 at the MAGED2 gene. The bar at top indicates where one arm of chrX was lost. (Red) HCC1954, (green) HMEC. (Bottom) Number of H3K36me3 reads belonging to haplotype 1 or 2. (F) Allelic H3K27me3 at the MGMT tumor suppressor. (Red) HCC1954, (green) HMEC. (Bottom) Number of H3K27me3 reads belonging to haplotype 1 or 2. (G) Allelic H3K9me3 near the CADM1 tumor suppressor. (Red) HCC1954, (green) HMEC. (Bottom) Number of H3K9me3 reads belonging to haplotype 1 or 2.
To assess if PMDs consist of one fully methylated allele and another fully unmethylated allele, we focused on the 15,309 partially methylated haplotype blocks with an allele-combined average %mCG between 40% and 60% (Fig. 4B). Plotting the %mCG of each phased allele illustrates a clear bias between the methylation status of each allele (Fig. 4B). Almost half (47%) of these partially methylated haplotype blocks exhibit significant allelic bias in DNA methylation (P ≤ 0.05, Fisher's exact test), with the majority (60%) having at least a 50% difference between %mCG on the two alleles (Fig. 4C). These results suggest that allele-specific methylation is prevalent and that PMDs frequently consist of two differentially methylated alleles.
To examine the status of histone modifications at the haplotype blocks identified above, we sequenced between 76.5 and 92.3 million ChIP-seq reads for H3K9me3, H3K27me3, and H3K36me3 in HCC1954 cells to gain adequate coverage and examined the allelic bias among the haplotype blocks that display allelic DNA methylation. By distinguishing the ChIP-seq reads corresponding to different haplotype blocks, we first identified 2100, 2479, and 2104 blocks that exhibit significant allelic bias for H3K9me3, H3K27me3, and H3K36me3, respectively (Fig. 4A) (P ≤ 0.05, Fisher's exact test). Overlapping these blocks with those showing allele-specific DNA methylation, we observe that 78% of overlapping H3K36me3 blocks are on the same allele as mCG (pH3K36me3 = 1.38 × 10−17, binomial). For example, ubiquitously expressed MAGED2 on chrX is located within a PMD, and DNA methylation is on the same allele as H3K36me3 (Fig. 4E). In contrast, the majority of overlapping H3K9me3 (75%) and H3K27me3 (79%) blocks are on the opposite allele from mCG (Fig. 4D) (pH3K9me3 = 1.75 × 10−12, pH3K27me3 = 2.11 × 10−15, binomial). For example, the MGMT tumor suppressor gene is partially methylated in HCC1954 and marked by H3K27me3, with these modifications belonging on opposite alleles (Fig. 4F). Similarly, a partially methylated haplotype block near the CADM1 tumor suppressor gene is marked by H3K9me3 on a different allele as mCG (Fig. 4G).
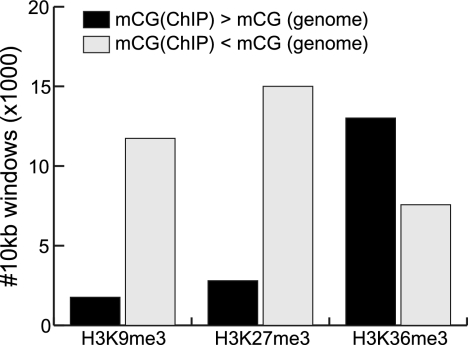
To further validate the mutual exclusiveness of H3K9me3 and H3K27me3 with mCG, we performed ChIP with these chromatin modifications, then determined the DNA methylation status of the resulting DNA by bisulfite sequencing (ChIP-methylC-seq). Of the 13,494 and 17,805 10-kb windows exhibiting significant differences in DNA methylation status between H3K9me3 and H3K27me3 ChIP-methylC-seq compared to methylC-seq reads (Fisher's exact test, P ≤ 0.01), 87.0% and 84.3% have less DNA methylation in the ChIP sample compared to the methylC-seq sample, respectively (pH3K9me3 < 10−100, pH3K27me3 < 10−100) (Fig. 5). In contrast, most (63.2%) of the 20,585 10-kb bins with differences in H3K36me3 ChIP-methylC-seq versus background are more enriched for mCG (pH3K36me3 < 10−100) (Fig. 5). Thus, in support of our observations, DNA enriched for H3K9me3 or H3K27me3 is depleted of mCG compared to DNA enriched for H3K36me3.
Figure 5.
Mutual exclusiveness of chromatin modifications and mCG by ChIP-methylC-seq. ChIP was performed for H3K9me3, H3K27me3, or H3K36me3 in HCC1954, followed by bisulfite conversion and sequencing. For 10-kb windows, mCG bias was compared to genomic background from methylC-seq (P ≤ 0.01, Fisher's exact test). Shown is the number of 10-kb windows showing significant differences, split by whether there is more or less mCG in the ChIP-methylC-seq sample.
A higher-order organization of H3K9me3 and H3K27me3
The mutually exclusive nature of H3K9me3 and H3K27me3 with mCG is one example of the organization of the HCC1954 epigenome. While H3K9me3 and H3K27me3 rarely overlapped (Supplemental Fig. S6), we also observed frequent examples of a higher-order organization between H3K9me3 and H3K27me3: These marks appear to be spatially organized such that large domains of H3K9me3 are flanked by regions of H3K27me3 enrichment. Examples include the DCC and DLC1 tumor suppressor genes (Fig. 6A,B).
Figure 6.
Large-scale organization of H3K9me3 and H3K27me3. (A) Hypomethylation near the DCC tumor suppressor coincides with H3K9me3, which is flanked by H3K27me3. (B) Hypomethylation near the DLC1 tumor suppressor coincides with H3K9me3, which is flanked by H3K27me3. (C) Enrichment of H3K9me3 within 2 Mb of H3K9me3 domains in HCC1954 and IMR90. (D) Enrichment of H3K27me3 within 2 Mb of H3K9me3 domains in HCC1954 and IMR90. (E) Enrichment of mCG within 2 Mb of H3K9me3 domains in HCC1954 and IMR90. (F) Fraction of H3K9me3 domains flanked by 0, 1, or 2 H3K27me3 domains.
To assess if this is a global phenomenon, we defined large domains of H3K9me3 by merging H3K9me3-enriched 10-kb windows, resulting in 376 domains spanning 589 Mb. To exclude H3K9me3 domains having internal enrichment of H3K27me3, we removed those domains with an average (H3K27me3 RPKM)/(input RPKM) ≥ 1.5, resulting in a final list of 322 H3K9me3 domains spanning 536 Mb of the human genome. We observed enrichment of H3K9me3 (Fig. 6C), depletion of mCG (Fig. 6E), and background levels of H3K27me3 within the bodies of these H3K9me3 domains (Fig. 6D). However, H3K27me3 was enriched in regions directly flanking the boundaries of H3K9me3 domains (Fig. 6D). Similar results are observed in IMR90 H3K9me3 domains, though the extent of H3K27me3 enrichment at H3K9me3 flanks is weaker (Fig. 6C–F). As H3K9me3 was not mapped in HMEC, we cannot assess this phenomenon directly in these cells. However, to identify potential H3K9me3 domains, we searched for regions depleted of DNA methylation that are simultaneously depleted of H3K27me3. These large domains, putatively similar to the H3K9me3 enriched domains found in HCC1956, also exhibit flanking peaks of H3K27me3 in HMEC, as in HCC1954 and IMR90 (Supplemental Fig. S7), suggesting that H3K27me3 flanking of H3K9me3 may be a common phenomenon.
A passive model of global hypomethylation
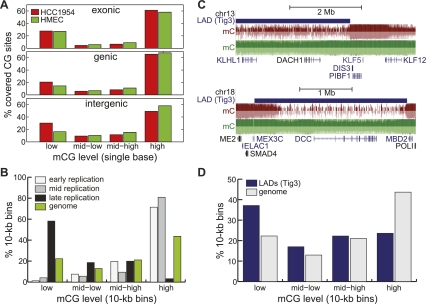
Thus far, we have focused on gross changes of DNA methylation across large 10-kb bins. To gain insight into which genomic features are more likely to undergo hypomethylation, we examined the methylation level at single cytosine residues spanning exons, intragenic regions, or intergenic regions. The distribution of cytosine methylation frequency at exons in HCC1954 is nearly indistinguishable from HMEC (Fig. 7A). At genic regions, defined to be the entire interval between transcription start site (TSS) and transcription terminal site (TTS), the frequency of lowly methylated cytosines is higher in HCC1954 than HMEC (Fig. 7A) (20.6% vs 14.5%, P < 10−16, binomial). However, hypomethylation is most pronounced in intergenic regions, where 30.2% of HCC1954 cytosines are lowly methylated compared to 16.2% in HMEC (Fig. 7A) (P < 10−16, binomial).
Figure 7.
A passive model of hypomethylation. (A) Distribution of HCC1954 %mCG for exonic (top), genic (middle), and intergenic (bottom) cytosine residues. (B) Distribution of HCC1954 %mCG for 10-kb regions that are consistently early-, middle-, and late-replicating in four cell types (Hansen et al. 2009), compared to the background genome (green). (C) Snapshots illustrating the overlap of hypomethylated regions with lamina-associated domains previously mapped in Tig3 cells (Guelen et al. 2008), at the DACH1 (top) and DCC (bottom) genes. (D) Distribution of HCC1954 %mCG for 10-kb regions that are found in Tig3 lamina-associated domains (blue), compared to the background genome (gray).
Two potential mechanisms can be envisioned to account for the global hypomethylation in cancer cells. One possibility is that the methyl group is actively removed from methylated cytosines. Several proteins, including AICDA (also known as AID), GADD45A, Tet family of proteins, ELP3, and TDG, have recently been implicated in demethylation of methylated cytosines (Barreto et al. 2007; Bhutani et al. 2010; Ito et al. 2010; Cortellino et al. 2011). None of these genes is significantly more expressed in HCC1943 than HMEC, nor do they harbor somatic mutations in HCC1954, suggesting that an active mechanism may not be responsible for global hypomethylation in HCC1954. Alternatively, DNA methylation may be gradually lost during replication of the cancer cells, which would be biased toward late-replicating regions of the genome. To test whether our data support this model, we compared the DNA methylation levels in HCC1954 to a compilation of regions that are consistently early-, middle-, and late- replicating in a panel of four cell lines (hESC, erythroid, lymphoid, and fibroblast cells) (Hansen et al. 2009). We find that 58.2% of consistently late-replicating regions have low methylation in HCC1954, significantly more than that expected by chance (22.3%, P < 10−16, binomial). In contrast, only 1.2% and 4.0% of consistently early- and middle-replicating regions, respectively, are lowly methylated. These results are consistent with a previous report (Aran et al. 2011) and the model that DNA methylation at late-replicating regions is gradually lost through many rounds of cell divisions. Further experiments are necessary to understand how late-replication timing leads to partial loss of DNA methylation.
DNA found near lamina-associated domains (LADs) has previously been shown to be late-replicating (Moir et al. 1994). As LADs have been shown to be stable across different cell types (Peric-Hupkes et al. 2010), to provide further support that hypomethylated domains in HCC1954 are late-replicating, we compared them to a previously published map of LADs in Tig3 human fibroblast cells (Guelen et al. 2008). We observe remarkable concordance between LADs in Tig3 cells and hypomethylated domains in HCC1954 (Fig. 7C). On a global scale, 37.2% of LADs are found in hypomethylated domains, compared to 22.3% expected by chance (P < 10−16, binomial), consistent with previous observations (Hansen et al. 2011).
Assessing the epigenetic contribution to increased transcription of repeat elements
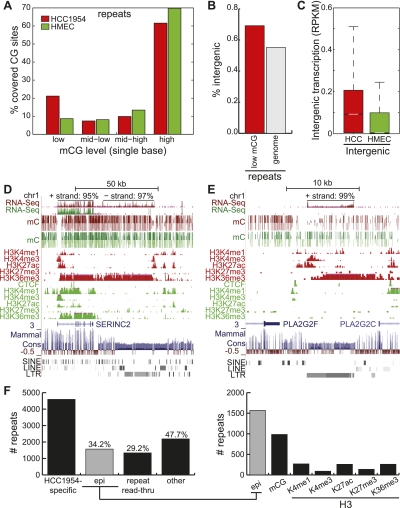
Increased expression of repetitive elements is a hallmark of cancer cells, and this aspect of cancer has frequently been attributed to global hypomethylation (Prak and Kazazian 2000). To assess the frequency of hypomethylation at repeats, we examined the methylation level at each cytosine residue contained within repeats. Dramatically, 21.2% of repeat-associated cytosines are lowly methylated in HCC1954, compared to 8.7% in HMEC (P < 10−16, binomial) (Fig. 8A). Lowly methylated repeats are also more frequently found in intergenic regions than expected by chance: while 55.2% of all repeat-associated cytosines are intergenic, the subset consisting of lowly methylated residues is 69.2% intergenic (P < 10−16, binomial) (Fig. 8B).
Figure 8.
Epigenetic contribution of increased repeat expression. (A) Distribution of %mCG of each repeat-associated cytosine residue spanned by at least 10 methylC-seq reads. (B) Percentage of all repeats (gray) and of lowly methylated repeats in HCC1954 (red) that are intergenic. (C) Distribution of intergenic repeat expression in HCC1954 and HMEC, expressed in RPKM. (White line) median, (box) 25th to 75th percentiles. (D) Snapshot of repeat expression and read-through with epigenetic signatures near the SERINC2 gene. RNA-seq strand distribution shown on top. (E) Snapshot of repeat expression and read-through with epigenetic signatures near the PLA2G2F gene. RNA-seq strand distribution shown on top. (F) Number of highly expressed repeats specific to HCC1954 and the number that can be explained by at least a 50% change in epigenetic modifications or read-through transcription from nearby repeats. (Epi) epigenetic.
To study the expression of repetitive elements, we focused on intergenic repeats, thereby avoiding ambiguities with genic features. The median expression level of intergenic repeats in HCC1954 (0.43 RPKM) is 5.2 times more than in HMEC (0.08 RPKM) (P < 10−16, rank sum test) (Fig. 8C), supporting previous observations of extensive repeat expression in cancer cells. Reminiscent of chromatin signatures at actively transcribed genes, we also observe hypomethylation at repeats concordant with peaks of H3K4me3 and H3K27ac (Fig. 8D,E). Strand-specific expression emanates from these peaks and coincides with enrichment of H3K36me3, extending to contiguous blocks of transcription spanning ∼31 kb and ∼7 kb near SERINC2 and PLA2G2F, respectively. Notably, these large blocks of repeat transcription span numerous adjacent repeat sequences and lack hypomethylation.
These observations suggest several explanations for repeat expression: (1) loss of a repressive epigenetic mark, (2) gain of an active chromatin mark, or (3) read-through transcription of neighboring repeats. To further explore these three possibilities, we quantified how much each component explains the expression of a set of 4592 intergenic repeats highly (RPKMHCC1954 ≥ 1) and specifically (RPKMHCC1954 ≥ 10 × RPKMHMEC) expressed in HCC1954. About one-third (34.2%) of these repeats exhibited at least a 50% change in an epigenetic modification. Of these, loss of DNA methylation is the most prominent epigenetic change, accounting for 62.7%. Gain of active chromatin modifications accounts for 42.1% of these repeats, while loss of H3K27me3 only makes up 8.7% (Fig. 8F).
To explain repeat expression by read-through of neighboring repeats, we first identified 6354 intergenic domains containing clustered and strand-specific RNA-seq reads (see Methods). While these domains only span 5.4% of all intergenic regions, they account for 31.0% of all repeats specifically expressed in HCC1954 (P < 10−16, binomial). As further evidence that these repeats are the result of read-through transcription, 94.0% are oriented in the dominant direction of RNA-seq reads in the domains, while the expectation is only 50% (P < 10−16, binomial). In total, these oriented repeats account for 29.2% of all HCC1954-specific repeats, and altogether about half (52.3%) of all repeats could be explained by either epigenetic changes or read-through repeat transcription (Fig. 8F).
Discussion
Global DNA hypomethylation is a hallmark in human cancer, but its functional consequences have been unclear (Hinshelwood and Clark 2008). By comparing the methylomes of a breast cancer line HCC1954 and the primary breast cell HMEC, we find a link between hypomethylation and the formation of repressive chromatin domains. Affected genes are frequently repressed in cancer cells and include tumor suppressors such as DNA repair gene MGMT (Esteller et al. 1999), the deleted in colorectal carcinoma gene, DCC (Fearon et al. 1990), and the deleted in liver cancer gene, DLC1 (Yuan et al. 1998). Thus, hypomethylation is not always an activating epigenetic change as previously assumed (Esteller et al. 2000; Esteller 2007, 2008). Rather, when it is accompanied by a gain of repressive chromatin, the result can be a decrease in expression.
The mechanism of global hypomethylation is a long-standing question in cancer epigenetics. Our data seem to support a passive mechanism whereby methylation is gradually lost over successive cell divisions, as opposed to an active mechanism involving demethylating enzymes (Wild and Flanagan 2010). Evidence for this passive mechanism includes a bias for hypomethylation at intergenic, late-replicating, and lamina-associated regions of the genome. Furthermore, given that global hypomethylation is a consistent feature of many diverse cancers (Gama-Sosa et al. 1983), the alternative hypothesis involving activation of demethylating enzymes appears less plausible. In light of the abnormal growth typical of cancer, a passive mechanism is more intuitive: Cancer cells grow faster than methylation can be copied from the replicating parental DNA, resulting in progressive loss of DNA methylation.
Increased expression of repetitive DNA sequences has been frequently observed in cancer cells, and this has often been linked to changes in DNA methylation or chromatin structure (Florl et al. 1999; Menendez et al. 2004; Schulz 2006; Ting et al. 2011). By integrating the transcriptome and epigenome data sets, our analysis suggests several possible mechanisms of repeat expression. Epigenetics can explain the expression of about one-third of HCC1954-specifically expressed repeats. Additionally, transcription read-through may be involved, as we observe that spatially clustered repeats are often expressed as a single transcriptional unit. One end of this unit is marked with the canonical epigenetic marks of transcription initiation (H3K4me3, H3K27ac, and hypomethylation), and the transcribed body with numerous repetitive elements contains the elongation mark H3K36me3. These repeats are potentially transcribed as a result of read-through transcription and span nearly a third of HCC1954-specifically expressed repetitive elements.
Partial DNA methylation is a recent observation made possible by whole genome shotgun bisulfite sequencing (Lister et al. 2009). Since PMDs are not observed in pluripotent stem cells but span about one-third of the genome in differentiated cell types such as fetal lung fibroblasts, the gradual loss of DNA methylation in PMDs during development may be interpreted as another form of global hypomethylation. However, the biological relevance and formative processes of PMDs has yet to be understood. We show that a significant fraction of PMDs correspond to allele-specifically methylated DNA. In these regions, the unmethylated allele is occupied by H3K9me3 and H3K27me3, while the hypermethylated allele is devoid of the repressive chromatin marks. This argues for a mutually exclusive relationship between DNA methylation and H3K9me3 or H3K27me3 at PMDs and is also in agreement with previous observations that highly methylated ES cells have much lower levels of H3K9me3 and H3K27me3 compared to differentiated cells (Lister et al. 2009; Hawkins et al. 2010). This exclusivity of H3K9me3 and DNA methylation is not incompatible with previous studies showing a close mechanistic tie between H3K9me3 and DNA methylation, particularly at constitutive heterochromatin, which is generally centromeric or telomeric (Schotta et al. 2004; Volkel and Angrand 2007). For example, H3K9me3 directs DNMT3B-dependent DNA methylation at pericentric satellite repeats in mouse ES cells (Lehnertz et al. 2003). As short-read sequencing technologies cannot map to these highly repetitive regions of the genome, our analyses are restricted to regions of facultative heterochromatin and euchromatin. Thus, at least in HCC1954 and IMR90, H3K9me3 appears to be exclusive of DNA methylation outside of constitutive heterochromatin.
Our observations suggest that DNA methylation changes in breast cancer cells may be mechanistically linked to the pathways responsible for H3K9me3 or H3K27me3. Interestingly, the genes encoding EZH2 and other PcG proteins responsible for H3K27me3 are frequently overexpressed in breast cancers (Kleer et al. 2003; Chang et al. 2011), and point mutations that lead to loss or gain of function have also been reported in these genes (Dalgliesh et al. 2010; Morin et al. 2010; Yap et al. 2010). The exclusive nature of repression, together with the observation that hypomethylation is coincident with an increase in repressive chromatin, suggests that these repressive mechanisms may be compensatory for each other and that therapies targeting one modification may not be sufficient to acquire gene activation.
Given the mutual exclusion of repressive epigenetic modifications, an unresolved question is whether global hypomethylation in cancer results in gain of H3K9me3/H3K27me3, or the other way around. Recently, it has been shown that DNA methylation prevents binding of the PRC2 complex to chromatin to deposit H3K27me3 (Lindroth et al. 2008; Bartke et al. 2010; Wu et al. 2010). Further, Komashko and Farnham observed that disruption of DNA methylation by 5-azacytidine in human embryonic kidney cells results in global increases of H3K9me3 and H3K27me3 (Komashko and Farnham 2010). Taken together, these data suggest that loss of DNA methylation in cancer cells may lead to the formation of repressive chromatin domains and silencing of tumor suppressor genes. We postulate, therefore, that inhibition of the H3K27me3 or H3K9me3 methyltransferases could be a new cancer therapeutic strategy. Further research is needed to identify the causes of DNA hypomethylation in these cells.
Methods
Cell culture
HCC1954 cells were grown in RPMI 1640 media supplemented with 10% FBS, 1× nonessential amino acids (Invitrogen 11140050), and 1× L-glutamine (Invitrogen 45000-676-1). Cells submitted to epigenetic analysis were between passage 34 and 42. Cryo-preserved HMECs at passage 6–7 were purchased from Lonza (CC-2551) and grown according to the manufacturer's instructions at 37°C/5% CO2. The cells were split two times before harvesting. Harvested HMECs highly express p16/CDKN2A, suggesting these cells have not reached stasis (Novak et al. 2008). Harvested HMECs express KLK6, COX7A1, EPCAM, KRT19, and PRDM1, all of which are hallmarks of early passage prestasis HMECs. Finally, harvested HMECs express TP53, indicating that they have not entered crisis (Garbe et al. 2009). See Supplemental Material for detailed descriptions.
Sequencing
Sonicated genomic DNA was submitted to Illumina paired-end library preparation according to the manufacturer's instructions. Libraries for mRNA-seq (Parkhomchuk et al. 2009), ChIP-seq (Hawkins et al. 2010), and methylC-seq (Lister et al. 2009) were created as described previously. ChIP-seq was performed with antibodies specific to H3K4me1, H3K4me3, H3K9me3, H3K27ac, H3K27me3, and H3K36me3. RNA-seq reads were mapped with TopHat (Trapnell et al. 2009), and other experiments were mapped with Bowtie (Langmead et al. 2009). Sequenced reads for RNA-seq and ChIP-seq were 36 bp, with methylC-seq at 100 bp, and genome sequencing being a mix of 36, 80, and 101 bp. See Supplemental Material for complete protocols.
Antibodies
ChIP-seq was performed using the following antibodies: H3K4me1 (Abcam ab8895-50, lot 720417), H3K4me3 (Millipore CS200580, lot DAM1612220), H3K9me3 (Abcam ab8898-100, lot 699671; Diagenode pAb-056-50, lot A93-0041), H3K27ac (Active Motif 39133, lot 19208002), H3K27me3 (Millipore 07-449, lot DAM161288), and H3K36me3 (Abcam ab9050-100, 707946). All antibodies used here were validated by peptide dot blot assays to ensure specificity to the correct histone modification (Egelhofer et al. 2011).
Read mapping
Reads from genome sequencing were mapped by the Bowtie program in two passes: first as paired-end mapping and then as single-end mapping of unmapped reads. ChIP-seq reads were also mapped by Bowtie. For both genome sequencing and ChIP-seq, only reads that mapped uniquely to hg18 with, at most, three mismatches were kept. MethylC-seq reads were mapped as previously described (Lister et al. 2009). RNA-seq reads were mapped by the TopHat program. For non-RNA libraries, PCR duplicates were removed with the Picard program (Picard 2011). All biological replicates were combined and compressed/indexed by the SAMtools suite into BAM format (Li et al. 2009). See Supplemental Material for detailed descriptions.
Quantifying RNA-seq expression
To quantify RPKM expression (reads per kilobase of model per million base pairs sequenced) at UCSC Known Genes (Hsu et al. 2006) and RefSeq genes (Pruitt et al. 2005), the Cufflinks program (Trapnell et al. 2010) was applied to the mapped TopHat reads. To allow for division between RPKM values, the lowest RPKM value was set to 5 × 10−6. To reduce redundancy of the gene lists, genes were merged and their expression values summed if (1) they had the same common gene name with the same TSS, or (2) they had the same common gene name and overlap each other. A gene is a member of a 10-kb DNA methylation domain category (low/mid-low/mid-high/high) if there is any overlap of the gene body with a member of the methylation category. Therefore, a gene can belong to multiple categories. To quantify expression of repetitive elements, we counted the number of reads mapping in each repeat and expressed the result in RPKM units, using only the subset of RNA-seq reads that are uniquely mapping to hg18.
Quantifying ChIP-seq enrichment
Enrichment of histone modifications in a specified region of the genome was quantified as log2 (ChIP RPKM/input RPKM). To avoid division by zero, a pseudocount was added depending on the depth of sequencing: Pseudocount = (# million base pairs mapped)/p, where p is the pseudocount factor, here set to 2. Thus, RPKM = (# reads in bin + pseudocount)/(# kb in model)/(# million base pairs mapped). This method of pseudocounts guarantees that bins having (1) no ChIP and input reads or (2) ChIP and input reads perfectly proportional to the number of reads sequenced will have log2 (ChIP RPKM/input RPKM) = 0.
Quantifying mCG enrichment
DNA methylation in the CG context was quantified as %mCG of all cytosine bases in the reference sequence. Specifically, for each of the Watson and Crick strands, all cytosine bases in the CG context in the reference sequence hg18 were identified. For all mapped methylC-seq reads in a given genomic interval, the number of cytosines in CG context that were called methylated as well as the number called unmethylated were counted. To reduce noise from sequencing error, only those bases with phred score ≥20, indicating sequencing confidence of at least 99%, were counted. Then %mCG = (# methylated CGs on both strands)/(# methylated CGs on both strands + # unmethylated CGs on both strands) × 100. This method assures that genomic intervals spanned by many methylC-seq reads devoid of mCG are accurately given a low %mCG value.
Defining haplotype blocks
To generate haplotypes, aligned reads were first used to determine SNPs in HCC1954 using the genotyping program bam2mpg (Teer et al. 2010). To safeguard against incorrect SNP calls due to sequencing error, only considered bases with phred score ≥20 were considered. Only genotypes with score ≥10 were kept, assuring that reported genotypes are at least exp(10) ≈ 22,000 times as probable as the next most probable genotype. With a list of SNPs and the sequence read information to link them together, previously developed error-correcting algorithms HASH and HapCUT were used to assemble haplotypes (Bansal and Vafna 2008; Bansal et al. 2008).
Assessing allele-specific epigenetic modifications
To assess allele-specific ChIP-seq enrichment at a given haplotype block, the number of ChIP reads landing in each of the two haplotypes hap1 and hap2 were counted. The background consisted of the number of occurrences of hap1 and hap2 from genome sequencing. Fisher's Exact Test was used to assess allele-specificity of ChIP-seq (P ≤ 0.05). To assess allele-specific DNA methylation at a given haplotype block, the number of methylated and unmethylated cytosines in CG context (given the reference genome sequence hg18 and with a phred score ≥20) were counted for hap1 and hap2. Fisher's Exact Test was used to assess allele-specificity of methylC-seq (P ≤ 0.05). See Supplemental Material for detailed descriptions.
To assess allele-specific DNA methylation from ChIP-methylC-seq data, the number of methylated and unmethylated cytosines in CG context (given the reference genome sequence hg18 and with a phred score ≥20) were counted for each 10-kb window using ChIP-methylC-seq and methylC-seq data, and Fisher's Exact test (P ≤ 0.01) was used to assess differential methylation.
Identifying large chromatin domains
To find large domains of H3K9me3 enrichment, log2 (ChIP RPKM/input RPKM) as described above was calculated for all 10-kb windows spanning the human genome. H3K9me3-enriched windows are defined as having (H3K9me3 RPKM)/(input RPKM) ≥ 1.5 . The binomial distribution with p = (# H3K9me3-enriched bins)/(# total bins) was used to find all domains of size 250 kb spanning these 10-kb windows that were significantly enriched for H3K9me3-enriched bins (P-value ≤ 0.01). To reduce redundancy of overlapping 250-kb domains, domains with no more than 50 kb of nonoverlapping regions were merged. As visual inspection indicated H3K27me3 domains were smaller than H3K9me3 domains, H3K27me3 domains were identified similarly but with a domain size of 100 kb rather than 250 kb.
Quantifying repeat-associated read-through transcription
We counted the number of reads mapping in each intergenic 1-kb bin and expressed the result in RPKM units, using only the subset of RNA-seq reads that are uniquely mapping to hg18. To reduce noise, we filtered for strand-specific bins containing at least three reads and having at least five times as many reads on one strand as on the other. To consolidate strand-specific transcribed intergenic clusters, bins within 5 kb on the same strand were merged, and merged regions smaller than 5 kb were discarded. Finally, repeats associated with read-through transcription were defined as repeats belonging to a merged region that are oriented in the same direction of transcription.
Data access
All the sequencing data generated here have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE29069.
Acknowledgments
This work was supported by the Ludwig Institute for Cancer Research and the Mary K. Chapman Foundation (J.R.E.). Work in the laboratory of J.R.E. is supported by the Howard Hughes Medical Institute and the Gordon and Betty Moore foundation. J.R.E. is a HHMI-GBMF Investigator. C.L. and V.B. were supported by an NSF fellowship, and grants 5RO1-HG004962 (NIH), and NSF-CCF-1115206 to V.B.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.125872.111.
References
- Alvarez-Díaz S, Valle N, García JM, Peña C, Freije JM, Quesada V, Astudillo A, Bonilla F, López-Otín C, Muñoz A 2009. Cystatin D is a candidate tumor suppressor gene induced by vitamin D in human colon cancer cells. J Clin Invest 119: 2343–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Toperoff G, Rosenberg M, Hellman A 2011. Replication timing-related and gene body-specific methylation of active human genes. Hum Mol Genet 20: 670–680 [DOI] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM 2009. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol 27: 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal V, Bafna V 2008. HapCUT: An efficient and accurate algorithm for the haplotype assembly problem. Bioinformatics 24: i153–i159 [DOI] [PubMed] [Google Scholar]
- Bansal V, Halpern AL, Axelrod N, Bafna V 2008. An MCMC algorithm for haplotype assembly from whole-genome sequence data. Genome Res 18: 1336–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, et al. 2007. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 445: 671–675 [DOI] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T 2010. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143: 470–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM 2010. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 463: 1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E JA, Stamatoyannopoulos A, Dutta R, Guigo TR, Gingeras EH, Margulies Z, Weng M, Snyder ET, Dermitzakis RE, Thurman et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, Woodward WA, Hsu JM, Hortobagyi GN, Hung MC 2011. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell 19: 86–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. 2011. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146: 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al. 2010. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463: 360–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: R60 doi: 10.1186/gb-2003-4-9-r60 [PubMed] [Google Scholar]
- Ding Y, Lee JF, Lu H, Lee MH, Yan DH 2006. Interferon-inducible protein IFIXalpha1 functions as a negative regulator of HDM2. Mol Cell Biol 26: 1979–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin AM, Huang TH, Toland AE 2009. Epigenetic alterations in the breast: Implications for breast cancer detection, prognosis, and treatment. Semin Cancer Biol 19: 165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhofer TA, Minoda A, Klugman S, Lee K, Kolasinska-Zwierz P, Alekseyenko AA, Cheung MS, Day DS, Gadel S, Gorchakov AA, et al. 2011. An assessment of histone-modification antibody quality. Nat Struct Mol Biol 18: 91–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M 2007. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet 8: 286–298 [DOI] [PubMed] [Google Scholar]
- Esteller M 2008. Epigenetics in cancer. N Engl J Med 358: 1148–1159 [DOI] [PubMed] [Google Scholar]
- Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG 1999. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res 59: 793–797 [PubMed] [Google Scholar]
- Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, Bussaglia E, Prat J, Harkes IC, Repasky EA, et al. 2000. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst 92: 564–569 [DOI] [PubMed] [Google Scholar]
- Everhard S, Tost J, El Abdalaoui H, Criniere E, Busato F, Marie Y, Gut IG, Sanson M, Mokhtari K, Laigle-Donadey F, et al. 2009. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro-oncol 11: 348–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, Hamilton SR, Preisinger AC, Thomas G, Kinzler KW, et al. 1990. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science 247: 49–56 [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B 1983. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301: 89–92 [DOI] [PubMed] [Google Scholar]
- Florl AR, Lower R, Schmitz-Drager BJ, Schulz WA 1999. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br J Cancer 80: 1312–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL 1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci 89: 1827–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, Ehrlich M 1983. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res 11: 6883–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe JC, Bhattacharya S, Merchant B, Bassett E, Swisshelm K, Feiler HS, Wyrobek AJ, Stampfer MR 2009. Molecular distinctions between stasis and telomere attrition senescence barriers shown by long-term culture of normal human mammary epithelial cells. Cancer Res 69: 7557–7568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951 [DOI] [PubMed] [Google Scholar]
- Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA 2009. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci 107: 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. 2011. Increased methylation variation in epigenetic domains across cancer types. Nat Genet 43: 768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. 2010. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6: 479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman A, Chess A 2007. Gene body-specific methylation on the active X chromosome. Science 315: 1141–1143 [DOI] [PubMed] [Google Scholar]
- Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, Fridlyand J, Sahin A, Agarwal R, Joy C, et al. 2009. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res 69: 4116–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, et al. 2007. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 8: R76 doi: 10.1186/gb-2007-8-5-r76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz JI, He X, Fan C, Perou CM 2008. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res 10: R75 doi: 10.1186/bcr2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibi K, Goto T, Mizukami H, Kitamura Y, Sakata M, Saito M, Ishibashi K, Kigawa G, Nemoto H, Sanada Y 2009a. MGMT gene is aberrantly methylated from the early stages of colorectal cancers. Hepatogastroenterology 56: 1642–1644 [PubMed] [Google Scholar]
- Hibi K, Sakata M, Yokomizo K, Kitamura YH, Sakuraba K, Shirahata A, Goto T, Mizukami H, Saito M, Ishibashi K, et al. 2009b. Methylation of the MGMT gene is frequently detected in advanced gastric carcinoma. Anticancer Res 29: 5053–5055 [PubMed] [Google Scholar]
- Hinshelwood RA, Clark SJ 2008. Breast cancer epigenetics: Normal human mammary epithelial cells as a model system. J Mol Med 86: 1315–1328 [DOI] [PubMed] [Google Scholar]
- Hoadley KA, Weigman VJ, Fan C, Sawyer LR, He X, Troester MA, Sartor CI, Rieger-House T, Bernard PS, Carey LA, et al. 2007. EGFR-associated expression profiles vary with breast tumor subtype. BMC Genomics 8: 258 doi: 10.1186/1471-2164-8-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R 1979. A new theory of carcinogenesis. Br J Cancer 40: 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D 2006. The UCSC Known Genes. Bioinformatics 22: 1036–1046 [DOI] [PubMed] [Google Scholar]
- Hu Z, Fan C, Livasy C, He X, Oh DS, Ewend MG, Carey LA, Subramanian S, West R, Ikpatt F, et al. 2009. A compact VEGF signature associated with distant metastases and poor outcomes. BMC Med 7: 9 doi: 10.1186/1741-7015-7-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41: 178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H, Baffa R, Numata SI, Murakumo Y, Rattan S, Inoue H, Mori M, Fidanza V, Alder H, Croce CM 1999. The FEZ1 gene at chromosome 8p22 encodes a leucine-zipper protein, and its expression is altered in multiple human tumors. Proc Natl Acad Sci 96: 3928–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal, and inner cell mass specification. Nature 466: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, et al. 2011. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331: 1199–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA 1999. The DNA methylation paradox. Trends Genet 15: 34–37 [DOI] [PubMed] [Google Scholar]
- Jones PA, Taylor SM, Wilson VL 1983. Inhibition of DNA methylation by 5-azacytidine. Recent Results Cancer Res 84: 202–211 [DOI] [PubMed] [Google Scholar]
- Jovanovic J, Ronneberg JA, Tost J, Kristensen V 2010. The epigenetics of breast cancer. Mol Oncol 4: 242–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminskas E, Farrell AT, Wang YC, Sridhara R, Pazdur R 2005. FDA drug approval summary: Azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist 10: 176–182 [DOI] [PubMed] [Google Scholar]
- Kelly TK, De Carvalho DD, Jones PA 2010. Epigenetic modifications as therapeutic targets. Nat Biotechnol 28: 1069–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. 2003. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci 100: 11606–11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohonen-Corish MR, Sigglekow ND, Susanto J, Chapuis PH, Bokey EL, Dent OF, Chan C, Lin BP, Seng TJ, Laird PW, et al. 2007. Promoter methylation of the mutated in colorectal cancer gene is a frequent early event in colorectal cancer. Oncogene 26: 4435–4441 [DOI] [PubMed] [Google Scholar]
- Komashko VM, Farnham PJ 2010. 5-azacytidine treatment reorganizes genomic histone modification patterns. Epigenetics 5: 229–240 [DOI] [PubMed] [Google Scholar]
- Laird PW 2003. The power and the promise of DNA methylation markers. Nat Rev Cancer 3: 253–266 [DOI] [PubMed] [Google Scholar]
- Laird PW 2010. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet 11: 191–203 [DOI] [PubMed] [Google Scholar]
- Lakshmikuttyamma A, Scott SA, DeCoteau JF, Geyer CR 2009. Reexpression of epigenetically silenced AML tumor suppressor genes by SUV39H1 inhibition. Oncogene 29: 576–588 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13: 1192 –1200 [DOI] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. 2010. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 363: 2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Park YJ, McLean CM, Dokshin GA, Persson JM, Herman H, Pasini D, Miro X, Donohoe ME, Lee JT, et al. 2008. Antagonism between DNA and H3K27 methylation at the imprinted Rasgrf1 locus. PLoS Genet 4: e1000145 doi: 10.1371/journal.pgen.1000145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, et al. 2011. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471: 68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. 2010. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466: 253–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez L, Benigno BB, McDonald JF 2004. L1 and HERV-W retrotransposons are hypomethylated in human ovarian carcinomas. Mol Cancer 3: 12 doi: 10.1186/1476-4598-3-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Montag-Lowy M, Goldman RD 1994. Dynamic properties of nuclear lamins:Lamin B is associated with sites of DNA replication. J Cell Biol 125: 1201–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. 2010. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 42: 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins M, Perreard L, Quackenbush JF, Gauthier N, Bayer S, Ellis M, Parker J, Perou CM, Szabo A, Bernard PS 2007. Agreement in breast cancer classification between microarray and quantitative reverse transcription PCR from fresh-frozen and formalin-fixed, paraffin-embedded tissues. Clin Chem 53: 1273–1279 [DOI] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10: 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak P, Jensen T, Oshiro MM, Watts GS, Kim CJ, Futscher BW 2008. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer Res 68: 8616–8625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DS, Troester MA, Usary J, Hu Z, He X, Fan C, Wu J, Carey LA, Perou CM 2006. Estrogen-regulated genes predict survival in hormone receptor-positive breast cancers. J Clin Oncol 24: 1656–1664 [DOI] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. 2009. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 27: 1160–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A 2009. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res 37: e123 doi: 10.1093/nar/gkp596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, et al. 2010. The genetic landscape of the childhood cancer medulloblastoma. Science 331: 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38: 603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreard L, Fan C, Quackenbush JF, Mullins M, Gauthier NP, Nelson E, Mone M, Hansen H, Buys SS, Rasmussen K, et al. 2006. Classification and risk stratification of invasive breast carcinomas using a real-time quantitative RT-PCR assay. Breast Cancer Res 8: R23 doi: 10.1186/bcr1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard2011. http://picard.sourceforge.net
- Prak ET, Kazazian HH Jr 2000. Mobile elements and the human genome. Nat Rev Genet 1: 134–144 [DOI] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM 2010. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 12: R68 doi: 10.1186/bcr2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR 2005. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts, and proteins. Nucleic Acids Res 33: D501–D504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolny C, Mazzone M, Tugues S, Laoui D, Johansson I, Coulon C, Squadrito ML, Segura I, Li X, Knevels E, et al. 2011. HRG inhibits tumor growth and metastasis by inducing macrophage polarization and vessel normalization through downregulation of PlGF. Cancer Cell 19: 31–44 [DOI] [PubMed] [Google Scholar]
- Ruike Y, Imanaka Y, Sato F, Shimizu K, Tsujimoto G 2010. Genome-wide analysis of aberrant methylation in human breast cancer cells using methyl-DNA immunoprecipitation combined with high-throughput sequencing. BMC Genomics 11: 137 doi: 10.1186/1471-2164-11-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 18: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz WA 2006. L1 retrotransposons in human cancers. J Biomed Biotechnol 2006: 83672 doi: 10.1155/JBB/2006/83672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274 [DOI] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. 2009. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit M, Riccardi L, Velasco P, Brown LF, Hawighorst T, Bornstein P, Detmar M 1999. Thrombospondin-2: A potent endogenous inhibitor of tumor growth and angiogenesis. Proc Natl Acad Sci 96: 14888–14893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia T, Smalley SV, Kohen P, Munoz A, Solis LM, Corvalan A, Faundez P, Devoto L, Camus M, Alvarez M, et al. 2008. Promoter hypermethylation of BRCA1 correlates with absence of expression in hereditary breast cancer tumors. Epigenetics 3: 157–163 [DOI] [PubMed] [Google Scholar]
- Teer JK, Bonnycastle LL, Chines PS, Hansen NF, Aoyama N, Swift AJ, Abaan HO, Albert TJ, Margulies EH, Green ED, et al. 2010. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res 20: 1420–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S, et al. 2011. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331: 593–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O'Meara S, Teague J, et al. 2009. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet 41: 521–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al. 2011. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469: 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkel P, Angrand PO 2007. The control of histone lysine methylation in epigenetic regulation. Biochimie 89: 1–20 [DOI] [PubMed] [Google Scholar]
- Weigelt B, Hu Z, He X, Livasy C, Carey LA, Ewend MG, Glas AM, Perou CM, Van't Veer LJ 2005. Molecular portraits and 70-gene prognosis signature are preserved throughout the metastatic process of breast cancer. Cancer Res 65: 9155–9158 [DOI] [PubMed] [Google Scholar]
- Wild L, Flanagan JM 2010. Genome-wide hypomethylation in cancer may be a passive consequence of transformation. Biochim Biophys Acta 1806: 50–57 [DOI] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. 2007. The genomic landscapes of human breast and colorectal cancers. Science 318: 1108–1113 [DOI] [PubMed] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE 2010. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329: 444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap DBJ, Chu T, Berg M, Schapira SW, Cheng A, Moradian RD, Morin AJ, Mungall B, Meissner M, Boyle et al. 2010. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117: 2451–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan BZ, Miller MJ, Keck CL, Zimonjic DB, Thorgeirsson SS, Popescu NC 1998. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res 58: 2196–2199 [PubMed] [Google Scholar]