Abstract
A subset of colorectal cancers was postulated to have the CpG island methylator phenotype (CIMP), a higher propensity for CpG island DNA methylation. The validity of CIMP, its molecular basis, and its prognostic value remain highly controversial. Using MBD-isolated genome sequencing, we mapped and compared genome-wide DNA methylation profiles of normal, non-CIMP, and CIMP colon specimens. Multidimensional scaling analysis revealed that each specimen could be clearly classified as normal, non-CIMP, and CIMP, thus signifying that these three groups have distinctly different global methylation patterns. We discovered 3780 sites in various genomic contexts that were hypermethylated in both non-CIMP and CIMP colon cancers when compared with normal colon. An additional 2026 sites were found to be hypermethylated in CIMP tumors only; and importantly, 80% of these sites were located in CpG islands. These data demonstrate on a genome-wide level that the additional hypermethylation seen in CIMP tumors occurs almost exclusively at CpG islands and support definitively that these tumors were appropriately named. When these sites were examined more closely, we found that 25% were adjacent to sites that were also hypermethylated in non-CIMP tumors. Thus, CIMP is also characterized by more extensive methylation of sites that are already prone to be hypermethylated in colon cancer. These observations indicate that CIMP tumors have specific defects in controlling both DNA methylation seeding and spreading and serve as an important first step in delineating molecular mechanisms that control these processes.
Promoter CpG island hypermethylation is a common strategy for cancer cells to silence tumor-suppressor gene expression (Jones and Baylin 2002). The notion of CpG island methylator phenotype (CIMP) arose from the initial observation that a subset of colorectal cancers has higher frequency of DNA hypermethylation at candidate tumor-suppressor promoters (Toyota et al. 1999). Thus far, the hypermethylated sites used to characterize CIMP have not been standardized, leading to inconsistencies in whether a colon cancer is appropriately described as CIMP (Yamashita et al. 2003; Issa 2004; Anacleto et al. 2005). Consequently, clinical studies attempting to correlate CIMP with prognosis and responsiveness to 5-fluorouracil chemotherapy have produced contradicting results (Van Rijnsoever et al. 2003; Shen et al. 2007; Barault et al. 2008; Lee et al. 2008; Kim et al. 2009; Ogino et al. 2009). Therefore, the uncertainty of CIMP's prognostic value may be linked to discrepancies in molecular classification. Objective validation of CIMP as an authentic genome-wide phenomenon and delineating a consistent set of molecular markers to classify tumors will be vital in determining its clinical relevance.
As a first step in unraveling the CIMP conundrum, we propose that a comparison of the DNA methylomes of normal colon, non-CIMP colon cancer, and CIMP colon cancer patient specimens is necessary. While genome-scale, array-based platforms may be sufficient in visualizing subgroups of colon cancers with a distinct preference for DNA methylation at loci present on the arrays, this type of approach cannot assess if (1) DNA methylation patterns in all genomic contexts are significantly different among the subgroups and are similar within each subgroup; (2) CpG islands are truly the preferential methylation targets in CIMP; and (3) additional molecular features, other than CpG island DNA methylation, can distinguish CIMP. Sequencing-based approaches, on the other hand, can objectively survey the genome to allow construction of individual DNA methylome profiles to answer the above questions. Currently, the most widely used marker panel for defining CIMP consists of CpG islands at CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1 (Weisenberger et al. 2006). In addition to defining DNA methylation markers for CIMP, this study also uncovered a close association between CIMP, BRAF mutation, and microsatellite instability (MSI). Here we use massively parallel sequencing technology to directly test, on the whole-genome level with minimum ascertainment bias, if these markers truly differentiate between subgroups of colon cancers with different propensities for CpG island DNA hypermethylation.
Results
CIMP markers differentiate between subgroups with distinct genome-wide DNA methylation patterns
We used MBD-isolated genome sequencing (MiGS) (Serre et al. 2010) to map the DNA methylomes of three sets of normal colon, non-CIMP colon cancers, and CIMP colon cancers. Briefly, we incubated randomly sheared genomic DNA from each sample with recombinant methyl CpG-biding domain (MBD) proteins to capture methylated DNA fragments for sequencing on the Illumina Genome Analyzer II. The CIMP tumors were selected based on DNA methylation at the five markers described by Weisenberger et al. and the presence of MSI and BRAF mutations (Weisenberger et al. 2006). The non-CIMP tumors have no methylation at the five markers, do not have MSI, and harbor KRAS mutations, which are mutually exclusive of BRAF mutations in colon cancer.
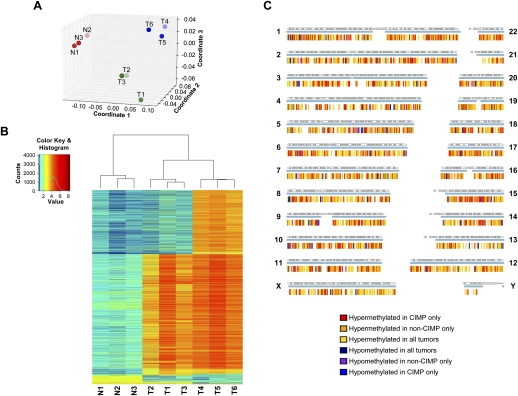
At a threshold of <5% false discovery rate (FDR), we assembled the DNA methylome of each specimen with the sequencing reads and visualized the nine samples based on their global DNA methylation profiles in a multidimensional scaling plot (Fig. 1A). The samples clustered to form three distinct groups consisting of normal (N1, N2, and N3), non-CIMP (T1, T2, and T3), and CIMP (T4, T5, and T6) specimens, respectively. This grouping pattern was independently verified by Principle Component Analysis and unsupervised hierarchical clustering using correlation distance (data not shown). This result suggests that our sample selection criteria robustly identified two populations of colon cancers that differ in their genome-wide DNA methylation profiles.
Figure 1.
Mapping and comparisons of the DNA methylomes of normal colon, non-CIMP colon cancer, and CIMP colon cancer specimens. (A) Multidimensional scaling plot of the nine samples using their individual genome-wide DNA methylation profiles. (Red dots) Normal colon samples; (green dots) non-CIMP colon cancer samples; (blue dots) CIMP colon cancer samples. (B) Heatmap and unsupervised hierarchical clustering of the nine samples based on their individual DNA methylation status at 6135 sharp yes/no differentially methylated sites. (N1, N2, N3) Normal colon specimens; (T1, T2, T3) non-CIMP cancer specimens; (T4, T5, T6) CIMP cancer specimens. (C) Spatial distribution of sharp yes/no differentially methylated sites in the genome. Each sharp yes/no differentially methylated site is represented by a vertical tick below each chromosome. Each differential methylation pattern is represented by one color according to the color key. The blue horizontal line under each chromosome represents regions of the chromosome where DNA methylation was detected in any of the nine samples.
CIMP tumors have significantly more CpG island DNA methylation on a genomic scale
Using the grouping structure revealed above, we tested for sites of differential DNA methylation among the three categories. While we could detect both quantitative and qualitative differential methylation in our data set, quantitative differences may be confounded by other genomic aberrations, such as copy number variations. Therefore, we focused our analysis on qualitative, sharp yes/no differential DNA methylation. We defined sharp yes/no differential DNA methylation as sites whose methylation pattern is consistent among samples in the same group but show a clear presence or absence of DNA methylation among the different groups. At <5% FDR, we identified 6135 differentially methylated sites between normal, non-CIMP, and CIMP (Table 1; Supplemental Table 1). Unsupervised clustering of the nine samples based on their methylation status at these 6135 sites corroborated the grouping structure observed using the whole-genome data (Fig. 1B). We detected sharp yes/no differential DNA methylation on all 24 chromosomes (Fig. 1C). Bisulfite sequencing performed on 15 randomly selected sites verified the MiGS data with 100% validation rate (examples in Supplemental Figs. 1, 2).
Table 1.
Summary of sharp yes/no differential DNA methylation sites
Consistent with previous reports, CIMP and non-CIMP colon cancers share a large number of hypermethylated sites (3780) when compared with normal colon (Table 1; Jones and Baylin 2002; Weisenberger et al. 2006). These commonly hypermethylated sites in all tumors are enriched for CpG islands but contain less repetitive family sequences than the genome average (Supplemental Table 2). These enrichment patterns are consistent with an existing paradigm of hotspots for DNA hypermethylation in colon cancers. In addition to the shared hypermethylation, CIMP tumors have an additional 2026 unique hypermethylated sites compared with only 49 sites that are unique to non-CIMP tumors. This difference (Chi-square test, p < 2.2 × 10−16) supports the hypothesis that CIMP tumors have higher levels of DNA methylation when compared with non-CIMP tumors. Furthermore, we tested select CIMP-specific hypermethylated loci in adjacent normal specimens using methylation-specific PCR (MSP) and verified that there is no basal-level DNA methylation at these sites in the normal colon, from which the CIMP tumors arose (Supplemental Fig. 3).
CIMP-specific hypermethylation is not restricted to any one genomic context, but CpG islands account for 80% of these sites (Table 1). Proportionally, promoter hypermethylation increased from 40% in hypermethylated sites shared by all tumors to 47% in CIMP-unique sites, and this increase is largely attributable to CpG island hypermethylation in the promoter context. By directly comparing the coverage of CpG island sequences by the CIMP-specific hypermethylated sites to that of the genome, we determined that this apparent enrichment is statistically significant (one-sided Binomial test, p < 2.23 × 10−308) (Supplemental Table 2). It is important to note that in CIMP tumors, the proportion of total detected methylation that did not overlap CpG islands was still significantly higher than methylation that did overlap CpG islands (83.8% vs. 16.2%), demonstrating that the enrichment for CpG island hypermethylation cannot be explained by an assay bias (Supplemental Fig. 4). These data together demonstrate, for the first time on a genome-wide scale, that CIMP cancers truly have a higher propensity for CpG island hypermethylation.
Genome-wide analysis reveals a novel methylation expansion phenotype in CIMP
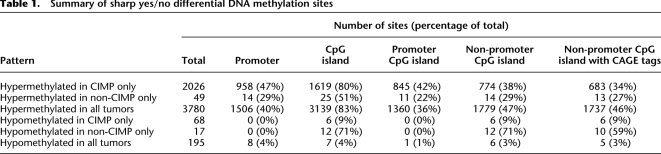
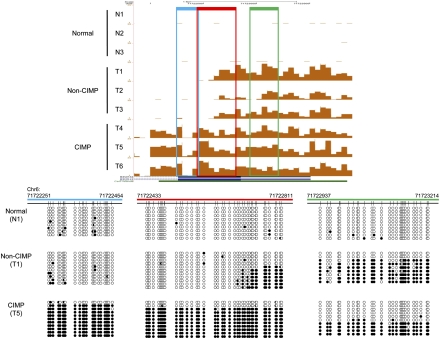
Further examination of differential methylation revealed that 506 CIMP-unique sites were contiguous with sites hypermethylated in all tumors. These sites represent CIMP-specific extension of tumor-specific methylation, and B3GAT2 is an example of a gene promoter where this phenomenon is observed (Fig. 2). Both non-CIMP and CIMP tumors showed hypermethylation of this gene, but CIMP tumors displayed additional hypermethylated sequences toward the 3′ end when compared with non-CIMP tumors. Such extensions of hypermethylated regions in CIMP tumors were seen in six possible arrangements at 488 regions throughout the genome (Fig. 3A; Supplemental Table 1). At these locations, the lengths of the methylated regions in CIMP were significantly larger (Welch t-test, p = 8.6 × 10−70) than those in non-CIMP tumors (Fig. 3B). This regional expansion is specific to these 488 sites and was not observed at other hypermethylated sites (Fig. 3C). The reverse phenomenon, extension of hypermethylated sites in non-CIMP tumors, was observed at only eight regions.
Figure 2.
Bisulfite sequencing of the B3GAT2 promoter. The B3GAT2 promoter is an example of a hypermethylation site shared by all tumors but is more extensively methylated in CIMP tumors. The UCSC Genome Browser screen capture shows the DNA methylation signal in each sample. The rectangles mark the regions validated by bisulfite sequencing and are color-coded to the corresponding bisulfite sequencing data. Each circle represents a CpG dinucleotide. (Black circles) Methylated cytosines; (white circles) unmethylated cytosines.
Figure 3.
Expansion of hypermethylation sites in CIMP. (A) Summary of the different types of CIMP-unique expansion of hypermethylation sites. Each colored rectangle denotes a positive methylation signal. (Yellow rectangles) DNA methylation regions shared by all tumors; (orange rectangles) DNA methylation regions observed only in CIMP tumors. (B) Boxplot comparing the sizes of hypermethylated regions in non-CIMP and CIMP tumors at the 488 sites with apparent expansion in CIMP. (Red box) Contains sites in non-CIMP tumors; (blue box) contains sites in CIMP tumors. The P-value for the difference detected is indicated. (C) Boxplot comparing the sizes of all other hypermethylated regions in non-CIMP and CIMP tumors. (Red box) Contains sites shared by all tumors (n = 3133); (blue box) contains sites unique to CIMP tumors (n = 1507). The P-value for the difference detected is indicated. (D) The size distribution of the expanded methylation regions in CIMP. The number of sites is tabulated for each length of the CIMP-specific expansions observed. (E) Motif analysis of all expansion regions (entire regions containing both the shared hypermethylation and the CIMP-specific expansions, n = 1180), unidirectional subset (CIMP-specific expansion that extends in only one direction from the shared hypermethylation, n = 423), and bidirectional subset (CIMP-specific expansion that extends both 5′ and 3′ from the shared hypermethylation, n = 22). The frequency of observing each motif within each subset of sequences is expressed as a percentage (% sites). The P-value of enrichment testing for individual motifs in each data set is indicated.
Recently, DNA methylation patterning has been linked to nucleosome positioning in human cells (Jeong et al. 2009; Chodavarapu et al. 2010), prompting us to investigate if this expansion phenotype in CIMP could be explained by nucleosome deposition. The sizes of the expanded regions were plotted on a histogram, but no distinct periodicity relating to nucleosome size was found (Fig. 3D). However, our approach relied on analyzing the genome using 50-bp windows and may not have the resolution needed to capture this phenomenon. Another possible mechanism for the spreading of DNA methylation in CIMP is targeted DNA methylation by specific DNA binding factors. Through sequence motif analysis, we identified 19 motifs that occurred frequently in these regions (Supplemental Table 3). Many of the factors that bind to these motifs, including SP1 and TFAP2A (AP2-α), are known to regulate CpG-island-containing promoters and, therefore, are not surprising to be associated with these hypermethylated regions. However, our analysis also identified unexpected candidates, such as ZBTB7B (CKROX) and CNOT3 (Fig. 3E), which warrant further empirical validation to establish a causal relationship to the observed DNA methylation spreading in CIMP.
Interestingly, unidirectional expansion of DNA methylation occurs in 86.7% of these regions as opposed to only 2.3% exhibiting bidirectional expansion (Fig. 3A). We performed motif analysis separately on these two subsets to determine if qualitative differences exist between them to explain the frequency disparity. While most identified motifs are common to both subsets, we did discover motifs unique to each subset that may suggest a more deliberate mechanism in dictating the pattern of DNA methylation spreading (Supplemental Table 3). For example, the binding motifs for transcription factors, WT1 and TEAD2 (ETF), were detected exclusively in all sequences within the bidirectional subset, while the binding motifs for ZNF148 (ZBP89) and RREB1 were found only in the unidirectional subset (Fig. 3E). Alternatively, the observed directionality could be a result of functional selection. The initial spreading of DNA methylation in CIMP may be bidirectional at most sites as sequelae of loss of boundary control. However, only the segments that confer functional advantages for the cancer cells were maintained through clonal expansion and observed in the resultant cancer. Supportive of this hypothesis, at least 67 regions containing the CIMP-specific extensions resulted in DNA methylation at either additional gene promoters or multiple annotated alternative transcription start sites (Supplemental Table 1). Of these, four regions displayed bidirectional expansions, while the remaining 63 showed unidirectional expansions.
Additional DNA hypermethylation in CIMP reinforces core cancer pathway changes
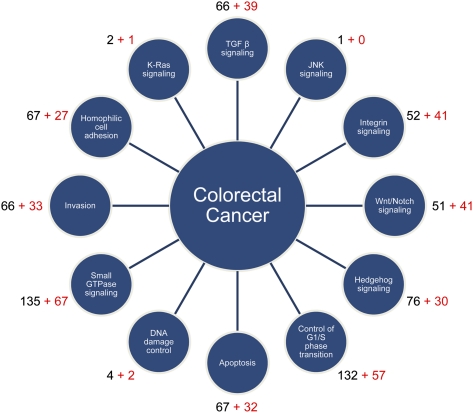
Of the 3975 sites differentially methylated between normal and tumor, 1514 sites overlapped with 1465 RefSeq gene promoters (Supplemental Table 1). Altered methylation of these gene promoters likely affect transcriptional activity, resulting in changes to cellular functions (Suzuki and Bird 2008). To understand how these genes can promote tumor development, we mapped them to the 12 core cancer signaling pathways (Fig. 4; Supplemental Table 4; Jones et al. 2008). Four hundred thirty hypermethylated genes shared by all tumors mapped to at least one pathway in this analysis. We performed a similar analysis for RefSeq gene promoters differentially methylated in CIMP cancers only and identified an additional 204 hypermethylated genes mapping to the core cancer pathways (Fig. 4). Using publicly available expression data sets (Rhodes et al. 2004; Parkinson et al. 2005), we were able to verify congruent differential gene expression in colon cancers for at least 137 and 57 genes in tumor-specific and CIMP-specific hypermethylated gene sets, respectively (Supplemental Table 5). Through this analysis, we could see that aberrant DNA methylation in colon cancers affects all 12 cancer signaling pathways. Furthermore, by separating the genes shared by all tumors from those unique to CIMP, additional epigenetic perturbation in almost all aspects of tumor biology is evident in CIMP (Fig. 4).
Figure 4.
Differentially methylated genes mapped to the 12 core cancer pathways. The 12 signaling pathways and processes that are most frequently altered in cancer are shown in satellite circles. The number of differentially methylated genes mapped to each of the 12 categories is indicated next to each circle. (Black numbers) The number of genes shared by all tumors; (red numbers) the number of additional genes unique to CIMP tumors.
Of note, this may still be an incomplete view of the effects of altered DNA methylation in colon cancers because non-promoter differential methylation, which was omitted in our pathway analysis, can also contribute to altered genome expression. For instance, non-promoter CpG islands have been demonstrated to represent cryptic alternative transcription start sites (Maunakea et al. 2010) and yet-to-be-characterized gene promoters (Illingworth et al. 2010). In fact, cap analysis of gene expression (CAGE) (Kodzius et al. 2006) sequence tags were detected at significant proportions of differentially methylated non-promoter CpG islands in our study (Table 1), providing evidence of possible transcription initiation at these sites. Furthermore, DNA methylation can modulate miRNA expression (Han et al. 2007; Lujambio et al. 2007; Toyota et al. 2008; Yan et al. 2011) and binding of regulatory proteins, such as CCCTC-binding factor (CTCF) (Bell and Felsenfeld 2000; Yang et al. 2003). Thus, non-promoter differential methylation overlapping these genomic elements can also alter cellular functions through differential expression of miRNAs and regulation of enhancer activities (Supplemental Table 1). In fact, differentially methylated sites shared by all tumors and unique to CIMP tumors both show enrichment for empirical CTCF binding sites when compared with the genome (Supplemental Table 2).
Unique sequence motifs identified in CIMP-specific hypermethylated sites
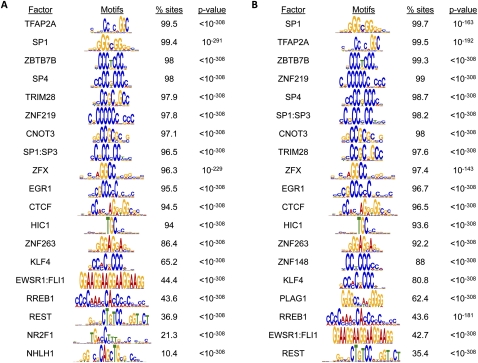
To begin addressing the mechanism behind increased DNA hypermethylation in CIMP tumors, we performed DNA sequence motif analysis for hypermethylated sites shared by all tumors and those unique to CIMP as separate data sets. The two sets of sequences share a large number of enriched motifs and show significant enrichment for binding sites of known CpG island-associated transcription factors, such as SP1, SP4, and TFAP2A (Fig. 5). Besides these expected motifs, we also identified some interesting common candidate transcription factors, such as TRIM28 and ZNF263, which are not readily associated with DNA methylation. Interestingly, TRIM28 is known to associate with SETDB1, a histone H3, lysine 9–specific methyltransferase, and HP1 to mediate heterochromatin formation (Schultz et al. 2002; Sripathy et al. 2006).
Figure 5.
Motif analysis for differentially methylated sites. Motif analysis for hypermethylated sites shared by all tumors (n = 3780) (A) and CIMP-specific hypermethylated sites (n = 2026) (B). The frequency of observing each motif within each subset of sequences is expressed as a percentage (% sites). The P-value of enrichment testing for individual motif in each data set is indicated.
More importantly, we identified the binding motifs for ZNF148 and PLAG1 to be uniquely enriched in the CIMP-specific hypermethylated subset (Fig. 5B). ZNF148 recognition motifs were found in 88% of the CIMP-specific hypermethylated sequences, while PLAG1 sites were detected in 62.4% of these same sequences. ZNF148, a Krüppel family zinc finger protein (Wang et al. 1993; Tommerup and Vissing 1995), binds to GC-rich DNA sequences and has been shown to recruit histone-modifying enzymes to modulate transcriptional activities at target promoters (Wu et al. 2007; Feng et al. 2009). PLAG1, an imprinted zinc finger protein, is oncogenic in salivary gland tumors and lipoblastoma (Kas et al. 1997; Astrom et al. 1999; Hibbard et al. 2000). While neither factor has been shown to directly associate with or target DNA methylation, these proteins may be reasonable candidates for future mechanistic studies. Their expression patterns in normal colon, non-CIMP cancers, and CIMP cancers as well as direct binding to CIMP-specific hypermethylated sequences will help clarify their roles in the initiation and propagation of aberrant DNA methylation in CIMP colon cancers.
Discussion
Aberrant DNA methylation patterns are a hallmark of human cancers. This phenomenon is particularly well studied in colorectal carcinoma, where global hypomethylation as determined by altered digestion patterns with methylation-sensitive restriction enzymes was first reported more than 25 years ago (Feinberg and Vogelstein 1983). Gene-specific hypermethylation was described several years later (de Bustros et al. 1988); and with the development of new techniques such as bisulfite genomic sequencing (Clark et al. 1994) and methylation-specific PCR (Herman et al. 1996), the number of hypermethylated loci characterized in colon cancer has skyrocketed. Upon closer examination of these sites, there was growing evidence for the presence of a subset of colon cancers that consistently showed hypermethylation of a specific set of genes. This subset of colon cancers was described as having a CpG island methylator phenotype. The original report used a candidate gene approach to establish that CIMP tumors frequently have the CDKN2A (also known as p16), THBS1, and MLH1 loci hypermethylated (Toyota et al. 1999). Clinical studies attempting to correlate CIMP status with clinical outcome have been confusing. CIMP tumors are typically associated with proximal tumor location, but whether it confers a good or poor prognosis is still unresolved and may depend on other markers such as BRAF mutation, KRAS mutation, and microsatellite instability (Van Rijnsoever et al. 2003; Barault et al. 2008; Ogino et al. 2009; Dahlin et al. 2010). These compounding variables along with the lack of a consistent, objective definition of CIMP have hindered the ability to determine its true clinical impact.
In light of this uncertainty regarding the behavior of CIMP tumors, we have characterized the DNA methylomes of CIMP and non-CIMP colon cancers using the definition of CIMP as proposed by Weisenberger et al. (2006). Specifically, the CIMP tumors we studied were hypermethylated at CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1, microsatellite unstable, and harbored BRAF mutations. The non-CIMP tumors we studied did not have any hypermethylation at those five loci, were microsatellite stable, and harbored KRAS mutations. Using these stringent definitions, the MiGS analysis established the presence of three well-defined groups according to their genome-wide DNA methylation patterns: normal colon, CIMP tumors, and non-CIMP tumors. This result clearly demonstrates that, as long as the definition of CIMP is strictly adhered to, CIMP and non-CIMP tumors are distinctly different. Thus, future studies using whole-genome DNA methylation analysis can use the clusters that we have generated to determine whether a tumor is more related to CIMP or non-CIMP. For example, tumors with varying numbers of positive CIMP markers, microsatellite stability, BRAF mutation, and KRAS mutation status can be placed on this multidimensional scaling plot to visualize how they ultimately cluster with respect to the CIMP and non-CIMP tumors. This will help objectively define the threshold for CIMP classification and identify additional distinct subgroups. Perhaps with this information, the problem of CIMP as a prognostic indicator can finally be solved.
One particularly striking result of our study is the discovery that CIMP tumors truly are prone to increased aberrant methylation. Moreover, CIMP tumors do not indiscriminately increase methylation in all regions of the genome. We showed that, of the hypermethylation sites that are unique to CIMP tumors, 80% of them occur in CpG islands and 42% map to promoter CpG islands. Thus, CIMP is truly an appropriate name for these tumors. MiGS is particularly suitable for making this observation since it is a technique that relies on next-generation sequencing and has only limited ascertainment bias (Bock et al. 2010; Harris et al. 2010). We have also described the functional consequence of this dramatic increase in CpG island hypermethylation by comparing methylation status with available gene expression data. All 12 core cancer signaling pathways are affected, and additional sites that are only hypermethylated in CIMP tumors may serve to reinforce the epigenetic changes to these pathways that typically occur in colorectal cancers. As epigenetic therapies gain traction, evaluation of the methylation status of these sites will be an important aspect of assessing treatment effect.
How abnormal methylation patterns develop in colon cancer is still unclear. There are some data suggesting that the expression of DNA methyltransferase 1, the enzyme responsible for maintaining DNA methylation in mammals, becomes progressively dysregulated during colorectal carcinogenesis (De Marzo et al. 1999), but the timing of this dysregulation with respect to the genesis of altered DNA methylation patterns remains to be clarified. Our detailed catalog of differentially methylated sites provides an attractive opportunity for generating new hypotheses for the origin of aberrant DNA methylation in colon cancer. In the present study, we approached the topic by analyzing these sequences for frequently occurring motifs of known DNA binding factors. While many of enriched motifs belong to transcription factors that are known to regulate CpG-island-containing promoters, we did discover some new candidates that may shed light on the mechanism behind abnormal DNA methylation. For instance, we identified a significant enrichment for binding motifs of TRIM28, also known as KAP-1, in the hypermethylated sites shared by all tumors. TRIM28 binds to the histone H3, lysine 9 (H3K9) methyltransferase, SETDB1, and HP1 to coordinate H3K9 methylation, heterochromatin formation, and gene silencing (Schultz et al. 2002; Sripathy et al. 2006; O'Geen et al. 2007; Takada et al. 2007). Furthermore, SETDB1 has been shown to directly interact with DNA methyltransferase 3A at epigenetically silenced promoters (Li et al. 2006). The close association between H3K9 methylation and DNA methylation suggests that TRIM28 may be involved in the initiation and maintenance of aberrant DNA methylation.
In addition to demonstrating that CIMP colon cancers indeed have exceptionally high capacity for CpG island hypermethylation, we also uncovered a novel characteristic of the CIMP methylation pattern. The increased DNA hypermethylation in both number and size of methylated regions suggest a universal perturbation in de novo DNA methylation in CIMP. The expansion of methylated regions in CIMP may also signify deregulation of epigenetic boundary control. Curiously, the spreading of DNA methylation in CIMP is most frequently unidirectional. We propose that this phenomenon can be achieved through at least two non-mutually exclusive mechanisms. First, the direction of DNA methylation spreading may be targeted by specific transcription factors. We demonstrated that this is a reasonable hypothesis by showing that binding sites of specific transcription factors, such as ZNF148 and RREB1, are preferentially enriched in the unidirectional subset when compared with the bidirectional subset. The second possibility for the observed pattern is a consequence of functional selective pressure. Many of the unidirectional CIMP-specific expansions extend to cover additional annotated transcriptional start sites or additional neighboring gene promoters. It is possible that the original CIMP-specific DNA methylation spreading was bidirectional at these sites, but only the portion conferring functional changes were selected for and maintained during cancer clonal expansion. Future studies focused on these CIMP-unique methylation patterns should prove useful in identifying the specific molecular mechanisms leading to these abnormalities. Such mechanistic understanding of the origin of CIMP will provide new therapeutic targets for the treatment of CIMP tumors.
Methods
Sample selection and preparation
Genomic DNA for three pairs of non-CIMP cancer and adjacent normal specimens and three pairs of CIMP cancer and adjacent normal specimens were extracted from tissues collected at the Cleveland Clinic Foundation under the appropriate IRB-approved protocol. All specimens were derived from Caucasian males. MiGS was performed for nine specimens: three normal specimens derived from the same patients providing the non-CIMP tumors, three non-CIMP tumors, and three CIMP tumors. The paired normal specimens to the CIMP tumors were characterized by MSP only. We classified CIMP using DNA methylation status at CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1 (Weisenberger et al. 2006). CIMP tumors must have DNA methylation at all five markers while non-CIMP tumors do not have DNA methylation at any of the five markers. We determined microsatellite stability using a panel of 10 markers (BAT25, BAT26, BAT40, BAT34C4, D5S346, D17S250, ACTC, D18S55, D10S197, and MYCL) (Lindor et al. 2006). Tumors containing >30% unstable markers were classified as microsatellite unstable while tumors containing zero unstable markers were deemed microsatellite stable. Mutation analysis for BRAF and KRAS was conducted as previously described (Sanchez et al. 2009). Additional clinical data for each cancer specimen are provided in Supplemental Table 6.
MBD-isolated genome sequencing (MiGS)
We captured and sequenced DNA methylated fractions of the genome as previously described (Serre et al. 2010). Briefly, genomic DNA was sheared randomly by sonication, and 5 μg of sheared genomic DNA was incubated with recombinant his-tagged methyl CpG binding domain proteins conjugated to magnetic beads. The bound fraction was eluted using Proteinase K digestion, purified using phenol:choloroform, and precipitated using isopropanol. We then prepared GAII ChIP-seq libraries from the isolated DNA according to the manufacturer's protocol (Illumina, IP-102-1001). Each library was sequenced on two lanes of the GAII to generate 36-bp single-end reads, and an average of ∼34 million sequencing reads was obtained for each sample.
Sequencing data analysis
Sequencing reads were mapped to the reference human genome (NCBI build 36.1, UCSC Hg18) using Bowtie (Langmead et al. 2009). Only uniquely mapped reads were retained for downstream analysis. If multiple identical reads were present, only one was retained for subsequent analysis. We assembled the individual DNA methylomes as previously described (Serre et al. 2010). Briefly, sequencing reads were extended to 120 bp, the average DNA fragment size in the sequencing library, and assigned to 50-bp non-overlapping windows to construct the methylation profile for each sample. Using the negative Binomial method, we determined that <5% FDR corresponds to windows containing ≥6 reads. The multidimensional scaling plot of the nine samples based on their genome-wide DNA methylation patterns was constructed using the R software.
Differential methylation analysis
Differential DNA methylation among the three categories was identified using our own signal deconvolution, pattern recognition followed by whole-genome scan, and differential testing approach (AH Ting, B Hu, Y Xu, in prep.). Briefly, methylation signals for each sample were decomposed into no-signal, background, and signal using a Bayesian Poisson Mixture model. Methylation sites were determined by consolidating methylated windows in close proximity on the genome and scanning based on the posterior probability map produced from the signal decomposition. For each identified methylation site, a test of differential methylation was performed using a linear mixed-effects model. The statistical significance of the differential methylation sites was assessed using the ANOVA models. For each site, the model contains fixed effects representing the mean methylation levels of the three categories. Random effects were introduced to account for the correlation structure of the data. For a CIMP sample, a random effect was specified to account for the correlation of the repeated windows on the same sample. Since a normal sample was paired with a non-CIMP sample in the sequencing study, a nested random effect was further specified. Finally, sites with <5% FDR were reported in Supplemental Table 1. We defined differential methylation as DNA methylation sites that are consistent among samples within the same group but statistically different among groups. Sharp yes/no methylation patterns refer to differential sites exhibiting clear presence or absence of DNA methylation among the three comparison groups.
Annotation of differential methylation sites
Each sharp yes/no differentially methylated site was annotated with five possible genomic contexts: RefSeq gene promoter, CpG island, CTCF binding site, miRNA coding region, and CAGE evidence (Supplemental Table 1). RefSeq gene promoters were defined as 1000 bp upstream and 500 bp downstream of an annotated RefSeq transcription start site. CpG islands were defined by the CpG island annotation from the reference human genome (NCBI build 36.1, UCSC Hg18) (Fujita et al. 2011). CTCF binding sites were compiled from all available CTCF ChIP-seq data sets (GM12878, H1-hESC, HepG2, HMEC, HSMM, HUVEC, K562, NHEK, and NHLF cells) deposited by the Broad Institute on the UCSC Genome Browser (Fujita et al. 2011). miRNA coding regions were defined as 500 bp upstream and downstream from an annotated miRNA. CAGE evidence was compiled from all available CAGE sequencing data sets (GM12878, H1hESC, HepG2, HUVEC, K562, NHEK, and prostate cells) deposited by the RIKEN Omics Science Center on the UCSC Genome Browser (Kodzius et al. 2006; Fujita et al. 2011). Hypermethylation sites that were expanded in CIMP tumors were noted in the column “CIMP-broad.” We defined sites located <500 bp apart as being in a contiguous region. Repetitive sequences in the differentially methylated sites were annotated using genomic coordinates provided by RepeatMasker. To test for enrichment of specific genomic context in a given differentially methylated data set, we summarized the proportion of 50-bp windows overlapping that genomic context in the data set and compared the frequency with that of the genome average using a one-sided Binomial test under the hypothesis that the frequency in our data set was greater than the genome average.
Bisulfite genomic sequencing and methylation-specific PCR (MSP)
One microgram of genomic DNA from each sample was treated with sodium bisulfite to convert unmethylated cytosines to uracils as previously described (Frommer et al. 1992). Following bisulfite conversion, PCR was performed for each of the selected validation targets using the primers and conditions listed in Supplemental Table 7. The PCR products were gel-purified and cloned into a pCR4-TOPO vector following the TOPO TA kit manual (Invitrogen, K4575-40). Individual clones were selected for sequencing using the M13R primer. MSP was performed for each of the selected targets using the primers and conditions listed in Supplemental Table 7. The PCR products were resolved on 2% agarose gels for visualization.
Motif analysis
Known sequence motifs for DNA binding factors were compiled by combining motif matrices from TRANSFAC (version 11.3) (Matys et al. 2003) and JASPAR (12-Oct-2009) (Sandelin et al. 2004). We used FIMO in the MEME package (version 4.6.0) (Bailey and Elkan 1994) to search for instances of known motifs in a given set of site sequences at a threshold of FDR < 0.01. Motifs are linked to their corresponding transcription factors through the metadata provided by TRASFAC and JASPAR. Enrichment of each motif in each data set was tested against a set of randomly selected sequences with similar length and GC-content distributions using a one-sided Binomial test. We generated such test data sets using the methylated sequences from our study that did not show differential DNA methylation. We reported motifs found in >10% of sites within each tested data set. Motif logos were created using the ceqlogo tool in the MEME package with the default stack algorithm (Schneider and Stephens 1990). If multiple motif matrices are linked to a transcription factor, only the logo associated with the motif with the maximum hits is presented in the figures.
Statistics
All statistical analyses were performed using the R software.
Pathway analysis
Differentially methylated RefSeq genes were analyzed in MetaCore (GeneGo) using Gene Ontology, Pathway Map, and Process Network terms defined for each of the 12 core cancer pathways described previously (Jones et al. 2008). Differentially methylated genes mapped to the 12 pathways are listed in Supplemental Table 4. The expression status for each gene mapped to the core pathways was verified in the Oncomine (Rhodes et al. 2004) and Arrayexpress databases (Parkinson et al. 2005). Genes showing consistent down-regulation in colon adenocarcinomas are listed in Supplemental Table 5.
Data access
The sequencing reads generated and used in this manuscript have been submitted to the NCBI Sequence Read Archive (SRA) (http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession number SRA029584.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.122788.111.
References
- Anacleto C, Leopoldino AM, Rossi B, Soares FA, Lopes A, Rocha JC, Caballero O, Camargo AA, Simpson AJ, Pena SD 2005. Colorectal cancer “methylator phenotype”: fact or artifact? Neoplasia 7: 331–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astrom AK, Voz ML, Kas K, Roijer E, Wedell B, Mandahl N, Van de Ven W, Mark J, Stenman G 1999. Conserved mechanism of PLAG1 activation in salivary gland tumors with and without chromosome 8q12 abnormalities: Identification of SII as a new fusion partner gene. Cancer Res 59: 918–923 [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Barault L, Charon-Barra C, Jooste V, de la Vega MF, Martin L, Roignot P, Rat P, Bouvier AM, Laurent-Puig P, Faivre J, et al. 2008. Hypermethylator phenotype in sporadic colon cancer: Study on a population-based series of 582 cases. Cancer Res 68: 8541–8546 [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485 [DOI] [PubMed] [Google Scholar]
- Bock C, Tomazou EM, Brinkman AB, Muller F, Simmer F, Gu H, Jager N, Gnirke A, Stunnenberg HG, Meissner A 2010. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol 28: 1106–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. 2010. Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Harrison J, Paul CL, Frommer M 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res 22: 2990–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlin AM, Palmqvist R, Henriksson ML, Jacobsson M, Eklöf V, Rutegård J, Oberg A, Van Guelpen BR 2010. The role of the CpG island methylator phenotype in colorectal cancer prognosis depends on microsatellite instability screening status. Clin Cancer Res 16: 1845–1855 [DOI] [PubMed] [Google Scholar]
- de Bustros A, Nelkin BD, Silverman A, Ehrlich G, Poiesz B, Baylin SB 1988. The short arm of chromosome 11 is a “hot spot” for hypermethylation in human neoplasia. Proc Natl Acad Sci 85: 5693–5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marzo AM, Marchi VL, Yang ES, Veeraswamy R, Lin X, Nelson WG 1999. Abnormal regulation of DNA methyltransferase expression during colorectal carcinogenesis. Cancer Res 59: 3855–3860 [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B 1983. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301: 89–92 [DOI] [PubMed] [Google Scholar]
- Feng Y, Wang X, Xu L, Pan H, Zhu S, Liang Q, Huang B, Lu J 2009. The transcription factor ZBP-89 suppresses p16 expression through a histone modification mechanism to affect cell senescence. FEBS J 276: 4197–4206 [DOI] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL 1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci 89: 1827–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. 2011. The UCSC Genome Browser database: update 2011. Nucleic Acids Res 39:D876–D882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Witmer PD, Casey E, Valle D, Sukumar S 2007. DNA methylation regulates microRNA expression. Cancer Biol Ther 6: 1284–1288 [DOI] [PubMed] [Google Scholar]
- Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, et al. 2010. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol 28: 1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB 1996. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci 93: 9821–9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbard MK, Kozakewich HP, Dal Cin P, Sciot R, Tan X, Xiao S, Fletcher JA 2000. PLAG1 fusion oncogenes in lipoblastoma. Cancer Res 60: 4869–4872 [PubMed] [Google Scholar]
- Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr AR, James KD, Turner DJ, Smith C, Harrison DJ, Andrews R, Bird AP 2010. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet 6:e1001134 doi: 10.1371/journal.pgen.1001134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP 2004. CpG island methylator phenotype in cancer. Nat Rev Cancer 4: 988–993 [DOI] [PubMed] [Google Scholar]
- Jeong S, Liang G, Sharma S, Lin JC, Choi SH, Han H, Yoo CB, Egger G, Yang AS, Jones PA 2009. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol 29: 5366–5376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB 2002. The fundamental role of epigenetic events in cancer. Nat Rev Genet 3: 415–428 [DOI] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. 2008. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321: 1801–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kas K, Voz ML, Roijer E, Astrom AK, Meyen E, Stenman G, Van de Ven WJ 1997. Promoter swapping between the genes for a novel zinc finger protein and β-catenin in pleiomorphic adenomas with t(3;8)(p21;q12) translocations. Nat Genet 15: 170–174 [DOI] [PubMed] [Google Scholar]
- Kim JH, Shin SH, Kwon HJ, Cho NY, Kang GH 2009. Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchows Arch 455: 485–494 [DOI] [PubMed] [Google Scholar]
- Kodzius R, Kojima M, Nishiyori H, Nakamura M, Fukuda S, Tagami M, Sasaki D, Imamura K, Kai C, Harbers M, et al. 2006. CAGE: cap analysis of gene expression. Nat Methods 3: 211–222 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cho NY, Choi M, Yoo EJ, Kim JH, Kang GH 2008. Clinicopathological features of CpG island methylator phenotype-positive colorectal cancer and its adverse prognosis in relation to KRAS/BRAF mutation. Pathol Int 58: 104–113 [DOI] [PubMed] [Google Scholar]
- Li H, Rauch T, Chen ZX, Szabo PE, Riggs AD, Pfeifer GP 2006. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J Biol Chem 281: 19489–19500 [DOI] [PubMed] [Google Scholar]
- Lindor NM, Smalley R, Barker M, Bigler J, Krumroy LM, Lum-Jones A, Plummer SJ, Selander T, Thomas S, Youash M, et al. 2006. Ascending the learning curve–MSI testing experience of a six-laboratory consortium. Cancer Biomark 2: 5–9 [DOI] [PubMed] [Google Scholar]
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Git A, et al. 2007. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res 67: 1424–1429 [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. 2003. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 31: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. 2010. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466: 253–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Geen H, Squazzo SL, Iyengar S, Blahnik K, Rinn JL, Chang HY, Green R, Farnham PJ 2007. Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet 3: e89 doi: 10.1371/journal.pgen.0030089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS 2009. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut 58: 90–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H, Sarkans U, Shojatalab M, Abeygunawardena N, Contrino S, Coulson R, Farne A, Lara GG, Holloway E, Kapushesky M, et al. 2005. ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res 33: D553–D555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A Chinnaiyan AM 2004. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JA, Krumroy L, Plummer S, Aung P, Merkulova A, Skacel M, DeJulius KL, Manilich E, Church JM, Casey G, et al. 2009. Genetic and epigenetic classifications define clinical phenotypes and determine patient outcomes in colorectal cancer. Br J Surg 96: 1196–1204 [DOI] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B 2004. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res 32: D91–D94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18: 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ III 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre D, Lee BH, Ting AH 2010. MBD-isolated genome sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res 38: 391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Catalano PJ, Benson AB III, O'Dwyer P, Hamilton SR, Issa JP 2007. Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil based chemotherapy. Clin Cancer Res 13: 6093–6098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripathy SP, Stevens J, Schultz DC 2006. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol 26: 8623–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A 2008. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 9: 465–476 [DOI] [PubMed] [Google Scholar]
- Takada I, Mihara M, Suzawa M, Ohtake F, Kobayashi S, Igarashi M, Youn MY, Takeyama K, Nakamura T, Mezaki Y, et al. 2007. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-γ transactivation. Nat Cell Biol 9: 1273–1285 [DOI] [PubMed] [Google Scholar]
- Tommerup N, Vissing H 1995. Isolation and fine mapping of 16 novel human zinc finger-encoding cDNAs identify putative candidate genes for developmental and malignant disorders. Genomics 27: 259–264 [DOI] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP 1999. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci 96: 8681–8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T 2008. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res 68: 4123–4132 [DOI] [PubMed] [Google Scholar]
- Van Rijnsoever M, Elsaleh H, Joseph D, McCaul K, Iacopetta B 2003. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin Cancer Res 9: 2898–2903 [PubMed] [Google Scholar]
- Wang Y-K, Kobori JA, Hood L 1993. The htβ gene encodes a novel CACCC box-binding protein that regulates T-cell receptor gene expression. Mol Cell Biol 13: 5691–5701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. 2006. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 38: 787–793 [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang X, Salmon M, Zehner ZE 2007. The zinc finger repressor, ZBP-89, recruits histone deacetylase 1 to repress vimentin gene expression. Genes Cells 12: 905–918 [DOI] [PubMed] [Google Scholar]
- Yamashita K, Dai T, Dai Y, Yamamoto F, Perucho M 2003. Genetics supersedes epigenetics in colon cancer phenotype. Cancer Cell 4: 121–131 [DOI] [PubMed] [Google Scholar]
- Yan H, Choi AJ, Lee BH, Ting AH 2011. Identification and functional analysis of epigenetically silenced microRNAs in colorectal cancer cells. PLoS ONE 6: e20628 doi: 10.1371/journal.pone.0020628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hu JF, Ulaner GA, Li T, Yao X, Vu TH, Hoffman AR 2003. Epigenetic regulation of Igf2/H19 imprinting at CTCF insulator binding sites. J Cell Biochem 90: 1038–1055 [DOI] [PubMed] [Google Scholar]