Abstract
The tumor microenvironment of colorectal carcinoma is a complex community of genomically altered cancer cells, nonneoplastic cells, and a diverse collection of microorganisms. Each of these components may contribute to carcinogenesis; however, the role of the microbiota is the least well understood. We have characterized the composition of the microbiota in colorectal carcinoma using whole genome sequences from nine tumor/normal pairs. Fusobacterium sequences were enriched in carcinomas, confirmed by quantitative PCR and 16S rDNA sequence analysis of 95 carcinoma/normal DNA pairs, while the Bacteroidetes and Firmicutes phyla were depleted in tumors. Fusobacteria were also visualized within colorectal tumors using FISH. These findings reveal alterations in the colorectal cancer microbiota; however, the precise role of Fusobacteria in colorectal carcinoma pathogenesis requires further investigation.
Malignant tumors are complex communities of oncogenically transformed cells with aberrant genomes, associated nonneoplastic cells including immune and stromal cells, and sometimes microbes, including bacteria and viruses. Several viruses that can integrate into the human genome directly cause cancer, such as human papillomavirus in cervical cancer (zur Hausen 2009) and Kaposi's sarcoma-associated herpesvirus in Kaposi's sarcoma (Chang et al. 1994). In other cases, micro-organisms lead indirectly to cancer through chronic inflammatory responses—a mechanism by which Helicobacter pylori contributes to both gastric cancer and MALT lymphoma (Cover and Blaser 2009; Polk and Peek 2010).
In the human distal gut, where microbial cells outnumber host cells nine-to-one (Goodman and Gordon 2010), the microbiome can impart both beneficial and detrimental effects on host physiology contributing to health or disease susceptibility. Gut microbial communities (microbiota) may also influence the development of colorectal carcinoma (Hope et al. 2005; Yang and Pei 2006; Rowland 2009). Sears and Pardoll have recently introduced the concept of the “alpha-bug”—wherein select members of a microbial community, in addition to possessing virulence and procarcinogenic features, are capable of remodeling the microbiome as a whole to drive proinflammatory immune responses and colonic epithelial cell transformation leading to cancer (Sears and Pardoll 2011).
We postulate that, if the microbiota play an active role in the pathogenesis of colorectal carcinoma, then these microbes will be found within the tumor microenvironment, and the composition of the tumor microbiome will differ from that of adjacent nonneoplastic tissue. We have undertaken unbiased, sequence-based approaches, followed by cytological analysis, to probe the differences in the microbial composition of the colorectal carcinoma tumor microenvironment relative to adjacent nonneoplastic tissue. We now report an association of Fusobacterium with the colonic mucosa of colorectal carcinoma.
Results
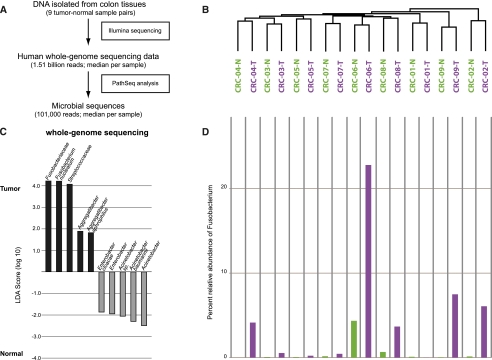
To determine the microbial composition of human colorectal cancer, we analyzed whole genome sequences of nine colorectal cancers and matched normal colons (Bass et al. 2011) using PathSeq, a computational subtraction pipeline that culls out candidate microbial sequences (Kostic et al. 2011). These presumed bacterial sequences were identified by alignment to known sequenced microbial genomes (Fig. 1A; Supplemental Fig. S1; Supplemental Table S1; Methods). PathSeq analysis also identified the presence of virus sequences in these specimens including human herpesvirus 7; however no significant differences in viral sequence levels were detected between tumor and normal DNA (Supplemental Table S2).
Figure 1.
Whole-genome sequencing analysis of the colorectal cancer microbiome. (A) Schematic of experimental and computational whole-genome sequencing analysis workflow. (B) Hierarchical clustering of phylotype relative abundance measurements demonstrates that microbial composition of tumor/normal pairs within individuals is more highly correlated than tumor/tumor pairs or normal/normal pairs from different individuals. (Green) Normal samples; (purple) tumors. (C) Linear discriminant analysis (LDA) coupled with effect size measurements identifies Fusobacterium as the most differentially abundant taxon in colon tumor versus normal specimens by whole-genome sequencing in nine individuals. Tumor-enriched taxa are indicated with a positive LDA score (black), and taxa enriched in normal tissue have a negative score (gray). Only taxa meeting an LDA significant threshold of 1.8 are shown. (D) Percent relative abundance for the genus Fusobacterium is depicted across all samples in the order of the labels in B, demonstrating a tumor-enrichment in most individuals.
Hierarchical clustering analysis of the species-specific relative abundances of microbial sequences revealed that the microbial communities of a tumor and matched noncancerous colon from a given patient were more similar to each other than are tumors or nonaffected colon samples from different patients (Fig. 1B). This finding suggests that a patient's intestinal ecosystem may be more significant in shaping the microbiota than the generic microenvironment of a colon tumor or normal colonic tissue.
To identify bacterial species whose sequences are more abundant in colorectal tumors than in the matched, noncancerous colorectal tissue, we applied a metagenomic biomarker discovery approach, LEfSe [linear discriminant analysis (LDA) coupled with effect size measurements], which performs a nonparametric Wilcoxon sum-rank test followed by LDA analysis to assess the effect size of each differentially abundant taxon (Segata et al. 2011). Using LEfSe, we found that Fusobacterium sequences were significantly enriched in the colorectal cancer metagenomes as were sequences from the family of Streptococcaceae (Fig. 1C,D; Supplemental Fig. S2). Other taxa were enriched in tumor or normal tissue, but their LDA enrichment scores were lower by two orders of magnitude or more (Fig.1C). The high abundance of Fusobacterium sequences, ranging over 20% of total bacterial sequences, is a feature of some but not all colorectal cancer genomes (Fig. 1D).
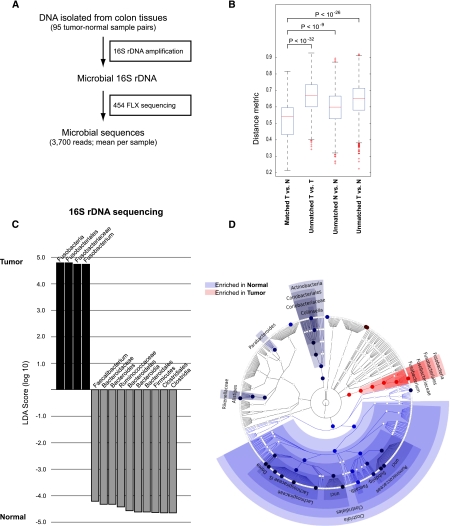
As our initial screen was performed on a sample size of nine cases, we next examined a larger cohort of 95 paired specimens of colon cancer and normal colonic DNA to survey the colon cancer microbiome and validate the tumor-specific enrichment of Fusobacterium. We amplified ribosomal 16S rDNA by PCR using consensus primers from 95 tumor/normal pairs, followed by pyrosequencing to assess the relative abundance of DNA from bacterial species (Fig. 2A). Overall, as was the case in our whole-genome sequence data, tumor/normal pairs from the same individual are much more highly correlated than tumor/tumor pairs or normal/normal pairs from different individuals (Fig.2B). Colorectal tumors were associated with broad phylum-level changes including the depletion (i.e., reduced relative abundance) of Firmicutes and Bacteroidetes, most prominently the Clostridia (Fig. 2C,D; Supplemental Fig. S3); however, the overall diversity in the tumors relative to adjacent tissue was not significantly different (Supplemental Fig. S4A,B). Consistent with our whole-genome sequencing results, the relative abundance of Fusobacterium was highly enriched in the population of tumor versus normal samples (Fig. 2C,D). However a tumor-enrichment for Streptococcaceae was not reproduced, most likely due to small sample-size in our initial whole-genome sequencing results. In addition, we analyzed patient metadata to identify correlations or possible confounding effects (including patient age, gender, ethnicity, tumor anatomic location, tumor purity, inflammation, necrosis, and vascularization) but only found a modest correlation with patient geographic location (Supplemental Fig. S5), as well as a correlation of higher microbial diversity with tumors of higher histological stage or grade (Supplemental Fig. S4C). The correlation of Fusobacteria abundance with geographic location may either indicate a real geographic effect or else a confounder introduced by slightly differing sample collection protocols at the collection sites, for example, time between surgery and freezing (see Methods).
Figure 2.
16S rDNA sequencing analysis of the colorectal cancer microbiome. (A) Schematic of experimental and computational 16S rDNA sequencing analysis workflow. (B) Beta-diversity distances calculated using phylotype relative abundance measurements between all pairs of samples demonstrate that the microbial composition of tumor/normal pairs within individuals is more highly correlated than tumor/tumor pairs, normal/normal pairs, or tumor/normal pairs from different individuals. (C) Linear discriminant analysis (LDA) coupled with effect size measurements identifies Fusobacterium as the most differentially abundant taxon in colon tumor versus normal specimens by 16S rDNA sequencing in 95 individuals. Tumor-enriched taxa are indicated with a positive LDA score (black), and taxa enriched in normal tissue have a negative score (gray). Only taxa meeting an LDA significant threshold of 4.2 are shown. (D) A cladogram representation of data in C. (Red) Tumor-enriched taxa; (blue) taxa enriched in normal tissue. The brightness of each dot is proportional to its effect size.
To determine if there was an absolute increase of Fusobacterium (as one possible explanation for enrichment is depletion of other bacterial species in the tumor microenvironment), we performed quantitative real-time PCR for Fusobacterium DNA relative to human DNA as a control. We found that Fusobacterium DNA was enriched in the tumor relative to normal in 98 paired samples (Supplemental Fig. S6).
As we have shown that Fusobacterium species are enriched in colorectal cancer DNA and tissue, we sought associations that might suggest that Fusobacteria are required for the survival or maintenance of colorectal cancer cells. Because Fusobacterium species can invade colonic epithelial cells (Strauss et al. 2011), we examined colorectal cancer cell lines and hepatic and lymph node metastases for evidence of fusobacterial DNA. Quantitative PCR analysis of 59 human colorectal cancer cell line DNAs revealed no significantly detectable Fusobacterium DNA; however, these in vitro passaged cell lines are often cultured in the presence of antibiotics (Supplemental Table S3). Strikingly, however, when we examined surgically resected colorectal cancer metastases, Fusobacterium was detected in two out of 11 cases (Supplemental Table S4).
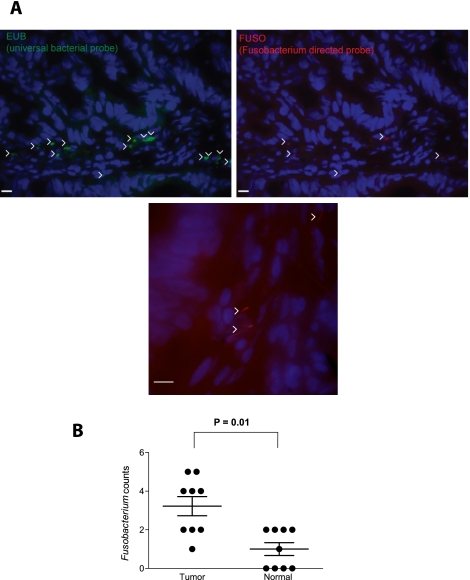
Given the increased abundance of Fusobacterium sequences in colon cancer DNA, we next asked whether Fusobacterium could be detected in histological sections of colon cancer and, if so, where. To address this question, we used 16S rDNA fluorescence in situ hybridization (FISH) oligonucleotide probes on colonic biopsy sections. Employing probeBase consortium 16S rDNA probes that detect the majority of bacteria (EUB338) and members of the genus Fusobacterium (FUSO) (Loy et al. 2008; Swidsinski et al. 2011), we performed FISH analysis on frozen (nine cases) and formalin-fixed paraffin- embedded (12 cases) tissue sections from colorectal cancer and normal colon (see Supplemental Table S5 for a summary of analyses performed on each sample). The Fusobacterium probes detected bacteria in the colorectal cancer and normal tissue sections and were quantitated within the lamina propria and mucus (Fig. 3A); z-section stacks suggest that some of the imaged bacteria may reside intracellularly (Supplemental Movies 1 and 2). Consistent with the analysis of Fusobacterium DNA described above, FISH-detected Fusobacteria were enriched in the colorectal cancer compared to the normal samples (Fig. 3B; Supplemental Fig. S7; see Supplemental Fig. S8 for a comparison of Fusobacterium quantitation across all four methods), in contrast to total bacteria counts which were more evenly distributed (Supplemental Fig. S7).
Figure 3.
Fluorescence in situ hybridization (FISH) detects enrichment of Fusobacteria in colorectal tumors. (A) FISH using an Oregon-Green 488-conjugated ‘‘universal bacterial'' 16S rDNA-directed oligonucleotide probe (EUB338, green) (top left); and Cy3-conjugated Fusobacterium (FUSO, red) (top right and bottom center) 16S rDNA-direct oligonucleotide probe demonstrates the presence of bacteria and Fusobacterium within the colonic mucosa of colorectal tumor samples. Representative images are shown with a 10-μm scale bar in the lower corner of each panel; white arrowheads mark bacteria. Epithelial cell nuclei were stained with DAPI. (B) To determine whether Fusobacterium was enriched in tumor versus normal pairs, three random 40× fields were chosen for scoring by an observer blind to tumor/normal status, using selection criteria of mucosal tissue depth and a minimum of five bacteria visualized by the EUB338 probe per field. Each dot represents data from either a tumor or normal sample from nine tumor/normal paired cases. The mean, SEM, and P-values (calculated by a Wilcoxon matched-pairs signed rank test) are shown.
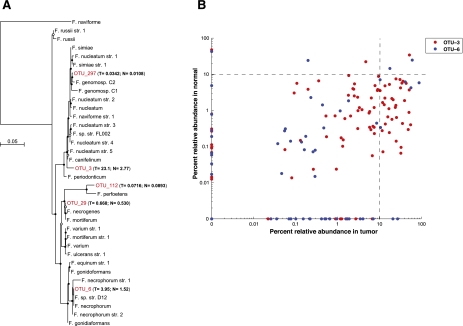
Finally, we sought to assess the specific Fusobacterium species that are enriched in colorectal carcinomas. Based on the 16S ribosomal DNA sequences, five out of a total of 409 operational taxonomic units (OTUs, a proxy for species) identified in our samples were classified as members of the Fusobacterium genus. By performing multiple sequence alignments using our five OTUs along with 16S rDNA sequences from a reference set of 31 Fusobacterium species and constructing maximum likelihood trees, the OTUs were identified as most closely related to Fusobacterium nucleatum, Fusobacterium necrophorum, Fusobacterium mortiferum, and Fusobacterium perfoetens (Fig. 4A). The percent relative abundance in colorectal tumors versus normal colons of the two most abundant OTUs is shown in Figure 4B, demonstrating that, for most patients, these OTUs are enriched in the tumor. Strikingly, only a subset of the cancers showed dramatic enrichment of Fusobacterium species, accounting for up to 89% of total bacterial DNA in some specimens; this result suggests that Fusobacteria may be uniquely related to pathogenesis of subsets of colorectal cancer. The OTU with the greatest similarity to F. nucleatum was the most dominant phylotype identified within cancers; however, some tumors contain more than one dominant species (Supplemental Fig. S9).
Figure 4.
Phylogenetic analysis identifies several Fusobacterium species in human colon cancer tissues. (A) Approximately-maximum-likelihood phylogenetic trees were constructed on the V3–V5 region of the 16S rDNA gene using 31 reference Fusobacterium species along with the five most prominent OTUs identified in colon cancer specimens (indicated in red). Nodes that have bootstrap support above 50% and 75% are indicated with a white and black dot, respectively. The mean percent relative abundance in tumor (T) and normal (N) of each OTU is indicated in parentheses. The full names of the reference strains appear in Supplemental Table S6. (B) The abundance of the indicated OTU relative to all other phylotypes in a given specimen is shown for the two most abundant Fusobacterium OTUs in tumors (x-axis) and normal colon tissue (y-axis); each point represents tumor and normal abundance data for a different individual. The lower-right quadrant of the graph highlights the substantial proportion of patients for whom the Fusobacterium abundance is >10% in tumors but <10% in the matched normal.
Discussion
In summary, genomic analysis of the microbiome of colorectal carcinomas reveals a significant enrichment of Fusobacterium species in these cancers, especially phylotypes with the greatest similarity to F. nucleatum, F. mortiferum, and F. necrophorum. This enrichment is confirmed by histological analysis of tumor tissue and also by the identification of Fusobacterium DNA in colon tumor metastases. Our analysis also reveals broader changes in the tumor environment, such as the depletion of the Bacteroidetes and Firmicutes phyla, most notably the order Clostridiales. Fusobacterium species may have a fitness advantage in the evolving tumor microenvironment, resulting in an altered microbiota in accordance with the “alpha-bug” hypothesis.
Interestingly, Fusobacterium species may be associated with inflammatory bowel diseases (IBD), including both ulcerative colitis and Crohn's disease (Neut et al. 2002; Ohkusa et al. 2002; Strauss et al. 2011), and IBD is a known risk factor, indeed, one of three highest risk factors, for colorectal cancer. Furthermore, consistent with our findings in colorectal carcinoma, others have reported that several Fusobacterium strains were associated with IBD; however, the majority (69%) were specifically associated with F. nucleatum (Strauss et al. 2011). Therefore, it is worth further exploration of a causal link between Fusobacterium spp. with inflammatory bowel disease and colorectal carcinoma pathogenesis.
F. nucleatum and other Fusobacterium species can elicit host proinflammatory response (Moore and Moore 1994) and possess virulence characteristics that promote their adhesiveness to host epithelial cells (Bachrach et al. 2005; Uitto et al. 2005) and their ability to invade into epithelial cells (Han et al. 2000; Strauss et al. 2011). Therefore, our findings of a tumoral enrichment of Fusobacterium spp. in colorectal carcinoma suggest the possibility that these organisms may contribute to tumorigenesis, perhaps in a limited subset of patients, most conceivably by an inflammatory-mediated mechanism. Alternatively, it is possible that Fusobacteria accumulate in the tumor microenvironment in the late stages of tumorigenesis and, therefore, do not have a significant role in tumor development. Our results do not prove a causal relationship between Fusobacterium and colorectal cancer; the establishment or repudiation of such a relationship will require further studies of colorectal cancer in both human subjects and animal models of the disease. Additionally, case-control studies comparing tumor microbiota to that of colonic epithelial tissues from healthy individuals will serve to demonstrate whether Fusobacterium species are more prevalent in individuals with colon cancer relative to the general population.
In summary, our findings reveal species-specific alterations in the colorectal cancer microbiota, which may lead to microbiota-directed prevention, diagnostic, prognostic, and treatment strategies for these cancers.
Methods
Sample collection and preparation
Colorectal adenocarcinoma and adjacent nonaffected tissue was obtained from the Vall d'Hebron University Hospital in Barcelona, Spain and Genomics Collaborative Inc. (GCI), using the sample collection protocols detailed below.
Vall d'Hebron University Hospital
All frozen tissue samples were collected following the Standard Operating Procedures at the site. Participants were enrolled in the study prior to surgery, and informed consent was obtained by the surgeon. After arterial ligation and surgical removal of the tissues, the specimens were immediately transferred from the operating room to the pathology suite and subsequently evaluated by the pathologist, and, if possible, one fragment of healthy tissue and one of tumor were chosen and placed in a cryotube and frozen immediately in liquid nitrogen (time interval between specimen removal and freezing: maximum = 30 min; median = 22 min). The frozen tissue was stored at −80°C until DNA extraction. All clinical data, captured on case report forms, was double-data entered into a clinical database.
Genomics Collaborative, Inc.
All frozen tissue samples were collected following the same Standard Operating Procedure at all collection sites, both within the United States and in Vietnam. Participants were enrolled in the study prior to surgery, and informed consent was obtained. After arterial ligation and surgical removal of the tissues, the tissue was transferred from the operating room to the pathology suite. The time taken from surgical removal of the specimen until the time it was received for processing in the pathology suite, as well as the temperature at which it was transferred (room temperature, or on ice) were recorded. The SOP required the tissue transmittal time (from removal to freezing of the sample) to ideally be from 30 to no more than 45 min. Upon dissection in the pathology suite, samples were cut into ∼1 g pieces and placed into prelabeled cyrovials supplied in the sample kits. Samples were immediately frozen in liquid nitrogen vapor phase in charged vapor-shippers located at each site. An H&E slide was cut from the adjacent face of each tissue sample and sent to GCI along with the frozen tissue samples. Upon receipt at GCI, the samples were qualified by pathologist review of the H&E slide, and all samples were stored in liquid N2 vapor freezers until requested for research. All clinical data, captured on case report forms, was double-data entered into a clinical database.
DNA extraction, whole genome sequencing, and analysis
DNA was extracted from colorectal carcinoma tumors and adjacent nonaffected tissues, and whole genome sequencing was performed as described previously (Bass et al. 2011). Initial alignments to the human reference genome were performed as described (Bass et al. 2011). All unaligned sequencing reads were (1) analyzed on PathSeq and (2) aligned to the complete set of fully sequenced bacterial and archaeal genomes (ftp://ftp.ncbi.nih.gov/genomes/Bacteria; downloaded 10-07-2010) by MegaBlast (Blast Tools version 2.2.23, word size 16, match reward 1, mismatch reward −2, gap open reward −5, gap extension reward −2). The top 30 sequence matches with >90% sequence identity and >90% query coverage were reported for each read (i.e., query). Classifications were performed at the domain, then phylum, then genus, then species level, requiring unique alignments (i.e., reads with equivalent E-values to multiple taxa were removed from analysis). At the species level, relative abundance (RA) for each organism was calculated as follows: RA = (# unique alignment positions in genome × 1,000,000) / (# total alignable reads × genome size). The RA values were then per-sample normalized such that the total relative abundance for each sample sums to one. The resulting normalized RA matrix was analyzed on LEfSe (Segata et al.2011).
Amplification and 454 sequencing of the 16S gene
The 16S gene data set consists of 454 FLX Titanium sequences spanning the V3 to V5 variable regions obtained for 190 samples (95 pairs). Detailed protocols used for 16S amplification and sequencing are available on the HMP Data Analysis and Coordination Center website (http://www.hmpdacc.org/tools_protocols/tools_protocols.php). In brief, genomic DNA was subjected to 16S amplifications using primers designed incorporating the FLX Titanium adapters and a sample barcode sequence, allowing directional sequencing covering variable regions V5 to partial V3 (Primers: 357F 5′-CCTACGGGAGGCAGCAG-3′ and 926R 5′-CCGTCAATTCMTTTRAGT-3′). Polymerase chain reaction (PCR) mixtures (25μl) contained 10 ng of template, 1x Easy A reaction buffer (Stratagene), 200 mM of each dNTP (Stratagene), 200 nM of each primer, and 1.25 U Easy A cloning enzyme (Stratagene). The cycling conditions for the V3–V5 consisted of an initial denaturation of 95°C for 2 min, followed by 25 cycles of denaturation at 95°C for 40 sec, annealing at 50°C for 30 sec, extension at 72°C for 5 min, and a final extension at 72°C for 7 min. Amplicons were confirmed on 1.2% Flash Gels (Lonza) and purified with AMPure XP DNA purification beads (Beckman Coulter) according to the manufacturer's instructions and eluted in 25 μL of 1× low TE buffer (pH 8.0). Amplicons were quantified on Agilent Bioanalyzer 2100 DNA 1000 chips (Agilent Technologies) and pooled in equimolar concentration. Emulsion PCR and sequencing were performed according to the manufacturer's specifications.
Processing of 16S sequence data
Resulting sequences were processed using a data curation pipeline implemented in mothur (Schloss et al. 2009), complimented by abundantOTU (Ye 2010), and custom PERL scripts. Sequences were removed from the analysis if they were <200 nt or >600 nt, had a read quality score <25, contained ambiguous characters, had a nonexact barcode match, or did show more than four mismatches to the reverse primer sequences (926R). Remaining sequences were assigned to samples based on barcode matches, after which barcode and primer sequences were trimmed, and reads were oriented such that all sequences begin with the 5′ end, according to standard sense-strand conventions. All sequences were aligned using a NAST-based sequence aligner to a custom reference based on the SILVA alignment (Pruesse et al. 2007; Schloss et al. 2009). Chimeric sequences were identified using the mothur implementation of the ChimeraSlayer algorithm (Haas et al. 2011). Quality filtered and chimera-free sequences were clustered into operational taxonomic units using abundantOTU (Ye 2010). Representative sequences per OTU were classified with the MSU RDP classifier v2.2 (Cole et al. 2009), using the taxonomy proposed by Garrity et al. (2007), maintained at the Ribosomal Database Project (RDP 10 database, version 6).
Quantitative PCR analysis
Quantitative real-time PCR was performed as described (Boutaga et al. 2005) using pan-Fusobacterium probe-primer sets as described (Boutaga et al. 2005). Fusobacterium quantitation was measured relative to human endogenous 18S [Applied Biosystems TaqMan Ribosomal RNA Control Reagents, Hs99999901_s1 (part number 4331182)].
Microbial FISH analysis
Frozen sections were fixed in Carnoy's solution overnight and embedded in paraffin, and 5-mm-thick sections were prepared and hybridized as previously described (Swidsinski et al. 2011). The sequences of the following FISH probes were obtained from probeBase (http://www.microbial-ecology. net/probebase/) (Loy et al. 2008): the ‘‘universal'' bacterial probe-EUB338 (pB-00159), Fusobacterium targeted probe (pB-00782). Slides were imaged on an Olympus B40 microscope and digitally photographed using IP Lab. Three random fields per sample were chosen by an observer blind to tumor/normal status, using selection criteria of mucosal tissue depth and a minimum of five bacteria visualized by the EUB338 probe per field. Composite z-stacks were assembled in IP Lab, and composite photomicrographs were assembled in Adobe Photoshop.
Data access
The 16S sequence data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession number SRP000383.
Acknowledgments
This work was funded by grants from the National Human Genome Research Institute (U54HG003067), The National Cancer Institute (P50CA127003; RC2CA148317; R01CA154426), and the Starr Cancer Consortium. A.D.K. was supported by a Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarship and an Albert J. Ryan Foundation Fellowship. A.I.O. was supported by the Rebecca Ridley Kry Fellowship of the Damon Runyon Cancer Research Foundation. We thank Jordi Barretina, Kristin Ardlie, Candace Guidicci, Lauren Ambrogio, and Susanna Hamilton of the Broad Institute and Emma Allen-Vercoe of the University of Guelph for assistance and advice.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.126573.111.
References
- Bachrach G, Ianculovici C, Naor R, Weiss EI 2005. Fluorescence-based measurements of Fusobacterium nucleatum coaggregation and of fusobacterial attachment to mammalian cells. FEMS Microbiol Lett 248: 235–240 [DOI] [PubMed] [Google Scholar]
- Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, Cibulskis K, Sougnez C, Voet D, Saksena G, Sivachenko A, et al. 2011. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. doi: 10.1038/ng.936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutaga K, van Winkelhoff AJ, Vandenbroucke-Grauls CM, Savelkoul PH 2005. Periodontal pathogens: A quantitative comparison of anaerobic culture and real-time PCR. FEMS Immunol Med Microbiol 45: 191–199 [DOI] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266: 1865–1869 [DOI] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, et al. 2009. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37: D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover TL, Blaser MJ 2009. Helicobacter pylori in health and disease. Gastroenterology 136: 1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity GM, Lilburn TG, Cole JR, Harrison SH, Tindall BJ 2007. Taxonomic outline of the Bacteria and Archaea, Release 7.7. http://www.taxonomicoutline.org/
- Goodman AL, Gordon JI 2010. Our unindicted coconspirators: Human metabolism from a microbial perspective. Cell Metab 12: 111–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, et al. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21: 494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YW, Shi W, Huang GT, Kinder Haake S, Park NH, Kuramitsu H, Genco RJ 2000. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun 68: 3140–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope ME, Hold GL, Kain R, El-Omar EM 2005. Sporadic colorectal cancer–role of the commensal microbiota. FEMS Microbiol Lett 244: 1–7 [DOI] [PubMed] [Google Scholar]
- Kostic AD, Ojesina AI, Pedamallu CS, Jung J, Verhaak RGW, Getz G, Meyerson M 2011. PathSeq: Software to identify or discover microbes by deep sequencing of human tissue. Nat Biotechnol 29: 4–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy A, Arnold R, Tischler P, Rattei T, Wagner M, Horn M 2008. probeCheck–a central resource for evaluating oligonucleotide probe coverage and specificity. Environ Microbiol 10: 2894–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WE, Moore LV 1994. The bacteria of periodontal diseases. Periodontol 2000 5: 66–77 [DOI] [PubMed] [Google Scholar]
- Neut C, Bulois P, Desreumaux P, Membre JM, Lederman E, Gambiez L, Cortot A, Quandalle P, van Kruiningen H, Colombel JF 2002. Changes in the bacterial flora of the neoterminal ileum after ileocolonic resection for Crohn's disease. Am J Gastroenterol 97: 939–946 [DOI] [PubMed] [Google Scholar]
- Ohkusa T, Sato N, Ogihara T, Morita K, Ogawa M, Okayasu I 2002. Fusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibody. J Gastroenterol Hepatol 17: 849–853 [DOI] [PubMed] [Google Scholar]
- Polk DB, Peek RM Jr 2010. Helicobacter pylori: Gastric cancer and beyond. Nat Rev Cancer 10: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO 2007. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland IR 2009. The role of the gastrointestinal microbiota in colorectal cancer. Curr Pharm Des 15: 1524–1527 [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. 2009. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears CL, Pardoll DM 2011. Perspective: Alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis 203: 306–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12: R60 doi: 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, Lynch T, Allen-Vercoe E 2011. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis 17: 1971–1978 [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Dorffel Y, Loening-Baucke V, Theissig F, Ruckert JC, Ismail M, Rau WA, Gaschler D, Weizenegger M, Kuhn S, et al. 2011. Acute appendicitis is characterized by local invasion with Fusobacterium nucleatum/necrophorum. Gut 60: 34–40 [DOI] [PubMed] [Google Scholar]
- Uitto VJ, Baillie D, Wu Q, Gendron R, Grenier D, Putnins EE, Kanervo A, Firth JD 2005. Fusobacterium nucleatum increases collagenase 3 production and migration of epithelial cells. Infect Immun 73: 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Pei Z 2006. Bacteria, inflammation, and colon cancer. World J Gastroenterol 12: 6741–6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y 2010. Fast and accurate identification and quantification of abundant species from pyrosequences of 16S rRNA by consensus alignment. In The Proceedings of BIBM, 153–157 Institute of Electrical and Electronics Engineers (IEEE), New York [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H 2009. Human papillomavirus and cervical cancer. Indian J Med Res 130: 209. [PubMed] [Google Scholar]