Abstract
An estimated 15% or more of the cancer burden worldwide is attributable to known infectious agents. We screened colorectal carcinoma and matched normal tissue specimens using RNA-seq followed by host sequence subtraction and found marked over-representation of Fusobacterium nucleatum sequences in tumors relative to control specimens. F. nucleatum is an invasive anaerobe that has been linked previously to periodontitis and appendicitis, but not to cancer. Fusobacteria are rare constituents of the fecal microbiota, but have been cultured previously from biopsies of inflamed gut mucosa. We obtained a Fusobacterium isolate from a frozen tumor specimen; this showed highest sequence similarity to a known gut mucosa isolate and was confirmed to be invasive. We verified overabundance of Fusobacterium sequences in tumor versus matched normal control tissue by quantitative PCR analysis from a total of 99 subjects (p = 2.5 × 10−6), and we observed a positive association with lymph node metastasis.
Few infectious agents have been unequivocally linked to cancer. Those that have, such as Human Papilloma Virus, Hepatitis B and C virus, and Helicobacter pylori, alone are responsible for an estimated 15% of the global cancer burden, based on strength of the association and prevalence of infection (Parkin 2006). Metagenomics methods developed over the past decade (Weber et al. 2002; Moore et al. 2011) provide a useful approach to identifying microbial sequence signatures in diseases that have a possible or suspected infectious etiology. There are variations on the method, but the basic approach involves shotgun sequencing bulk DNA or RNA isolated from disease tissue, computational subtraction of all sequence reads recognized as human, and comparison of the residual reads to databases of known microbial sequences in order to identify microbial species present in the initial specimen. The method is complementary to traditional culture and histolology–based protocols, and new massively parallel sequencing technologies impart high sensitivity. At present the power of the method remains restricted by the content of microbial sequence databases, but with our increasing reach into microbial sequence space, the comprehensiveness of these data resources continues to improve. In oncology, the identification of a novel polyomavirus in Merkel Cell carcinoma (Feng et al. 2008) is a recent demonstration of the utility of a metagenomics approach.
Colorectal carcinoma (CRC) is the fourth leading cause of cancer deaths, responsible for approximately 610,000 deaths per year worldwide (World Health Organization 2011). It is also one of the first and best genetically characterized cancers, and specific somatic mutations in oncogenes and tumor suppressor genes have been found that are associated with progression from adenomatous lesions (polyps) to invasive carcinoma (Vogelstein et al. 1988). The root cause of CRC is unclear, but inflammation is a well-recognized risk factor (Wu et al. 2009; McLean et al. 2011). Given the link between H. pylori–mediated inflammation and gastric cancer (Marshall and Warren 1984), we asked if inflammatory microorganisms are associated with other gastrointestinal (GI) cancers. We began to address this question by undertaking a metagenomic survey of colorectal carcinoma.
Results
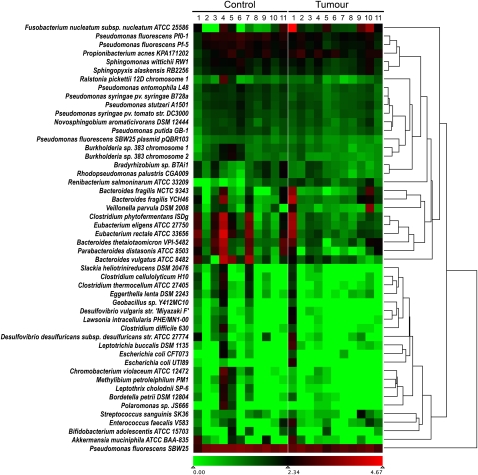
Total RNA was isolated from frozen sections of 11 matched pairs of colorectal carcinoma and adjacent normal tissue specimens. RNA was purified by host ribosomal sequence depletion, rather than poly(A) selection, in order to retain non-polyadenylated sequences of potential microbial origin. In our screen, we analyzed RNA rather than DNA in order to detect active, transcribing microorganisms and to allow for the detection of RNA viruses that may be present. Illumina RNA-seq libraries were constructed, barcoded, and pooled, and two lanes of paired-end sequencing data were obtained using the Illumina GAIIx platform. Reads were filtered for base quality and low complexity, then aligned pairwise to human rRNA and cDNA (Flicek et al. 2011) and genome (hg18) reference sequences using Burrows-Wheeler Aligner (BWA) (Li and Durbin 2010), as previously described (Moore et al. 2011). Aligned reads were removed from the data set, leaving 34.9 million pairs (Supplemental Table S1). These residual read-pairs were then used to search a custom database containing accessions for all RefSeq bacterial and viral genomes, using Novoalign (http://novocraft.com), which is a slower but more permissive aligner than BWA. Our analysis was alignment-based, because the abundance of candidate organisms can be inferred more directly from alignments than from de novo assemblies. For accuracy, we tallied only unambiguous alignments where the best match to both the forward and reverse mate pair was to the same genome accession. These alignments identified a total of 670 distinct genome accessions, representing 415 species (Supplemental Table S1). These were predominantly (97%) bacterial, although several herpes virus sequences were detectable at low levels, and one tumor showed over-abundance (142 raw read-pairs) of human papillomavirus type 107 (GenBank accession EF422221.1). A wide distribution of bacterial species abundance was apparent, with 30 species representing 95% of the sequence data (Supplemental Fig. S1). Of the 670 distinct genome accessions hit, 63% were found in both tumor and normal specimens. Alignments specific to only tumor or only control specimens were due to rare sequences, and, therefore, the representation in one group or the other may simply reflect sampling bias. The only alignments we obtained that were markedly disproportionate between tumor and control were to the genome of Fusobacterium nucleatum subsp. nucleatum (American Type Culture Collection [ATCC] 25586), a Gram-negative anaerobe. F. nucleatum was the organism with the highest number of hits overall (21% of all alignments), and nine of the 11 subjects showed at least twofold higher read counts in tumor relative to corresponding control tissue (Fig. 1). Differential abundance ranged from 0.1-fold to 256-fold, with a mean over-abundance of 79-fold. The majority of the hits were to highly abundant F. nucleatum ribosomal transcripts, but other non-ribosomal F. nucleatum gene products were also detected (Supplemental Fig. S2).
Figure 1.
Relative abundance of microbial genomes in tumor and control specimens. Numbers of read-pairs that matched known microbial sequences were normalized according to sequencing depth for both tumor and matched normal samples. The abundance of normalized bacterial read-pairs ranged from zero to a maximum of 66,896 represented by a transition from green to red on a log10 scale. F. nucleatum sequences were present in the tumor samples at levels twofold or greater than in normal samples in nine out of the 11 subjects. The mean over abundance across all subjects was 79-fold.
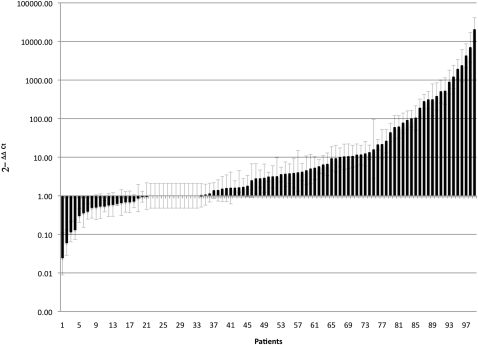
To explore further the observation of disparate F. nucleatum read counts between tumor and matched normal samples in our RNA-seq data set, we developed a targeted quantitative real-time polymerase chain reaction (qPCR) assay to interrogate additional samples. To design the qPCR primers and probe, we gathered the 51,677 read-pairs from tumor sample 1 that matched F. nucleatum and performed a local de novo assembly using SSAKE (Warren et al. 2007) to obtain 861 total contigs, ranging in length from 100 to 1433 bp. The majority of these contigs matched genes encoding F. nucleatum ribosomal RNAs and proteins, but we also obtained 82 contigs that gave BLASTN (Basic Local Alignment Search Tool) alignments of 80% or greater sequence identity to other F. nucleatum protein-coding genes. A 161-bp contig that returned a high-quality BLAST match (95% identity) to the nusG gene (GenBank accession AAL94126.1) of F. nucleatum and no match to any gene of any other species, was used as the target for designing a qPCR (Taqman, ABI) primer/probe set. The initial metagenomics screen described above involved interrogation of expressed genes; however, once we established F. nucleatum as a candidate pathogen, we switched to analysis of gDNA because a larger amount of high-quality DNA than RNA was obtainable from the frozen tissue sections. We conducted qPCR on gDNA isolated from an additional 88 colorectal carcinomas and matched normal specimens and confirmed an over-representation of F. nucleatum in tumor versus matched normal specimens (p = 2.5 × 10−6, two-tailed ratio t-test) (Fig. 2). The Fusobacterium abundance measured by qPCR correlated with that measured from the RNA-seq data (Pearson's r = 0.97). The mean overall abundance of Fusobacterium was found to be 415 times greater in the tumor samples (n = 99) than in the matched normal samples (n = 99) (Fig. 2).
Figure 2.
Relative abundance of Fusobacterium in tumor versus normal colorectal carcinoma biopsies. Relative amounts of Fusobacterium DNA were determined between tumor and matched normal biopsies in 99 subjects, using quantitative real-time PCR (qPCR). The cycle threshold (Ct) values for the normal samples had a Ct range of 25.5 to 40, and the Ct range for the tumor samples was between 21.4 and 40. The data shown are mean values from two independent experiments. Fusobacterium load, as determined by qPCR, was found to be significantly higher in the tumor samples versus the matched control samples (two-tailed ratio t-test, p = 2.52 × 10−6).
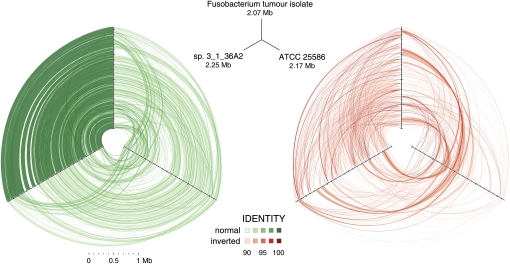
We attempted to culture Fusobacteria anaerobically, directly from 12 of the frozen tumor sections that showed high abundance by qPCR, and we obtained a single isolate (CC53). We purified high-molecular-weight (HMW) gDNA from this culture, constructed and sequenced a WGS (whole-genome shotgun) library using the Illumina HiSeq platform, and obtained an excessive number (64,819,156) of quality filtered, paired 100-nt reads. These reads were aligned to the F. nucleatum type strain ATCC 25586 (GenBank accession NC_003454.1) sequence, covering 76% of this reference genome with 2661-fold mean depth and 95.6% ± 2.0% (mean ± SD) identity. Furthermore, we aligned reads from CC53 to 483 additional draft genome sequences available from the Human Microbiome Project (HMP) (Nelson et al. 2010) including 16 as-of-yet incomplete Fusobacterium genomes. CC53 aligned with highest identity to Fusobacterium sp. 3_1_36A2, covering 91.6% of the 12-supercontig draft assembly with 99.5 ± 1.2% (mean ± SD) sequence identity. Three-way analysis among these strains using cross_match Smith-Waterman alignments confirmed that CC53 is closest to Fusobacterium sp. 3_1_36A2 (Fig. 3). Some notable differences were apparent, however. We observed 19 segments from strain 3_1_36A2 that were missing from CC53. The majority (156/206) of the predicted coding sequences (CDS) on these segments from strain 3_1_36A2 had unknown function, but there were numerous sequences indicative of prophage content, including genes encoding putative helicase, integrase, recombinase, terminase, and topoisomerase activity (Supplemental Table S2). De novo assembly of unmapped CC53 reads yielded 82 kb of sequence in 67 contigs ≥500 nt. These contigs aligned with variable sequence identity to one of the 16 Fusobacterium genome assemblies or the ATCC type strain. BLASTX (Altschul et al. 1997) searches of GenBank-nr identified 99 coding sequences (Supplemental Table S3), the most recurrent of which was hemolysin, a bacterial endotoxin. Most, however, had no predicted function (Supplemental Table S3). Although we were able to culture Fusobacterium from only a single tumor section, we used primer walking to interrogate an additional four samples where qPCR-predicted levels of Fusobacterium were high. Sanger sequences from these amplicons comprised 68,694 total base pairs and each aligned with highest sequence similarity (93%–100%) to one of the various Fusobacterium draft genomes, although we could not assign unambiguously a specific best matching strain to any of these samples, due perhaps to within-sample strain heterogeneity.
Figure 3.
Hive plots showing alignment of three Fusobacterium genomes. Approximately 32 million high-quality WGS Illumina HiSeq reads (≥99 consecutive Q30 bases) from Fusobacterium tumor isolate CC53 were assembled with SSAKE (v3.7, default options) into 379 contigs. The contigs were aligned using cross_match (-minmatch 29 -minscore 59 -masklevel 101) to the complete F. nucleatum susb. nucleatum ATCC 25586 genome and, independently, to the 12-contig HMP Fusobacterium sp. 3_1_36A2 assembly, respectively; and ordered/oriented based on the highest identity to the latter sequence. Three-way cross_match (http://www.phrap.org) alignments between each Fusobacterium genome were performed and represented visually using hive plots (http://www.hiveplot.com). For each, the top, left, and right axes are proportional to genome size and represent the Fusobacterium tumor isolate CC53 (2.07 Mb), the HMP sp. 3_1_36A2 (2.25 Mb), and the ATCC 25586 type strain (2.17 Mb), in that order. Synteny between the isolates is depicted by green and red links that show direct and inverted alignments, respectively. Sequence similarity and synteny is highest between CC53 and sp. 3_1_36A2, as evidenced by a greater density of high similarity sequence matches between them, relative to ATCC 25586, and shared patterns of inversions compared to this reference strain. Three regions of sequences present in sp. 3_1_36A2 but absent from CC53 are apparent as conspicuous gaps on the sp. 3_1_36A2 axis. Sequence segments unique to CC53 are not visible at this scale.
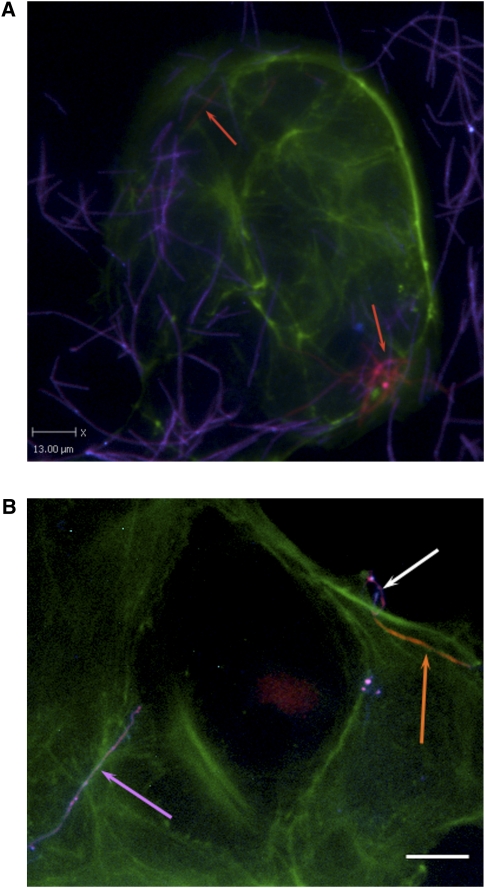
We were interested to determine if CC53 would demonstrate invasiveness in human colonic epithelial cells. We used immunofluorescence and an antibody-based differential staining method, described previously (Strauss et al. 2011), to measure invasion of cultured colonic adenocarcinoma-2 (Caco-2) cells by the Fusobacterium tumor isolate. We identified two previous Fusobacterium polyclonal antibodies, one rabbit (EAV_AS1) and one rat (EAV_AS2), which reacted positively to CC53. Caco-2 cells were grown on glass coverslips, infected with CC53 culture (at a multiplicity of infection of 100:1), and then differentially stained with anti-Fusobacterium antibodies conjugated to different fluorophores before and after Caco-2 cell permeabilization. We confirmed the invasiveness of CC53 in this model system (Fig. 4).
Figure 4.
Representative differentially stained immunofluorescence image showing strain CC53 invading Caco-2 cells. (A) The differential staining method allows for delineation between bacteria that have penetrated the host cells (labeled for actin in green) to reside within them (orange, also indicated with orange arrows), and bacteria present on the outside of the cell (purple). CC53 shows a very long, fine, thread-like cell morphology. (B) Detail of CC53 invasion. (Top right) A representative CC53 cell in the process of invading the Caco-2 host cell (image is differentially stained as for 4A). The long, thread-like cells appear to penetrate host cells pole first. (Orange arrow) The CC53 cell that is internalized; (white arrow) the external portion of the same bacterial cell that has looped around on itself, demonstrating apparent flexibility. (Purple arrow) A single CC53 cell that has not invaded the host cells, for contrast. Bar, 15 μm. In the immunofluorescence micrograph, CC53 shows a very long, flexible cell morphology. (Green) Actin (Caco-2 cells); (orange) invasive and internalized bacteria; (purple) bacteria external to the cell.
We explored clinical correlates of Fusobacterium overabundance but did not observe any association with tumor stage, tumor site, history of treatment, patient age, or survival. To explore histopathological correlates, a hematoxylin-and-eosin (H&E) stained section from a representative cross-section clinical block from each tumor was scored for lymphocytic infiltrates, myeloid/neutrophil infiltrates, circumferential involvement, and luminal or geographic necrosis, and these scores were compared to Fusobacterium relative abundance (tumor vs. control). Fusobacterium showed higher relative abundance in tumors with >50% circumferential involvement (unpaired, two-tailed t-test, p = 0.0023). In addition, we found that subjects with high-relative-abundance Fusobacterium in tumor relative to matched control tissue were significantly more likely to have regional lymph node metastases, as determined by their TNM (tumor, node, metastases) scores (one-tailed Fisher's exact test, p = 0.0035) (Supplemental Fig. S3). Specifically, lymph node metastases were present in 29/39 patients in the high-abundance Fusobacterium group versus 26/58 in the low-abundance group.
Discussion
F. nucleatum is an invasive (Han et al. 2000; Swidsinski et al. 2011), adherent (Weiss et al. 2000), and pro-inflammatory (Krisanaprakornkit et al. 2000; Peyret-Lacombe et al. 2009) anaerobic bacterium. It is common in dental plaque (Bolstad et al. 1996; Ximenez-Fyvie et al. 2000), and there is a well-established association between F. nucleatum and periodontitis (Signat et al. 2011). Anecdotally, F. nucleatum has been found to cause cerebral abscesses (Kai et al. 2008) and pericarditis (Han et al. 2003), and it is one of the Fusobacterium species responsible for Lemierre's syndrome, a rare form of thrombophlebitis (Weeks et al. 2010). More recently, various Fusobacteria including F. nucleatum have been implicated in acute appendicitis, where they have been found by immunohistochemistry (IHC) as epithelial and submucosal infiltrates that correlate positively with severity of disease (Swidsinski and Ismail 2011). Furthermore, when isolated from human intestinal biopsy material, F. nucleatum has been found to be more readily culturable from patients with GI disease than healthy controls, and the strains grown from inflamed biopsy tissue tend to display a more invasive phenotype (Strauss et al. 2008, 2011).
Our observation of a highly significant over-representation of F. nucleatum in colorectal tumor specimens was largely unexpected, given that it is generally regarded as an oral pathogen—it is not an abundant constituent of the normal gut microbiota (Qin et al. 2010). A new report of rRNA sequences from a small number of colorectal tumor and control samples (Marchesi et al. 2011) highlighted a trend toward elevated Coriobacteria in tumors, but the data were also suggestive of an abundance of Fusobacteria. We are also aware of an independent report of Fusobacterium in colorectal cancer copublished in this issue (Kostic et al. 2012). Thus, there is increasing evidence that Fusobacterium infection is common in colorectal carcinoma, but it remains to be determined if there is any involvement of Fusobacterium in tumorogenesis. The presence of this bacterium may simply represent an opportunistic infection at an immuno-compromised site, but the possibility of a role in tumor etiology, perhaps through pro-inflammatory mechanisms, deserves further scrutiny. Analysis of colorectal carcinomas arising in association with chronic inflammatory bowel disease would be of interest. Furthermore, since colorectal carcinoma typically arises from earlier-stage adenomatous lesions (polyps), it would also be useful to screen early-stage lesions for the presence of this bacteria in a prospective study. In the present study, three of the 99 tumor sections were composed entirely of the adenomatous component of adenocarcinoma specimens. By qPCR, two out of these three had very high Fusobacterium content, and, in fact, one of these gave the highest tumor normal ratio of all samples. This supports the notion that a comprehensive study of early-stage lesions may help determine whether Fusobacterium infection is related to the early stages of tumor progression. If so, it may be an appropriate target for vaccination and/or antimicrobial therapy. Finally, should an etiological link prove to be absent, the highly significant association of F. nucleatum with colorectal carcinoma may still be of clinical utility. For example, it may be possible to exploit this association for screening purposes. Although we did not find F. nucleatum tumor abundance to be elevated in all subjects in the present study, we did not characterize the fecal microbiota in these patients, and we have not yet obtained data from healthy subjects to compare to our findings from CRC patients. Further work of this nature may reveal F. nucleatum–related biomarkers that are informative with respect to CRC presence or CRC risk, although this remains entirely speculative at the present time. There is also renewed interest in the potential utility of bacterial cancer therapy (Forbes 2010), an approach that relies fundamentally on the availability of microbes with natural tumor specificity.
Methods
Clinical specimens
For all cases, fresh CRC samples were obtained with informed consent by the BC Cancer Agency Tumor Tissue Repository (BCCA-TTR) (Watson 2010), which operates as a dedicated biobank with approval from the University of British Columbia–British Columbia Cancer Agency Research Ethics Board (BCCA REB). The BCCA-TTR platform is governed by Standard Operating Procedures (SOPs) that meet or exceed the recommendations of international best practice guidelines for repositories (NCI Office of Biorepositories and Biospecmen Research, NCI Best Practices for Biospecimen Resources). Specimens are handled with very close attention to maintaining integrity and isolation. Overall average collection time (time from removal from surgical field to cryopreservation in liquid nitrogen) for all colorectal cases in the BCCA-TTR is 31 min. For this study, biospecimens were held briefly at −20°C during frozen sectioning, using 100% ethanol to clean the blade between all samples. Clinical-pathological and outcomes data were obtained from the BC Cancer Agency clinical chart including tumor features reported according to the American College of Pathologists criteria and the “Protocol for Examination of Specimens from Patients with Primary Carcinoma of the Colon and Rectum.” This included histological features indicative of inflammatory and immune response (lymphoid and myeloid cell infiltrates), which were assessed as none, mild–moderate, or marked using semiquantitative scoring as well as the percent area of tumor involved by necrosis, by a pathologist in a representative tumor cross-section.
Metagenomic library construction and sequencing
Eleven colorectal tumor samples and 11 matched normal samples were processed, as detailed previously (Moore et al. 2011), using an RNeasy Plus mini kit (QIAGEN) to purify total RNA or an AllPrep DNA/RNA mini kit (QIAGEN) to purify both DNA and RNA. RNA quality and concentration were assessed using Agilent Bioanalyzer 2000 RNA Nanochips. Ribosomal RNAs were depleted from 1 μg of total RNA using the manufacturer's protocol for the RiboMinus Eukaryote Kit for RNA-seq (Invitrogen). Depletion was assessed using Agilent Bioanalyzer 2000 RNA Nanochips. All samples were found to have ≤10% residual ribosomal RNA contamination and were processed as described previously (Shah et al. 2009; Morin et al. 2010) for the construction of Illumina libraries, with the following modifications: Each paired-end library was PCR amplified for 15 cycles using the standard Illumina PE1 PCR primer plus one of 12 modified PE2 primers, each including a unique 6-base insertion as an index sequence. Libraries prepared using indexed primers were then combined in pools of 11 each (one tumor pool, one control pool), gel-purified, and then sequenced on the Illumina GAIIx platform. One lane of 75-bp paired-end sequence was obtained for each of the two pools.
Bioinformatics analysis
Paired-end sequence reads from indexed tumor and adjacent normal sample libraries were processed as described (Moore et al. 2011). Briefly, corresponding human RNA-seq libraries were aligned with BWA (version 0.5.4 [sample -o 1000, default options]) (Li and Durbin 2010) sequentially against human rRNA, cDNA, and genome reference sequences (Flicek et al. 2011). Pairs aligning logically or containing reads having either an average base quality below phred 20 (Ewing et al. 1998), and/or more than 20 consecutive homopolymeric bases were subtracted from the original data. Read-pairs that remained unaligned to any of the human sequence databases were used to interrogate a custom-built sequence collection of well-characterized bacterial and viral genes and genomes using novoalign (version 2.05.20 [-o SAM -r A -R 0, default options]). Alignments were run on a single 3-GHz 8-CPU Intel Xeon 64-bit 61-GB RAM computer running CentOS release 5.4. Multiplexed reads from the tumor and normal libraries were deconvoluted according to sequence tags (i.e., barcodes), and the number of read-pairs that mapped unambiguously to a single location were tallied for each indexed sample and normalized against the sample read count. Ultimately, read-pair count was reported for each GenBank accession in our microbial genome database, sorted in decreasing order by the sum of unambiguous pairs, and Perl scripts were developed to mine these data. Read counts were graphically visualized first by clustering common accession reads using UPGMA (Sokal and Michener 1958) and then displayed as a heat map (log10 scale) using the Mayday package (http://www-ps.informatik.uni-tuebingen.de/mayday/wp/).
Quantitative PCR
A custom TaqMan primer/probe set was designed to amplify F. nucleatum DNA that matched the contiguous sequence from the WTSS experiment. The cycle threshold (Ct) values for Fusobacterium were normalized to the amount of human biopsy gDNA in each reaction by using a primer/probe set for the reference gene, prostaglandin transporter (PGT), as previously described (Wilson et al. 2006). The reaction efficiency for the Fusobacterium assay and the PGT assay were found to be 97% and 98%, respectively. The fold difference (2−ΔΔCt) in Fusobacterium abundance in tumor versus normal tissue was calculated by subtracting ΔCttumor from ΔCtnormal, where ΔCt is the difference in threshold cycle number for the test and reference assay. Isolated biopsy DNA was quantified by PicoGreen Assay (Invitrogen) on a Wallac Victor spectrophotometer (Perkin Elmer). Each reaction contained 5 ng of DNA and was assayed in duplicate in 20 μL reactions containing 1× final concentration TaqMan Universal Master Mix (ABI part number 4304437), 18 μM each primer, and 5 μM probe and took place in a 384-well optical PCR plate. Amplification and detection of DNA was performed with the ABI 7900HT Sequence Detection System (Applied Biosystems) using the following reaction conditions: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 sec at 95°C and 1 min at 60°C. Cycle thresholding was calculated using the automated settings for SDS 2.2 (Applied Biosystems). The primer and probe sequences for each assay were as follows: Fusobacteria forward primer, 5′-CAACCATTACTTTAACTCTACCATGTTCA-3′; Fusobacteria reverse primer, 5′-GTTGACTTTACAGAAGGAGATTATGTAAAAATC-3′; Fusobacteria FAM probe, 5′-GTTGACTTTACAGAAGGAGATTATGTAAAAATC-3′; PGT forward primer, 5′-ATCCCCAAAGCACCTGGTTT-3′; PGT reverse primer, 5′-AGAGGCCAAGATAGTCCTGGTAA-3′; PGT FAM probe, 5′-CCATCCATGTCCTCATCTC-3′. The entire qPCR experiment was performed a second time using the same samples and methods as outlined above, for the purpose of replication, and very similar results were obtained.
Fusobacterium culture
Frozen tumor sections were thawed and immediately placed into 500 μL of pre-reduced phosphate-buffered saline, and the tissue was agitated and gently broken up using a pipette fitted with a sterile, wide-bore, plugged tip. One hundred milliliter aliquots of this suspension were directly spread onto pre-reduced fastidious anaerobe agar (FAA) plates supplemented with 5% defibrinated sheep blood (DSB), and incubated for 10 d in a humidified anaerobe chamber (Ruskinn Bug Box). Plates were inspected every 2 d for growth, and all colonies were picked and streak-purified on further pre-reduced FAA + 5% DSB plates. Single colonies were examined by phase microscopy using a Leica ICC50 microscope fitted with a 100× oil immersion objective, looking for slender rods or needle-shaped cells characteristic of F. nucleatum. gDNA was isolated from positively identified isolates using a Maxwell 16 instrument with cell DNA cartidges, and aliquots were used as template in PCR with primers and conditions as described by Kim et al. (2004). A product size of 495 nt confirmed that the isolate belonged to the Fusobacterium genus, and a further PCR to partially amplify 16S rRNA gene was performed using the same DNA template using primers and conditions as defined by Ben-Dov et al. (2006). This product was sent for Sanger sequence analysis to MWG Operon, and obtained traces that confirmed F. nucleatum as the species. In total, three clones of the isolated strain were obtained from the tumor specimen from patient number 53, and named CC53 F, G, and H, respectively. All strains were stored at −80°C in cryoprotectant media (12% [w/v] skim milk powder, 1% [v/v] dimethyl sulfoxide, and 1% [v/v] glycerol).
Primer walking
PCR primers were designed using primer 3.0 and the F. nucleatum types strain (ATCC 25586) genome as reference. For PCR, 1 ng of extracted gDNA was used as template, and Phusion polymerase (NEB) and buffers were used for the PCR. Cycling conditions were as follows: 2 min at 94°C, then 30 sec at 94°C, 30 sec at 67°C, and 30 sec at 72°C for 30 cycles. PCR products were purified using Ampure magnetic beads. Sequencing reactions were done using BigDye 3.1, and reaction products were run on AB 3730xl. Phred quality 30 trimmed sequences were used in a BLASTN alignment against the HMP reference genome data, keeping the hit with the highest sequence identity.
Whole-genome sequencing of a representative strain
Fusobacterium genomic DNA was sonicated, and size fractions between 175 and 200 bp and between 400 and 450 bp were isolated following PAGE. WGSS paired end Illumina libraries were prepared from each size fraction as described previously with the following modifications: The final PCR amplification was increased to 15 cycles and contained the standard Illumina PE1 PCR primer and an indexed PE2 primer as detailed above for RNA-seq library construction. A total of 92.0 million paired 100-nt reads were obtained from a single lane of the Illumina HiSeq instrument. After quality filtering, keeping only pairs with an average base quality of Q30 or higher, 64.8 million paired reads were aligned with novoalign (http://www.novocraft.com) (-o SAM -r A -R 0) onto the F. nucleatum subsp. nucleatum ATCC 25586 (GenBank accession NC_003454.1) and Fusobacterium sp. 3_1_36A2 genome sequences (HMP accessions GG698790-GG698801), respectively. Paired read alignments were processed using custom Perl scripts that tracked genome sequence coverage, depth of coverage, and average sequence identity of mapped pairs. Annotation of strain sp. 3_1_36A2 regions devoid of read alignments was performed by extracting the coordinates of alignment gaps 1 kb or larger and mining the HMP GenBank-format file for existing gene annotations (http://www.hmpdacc.org/data_genomes.php). Reads that did not align onto the sp. 3_1_36A2 genome assembly were quality-trimmed to only include those having 70 or more consecutive Q30 bases and assembled with SSAKE (v3.7 [-p 1 -m 20 -o 2 -r 0.7]) in 67 contigs (mean size = 1225 bp; max size = 6018 bp; total bases = 82,076 bp; N50 = 1359 bp). The contigs were annotated using BLASTX (v2.2.25), reporting the best hit for each high-scoring pair and manually inspecting each alignment.
In a separate analysis, the 64.8 million paired quality control (QC) reads were filtered further, leaving only sequences having 99 or 100 consecutive Q30 bases. This aggressive filter yielded ∼32 million total reads, including 4.5 million paired and 22.9 million unpaired reads, and assembled with SSAKE (v3.7 -p 1 -m 20 -o 2 -r 0.7) into 379 contigs (mean size = 5460 bp; max size = 31,878 bp; total bases = 2,069,558 bp; N50 = 8680 bp). The Fusobacterium sp. 3_1_36A2 genome assembly was aligned onto the type strain using cross_match (http://www.phrap.org [-minmatch 29 -minscore 59 -masklevel 101]) and ordered/oriented based on the latter. Fusobacterium tumor isolate contigs were, in turn, aligned onto the reordered Fusobacterium sp. 3_1_36A2 HMP genome assembly and ordered/oriented according to that genome sequence, using the same cross_match parameters. Three-way cross_match alignments between the ordered Fusobacterium genomes were performed and plotted using hive plots (http://www.hiveplot.com).
Epithelial cell invasion assays
Caco-2 cell invasion assays with CC53 were carried out in triplicate using a differential staining immunofluorescence procedure as previously described (Strauss et al. 2011). Briefly, bacterial cultures were grown to late log phase according to predetermined growth-curve data and normalized for cell number using McFarland standards. Caco-2 cells were grown to 80% confluence on glass coverslips in 24-well plates and infected at a multiplicity of infection of 100:1 (bacterial cells:intestinal cells). Infected cells were maintained for 4 h at 37°C, 5% CO2 following infection, after which time cells were washed with PBS to remove non-adherent bacteria and then fixed with 2.5% paraformaldehyde, and blocked in 10% (v/v) normal goat serum. Prepared polyclonal antibodies were diluted to 1/500, applied to coverslips, and incubated for 1 h at 37°C. Coverslips were then incubated with donkey anti-rabbit (EAV_AS1) or anti-rat (EAV_AS2) Alexa 350 (1/100) (Molecular Probes), permeabilized by the addition of 0.1% Triton X-100, and then re-incubated with prepared polyclonal antibodies, as above. Following this, cells were labeled with donkey anti rat or anti-rabbit Cy3 (1/500) for 30 min at 37°C, as well as Alexa 488 Phalloidin (Molecular Probes) (1/200). Coverslips were mounted onto glass slides and examined at 40× magnification using a Leica DMIREB2 microscope and an ORCA-ER digital camera. Images were captured using Volocity (Improvision) software. Using this protocol, bacteria external to the host cell were labeled with both Cy3 and Alexa 350 (and appeared purple when channels were merged), whereas bacteria inside the cells were labeled with Cy3 only (appearing only orange when channels were merged). Each invasion assay was carried out on three separate occasions using freshly prepared Caco-2 cells and bacterial inocula.
Data access
The sequencing data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRP007584.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research, Genome British Columbia, and the Crohn's & Colitis Foundation of Canada. We thank Michelle Daigneault, Yongjun Zhao, and Michael Mayo for technical support. We thank Dr. Joanne Johnson for project management assistance.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.126516.111.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Dov E, Shapiro OH, Siboni N, Kushmaro A 2006. Advantage of using inosine at the 3′ termini of 16S rRNA gene universal primers for the study of microbial diversity. Appl Environ Microbiol 72: 6902–6906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad A, Jensen H, Bakken V 1996. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin Microbiol Rev 9: 55–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319: 1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. 2011. Ensembl 2011. Nucleic Acids Res 39: D800–D806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes N 2010. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer 10: 784–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YW, Shi W, Huang GT, Kinder Haake S, Park NH, Kuramitsu H, Genco RJ 2000. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun 68: 3140–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han XY, Weinberg JS, Prabhu SS, Hassenbusch SJ, Fuller GN, Tarrand JJ, Kontoyiannis DP 2003. Fusobacterial brain abscess: a review of five cases and an analysis of possible pathogenesis. J Neurosurg 99: 693–700 [DOI] [PubMed] [Google Scholar]
- Kai A, Cooke F, Antoun N, Siddharthan C, Sule O 2008. A rare presentation of ventriculitis and brain abscess caused by Fusobacterium nucleatum. J Med Microbiol 57: 668–671 [DOI] [PubMed] [Google Scholar]
- Kim M-K, Kim H-K, Kim B-O, Yoo SY, Seong J-H, Kim D-K, Lee SE, Choe S-J, Park J-C, Min B-M, et al. 2004. Multiplex PCR using conserved and species-specific 16S rDNA primers for simultaneous detection of Fusobacterium nucleatum and Actinobacillus actinomycetemcomitans. J Microbiol Biotechnol 14: 110–115 [Google Scholar]
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, et al. 2012. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res (this issue). doi: 10.1101/gr.126573.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisanaprakornkit S, Kimball JR, Weinberg A, Darveau RP, Bainbridge BW, Dale BA 2000. Inducible expression of human β-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: Multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect Immun 68: 2907–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, Tjalsma H 2011. Towards the Human Colorectal Cancer Microbiome. PLOS ONE 6: e20447 doi: 10.1371/journal.pone.0020447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall B, Warren J 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic-ulceration. Lancet 1: 1311–1315 [DOI] [PubMed] [Google Scholar]
- McLean MH, Murray GI, Stewart KN, Norrie G, Mayer C, Hold GL, Thomson J, Fyfe N, Hope M, Mowat NA, et al. 2011. The inflammatory microenvironment in colorectal neoplasia. PLoS ONE 6: e15366 doi: 10.1371/journal.pone.0015366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RA, Warren RL, Freeman JD, Gustavsen JA, Chénard C, Friedman JM, Suttle CA, Zhao Y, Holt RA 2011. The sensitivity of massively parallel sequencing for detecting candidate infectious agents associated with human tissue. PLoS ONE 6: e19838 doi: 10.1371/journal.pone.0019838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. 2010. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 42: 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, Rusch DB, Mitreva M, Sodergren E, et al. 2010. a catalog of reference genomes from the human microbiome. Science 328: 994–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin D 2006. The global health burden of infection-associated cancers in the year 2002. Int J Cancer 118: 3030–3044 [DOI] [PubMed] [Google Scholar]
- Peyret-Lacombe A, Brunel G, Watts M, Charveron M, Duplan H 2009. TLR2 sensing of F. nucleatum and S. sanguinis distinctly triggered gingival innate response. Cytokine 46: 201–210 [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, et al. 2009. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461: 809–813 [DOI] [PubMed] [Google Scholar]
- Signat B, Roques C, Poulet P, Duffaut D 2011. Role of Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol 13: 25–35 [PubMed] [Google Scholar]
- Sokal R, Michener C 1958. A statistical method for evaluating systematic relationships. Univ Kans Sci Bull 28: 1409–1438 [Google Scholar]
- Strauss J, White A, Ambrose C, McDonald J, Allen-Vercoe E 2008. Phenotypic and genotypic analyses of clinical Fusobacterium nucleatum and Fusobacterium periodonticum isolates from the human gut. Anaerobe 14: 301–309 [DOI] [PubMed] [Google Scholar]
- Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, DeVinney R, Lynch T, Allen-Vercoe E 2011. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis 17: 1971–1978 [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Dörffel Y, Loening-Baucke V, Theissig F, Rückert JC, Ismail M, Rau WA, Gaschler D, Weizenegger M, Kühn S, et al. 2011. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut 60: 34–40 [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL 1988. Genetic alterations during colorectal-tumour development. N Engl J Med 319: 525–532 [DOI] [PubMed] [Google Scholar]
- Warren RL, Sutton GG, Jones SJ, Holt RA 2007. Assembling millions of short DNA sequences using SSAKE. Bioinformatics 23: 500–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PH 2010. Canadian Tumour Repository Network. Biopreserv Biobank 8: 181–185 [DOI] [PubMed] [Google Scholar]
- Weber G, Shendure J, Tanenbaum DM, Church GM, Meyerson M 2002. Identification of foreign gene sequences by transcript filtering against the human genome. Nat Genet 30: 141–142 [DOI] [PubMed] [Google Scholar]
- Weeks DF, Katz DS, Saxon P, Kubal WS 2010. Lemierre syndrome: report of five new cases and literature review. Emerg Radiol 17: 323–328 [DOI] [PubMed] [Google Scholar]
- Weiss EL, Shaniztki B, Dotan M, Ganeshkumar N, Kolenbrander PE, Metzger Z 2000. Attachment of Fusobacterium nucleatum PK1594 to mammalian cells and its coaggregation with periodontopathogenic bacteria are mediated by the same galactose-binding adhesin. Oral Microbiol Immunol 15: 371–377 [DOI] [PubMed] [Google Scholar]
- Wilson G, Flibotte S, Chopra V, Melnyk B, Honer W, Holt R 2006. DNA copy-number analysis in bipolar disorder and schizophrenia reveals aberrations in genes involved in glutamate signaling. Hum Mol Genet 15: 743–749 [DOI] [PubMed] [Google Scholar]
- World Health Organization 2011. Fact sheet no. 297. World Health Organization, Geneva, Switzerland. http://www.who.int/mediacentre/factsheets/fs297/en/ [Google Scholar]
- Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, et al. 2009. A human colonic commensal promotes colon tumourigenesis via activation of T helper type 17 T cell responses. Nat Med 15: 1016–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ximenez-Fyvie LA, Haffajee AD, Socransky SS 2000. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J Clin Periodontol 27: 648–657 [DOI] [PubMed] [Google Scholar]