Abstract
Most of the studies characterizing DNA methylation patterns have been restricted to particular genomic loci in a limited number of human samples and pathological conditions. Herein, we present a compromise between an extremely comprehensive study of a human sample population with an intermediate level of resolution of CpGs at the genomic level. We obtained a DNA methylation fingerprint of 1628 human samples in which we interrogated 1505 CpG sites. The DNA methylation patterns revealed show this epigenetic mark to be critical in tissue-type definition and stemness, particularly around transcription start sites that are not within a CpG island. For disease, the generated DNA methylation fingerprints show that, during tumorigenesis, human cancer cells underwent a progressive gain of promoter CpG-island hypermethylation and a loss of CpG methylation in non-CpG-island promoters. Although transformed cells are those in which DNA methylation disruption is more obvious, we observed that other common human diseases, such as neurological and autoimmune disorders, had their own distinct DNA methylation profiles. Most importantly, we provide proof of principle that the DNA methylation fingerprints obtained might be useful for translational purposes by showing that we are able to identify the tumor type origin of cancers of unknown primary origin (CUPs). Thus, the DNA methylation patterns identified across the largest spectrum of samples, tissues, and diseases reported to date constitute a baseline for developing higher-resolution DNA methylation maps and provide important clues concerning the contribution of CpG methylation to tissue identity and its changes in the most prevalent human diseases.
Epigenetics encompasses a large number of mechanisms underlying embryonic development, differentiation, and cell identity, including DNA methylation and histone modifications (Bernstein et al. 2007; Hemberger et al. 2009). The existence of distinct epigenomes might explain why the same genotypes generate different phenotypes, such as those seen in Agouti mice (Michaud et al. 1994), cloned animals (Humpherys et al. 2001), and monozygotic twins (Fraga et al. 2005; Kaminsky et al. 2009). Most importantly, epigenetic alterations are increasingly recognized as being involved in human diseases (Das et al. 2009), such as cancer (Jones and Baylin 2007; Esteller 2008) and imprinting (Feinberg 2007), neurological (Urdinguio et al. 2009), cardiovascular (Gluckman et al. 2009), and autoimmune (Richardson 2007) disorders, among others. For the first time, it is possible to define whole epigenomes, which represent all epigenetic marks in a given cell type, thanks to the development of powerful new genomics technologies (Bernstein et al. 2007; Esteller 2007; Jones and Baylin 2007; Bonetta 2008; Lister and Ecker 2009). Furthermore, coordinated epigenomic projects are starting to be launched (Jones et al. 2008; Abbot 2010).
One of the earliest studied epigenetic marks in eukaryotes is cytosine DNA methylation, which acts as a stably inherited modification affecting gene activity and cellular biology. Determining the complete DNA methylome entails describing all the methylated nucleotides in an organism. The gold standard technique for analyzing the methylation state of individual cytosines is bisulfite sequencing in which unmethylated cytosines are converted to uracils and read as thymines, while methylated cytosines are protected from conversion. Bisulfite sequencing yields precise nucleotide resolution data, but this method has been limited to relatively small genome coverage (Rakyan et al. 2004; Eckhardt et al. 2006; Frigola et al. 2006; Zhang et al. 2009), although it has proved useful for analyzing viral DNA methylomes (Fernandez et al. 2009). Alternative approaches involve the isolation of methylated fractions of the genome by methylation-sensitive restriction (Lippman et al. 2005; Irizarry et al. 2008), immunoprecipitation with a methylcytosine (Weber et al. 2005; Keshet et al. 2006; Weber et al. 2007; Down et al. 2008) or methyl-CpG binding domain antibody (Ballestar et al. 2003; Rauch et al. 2009), combined with hybridization to genomic microarrays or ultrasequencing. This is exemplified by the recent DNA methylation analyses of the Arabidopsis genome (Zhang et al. 2006; Vaughn et al. 2007; Zilberman et al. 2007), which are further expanded by using sequencing-by-synthesis (MethylC-Seq) technology (Lister et al. 2008) and shotgun bisulfite genomic sequencing (Cokus et al. 2008). In representing mouse pluripotent and differentiated cells, bisulfite sequencing has covered roughly 1 million distinct CpG dinucleotides (4.8% of all CpGs) (Meissner et al. 2008), and two human cell lines (one each from embryonic stem cells and fetal fibroblasts) have been analyzed using MethylC-Seq, including 94% of the cytosines in the genome (Lister et al. 2009). Using whole-genome bisulfite sequencing, the DNA methylome analysis of peripheral blood mononuclear cells from a single case has also been recently reported (Li et al. 2010).
Only a small number of base-resolution DNA methylomes have been described so far. Nevertheless, even with the enormous advantages that genetic sequencing has over DNA methylation characterization with respect to time and technology, very few full genomes have been reported, either. From the genetic standpoint, this current shortage of information is being tackled through the development of efforts such as the 1000 Genomes Project (Kuehn 2008; Siva 2008) or by genome-wide association scan (GWAS) studies in which an association with a phenotype or a disease can be established if we limit the number of nucleotides assessed and thus the extent of coverage of the genome (Cantor et al. 2010; Ku et al. 2010). We decided to combine these two approaches—extremely extensive analyses of hundreds of normal and disease-associated cells and tissues with intermediate coverage of CpG dinucleotides—to obtain a DNA methylation fingerprint of 1628 human samples corresponding to healthy individuals and in those affected by the diseases most commonly associated with death in the Western world, such as cancer, neurological disorders, and cardiovascular disease.
Results
Description of 1628 samples and analysis of 1505 CpG sites
We first studied the genomic DNA from 1628 human samples corresponding to 424 normal tissues (180 leukocytes, 97 colon mucosa, and 227 other normal samples), 1054 tumorigenic samples (premalignant lesions, primary tumors, and metastases), and 150 non-cancerous disorders, such as brain lesions from Alzheimer's disease, dementia with Lewy bodies, aortic atherosclerotic lesions, myopathies, and autoimmune disorders. Supplemental Table 1 shows the complete list of samples studied. The age of donors ranged from 6 mo to 102 yr, with an average age of 57 yr. Forty percent (n = 648) were men, and 38% (n = 623) were women, the gender of the remaining 22% (n = 357) not being known. Eighty-seven percent (n = 1421) of the samples were from European volunteers and patients, while 4% (n = 59) and 2% (n = 36) were from Asian and North American populations, respectively; the origin was not known for 7% (n = 112) of cases. Finally, 93% (n = 1512) of the samples were primary tissues obtained at the time of the clinically indicated procedures, while 7% (n = 116) were obtained from established cell lines. Supplemental Figure 1 summarizes the described sample distribution. For all these samples, we obtained the DNA methylation fingerprints defined by the status of 1505 CpG sites located from −1500 bp to +500 bp around the transcription start sites (Supplemental Fig. 2) of 808 selected genes using the GoldenGate DNA methylation BeadArray (Illumina, Inc.) assay (Bibikova et al. 2006; Byun et al. 2009; Christensen et al. 2009). The panel of genes includes oncogenes and tumor-suppressor genes, imprinted genes, genes involved in various signaling pathways, and those responsible for DNA repair, cell cycle control, metastasis, differentiation, and apoptosis (Bibikova et al. 2006; Byun et al. 2009; Christensen et al. 2009). Sixty-nine percent (n = 1044) of the 1505 CpG sites studied are located within a canonical CpG island (Takai and Jones 2002), while 31% (n = 461) are situated outside CpG islands (Supplemental Fig. 2). All human chromosomes, except the Y chromosome, are represented among the CpG sites analyzed (Supplemental Fig. 2). CpG sites in “CpG island shores,” regions of comparatively low CpG density within 2 kb of CpG islands, are not printed in the array used, and their biological relevance has already been extensively studied (Doi et al. 2009; Irizarry et al. 2009). Briefly, in our case, four probes were designed for each CpG site: two allele-specific oligos (ASOs) and two locus-specific oligos (LSOs). Each ASO–LSO oligo pair corresponded to either the methylated or unmethylated state of the CpG site. After bisulfite treatment conversion, the remaining assay steps were identical to those of the GoldenGate genotyping assay using Illumina-supplied reagents and conditions, and the arrays were imaged using a BeadArray Reader (Illumina, Inc.). Each methylation data point was represented by fluorescent signals from the M (methylated) and U (unmethylated) alleles. Before analyzing the CpG methylation data, we excluded possible sources of technical biases that could have influenced the results. Every beta value in the GoldenGate platform is accompanied by a detection P-value, and we observed that a threshold P-value above 0.01 indicated unreliable beta values (130 CpGs). X-chromosome CpG sites with female-specific DNA methylation (Reik and Lewis 2005) were also excluded (44 CpGs). Finally, nine CpG sites that were unmethylated in all normal and disease-associated samples were also excluded. Using these filters, 1322 CpGs proved to be reliable and were used subsequently in the study. Further technical information is provided in the Supplemental Methods. The precise DNA methylation status of every CpG dinucleotide analyzed in each of the 1628 samples studied is freely available by downloading from the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE28094.
DNA methylation fingerprint of human normal tissues
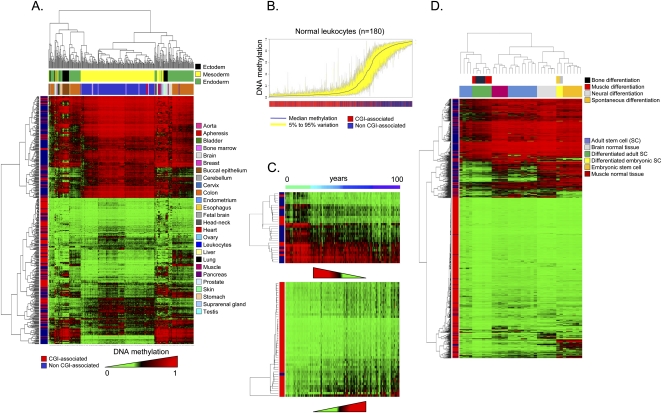
We analyzed first the DNA methylation fingerprints for 424 human normal tissues. Of the 424 normal tissues studied, only 1% (n = 17) of CpGs (corresponding to 14 genes) were methylated in all the samples studied (Supplemental Table 2). These exclusively methylated CpG dinucleotides were preferentially located outside CpG islands (82%; Fisher's exact test, p = 1.97×10−5). Conversely, 37% (n = 488) of the CpGs, corresponding to 359 5′ ends of genes, were exclusively unmethylated in every normal tissue studied (Supplemental Table 3). These always-unmethylated CpG dinucleotides were almost exclusively located within CpG islands (98%; Fisher's exact test, p = 2.20×10−85) and were associated with housekeeping expression genes (Fisher's exact test, p = 1.13×10−4) (Supplemental Methods). Most importantly, significant differential DNA methylation (Kruskal-Wallis rank-sum test, p < 2.21×10−16) was encountered between different normal samples of 511 CpG dinucleotides using elastic net classifiers, which enabled their distinction on the basis of tissue type using an unsupervised hierarchical clustering approach (Fig. 1A). The 511 CpG sites described correspond to 359 genes and, providing further validation to the data, 220 genes (61%; 220) and 137 (38%) were previously identified as genes with tissue-specific DNA methylation using the same 1505 CpG platform (Byun et al. 2009) or a 27,000-CpG microarray (Nagae et al. 2011), respectively. Illustrative examples of genes found in the three sets, and also confirmed by bisulfite genomic sequencing in another independent study (Eckhardt et al. 2006), include TBX1 (T-box 1), OSM (oncostatin M), and GP1BB (glycoprotein Ib [platelet] beta polypeptide). Examples of tissue-specific CpG methylation further validated by pyrosequencing (“technical replicates”) are shown in Supplemental Figure 3.
Figure 1.
DNA methylation fingerprints for human normal tissues. (A) Unsupervised hierarchical clustering and heatmap including CpG dinucleotides with differential DNA methylation encountered between different normal primary samples. Tissue type and development layers are displayed in the different colors indicated in the figure legends. Average methylation values are displayed from 0 (green) to 1 (red). (B) Deviation plot for the 1322 CpG sites studied in leukocyte samples showing that little CpG methylation heterogeneity (yellow area) occurs overall at CpG sites within CpG islands (red lines in the track below), while more differences in CpG methylation are observed outside CpG islands (blue lines in the track below). (C) Unsupervised hierarchical clustering and heatmap including sets of genes with high correlation values between hypomethylation (up) and hypermethylation (down) with aging. (D) Unsupervised hierarchical clustering and heatmap showing the DNA methylation patterns of embryonic and adult stem cells, comparing them with corresponding normal and differentiated tissues (muscle, bone, and neuron; and muscle and brain, respectively).
For our 359 genes with tissue-type-specific CpG methylation, their expression patterns in the 21 normal tissues are known (GEO Expression Omnibus, GEO; http://www.ncbi.nlm.nih.gov/geo/) (Supplemental Methods). Unsupervised clustering analysis of the expression of these 359 genes discriminates each normal tissue type, as the CpG methylation did, reinforcing the association between DNA methylation and transcriptional silencing of the neighboring gene for these targets (Supplemental Fig. 3). Strikingly, the CpG sites for which methylation status was the most valuable for discriminating between tissue types were those located in non-CpG-island 5′ ends (Fisher's exact test, p = 5.85×10−49). These data support the long-standing hypothesis that most housekeeping genes contain CpG islands around their transcription start sites, while half of the tissue-specific genes have a CpG island at their 5′ ends, and the other half are 5′-CpG-poor (Illingworth and Bird 2009). The top-scoring genes with defined organ-specific DNA methylation are listed in Supplemental Table 4. The tissue-type-specific DNA methylation patterns, which are in line with previous observations in humans (Eckhardt et al. 2006; Shen et al. 2007; Byun et al. 2009; Christensen et al. 2009), also match the developmental layers in which the tissues originated (endoderm, mesoderm, or ectoderm) (Fig. 1A), implying the existence of germ-layer-specific DNA methylation (Sakamoto et al. 2007). Interestingly, 49 CpG sites corresponding to 26 imprinted genes were also included in the assay (Supplemental Fig. 4). We observed that CpG sites located outside differentially methylated regions (DMRs) (Dindot et al. 2009; Monk 2010) behaved like the CpGs of non-imprinted genes in normal tissues: CpGs located within and outside CpG islands were unmethylated and methylated, respectively (Supplemental Fig. 4). However, CpGs within DMRs were 50% methylated in all normal tissue types studied (Supplemental Fig. 4).
Within the same tissue type, interindividual DNA methylation differences were minimal. For example, the DNA methylation deviation plot for the 1322 CpG sites studied in leukocyte samples from 180 healthy donors showed little heterogeneity (Fig. 1B). However, it is interesting to note that the main DNA methylation differences between individuals occurred at CpG sites located outside CpG islands in comparison to CpG-island-associated CpG dinucleotides (Wilcoxon test, p = 3.52×10−39) (Fig. 1B). One interesting issue concerned the putative impact of aging on the DNA methylation patterns of normal tissues in humans (Christensen et al. 2009; Rakyan et al. 2010; Teschendorff et al. 2010) and mice (Maegawa et al. 2010). Our analysis of the leukocyte samples from the 180 healthy donors (Fig. 1B) revealed sets of genes that were significantly hypermethylated (n = 43) or hypomethylated (n = 25) during the normal aging process (Fig. 1C; Supplemental Table 5). Examples of age-specific CpG methylation further validated by pyrosequencing are shown in Supplemental Figure 4. It is encouraging to note that there are genes with age-related methylation found in our study that were also identified in the mentioned previous reports using the same 1505 CpG platform (Christensen et al. 2009) or the 27,000 CpG microarray (Rakyan et al. 2010; Teschendorff et al. 2010). Among these, we can underline for the age-hypermethylated genes MYOD1 (myogenic differentiation 1), and for the age-hypomethylated genes representative examples include NOD2 (also known as CARD15, caspase recruitment domain-containing protein 15), ACVR1 (activin A receptor type I), and SOD3 (Superoxide Dismutase 3). Furthermore, we also found that the CpG hypermethylation events in aging were significantly more likely to occur in the promoters of those genes with enriched Polycomb occupancy (Fisher's exact test, p = 3.83×10−8; permutation P-value = 0.0014) and the presence of the bivalent histone domain (3mK4H3 + 3mK27H3) (Fisher's exact test, p = 9.03×10−4; permutation P-value = 0.0354) in embryonic stem cells (Supplemental Fig. 4), as was recently suggested (Rakyan et al. 2010; Teschendorff et al. 2010).
In addition to the tissue-type-specific DNA methylation patterns, one group of normal cells had distinctive DNA methylation profiles: embryonic and adult stem cells (Fig. 1D). Adult and embryonic stem cells both had DNA methylation fingerprints that did not resemble any of the differentiated primary normal tissues studied (Fig. 1D). Furthermore, we confirmed that the previously studied samples from multipotent adult stem cells (Aranda et al. 2009) had different DNA methylation fingerprints from pluripotent embryonic stem cells (Fig. 1D). Herein, we went further to show that induction of differentiation of both types of stem cells through different lineages produced DNA methylation fingerprints that resembled those present in the corresponding normal differentiated tissues, such as muscle or neuron (Fig. 1D). Interestingly, in vitro–differentiated material from adult and embryonic stem cells did not completely recapitulate the DNA methylation patterns present in the corresponding primary differentiated tissues, and there were always deficiently methylated CpG sites. Supplemental Table 6 provides examples of these in muscle and neuronal tissues. Supplemental Figure 5 shows examples of tissue-specific CpG methylation, unachieved upon in vitro differentiation of stem cells and validated by pyrosequencing analysis.
DNA methylation fingerprint of human cancer
We next studied the DNA methylation fingerprints for 1054 human tumorigenesis samples. Genetic and epigenetic alterations both contribute to cancer initiation and progression (Jones and Baylin 2007; Esteller 2008). One of the first epigenetic alterations found in human cancer was the global low level of DNA methylation in tumors compared with healthy tissue counterparts. Global DNA hypomethylation is accompanied by hypermethylation of CpG islands at specific promoter regions. Nowadays, hypermethylation of the CpG islands in the promoter regions of tumor-suppressor genes is also recognized as a major event in the origin of many cancers (Jones and Baylin 2007; Esteller 2008). Tumor-suppressor genes disrupted by DNA methylation-associated transcriptional silencing in sporadic tumors include the retinoblastoma tumor suppressor gene (RB1), VHL (associated with von Hippel-Lindau disease), the cell cycle inhibitor CDKN2A (also known as p16INK4a), MLH1 (a homolog of Escherichia coli mutl), and BRCA1 (breast-cancer susceptibility gene 1) (Jones and Baylin 2007; Esteller 2008). Using candidate gene approaches and early epigenomics technologies, a CpG-island hypermethylation profile of human primary tumors emerged that suggested that a defining DNA hypermethylome could be assigned to each tumor type (Costello et al. 2000; Esteller et al. 2001; Ballestar and Esteller 2008). Herein, we have analyzed the DNA methylation fingerprints of 1054 human tumorigenesis samples, including 855 primary malignancies (611 solid tumors from 19 tissue types and 244 hematological malignancies), 50 metastatic lesions, 25 premalignant lesions, 82 cancer cell lines, and 42 cancers of unknown primary origin (CUPs) (Supplemental Table 1). The DNA methylation map that emerges shows a tumor-type-specific profile characterized by the progressive gain of CpG methylation within CpG-island-associated promoters and a cumulative loss of CpG methylation outside CpG islands in the different steps of tumorigenesis.
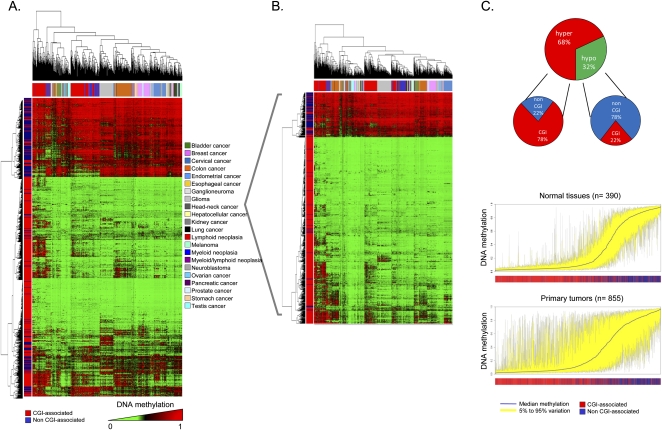
First, unsupervised clustering of the DNA methylation profiles obtained from the 855 primary tumors demonstrated that each type of malignancy had its own aberrant DNA methylation landscape (Fig. 2A). From a quantitative standpoint, 1003 CpG sites (76% of the 1322 validated CpGs) had significantly different methylation levels between tumor types (Kruskal-Wallis rank-sum test, p < 2.2×10−16). The distinction of primary tumors by their tissue of origin was maintained even when we subtracted the tissue-type-specific DNA methylation described above (511 CpG sites) (Supplemental Table 4) from the analysis of the DNA methylation profiles for each normal tissue (Fig. 2B). Comparing each tumor type with its corresponding normal tissue, 729 CpG sites (55% of the 1322 CpGs) showed differential DNA methylation. Using these tumor/normal differentially methylated CpG sites, overall human primary tumors were characterized by increased levels of CpG dinucleotide methylation: 68% (n = 496) were hypermethylated and 32% (n = 233) were hypomethylated (t-test, p = 3.521×10−5) (Fig. 2C). Most importantly, the location of these DNA methylation events differed: CpG dinucleotide hypermethylation occurred within CpG islands (78%), while CpG hypomethylation was present in 5′ ends of non-CpG-island genes (78%; Fisher's exact test, p = 2.59×10−47; permutation P-value < 0.001) (Fig. 2C). A DNA methylation deviation plot for the 1322 CpG sites studied in all normal primary tissues (n = 390) versus all primary tumors (n = 855) shows the hypermethylated CpG sites within CpG islands and hypomethylated CpG sites outside CpG islands observed in the malignancies (Fig. 2C) (Paired Wilcoxon test, p < 2.2×10−16). CpG sites with cancer-specific differential methylation according to tumor type in comparison with their corresponding normal tissue are provided in Supplemental Table 7. Examples of cancer-type-specific CpG methylation further validated by pyrosequencing are shown in Supplemental Figure 6. Those CpG sites with highly specific methylation changes occurring only in one tumor type are shown in Supplemental Table 8. Interestingly, we also confirmed the previous observation (Ohm et al. 2007; Schlesinger et al. 2007; Widschwendter et al. 2007) that the CpG hypermethylation events in cancer were significantly more likely to occur in the promoters of those genes with enriched Polycomb occupancy (Fisher's exact test, p = 5.03×10−6; permutation P-value = 0.0012) and the presence of bivalent histone domains (3mK4H3 + 3mK27H3) (Fisher's exact test, p = 5.97×10−4; permutation P-value = 0.0278) in embryonic stem cells (Supplemental Fig. 6). We also found evidence to reinforce the link between the 5′-end CpG methylation and transcriptional silencing (Jones and Baylin 2007; Esteller 2008) by developing expression microarray studies (Supplemental Methods) in the 19 primary colorectal tumors from which we had obtained the DNA methylation profiles. We observed that the median expression of all the CpG hypermethylation-associated genes was significantly lower than in those CpG hypomethylation-linked genes (Kruskal-Wallis test, p = 1.56×10−8) (Supplemental Fig. 6).
Figure 2.
DNA methylation fingerprint of human cancer. (A) Unsupervised hierarchical clustering and heatmap showing distinction of primary tumor DNA methylation fingerprints according to the tissue of origin. (B) Unsupervised hierarchical clustering and heatmap of primary tumors excluding CpG sites with tissue-specific methylation. (C, above) Pie charts displaying the percentage of hypermethylated CpG sites (red) and hypomethylated CpG sites (green) in human malignancies, and their distribution in CpG islands (CGI in red) and outside CpG islands (non-CGI in blue). (Below) Deviation plot for the 1322 CpG sites showing the great methylation heterogeneity (yellow area) of primary tumors in comparison with normal primary tissues.
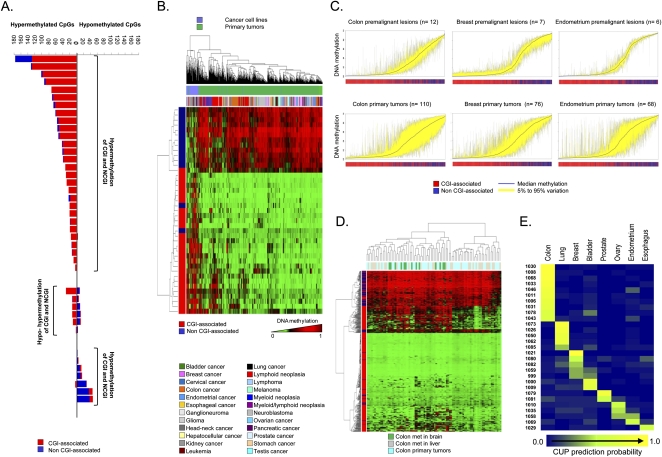
For our largest set of samples with paired normal–tumor tissues from the same patient (41 cases of colorectal cancer), we observed that of the 1322 CpG sites studied, CpG dinucleotides within CpG-island promoters became significantly more DNA-methylated in 79% of cases (34 of 43 normal/tumor pairs; Wilcoxon test, p = 2.47×10−7), while CpGs located in non-CpG-island promoters more commonly underwent DNA hypomethylation events, in 51% of cases (22 of 43 normal/tumor pairs; Wilcoxon test, p = 0.001). If we consider the colorectal tumor population as a whole, in 68% of cases (28 of 41) the primary malignancy gained CpG dinucleotide methylation within promoter CpG islands and non-CpG-island promoters, while in 15% of tumors (six of 41) the gain of CpG island methylation occurred in a context of loss of promoter non-CpG-island methylation (Fig. 3A). Interestingly, 17% of cases (seven of 41) featured a loss of methylation in both promoter CpG islands and non-CpG-island promoters (Fig. 3A). Thus, the presence of hypermethylation of promoter CpG islands appears to be a common hallmark of human tumors, but there are subsets of cancers that present other DNA methylation profiles at promoter CpG sites that suggest additional and complex aberrant DNA methylation pathways in tumorigenesis. For example, the possibility that DNA hypomethylation events at CpGs located in non-CpG-island promoters, typical of genes with restricted tissue-specific expression (Illingworth and Bird 2009), can cause a loss of cellular identity in transformed cells is worth further investigation.
Figure 3.
Scenarios of DNA methylation changes in human tumorigenesis. (A) Bart plot showing the CpG hypermethylation or hypomethylation changes observed when comparing paired normal–tumor tissues from the same colorectal cancer patient. They can be distinguished if the methylation change occurs in CpG island (CGI) or non-CpG island (non-CGI)–associated CpG. (B) Unsupervised hierarchical clustering and heatmap including a set of specific CpG sites that undergo differential DNA methylation only in cancer cell lines. (C) Deviation plot for the 1322 CpG sites shows greater CpG methylation heterogeneity (yellow area) in established tumors (colon, breast, and endometrial cancers) than in their corresponding premalignant lesions. (D) DNA methylation unsupervised clustering analyses and heatmap of primary tumors, local liver metastases, and distant brain metastases from the same colorectal cancer patient. A CpG methylation-specific pattern for brain metastases (green lanes) is observed. (E) CpG methylation prediction heatmap showing the CUP classification to a specific tumor type.
As cancer cell lines are a major tool in biomedical research, we next examined how the DNA methylation profiles of cell lines differ from those of the primary tumor types. The analyses of the DNA methylation fingerprints of 82 human cancer cell lines representing 14 tumor types (Supplemental Table 1) showed that, overall, they preserved their original cancer-type-specific profile and underwent an increase in the levels of CpG dinucleotide methylation in comparison with the corresponding normal tissues (Paired Wilcoxon test, p < 2.2×10−16) (Supplemental Fig. 7), as occurs with most primary tumors. Examples of CpG methylation in cancer cell lines further validated by pyrosequencing are shown in Supplemental Figure 7. In the same line as primary malignancies, the hypermethylated CpG sites in cancer cell lines occurred significantly more often within CpG islands (Supplemental Fig. 7), while CpG hypomethylation events mainly happened around transcription start sites that did not contain a CpG island (Paired Wilcoxon test, p < 2.2×10−16) (Supplemental Fig. 7). However, there were qualitative and quantitative differences. First, human cancer cell lines had significantly greater hypermethylation of promoter CpG islands and non-CpG-island promoters (Paired Wilcoxon test, p < 2.2×10−16) (Supplemental Fig. 7). At this stage, we cannot distinguish whether these greater changes are associated with the in vitro growth of these cells over many years, or if the DNA methylation changes were more detectable because there was no contaminating normal tissue, as is the case in primary tumors. Second, there are a set of specific CpG sites that only undergo differential DNA methylation in cancer cell lines (Supplemental Table 9), which enable them to be classified into a distinct clustering arm in the unsupervised analysis (Fig. 3B). We further tested the association between hypermethylated CpGs at the 5′ ends and transcriptional silencing of the corresponding gene by treating five cancer cell lines (SW480, HN-011A, HN-011B, IGR37, and IGR39) with the DNA demethylating agent 5′-aza-2′-deoxycytidine, followed by gene expression microarray analysis (Supplemental Methods). We observed that while genes with associated hypermethylated CpGs had a low median expression compared with their corresponding normal tissues, upon treatment with the hypomethylating agent, their expressions were restored (Supplemental Fig. 8).
The comprehensive collection of human tumorigenesis samples studied here allowed us to address two other interesting aspects of cancer epigenetics: timing and progression. For genetic changes, it is well known that there is an accumulation of genetic events that drive the carcinogenesis process from the healthy tissue to early premalignant lesions and finally to established full-blown tumors and metastasis, as exemplified by colorectal tumorigenesis (Fearon and Vogelstein 1990). Candidate gene approaches and limited epigenomics strategies have also indicated that this could be a pathway leading to aberrant DNA methylation changes (Fraga et al. 2004). Our analysis of the DNA methylation signatures in progressive samples of three different tumorigenesis pathways (colon, breast, and endometrial cancers) demonstrated the increasing degree of CpG dinucleotide methylation within promoter CpG islands and a loss of CpG methylation outside CpG islands in consecutive steps (Fig. 3C). The DNA methylation deviation plot for the 1322 CpG sites in colorectal adenomas versus primary colorectal tumors, breast hyperplasias versus primary breast tumors, and endometrial hyperplasias versus primary endometrial carcinomas demonstrated that the full-blown tumors had significantly greater hypermethylation of promoter CpG islands in association with the loss of CpG methylation in non-CpG islands than their corresponding premalignant lesions (Paired Wilcoxon test, p < 2.2×10−16) (Fig. 3C). Most importantly, for colorectal tumors where we had DNA from brain metastasis available, these distant metastasis lesions achieved higher levels of promoter CpG-island hypermethylation and lower levels of non-CpG-island methylation than the primary colon malignancies (Paired Wilcoxon test, p < 2.2×10−16), suggesting that these pathological entities are the final stages of the disease. In fact, the DNA methylation unsupervised clustering analyses of primary tumors, local liver metastases, and distant brain metastases from the same colorectal cancer patient showed that there were specific hypermethylated CpGs in the brain metastases (Fig. 3D; Supplemental Table 10). Examples of specific CpG methylation in the brain metastasis of colorectal tumors validated by pyrosequencing are shown in Supplemental Figure 8. Ninety percent of cancer deaths are attributable to the development of metastasis (Mehlen and Puisieux 2006), thus these findings might have a translational value for the prediction of the metastatic capacity of a particular tumor, as has recently been shown for hypermethylated microRNA loci, and it might be a useful molecular marker in the decision process for medical and surgical intervention in the disease.
The DNA methylation fingerprints of human cancer obtained in our study can also provide additional important molecular diagnostic and prognostic biomarkers for the management of neoplasias. One example we have assessed is the case of the clinical entities classified as cancers of unknown primary origin (CUPs). These are patients who present metastatic diseases for which the primary site cannot be found despite standard investigation. The median survival in randomized studies of these patients is extremely poor (Abbruzzese et al. 1995), but if it were possible to predict the primary tumor site, the patient could be treated with a site-specific program, potentially resulting in better survival than that provided by non-specific treatment, for which the current median is only 7 mo (Greco and Pavlidis 2009). We have analyzed the DNA methylation fingerprints of 42 CUPs and compared the DNA methylation landscapes obtained with those from the aforementioned human malignancy collection where the original tissue type was known. We were able to assign a given tumor type for these CUPs in 69% (29 of 42) of cases using L1-regularized logistic regression with misclassification (R, version 2.10) to create a prediction heatmap (Fig. 3E). A proposed foster primary in these 29 cases was also achieved by conventional clustering analysis (Supplemental Fig. 8). Most importantly, the tumor type prediction of the CUPs based on the DNA methylation analyses was fully confirmed in 78% of cases (seven of nine) for which detailed pathological analysis developed at a later stage in a blind fashion was able to provide a diagnosis. We might also conclude that the remaining 31% (13 of 42) of the studied CUP cases did not represent any of the 19 tumor types included in our analysis (Supplemental Table 1). The three most common tumor types present in the DNA methylation-assigned CUPs were colorectal cancer (34%, 10 of 29), non-small-cell lung cancer (17%, five of 29), and breast tumors (17%, five of 29). These cases are particularly interesting because the introduction of targeted therapies, such as treatment with epidermal growth factor receptor (EGFR) antibodies in colorectal cancer, small-molecule inhibitors for EGFR mutations in lung adenocarcinoma, and more personalized chemotherapy options for breast cancer as a function of the hormonal and ERBB2 receptor status have improved the outcome of these patients (Harris and McCormick 2010). Thus, it is tempting to propose that the prediction of a foster primary site for CUPs based on the DNA methylation profiles might identify a more specific treatment regimen for these patients that would improve their quality of life and survival.
DNA methylation fingerprint of non-cancerous human diseases
We also analyzed the DNA methylation profiles for 150 non-cancerous human diseases. Although most of the aberrant DNA methylation patterns described in human disease have been reported for cancer, there is no reason to believe that disrupted DNA methylation signatures are not present, and might drive other common human diseases (Feinberg 2007), such as neurological (Urdinguio et al. 2009), cardiovascular (Gluckman et al. 2009), and autoimmune (Richardson 2007) disorders. The data on DNA methylation changes outside cancer are still scarce, but this could be more likely because of the small number of studies devoted to these pathologies than because DNA methylation disruption is genuinely of little importance in the origin and progression of these diseases. To address this issue, we analyzed the corresponding target tissues of 150 non-cancerous human diseases, including cerebral cortex lesions from Alzheimer's (n = 11) and dementia with Lewy bodies (n = 13), atherosclerotic lesions from the aorta (n = 18), skeletal muscle from myopathies (n = 17), leukocytes from autoimmune disorders (n = 21), and other non-tumoral diseases and tissues (n = 70) (Supplemental Table 1).
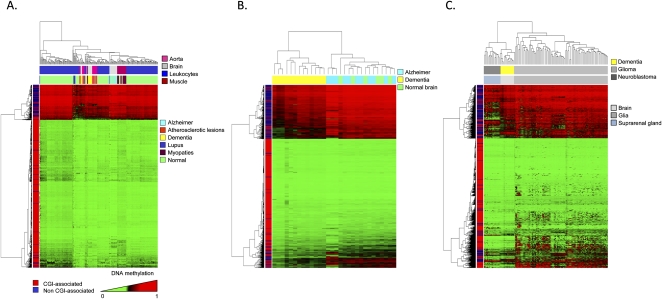
One of the most striking observations was that the described non-tumoral diseases in an unsupervised clustering had a distinct DNA methylation pattern, even if the tissue-specific CpG methylated sites were not included in the analysis (Fig. 4A). In the cases of dementia with Lewy bodies (Fig. 4B) and systemic lupus erythematosus (Supplemental Fig. 9), the DNA methylation patterns obtained from the 1322 CpG sites distinguished them from their corresponding normal tissues. Most importantly, the corresponding distinctions between brain samples of dementia with Lewy bodies versus normal brain and leukocytes of lupus patients versus healthy donor samples were exclusively associated with CpG hypomethylation events in the disease tissue (Supplemental Table 11). Examples of dementia with Lewy bodies–specific CpG hypomethylation further validated by pyrosequencing are shown in Supplemental Figure 9. Interestingly, the sequestration of DNA methyltransferase 1 (DNMT1) in the cytoplasm of neurons from patients affected by dementia with Lewy bodies has been recently described (Desplats et al. 2011), a mechanism that could explain the hypomethylation events observed in this disease using our approach. Related to the lupus patients, it is noteworthy to consider that these samples were also previously analyzed using the same 1505 CpG array to search for DNA methylation differences between monozygotic twins (Javierre et al. 2010). Herein, they were studied in a more stringent manner because they were compared to a new large set of normal leukocytes (n = 180) and with a higher cutoff value for methylation. Among the lupus-common genes derived from both studies, it is relevant to mention the hypomethylation event targeting PI3 (Proteinase Inhibitor 3), a protein that has been involved in psoriasis with an autoimmune component (Tjabringa et al. 2008). With the CpG array used, we were unable to find any significant difference between brain samples from Alzheimer's patients (Fig. 4B), aorta samples from atherosclerotic lesions (Supplemental Fig. 9), myopathies (data not shown), and their respective normal tissues.
Figure 4.
DNA methylation fingerprint in non-tumoral human diseases. (A) Unsupervised hierarchical clustering and heatmap of several non-tumoral diseases showing distinct DNA methylation profiles. (B) Unsupervised hierarchical clustering and heatmap showing significant differences between the DNA methylation patterns of dementia with Lewy bodies and normal controls. The CpG methylation platform used was unable to detect significant differences in the case of Alzheimer's versus healthy brain tissues. (C) Unsupervised hierarchical clustering and heatmap showing differences between dementia with Lewy bodies and neuroectodermal tumors (glioma and neuroblastoma).
The DNA methylation profiles obtained from the aforementioned non-cancer disorders were distinct from those observed in tumors originating from the same cell type. Dementia with Lewy bodies’ patients had CpG-site methylation patterns that distinguished them not only from normal brain (Fig. 4B), but also from neuroectodermal tumors, such as glioma and neuroblastoma (Fig. 4C). Interestingly, brain samples from dementia with Lewy bodies’ patients were closer, from a DNA methylation fingerprint perspective, to neuroblastomas than to gliomas (Fig. 4C), a characteristic that might be associated with the different cell biology of the disorders. Although in dementia associated with Alzheimer's disease there is a high grade of neuronal cell death that causes an over-representation of glia cells in the studied samples (gliosis) (Jellinger and Stadelmann 2001; Teaktong et al. 2003), in the dementia with Lewy bodies’ brain there is not such massive neuronal cell death (Jellinger and Stadelmann 2001; Teaktong et al. 2003), and the DNA methylation profiles observed resembled those found in neuron-enriched samples, such as neuroblastomas. In this regard, the existence of different DNA methylation patterns among brain regions with different cell composition has also been suggested (Ladd-Acosta et al. 2007). Distinct DNA methylation profiles for non-malignant and malignant disorders originating from the same cell type also occur for leukocytes of lupus patients displaying DNA methylation profiles that are different from those present in healthy donors or in leukemias (Supplemental Fig. 9).
Overall, these findings suggest that few specific DNA methylation changes in non-cancerous human diseases could be responsible for the observed phenotypes of these entities; they nevertheless merit further attention. Most importantly, the specific DNA methylation changes found in the described disorders occurred in clear contrast to human cancer, where the DNA methylation profile undergoes a wide-ranging, global change characterized by the gain of promoter CpG-island methylation and loss of non-CpG-island methylation. These results underlie the multifactorial nature of human cancer that involves epigenetic “hits” in almost all known cellular pathways, exemplified by the aberrant DNA methylation fingerprints obtained here.
Discussion
Disruption of the DNA methylation patterns is emerging as a common feature of human disease (Portela and Esteller 2010), where cancer is the disorder on which most of the studies have been focused (Jones and Baylin 2007; Esteller 2008). From the initial studies looking at a single locus, we have now available a wide range of epigenomics techniques to study multiple CpG sites in the human genome. In addition to methods that isolate methylated fractions of the genome by methylation-sensitive restriction (Lippman et al. 2005; Irizarry et al. 2008), immunoprecipitation with a methylcytosine (Weber et al. 2005, 2007; Keshet et al. 2006; Down et al. 2008) or methyl-CpG binding domain antibody (Ballestar et al. 2003; Rauch et al. 2009) and the genome-wide bisulfite genomic sequencing approaches (Li et al. 2010; Lister et al. 2009), it is worthwhile to highlight DNA methylation bead microarrays (Bibikova et al. 2006). This approach has the advantage that it can be used in a common standard manner by different laboratories around the world with similar bioinformatics packages, and the raw data can be user-friendly, deposited, and shared. Herein, using the first version of the DNA methylation bead microarray, which included 1505 CpG sites corresponding to 808 genes, we have studied the largest collection of human samples to date, 1628, that included 424 normal tissues, 1054 tumorigenic samples, and 150 non-cancerous disorders. Our data provide new clues about the DNA methylation profiles present in normal and disease-associated tissues and also expand and confirm previous reports in this area obtained using the same platform (Aranda et al. 2009; Byun et al. 2009; Christensen et al. 2009; Javierre et al. 2010) or a second DNA methylation bead microarray that includes 27,000 CpG sites (Rakyan et al. 2010; Teschendorff et al. 2010; Nagae et al. 2011). In normal cells, the derived picture reinforces the role of methylation in non-CpG-island 5′ ends to determine tissue-specific expression, the shift in the DNA methylation landscape from pluripotent to differentiated cells, and the existence of a DNA methylation drift associated with aging. For transformed cells, the study demonstrates that tumors undergo mostly a progressive CpG hypermethylation within CpG islands, while CpG hypomethylation occurs in 5′ ends of non-CpG-island genes. For other human disorders, such as dementia with Lewy bodies and lupus, we show that they also possess a particular DNA methylation fingerprinting that is mainly characterized by CpG hypomethylation events. One extra value of the present study is that it not only provides new DNA methylation markers for all the described normal and pathological settings, but it also validates previous results in aging (Christensen et al. 2009; Rakyan et al. 2010; Teschendorff et al. 2010), tissue specificity (Eckhardt et al. 2006; Byun et al. 2009; Christensen et al. 2009), or lupus (Javierre et al. 2010). Furthermore, the deposited data for the 1628 human samples (http://www.ncbi.nlm.nih.gov/geo; accession number GSE28094) can be a value resource for further biocomputational and meta-analysis studies.
Overall, the goal of the research described here was to examine human DNA methylation profiles comprehensively from an extremely extensive range of samples that covers physiological changes (across different tissue types, sex, age, geography, differentiation vs. stemness, primary vs. cell culture, etc.) and human diseases (cancer and common non-tumoral diseases, such as neurological, cardiovascular, and autoimmune disorders). The results obtained indicate that different DNA methylation fingerprints are observed in most of the described conditions, cancer samples being the result of the most extreme type of DNA methylation change observed, in which a profile of an increased degree of CpG dinucleotide methylation within promoter CpG islands and a loss of CpG methylation outside CpG islands is a common hallmark, as described above. A DNA methylation signature that becomes more distorted as the disease progresses can provide potentially relevant clues for improving disease management for these patients, such as we have demonstrated for the CUP cases.
We would like to underscore the relevance of the CUP DNA methylation fingerprints. Despite the increasing sophistication in the diagnostic tools for malignancies, deaths due to CUP were estimated to be 45,230 in 2007 in the United States (American Cancer Society 2007). CUPs have an incidence of 6% among all malignancies, and in 25% of cases, the primary site cannot be identified even upon postmortem examination (American Cancer Society 2007). The inability to identify the primary site of the cancer and the impossibility to provide the right treatment has a large impact on the expected clinical outcome of these patients. Herein, the acquisition of DNA methylation fingerprints for 1054 tumorigenic samples allowed the classification according to cancer type of almost 70% of the studied CUPs, a result that can make a difference in the prognosis of these patients. This is just an example of the possible translational use of the DNA methylation profiles provided. Other uses might follow, and they will require further development, such as our finding of a distinct DNA methylation fingerprint between local liver metastases and distant brain metastases derived from colorectal tumors that might suggest the use of DNA methylation patterns to predict the metastatic spectrum of a given cancer. We would also like to highlight another promising step in the clinical-benefits direction by the recent finding of 27,000 CpG-site DNA methylation profiles in blood that are associated with bladder cancer risk (Marsit et al. 2011).
One obvious limitation of our approach is the level of resolution, since only 1505 CpG sites were interrogated. The increasing number of studies developed and under way using the 27,000-CpG-site platform and the future reports using the new 450K-CpG-site microarray will be useful to further validate and complement the DNA methylation profiles obtained. We can only imagine how the firm, automatic, and affordable establishment of whole-genome sequencing of complete human DNA methylomes (Lister et al. 2009; Li et al. 2010) will yield further knowledge about the role of DNA methylation in cellular identity and its loss in disease. Even so, the 1628 DNA methylation fingerprints described herein, and displayed by tissue type and disease in Figure 5, are a promising starting point for understanding the variation of human DNA methylation over a range of normal and pathological conditions.
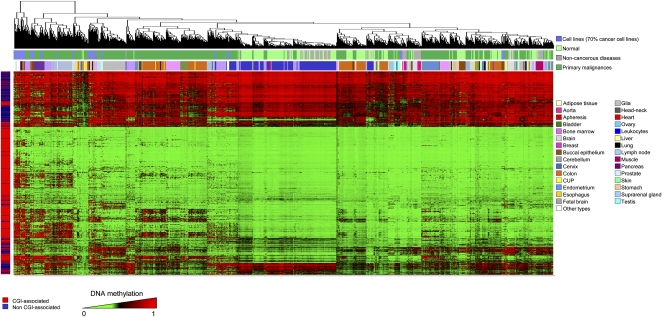
Figure 5.
A DNA methylation fingerprint of 1628 human samples. Unsupervised hierarchical clustering and heatmap of all the CpG methylation maps obtained in the study, by tissue and disease type.
Methods
Filtering of probes and samples
Although the GoldenGate Assay by Illumina is an established, highly reproducible method for DNA methylation detection, there is currently no standard procedure for post-filtering of probes and samples commonly used. Before analyzing the methylation data, we explored several ways of excluding possible sources of biological and technical biases that could have affected and improved the accuracy of the results. Every beta value in the GoldenGate platform is accompanied by a detection P-value. We based the criteria of filtering on these P-values reported by the assay. We examined two aspects of filtering out probes and samples based on the detection P-values, selecting a threshold and a cutoff. Our analyses indicated that a threshold value of 0.01 allows a clear distinction to be made between reliable and unreliable beta values. We selected the cutoff value as 5%. Following this criterion, we first removed all probes with detection P-values >0.01 in 5% or more of the samples. As a second step, we removed all samples with detection P-values >0.01 in 5% or more of their (remaining) probes. In total, 130 probes and 87 samples were removed. We also checked for and removed consistently unmethylated and methylated probes. We ignored all cell line samples and focused on the remaining 1521 (primary tissue) samples. All probes exhibiting a degree of methylation <0.25 for all primary tissue samples were considered to be consistently unmethylated. Similarly, probes with a degree of methylation >0.75 for all primary tissue samples were considered to be consistently methylated. We identified nine consistently unmethylated probes; none of the probes fit our definition for being consistently methylated. A known biological factor is that one copy of chromosome X is methylated in women (Reik and Lewis 2005), and, therefore, we decided to identify and remove all probes with prominent gender-specific methylation, to avoid hidden bias in the subsequent analyses. We considered the set of 1271 samples with gender information; approximately half of them were female. We defined a probe to be gender-specific if (1) the probe showed a significant differential methylation between the two sample groups, as determined by the Mann-Whitney U-test with FDR correction; and (2) the mean methylation degrees of females and males for this probe differed by at least 0.17 (a limitation of the GoldenGate assay). After excluding 130 probes that were not of sufficient quality, nine that were consistently unmethylated and 44 that were gender-specific, 1322 probes were available for further statistical analyses.
Analysis of differentially methylated probes
The large cohort of heterogeneous methylation profiles allows us to identify differentially methylated probes under a variety of scenarios. We analyzed different groups of tissue samples separately (normal primary tissues, cancerous and non-cancerous diseases, and cancer cell lines). We performed all statistical analyses using the R environment for statistical computing (version 2.10; http://www.R-project.org). Further explanation about detection of differentially methylated probes and genes in each scenario, statistical analyses, and graphical representations are provided in the Supplemental Methods.
Pyrosequencing
Pyrosequencing assays were designed to analyze and validate the results obtained from the array under different scenarios. Sodium bisulfite modification of 0.5 μg of genomic DNA isolated from different tissues was carried out with the EZ DNA Methylation Kit (Zymo Research Corporation) following the manufacturer's protocol. Bisulfite-treated DNA was eluted in 15-μL volumes with 2 μL used for each PCR. The set of primers for PCR amplification and sequencing were designed with a specific program (PyroMark assay design version 2.0.01.15). Primer sequences were designed to hybridize with CpG-free sites to ensure methylation-independent amplification. PCR was performed with primers biotinylated to convert the PCR product to single-stranded DNA templates. We used the Vacuum Prep Tool (Biotage) to prepare single-stranded PCR products according to the manufacturer's instructions. Pyrosequencing reactions and quantification of methylation were performed in a PyroMark Q24 System version 2.0.6 (QIAGEN). Graphs of methylation values show bars identifying CpG sites with values from 0% (white) to 100% (black).
Classification of CUPs
We used the advanced method L1-regularized logistic regression with misclassification to classify the 42 CUP samples in our data set into one of the known cancer types. By classifying a CUP, this classifier gives probabilities (values between 0 and 1) for every known cancer type. A CUP prediction heatmap was derived in R (version 2.1.0) (Fig. 3E). The CUP samples were selected on the basis of having a >30% probability of being ascribed to a specific tumor type. The arrangement of the samples in the heatmap was established by (1) ordering the tumor types by the number of CUPs ascribed to each one; and (2) within each tumor type, ranking the CUPs from the highest to lowest probability of ascription.
Expression data analysis
CEL files containing normal tissue gene expression data were downloaded from the GEO database. Data series, samples, and analysis procedures are detailed in the Supplemental Methods.
Enrichment of PcG-marks and bivalent domains in different methylation groups
The presence of PcG-marks and bivalent domains in different methylation groups was compared using a Fisher's exact test. In addition to a Fisher's exact test, we calculated permutation-based P-values to account for interdependencies between the methylation states of different CpGs. Briefly, we performed a Fisher's exact test in 104 random reassignments of the studied samples and calculated the proportion of resulting P-values that is lower than or equal to the originally obtained one. A genome-wide map of Polycomb target genes and 3mK4H3/3mK27H3-enriched genes in ESCs is available as supplemental material of the articles by Lee et al. (2006) and Pan et al. (2007), respectively.
Human cancer cell lines and expression upon 5-aza-2′-deoxycytidine treatment
Five cancer cell lines—SW480 (colon), HN-011A and HN-011B (esophagus), and IGR37 and IGR39 (melanoma)—were grown in DMEM medium supplemented with 4 mM glutamine, 10% FBSm and 100 units/mL penicillin/streptomycin at 37°C/5% CO2. All cell lines were treated with 1 μM 5-aza-2′-deoxycytidine (Sigma) for 72 h. Total RNA was isolated from all cell lines before and after 5-aza-2′-deoxycytidine treatment by TRIzol extraction (Invitrogen), and 5 μg was hybridized on the Human GeneChip U133 Plus 2.0 expression array (Affymetrix). Expression data were normalized and analyzed following the same procedures described in the Supplemental Methods.
Data access
The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE28094.
Acknowledgments
This work was supported by European Grants CANCERDIP HEALTH-F2-2007-200620, LSHG-CT-2006-018739-ESTOOLS, LSHC-CT-2006-037297- MCSCs, the Dr. Josef Steiner Cancer Research Foundation Award, the Fondo de Investigaciones Sanitarias Grant PI08-1345, Consolider Grant MEC09-05, the Spanish Association Against Cancer (AECC), Spanish Ministry of Education and Science (SAF2009-07319), the Lilly Foundation Biomedical Research Award, Fundacio Cellex, and the Health Department of the Catalan Government (Generalitat de Catalunya). R.S. is supported by BMBF and Deutsche Krebshilfe. J.B. is supported in part by the University College London Cancer Institute Experimental Cancer Medicine Centre and the University College London Hospitals and University College London Comprehensive Biomedical Research Centre. M.E. is an Institucio Catalana de Recerca i Estudis Avançats (ICREA) Research Professor.
Authors’ contributions: A.F.F., Y.A., C.B., and M.E. conceived and designed the experiments. All authors analyzed the data. A.F.F., Y.A., C.B., and M.E. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.119867.110.
References
- Abbott A 2010. Project set to map marks on genome. Nature 463: 596–597 [PubMed] [Google Scholar]
- Abbruzzese JL, Abbruzzese MC, Lenzi R, Hess KR, Raber MN 1995. Analysis of a diagnostic strategy for patients with suspected tumors of unknown origin. J Clin Oncol 13: 2094–2103 [DOI] [PubMed] [Google Scholar]
- American Cancer Society 2007. Statistics for 2007. American Cancer Society Statistics; Available at http://www.cancer.org/docroot/stt/stt_0.asp [Google Scholar]
- Aranda P, Agirre X, Ballestar E, Andreu EJ, Román-Gómez J, Prieto I, Martín-Subero JI, Cigudosa JC, Siebert R, Esteller M, et al. 2009. Epigenetic signatures associated with different levels of differentiation potential in human stem cells. PLoS ONE 4: e7809 doi: 10.1371/journal.pone.0007809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestar E, Esteller M 2008. SnapShot: The human DNA methylome in health and disease. Cell 135: 1144–1144.e1 [DOI] [PubMed] [Google Scholar]
- Ballestar E, Paz MF, Valle L, Wei S, Fraga MF, Espada J, Cigudosa JC, Huang TH, Esteller M 2003. Methyl-CpG binding proteins identify novel sites of epigenetic inactivation in human cancer. EMBO J 22: 6335–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES 2007. The mammalian epigenome. Cell 128: 669–681 [DOI] [PubMed] [Google Scholar]
- Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, et al. 2006. High-throughput DNA methylation profiling using universal bead arrays. Genome Res 16: 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetta L 2008. Epigenomics: Detailed analysis. Nature 454: 795–798 [DOI] [PubMed] [Google Scholar]
- Byun HM, Siegmund KD, Pan F, Weisenberger DJ, Kanel G, Laird PW, Yang AS 2009. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet 18: 4808–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor RM, Lange K, Sinsheimer JS 2010. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am J Hum Genet 86: 6–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, et al. 2009. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet 5: e1000602 doi: 10.1371/journal.pgen.1000602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC, et al. 2000. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet 24: 132–138 [DOI] [PubMed] [Google Scholar]
- Das R, Hampton DD, Jirtle RL 2009. Imprinting evolution and human health. Mamm Genome 20: 563–572 [DOI] [PubMed] [Google Scholar]
- Desplats P, Spencer B, Coffee E, Patel P, Michael S, Patrick C, Adame A, Rockenstein E, Masliah E 2011. Alpha-synuclein sequesters Dnmt1 from the nucleus: A novel mechanism for epigenetic alterations in Lewy body diseases. J Biol Chem 286: 9031–9037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dindot SV, Person R, Strivens M, Garcia R, Beaudet AL 2009. Epigenetic profiling at mouse imprinted gene clusters reveals novel epigenetic and genetic features at differentially methylated regions. Genome Res 19: 1374–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. 2009. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet 41: 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, et al. 2008. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol 26: 779–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. 2006. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet 38: 1378–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M 2007. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet 8: 286–298 [DOI] [PubMed] [Google Scholar]
- Esteller M 2008. Epigenetics in cancer. N Engl J Med 358: 1148–1159 [DOI] [PubMed] [Google Scholar]
- Esteller M, Corn PG, Baylin SB, Herman JG 2001. A gene hypermethylation profile of human cancer. Cancer Res 61: 3225–3229 [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B 1990. A genetic model for colorectal tumorigenesis. Cell 61: 759–767 [DOI] [PubMed] [Google Scholar]
- Feinberg AP 2007. Phenotypic plasticity and the epigenetics of human disease. Nature 447: 433–440 [DOI] [PubMed] [Google Scholar]
- Fernandez AF, Rosales C, Lopez-Nieva P, Grana O, Ballestar E, Ropero S, Espada J, Melo SA, Lujambio A, Fraga MF, et al. 2009. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res 19: 438–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Herranz M, Espada J, Ballestar E, Paz MF, Ropero S, Erkek E, Bozdogan O, Peinado H, Niveleau A, et al. 2004. A mouse skin multistage carcinogenesis model reflects the aberrant DNA methylation patterns of human tumors. Cancer Res 64: 5527–5534 [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, et al. 2005. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci 102: 10604–10609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark SJ 2006. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet 38: 540–549 [DOI] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS 2009. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat Rev Endocrinol 5: 401–408 [DOI] [PubMed] [Google Scholar]
- Greco FA, Pavlidis N 2009. Treatment for patients with unknown primary carcinoma and unfavorable prognostic factors. Semin Oncol 36: 65–74 [DOI] [PubMed] [Google Scholar]
- Harris TJ, McCormick F 2010. The molecular pathology of cancer. Nat Rev Clin Oncol 7: 251–265 [DOI] [PubMed] [Google Scholar]
- Hemberger M, Dean W, Reik W 2009. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat Rev Mol Cell Biol 10: 526–537 [DOI] [PubMed] [Google Scholar]
- Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout WM III, Biniszkiewicz D, Yanagimachi R, Jaenisch R 2001. Epigenetic instability in ES cells and cloned mice. Science 293: 95–97 [DOI] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP 2009. CpG islands–‘a rough guide.’ FEBS Lett 583: 1713–1720 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, Wen B, Feinberg AP 2008. Comprehensive high-throughput arrays for relative methylation (CHARM). Genome Res 18: 780–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41: 178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O'Hanlon TP, Rider LG, et al. 2010. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res 20: 170–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellinger KA, Stadelmann C 2001. Problems of cell death in neurodegeneration and Alzheimer's disease. J Alzheimers Dis 3: 31–40 [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB 2007. The epigenomics of cancer. Cell 128: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Archer TK, Baylin SB, Beck S, Berger S, Bernstein BE, Carpten JD, Clark SJ, Costello JF, Doerge RW, et al. ; The American Association for Cancer Research Human Epigenome Task Force & European Union, Network of Excellence, Scientific Advisory Board 2008. Moving AHEAD with an international human epigenome project. Nature 454: 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GH, Wong AH, Feldcamp LA, Virtanen C, Halfvarson J, Tysk C, et al. 2009. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet 41: 240–245 [DOI] [PubMed] [Google Scholar]
- Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, et al. 2006. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet 38: 149–153 [DOI] [PubMed] [Google Scholar]
- Ku CS, Loy EY, Pawitan Y, Chia KS 2010. The pursuit of genome-wide association studies: where are we now? J Hum Genet 55: 195–206 [DOI] [PubMed] [Google Scholar]
- Kuehn BM 2008. 1000 Genomes Project promises closer look at variation in human genome. JAMA 300: 2715. [DOI] [PubMed] [Google Scholar]
- Ladd-Acosta C, Pevsner J, Sabunciyan S, Yolken RH, Webster MJ, Dinkins T, Callinan PA, Fan JB, Potash JB, Feinberg AP 2007. DNA methylation signatures within the human brain. Am J Hum Genet 81: 1304–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhu J, Tian G, Li N, Li Q, Ye M, Zheng H, Yu J, Wu H, Sun J, et al. 2010. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol 8: e1000533 doi: 10.1371/journal.pbio.1000533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Colot V, Martienssen R 2005. Profiling DNA methylation patterns using genomic tiling microarrays. Nat Methods 2: 219–224 [DOI] [PubMed] [Google Scholar]
- Lister R, Ecker JR 2009. Finding the fifth base: Genome-wide sequencing of cytosine methylation. Genome Res 19: 959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegawa S, Hinkal G, Kim HS, Shen L, Zhang L, Zhang J, Zhang N, Liang S, Donehower LA, Issa JP 2010. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res 20: 332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit CJ, Koestler DC, Christensen BC, Karagas MR, Houseman EA, Kelsey KT 2011. DNA methylation array analysis identifies profiles of blood-derived DNA methylation associated with bladder cancer. J Clin Oncol 29: 1133–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlen P, Puisieux A 2006. Metastasis: a question of life or death. Nat Rev Cancer 6: 449–458 [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud EJ, van Vugt MJ, Bultman SJ, Sweet HO, Davisson MT, Woychik RP 1994. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev 8: 1463–1472 [DOI] [PubMed] [Google Scholar]
- Monk D 2010. Deciphering the cancer imprintome. Brief Funct Genomics 9: 329–339 [DOI] [PubMed] [Google Scholar]
- Nagae G, Isagawa T, Shiraki N, Fujita T, Yamamoto S, Tsutsumi S, Nonaka A, Yoshiba S, Matsusaka K, Midorikawa Y, et al. 2011. Tissue-specific demethylation in CpG-poor promoters during cellular differentiation. Hum Mol Genet doi: 10.1093/hmg/ddr170 [DOI] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. 2007. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 39: 237–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA 2007. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 1: 299–312 [DOI] [PubMed] [Google Scholar]
- Portela A, Esteller M 2010. Epigenetic modifications and human disease. Nat Biotechnol 28: 1057–1068 [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Hildmann T, Novik KL, Lewin J, Tost J, Cox AV, Andrews TD, Howe KL, Otto T, Olek A, et al. 2004. DNA methylation profiling of the human major histocompatibility complex: A pilot study for the human epigenome project. PLoS Biol 2: e405 doi: 10.1371/journal.pbio.0020405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Maslau S, Andrew T, Yang TP, Beyan H, Whittaker P, McCann OT, Finer S, Valdes AM, et al. 2010. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res 20: 434–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP 2009. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci 106: 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Lewis A 2005. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nat Rev Genet 6: 403–410 [DOI] [PubMed] [Google Scholar]
- Richardson B 2007. Primer: epigenetics of autoimmunity. Nat Clin Pract Rheumatol 3: 521–527 [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Suzuki M, Abe T, Hosoyama T, Himeno E, Tanaka S, Greally JM, Hattori N, Yagi S, Shiota K 2007. Cell type-specific methylation profiles occurring disproportionately in CpG-less regions that delineate developmental similarity. Genes Cells 12: 1123–1132 [DOI] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 39: 232–236 [DOI] [PubMed] [Google Scholar]
- Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa JP 2007. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet 3: 2023–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siva N 2008. 1000 Genomes project. Nat Biotechnol 26: 256 doi: 10.1038/nbt0308-256b [DOI] [PubMed] [Google Scholar]
- Takai D, Jones PA 2002. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci 99: 3740–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teaktong T, Graham A, Court J, Perry R, Jaros E, Johnson M, Hall R, Perry E 2003. Alzheimer's disease is associated with a selective increase in alpha7 nicotinic acetylcholine receptor immunoreactivity in astrocytes. Glia 41: 207–211 [DOI] [PubMed] [Google Scholar]
- Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell CG, Maxwell AP, et al. 2010. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res 20: 440–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjabringa G, Bergers M, van Rens D, de Boer R, Lamme E, Schalkwijk J 2008. Development and validation of human psoriatic skin equivalents. Am J Pathol 173: 815–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdinguio RG, Sanchez-Mut JV, Esteller M 2009. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol 8: 1056–1072 [DOI] [PubMed] [Google Scholar]
- Vaughn MW, Tanurdzic M, Lippman Z, Jiang H, Carrasquillo R, Rabinowicz PD, Dedhia N, McCombie WR, Agier N, Bulski A, et al. 2007. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol 5: e174 doi: 10.1371/journal.pbio.0050174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37: 853–862 [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. 2007. Epigenetic stem cell signature in cancer. Nat Genet 39: 157–158 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Rohde C, Tierling S, Jurkowski TP, Bock C, Santacruz D, Ragozin S, Reinhardt R, Groth M, Walter J, et al. 2009. DNA methylation analysis of chromosome 21 gene promoters at single base pair and single allele resolution. PLoS Genet 5: e1000438 doi: 10.1371/journal.pgen.1000438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S 2007. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39: 61–69 [DOI] [PubMed] [Google Scholar]