Abstract
Enhancer of rudimentary, e(r), encodes a small nuclear protein, ER, that has been implicated in the regulation of pyrimidine metabolism, DNA replication and cell proliferation. In Drosophila melanogaster, a new recessive Notch allele, Nnd-p, was isolated as a lethal in combination with an e(r) allele, e(r)p2. Both mutants are viable as single mutants. Nnd-p is caused by a P-element insertion in the 5′ UTR, 378-bp upstream of the start of translation. Together the molecular and genetic data argue that Nnd-p is a hypomorphic allele of N. The three viable notchoid alleles, Nnd-p, Nnd-1 and Nnd-3, are lethal in combination with e(r)− alleles. Our present hypothesis is that e(r) is a positive regulator of the Notch signaling pathway and that the lethality of the N e(r) double mutants is caused by a reduction in the expression of the pathway. This is supported by the rescue of the lethality by a mutation in Hairless, a negative regulator of N, and by the synthetic lethality of dx e(r) double mutants. Further support for the hypothesis is a reduction in E(spl) expression in an e(r)− mutant. Immunostaining localizes ER to the nucleus, suggesting a nuclear function for ER. A role in the Notch signaling pathway, suggests that e(r) may be expressed in the nervous system. This turns out to be the case, as immunostaining of ER shows that ER is localized to the developing CNS.
Keywords: enhancer of rudimentary, Notch, deltex, neurogenesis, transcriptional regulation
Introduction
The Drosophila enhancer of rudimentary, e(r), gene encodes ER, a small protein of 104 amino acids.1,2 An examination of the amino acid sequence does not reveal any motifs that indicate a putative function or mode of regulation of the protein other than two conserved casein kinase II sites.3 A mutagenic analysis of these sites indicate that they are important in the regulation of the activity of ER.3 ER is a highly conserved protein that has been found in plants, animals and protists, but has yet to be found in fungi.2 ER is 76% identical to the ER homologue, ERH, of vertebrates, 49% identical to the C. elegans ERH and 40% identical to the plant (Arabidopsis) ERH.2 The vertebrate ER homologues are very highly conserved. The human and mouse proteins are identical and differ from the zebrafish ERH by a single conservative amino acid change.
While the protein is highly conserved, the biochemical and cellular function of ER has yet to be determined. Genetic and biochemical data argue for the involvement of ER in a number of cellular processes such as pyrimidine metabolism, cell cycle regulation and cellular proliferation. Mutations in e(r) were first identified by their ability to enhance the mutant wing phenotype of rudimentary, r, mutants.1 Rudimentary encodes a large multifunctional protein containing the first three enzymatic activities of pyrimidine biosynthesis.4–6 These studies led to the proposal that e(r) was involved in the regulation of pyrimidine metabolism, but not directly by regulating r expression. Pyrimidine synthesis is high in proliferating cells and has been examined as an avenue to treat cancer cells.7–9 In this vein, ER has also been shown to be expressed at high levels in certain human cancers, and has been proposed as a target for drug therapy for certain cancers.10 An ER-GFP fusion protein has been shown to be nuclearly localized in human cells,11 which argues for a nuclear function of the protein. A number of yeast two-hybrid screens have identified potential binding partners with ER which have nuclear functions. These include a transcription factor, DCoH/PCD in Xenopus,12 a protein involved in the regulation of DNA replication in humans, PDIP46/SKAR,11 and a nuclear zinc-finger protein that may regulate DNA synthesis and cyclin dependent kinases in humans, Ciz1.13,14 In addition ER has been shown to co-immunoprecipitate with SPT3, a transcription elongation factor15 and with FCP1, a TFIIF-associating component of CTD phosphatase.16 All of these studies suggest that ER may have a number of general nuclear functions involving DNA replication and transcription and are consistent with a role for ER in cell proliferation.
The high conservation of ER and its unique protein structure argue for an important non-redundant function for ER, and suggest that ER performs a vital function for the cell. Given these observations, it was initially surprising that deletions of e(r) are viable. However these mutants have low viability and low female fertility, which reveal that e(r) has a necessary function and explain the high conservation of the protein.17 The expression of e(r) in the ovaries and the deposition of the mRNA and protein into the developing oocyte are consistent with a role in oogenesis.17 In the present study we present genetic and expression data that reveal a role of e(r) as a positive regulator of the Notch signaling pathway. All of the data indicate that in the absence of ER, the Notch signaling pathway is downregulated.
Results
Synthetic lethality between N and e(r) mutations.
Hypomorphic mutants of e(r) are viable.1 One such mutant, e(r)p2, is caused by the insertion of a small, unmarked P element into the 5′ control region of e(r).20 In a screen to isolate X-linked lethal mutations caused by the mobilization of the P element in e(r)p2, one lethal mutation out of 10,024 chromosomes was isolated (Fig. 1). Since e(r)p2 was caused by a P-element insertion, it was thought that the lethality might have been caused by a deletion generated by the imprecise excision of the P element. However, when the e(r) gene from this chromosome was analyzed by PCR, it was shown that the original P element of e(r)p2 was still intact and that a deletion within the e(r) region had not been generated. Additional genetic analyses showed that the lethality required the presence of e(r)p2 and a mutation that mapped just to the right of white, w. This fact and the fact that the few male escapers had severely notched wings (Fig. 2A) suggested that the second mutation might be a N allele.
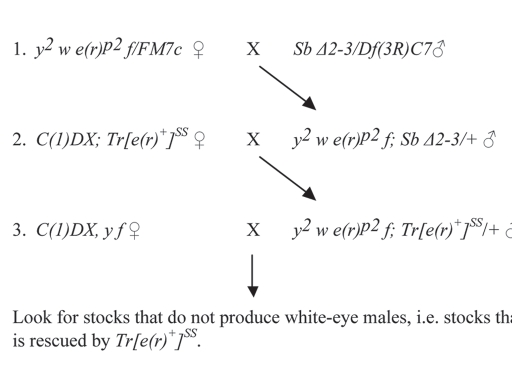
Figure 1.
Screen for hybrid-dysgenesis-induced X-linked lethals in an e(r)p2-containing X chromosome. In this scheme, the only mobile P element is the one in e(r)p2. This element is mobilized by the introduction of the active but immobilized P element, Δ2–3. Tr[e(r)+]SS is a construct carrying e(r)+ and w+.1 In cross 3, males were individually mated to 5 C(1)DX, y f females, since each male represents a single mutagenized X chromosome. Crosses which produced red-eye males, but no white-eye males were kept as possible X-linked lethal stocks. From 10,024 stocks, one lethal stock was isolated.
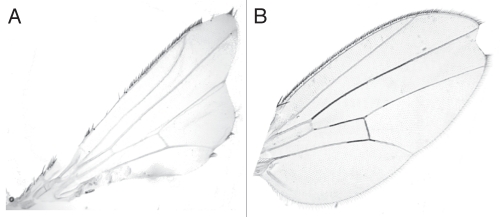
Figure 2.
Wings of Nnd-p e(r)p2 and Nnd-p e(r)+ males. (A) Wing of a Nnd-p e(r)p2 escaper male. (B) Wing of Nnd-p e(r)p2; Tr[e(r)+]SS/+ male. The presence of Tr[e(r)+]SS rescues both the lethality (Table 2) and the severe wing notching of Nnd-p e(r)p2. This wing phenotype is similar to what is seen in some Nnd-p males.
The putative new N allele was separated from e(r)p2 by recombination and the single mutant flies were viable and fertile (Table 1). The large majority of the mutant flies look wild-type, but occasionally apical wing nicks similar to those seen in recessive notchoid alleles are seen (Fig. 2B). Complementation crosses indicate that the mutation does not complement two recessive notchoid alleles of N, Nnd-1 and Nnd-3, but does complement recessive eye-facet alleles of N, Nspl-1, Nfa-g and Nfa-swb (Tsubota SI, unpublished results). In addition like Nnd-3, the new allele is lethal as a heterozygote with lethal null N alleles. Using a number of different N loss-of-function alleles, no heterozygotes were seen as compared to 1,730 control females (Table 3). All of the data support the conclusion that the new mutation is a hypomorphic N allele, and thus we have named the allele, Nnd-p.
Table 1.
Viability of N, dx and e(r) double-mutant combinations
| X chromosome tested | Heterozygous females | Hemizygous tester males |
| Single mutants | ||
| Nnd-p | 351 | 276 |
| Nnd-1 | 153 | 158 |
| Nnd-3 | 159 | 213 |
| e(r)p2 | 344 | 333 |
| e(r)27-1 | 734 | 243 |
| e(r)37-6 | 1137 | 530 |
| dx | 185 | 197 |
| Double mutants | ||
| Nnd-p e(r)p2 | 2835 | 32 |
| Nnd-p e(r)27-1 | 4209 | 0 |
| Nnd-p e(r)37-6 | 2426 | 0 |
| Nnd-1 e(r)p2 | 304 | 0 |
| Nnd-1 e(r)27-1 | 1741 | 1 |
| Nnd-3 e(r)p2 | 700 | 3 |
| Nnd-3 e(r)27-1 | 258 | 0 |
| Nfa-g e(r)27-1 | 1021 | 587 |
| Nspl-1 e(r)27-1 | 471 | 95 |
| Nfa-swb e(r)27-1 | 1010 | 305 |
| dx e(r)27-1 | 1481 | 0 |
All of the mutations are on the X chromosome, so viability was measured as the number of hemizygous males vs. the number of heterozygous sisters as a control. If there are no viability differences between the two groups, the numbers should be roughly equal. The data for the single mutants e(r)p2, e(r)27-1 and e(r)27-1 have been previously reported in reference 17 and are used here as controls.
Table 3.
Lethality of N−/Nnd-p heterozygotes
| Tester N allele | Tester N allele/Nnd-p | FM7c/Nnd-p |
| N55e11 | 0 | 173 |
| Df(1)N81k1 | 0 | 649 |
| Nco | 0 | 466 |
| N55e11 | 0 | 442 |
| Grand total | 0 | 1730 |
All of the tester N alleles are null mutations. The viability of the N−/Nnd-p females was compared to their FM7c/Nnd-p sisters. If there are no differences in viability, then the two groups should have similar numbers.
Given that the mutation was generated in a hybrid-dysgenesis screen and that P element insertions into the 5′ end of the N gene had previously been described in reference 21, PCR was used to examine the N gene for insertions. A 1.1-kb insertion was found in the 5′ end of the N gene. DNA sequencing revealed that this element was indeed a P element and that the insertion site was within the region encoding the 5′ UTR, 378-bp upstream of the start of translation (Fig. 3A). The position of the P element and the hypomorphic nature of the mutant phenotype suggest that Nnd-p is a hypomorphic mutation caused by either a decrease in the transcription of the N gene or a decrease in the translation of the N mRNA.
Figure 3.
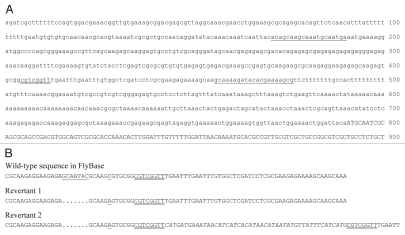
DNA sequences of Nnd-p and its hybrid-dysgenesis-induced revertants. (A) Nucleotide sequence of the P-element insertion site in Nnd-p. The first 900 bp of the first N exon are shown. The protein-coding region is in uppercase letters. The 8-bp site duplication created by the insertion of the P element is double underlined. The positions of the two primers used to amplify the revertant fragments for DNA sequencing are underlined. (B) Partial sequence of the two revertants of Nnd-p. A partial sequence of the wild-type sequence of N reported in FlyBase (CG3936, FBgn0004647) is given as a reference. It contains a seven-base-pair sequence that is not found in Nnd-p or the two revertants and a C/A polymorphism. These sequences are unde-lined. The eight-base-pair sequence that was duplicated during the insertion of the P element in Nnd-p is double underlined in all of the sequences. The first revertant is a precise excision of the P element. The second revertant is an imprecise excision. A forty-eight base-pair sequence derived from the P element is located between the site duplications in this revertant.
Given the novelty of the proposed lethal interaction between N and e(r) mutations, it was important to verify that the synthetic lethality of the double-mutant X chromosomes requires both notchoid alleles of N and mutations of e(r). Various double mutant combinations were constructed with N mutations (Nnd-p, Nnd-1, Nnd-3) and mutations of e(r)—one hypomorphic alleles, e(r)p2 and two null alleles, e(r)27-1 and e(r)37-6. While each of the single mutants has good viability, all of the double mutants have greatly decreased viability (Table 1). To further verify the specificity of this lethal interaction, duplications containing the wild-type alleles of either N or e(r) were tested for their abilities to rescue the lethality. In the case of e(r), Tr[w+ e(r)+]SS, a transgene consisting of a 6.1-kb SalI fragment containing e(r)+, was used.1 This transgene rescues the lethality of Nnd-p e(r)p2 and Nnd-p e(r)37-6 (Table 2). For N, Dp(1;3)DC109, a 93-kb genomic fragment that contains the entire N gene inserted into the third chromosome, was used in the rescue experiments. No other genes are contained within this duplication. Dp(1;3)DC109 rescues the lethality of Nnd-p e(r)37-6 males (Table 2). Together the rescue experiments demonstrate that the lethality of N e(r) double mutants requires mutations at both genes and rules out the possibility that the synthetic lethality is caused by an interaction with an unidentified mutation.
Table 2.
Rescue of double-mutant lethality by a wild-type transgene of dx, e(r) or N
| Genotype tested | Heterozygous females | Hemizygous tester males |
| Nnd-p e(r)p2; Tr[e(r)+]SS | 467 | 205 |
| Nnd-p e(r)37-6; Tr[e(r)+]SS | 359 | 159 |
| dx e(r)27-1; Tr[e(r)+]SS | 95 | 71 |
| Nnd-p e(r)37-6; Dp(1;3)DC109 | 130 | 70 |
| dx e(r)27-1; Dp(1;3)DC157 | 166 | 118 |
| dx e(r)27-1; Dp(1;3)DC109 | 173 | 57 |
Females that were heterozygous for the double-mutant combination were crossed to males that were homozygous for a transformation construct carrying the wild-type allele of either dx, e(r) or N. Each of the three transgenes is located on the third chromosome. As in Table 1, viability was measured as the number of hemizygous males vs. the number of heterozygous sisters as a control. All individuals were heterozygous for one of the transgene constructs (for dx - Dp(1;3)DC157, for e(r) - Tr[e(r)+]SS, or for N - Dp(1;3)DC109. Rescue was seen as the presence of hemizygous males. Compare these results to those in the absence of the transgene (Table 1). Note that the lethality of the e(r) Nnd-p double mutants is rescued by either an e(r) or a N transgene and that the lethality of the e(r) dx double mutant is resculed by either an e(r) or a dx transgene. Also note that the lethality of the e(r) dx double mutant is also rescued by a N transgene, which argues that the lethality is caused by a decrease in N expression.
To further confirm that the lethal interaction with e(r) requires the Nnd-p mutation and to confirm that the P element inserted into the N gene is responsible for the Nnd-p mutation, two hybrid-dysgenesis-induced revertants of Nnd-p were isolated and their DNA sequences determined. The first revertant was isolated as a revertant of the lethal interaction between Nnd-p and a deletion of e(r), e(r)27-1. This revertant is a precise excision of the P element associated with Nnd-p (Fig. 3B). The second revertant was isolated as a revertant of the lethality of N8 /Nnd-p heterozygotes. This revertant is an imprecise excision of the P element. Left behind are the two copies of the 8-bp site duplication with forty-eight base pairs of P-element related sequence between them (Fig. 3B). This second reversion also reverts the synthetic lethality with e(r) mutations. Thus, these two revertants verify both that the P element in the N 5′ UTR is responsible for the Nnd-p allele and that the synthetic lethality with e(r) mutations requires the Nnd-p allele.
The DNA sequencing also revealed a polymorphism in the N gene. Nnd-p and the two revertants have sequence differences with the N sequence in Genbank and FlyBase (CG3936, FBgn0004647). Each of the sequenced alleles has a 7-bp deletion and a C to A transversion (Fig. 3). While these sequence differences are not responsible for the Nnd-p mutation, they are useful in verifying the common origin of the three alleles.
Lethal interactions are seen between the recessive notchoid N alleles and e(r)− alleles. To determine if this lethal interaction extends to other recessive N alleles, e(r)27-1 was combined with three recessive eye-phenotype alleles, Nfa-g, Nfa-swb and Nspl-1. These alleles did not show a lethal interaction with e(r) null alleles (Table 1), nor did they show an enhancement in the mutant eye phenotypes (data not shown). It appears that the N-e(r) interaction is allele specific. This may indicate temporal and/or tissue specificity in the interaction.
Interactions between e(r) and known regulators of the Notch signaling pathway.
It is likely that the N-e(r) interactions are acting through the Notch signaling pathway and that the lethal interactions are the result of the downregulation of the pathway. To test this possibility we examined the ability of a Hairless, H, mutation to suppress the lethality of the Nnd-p e(r)p2 double mutant. H encodes a transcriptional co-repressor that downregulates the Notch signaling pathway.22,23 Reducing Hairless activity by 50% with the H mutation results in the upregulation of the Notch signaling pathway and the suppression of some of the visible phenotypes associated with N mutations.22 If the lethality of Nnd-p e(r)p2 is caused by a downregulation of the Notch signaling pathway by e(r)p2, then one might expect that a H mutation would upregulate the pathway and suppress the lethality. This is indeed the case (Table 4). Nnd-p e(r)p2 /Y; H/+ males are viable and do not display the severe wing-notching seen in Nnd-p e(r)p2/Y; +/+ escapers.
Table 4.
Rescue of synthetic lethality with a H loss-of-function mutation
| Nnd-p e(r)p2/+ females | Nnd-p e(r)p2/Y males | ||
| In(3RC)/+ | H/+ | In(3RC)/+ | H/+ |
| 62 | 60 | 0 | 44 |
Nnd-p e(r)p2/FM7C females were crossed to H/In(3R)C males. The ability of a H mutation to rescue the lethality of Nnd-p e(r)p2 was seen as the presence of these males in a H background but not in an In(3RC) background. The females serve as a control for the viability of flies carrying either H or In(3RC).
Another interaction with the Notch signaling pathway is shown with deltex, dx. The dx gene encodes Dx, a protein that positively regulates N through binding to the Ankyrin repeats of the intracellular domain of N.24 If both dx and e(r) are acting as positive regulators of the Notch signaling pathway, then mutations in both genes may produce a severe reduction in N activity and thus result in lethality. The results are consistent with this proposal. While dx and e(r)27-1 are both viable as single mutants, the double mutant is lethal (Table 1). This lethality is rescued by both an e(r)+ transgene and a dx+ transgene, which confirms the specificity of the lethal interaction (Table 2). The hypothesis is that the dx e(r)27-1 double mutants are dying because of reduced N activity due to mutations in these two putative positive regulators of N. If this is the case, then the lethality should be rescued by a N duplication. This is exactly what is seen (Table 2). Double mutant dx e(r)27-1 males in the presence of a N duplication show good viability. This supports the conclusion from the lethal interactions between N and e(r), that e(r) is a positive regulator of N and necessary for normal levels of N activity.
Delta, Dl and Serrate, Ser, encode the known ligands of N in Drosophila. If e(r) is positively regulating the Notch signaling pathway, then reducing both the levels of ER and either Dl or Ser, may further compromise the pathway. This could be seen as an interaction between mutations in e(r) and Dl and Ser. Two loss-of-function alleles of Dl were examined. Both are homozygous lethal and as heterozygotes display the dominant vein-thickening of L2. The wing phenotype and viability of the heterozygotes were examined in the background of e(r)+ and e(r)−. The Dl wing phenotype was not affected, however the viability was dramatically decreased in the absence of e(r) (Table 5). In the background of e(r)27-1, an e(r) null mutation, viability of Dl6B was reduced to 49% and the viability of Dl3 was reduced to 21%. Again, these interactions are consistent with a reduction in the activity of the Notch signaling pathway in the absence of e(r). No interactions were seen in the Ser e(r)27-1 double mutants. The Ser wing phenotypes were not suppressed or enhanced and the viability of the Ser/+ males was not affected by e(r)27-1. While these are preliminary results, it is conceivable that e(r) may be interacting with the Dl-N signaling pathway but not with the Ser-N signaling pathway.
Table 5.
Viability of e(r)27-1 Dl and e(r)27-1 Ser double mutants
| e(r)27-1/+ females | e(r)27-1 males | *FM7C males | |||
| Dl6B/+ | +/+ | Dl6B/+ | +/+ | Dl6B/+ | +/+ |
| 274 | 296 | 41 | 84 | 99 | 65 |
| e(r)27-1/+ females | e(r)27-1 males | *FM7C males | |||
| Dl3/+ | +/+ | Dl3/+ | +/+ | Dl3/+ | +/+ |
| 300 | 379 | 42 | 200 | 128 | 160 |
| e(r)27-1/+ females | e(r)27-1 males | ||||
| Ser/+ | +/+ | Ser/+ | +/+ | ||
| 137 | 174 | 42 | 46 | ||
Crosses were performed to determine if the Dl or Ser mutations affected the viability of e(r)27-1. Viability of flies heterozygous for the Dl or Ser mutation was compared to viability of flies carrying two wild-type alleles. Heterozygous females and FM7C males from the crosses were used as controls.
Not all of the vials were scored for FM7C males, so the absolute numbers cannot be compared to those of e(r)27-1 males, however, the data show that the decrease in viability seen in the Dl e(r)27-1 males was not a general effect of Dl mutations on all males.
Regulation of the Notch signaling pathway by e(r).
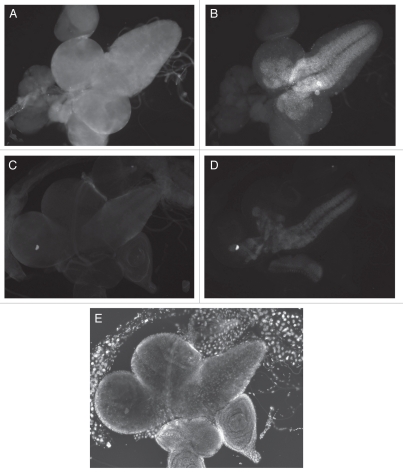
All of the genetic data indicate that e(r) is a positive regulator of the Notch signaling pathway and that the lethality between Nnd-p and e(r) mutations and between dx and e(r)27-1 is caused by a reduction in the activity of the pathway. The fact that in each case the lethality is rescued by a N duplication suggests that the lethality is caused by a reduction in N activity and that N activity is decreased in an e(r) null mutant. To examine this hypothesis, the effect of an e(r) deletion on E(spl) expression was measured. N is a transcription activator of E(spl), making E(spl) a convenient reporter for N activity.25 The e(r) null allele used in this assay was e(r)27-1. The effect of e(r) activity on E(spl) expression was examined in third-instar larval CNS and optic lobes of e(r)+ and e(r)27-1 males. In e(r)+ males, ER is localized throughout the larval CNS and optic lobes (Fig. 4A), while, E(spl) is localized in a subset of these cells (Fig. 4B). This makes it easy to examine E(spl) expression in the absence of e(r) expression. In the brain and CNS of the e(r)27-1 males, the level of ER is vastly reduced (Fig. 4C). What staining is present is background staining only seen at high exposures. E(spl) expression is considerably reduced in these same tissues (Fig. 4D). In order to see any E(spl) expression in the e(r)27-1 mutant background, the exposure times of the image captures had to be increased 2.5 fold over those of e(r)+. In e(r)27-1 the reduced expression of E(spl) is seen in the same spatial pattern as seen in the wild-type larvae. These data indicate that e(r) is necessary for normal levels of expression of E(spl), but not for the normal spatial pattern of E(spl) expression. These data are in agreement with the genetic data that show that e(r) is a positive regulator of the Notch signaling pathway. The downregulation of E(spl) expression also indicates that e(r) is acting upstream of E(spl) in the Notch signaling pathway.
Figure 4.
Immunolocalization of ER and E(spl) in the 3rd instar larval CNS and optic lobes. (A and B) e(r)+ male, exposure 100 msec. (C and D) e(r)27-1 male, exposure 250 msec. (E) DAPI staining of (C and D). This aids in the identification of the tissues. (A and C) ER immunostaining. (B and D) E(spl) immunostaining. (A) ER immunostaining is seen throughout the larval CNS and optic lobes, (B) E(spl) is seen in a subset of cells in the larval CNS. (C) ER is vastly reduced in an e(r)27-1 mutant. (D) In the same e(r)27-1 mutant, E(spl) staining is also clearly reduced when compared to wild-type, even with 2.5 times the exposure.
Immunolocalization of ER in embryos and Drosophila Schneider 2 cells.
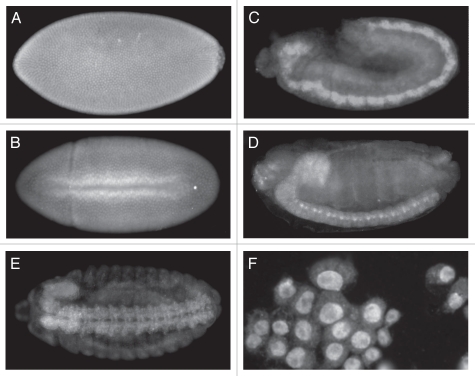
The data indicating that e(r) regulates the Notch signaling pathway suggest that e(r) is expressed in the developing nervous system. To examine this possibility wild-type embryos were immunostained with the anti-ER antibody. Around stage 11, ER can clearly be seen concentrated in the developing CNS (Fig. 5C). Staining is seen in the developing brain and ventral nerve chord. Expression is also diffuse, showing lower expression of ER throughout the embryo. By stage 14, ER can clearly be seen in the ventral nerve cord of the CNS and the brain (Fig. 5D and E). The ventral view, clearly show ER in the brain and the two tracks of the ventral nerve cord of the CNS. Although less intense, ER also appears to be localized to the peripheral nervous system (Fig. 5E). These data along with the genetic interactions with N and dx and the downregulation of E(spl) expression in an e(r) null background, point to a role of e(r) in the Notch signaling pathway in the developing nervous system.
Figure 5.
Immunolocalization of ER in wild-type embryos and Schneider 2 cells. Anterior is to the left and posterior is to the right for the embryos. (A) Stage 5-Syncytial blastoderm showing pole cells on the right. ER is localized to the nuclei. (B) Stage 7-ventral mesoderm invagination. ER can still be seen localized primarily to the nuclei. (C) Stage 11. ER shows a diffuse general localization, but much higher levels in the developing CNS. (D) Stage 14. Staining is seen in the brain lobe on the anterior dorsal side of the embryo and in the ventral nerve cord. (E) Stage 14, ventral view. Staining is seen in the brain lobe and the two tracks of the ventral nerve cord. Staining also appears in the peripheral nervous system, although at lower levels. (F) Schneider 2 cells. Staining is clearly localized to the nuclei.
The immunolocalization of ER in early embryos shows a strong localization to the nucleus (Fig. 5A and B). Initially maternal ER is distributed throughout the unfertilized egg.17 After nuclear cleavage at the syncytial blastoderm stage, ER localizes to the nucleus (Fig. 5A). This nuclear localization persists and is evident during gastrulation at the ventral furrow formation (Fig. 5B). Strong localization of ER can also be seen in Drosophila Schneider 2 (S2) cells (Fig. 5F). These results are in agreement with studies in mammalian cells that used a ER-GFP fusion to demonstrate nuclear localization.11 If e(r) is indeed a positive regulator of the Notch signaling pathway, then the nuclear localization suggests that e(r) is regulating the Notch signaling pathway at the level of transcription.
Discussion
Nnd-p is a P element insertion into the first exon of the N gene, upstream of the start of translation. Both revertants of Nnd-p have lost this element, verifying that the P element is the cause of the mutation. The very weak mutant phenotype of Nnd-p flies and the inability of Nnd-p to complement the lethality of N deletions argue that Nnd-p results in a partial reduction in N activity. An earlier report analyzed 13 hybrid-dysgenesis-induced N mutations.21 Like Nnd-p, all of the mutations were P element insertions in the first exon, upstream of the start of translation. Twelve of the mutations were recessive lethals, indicative of a large reduction in N expression and one was a recessive, hypomorphic, notchoid-like mutation similar in phenotype to Nnd-p. It is clear that that P elements insert preferentially into the 5′ end of the N gene. What is also clear is that there is a range in the reduction in N activity by the insertions. The two notchoid-like alleles, Nnd-p and Nnd3.1072, reduce expression the least, followed by five recessive-lethal alleles that do not show the wing nicking as heterozygotes, to seven recessive-lethal alleles that show the wing nicking as heterozygotes. This last group is displays the phenotype indicative of a N null mutation.
The body of evidence from studies in Drosophila and vertebrates suggests that e(r) is involved in a number of different biochemical and developmental pathways. Genetically it was first shown to be involved in pyrimidine metabolism through its interactions with rudimentary, a gene encoding the first three enzymatic activities in pyrimidine biosynthesis.4–6 In Drosophila, it is expressed maternally during oogenesis and deposited into the mature oocyte, consistent with the decreased fertility of e(r) null females.17 In vertebrates, a role for e(r) in cell proliferation is suggested by its increased expression in certain human cancer cells.10 Another piece of evidence linking ER to DNA replication is the identification of PDIP46/SKAR as a potential binding partner.11 This protein interacts with DNA polymerase γ, one of the major polymerases in DNA replication. This study also demonstrated that a human ER-GFP fusion protein was nuclearly localized, consistent with a nuclear function for ER.11 ER is also a binding partner of Ciz1,13 a nuclear protein that has been shown to activate DNA synthesis and entry into S phase in vertebrates.14 Finally, ER has been shown to associate with a number of nuclearly localized and functionally distinct proteins involved in transcription.12,15,16 One of these interacting proteins is DCoH/PCD, a dimerization cofactor for the transcription factor, hepatocyte nuclear factor 1.12 The authors conclude that in Xenopus, ER is acting as a transcription co-repressor by interfering with the activation of hnf-1. The other studies show that ER copurifies with SPT5,15 a transcription elongation factor and with FCP1,16 a phosphatase associated with the CTD of the RNA polymerase II. Together these data suggest that ER may have a role in transcription elongation. The authors propose that ER is acting to dampen transcription by counteracting the positive effects of SPT5 and FCP1.16 Thus the vertebrate data argue that ER can negatively regulate transcription at a number of different steps.
The data with the vertebrate ER all indirectly indicate that ER is localized to the nucleus. This has been done with protein-interaction assays and an ER-GFP fusion protein. In the present study, we utilize immunolocalization to show that the endogenous Drosophila ER is nuclearly localized in early embryos and in S2 cells. This is the first demonstration that the endogenous ER is nuclearly localized and supports the proposed nuclear functions of ER. The association of ER with nuclear proteins of disparate functions is not easily explained. It suggests that ER may have a general nuclear function that requires it to interact with many different proteins and many different pathways. This could explain the fact that a null mutants of e(r), while viable and without any noticeable morphological defects, have low viability.17
The present study suggests a new function to e(r) in the Notch signaling pathway possibly in neurogenesis. Our data indicate that e(r) is a positive regulator of the Notch signaling pathway and is acting upstream of E(spl) in the pathway. In embryos, ER is localized to the developing nervous system and is nuclearly localized in embryos and S2 cells. Given the nuclear localization of ER in vertebrates11 and in Drosophila and the proposed roles of ER as a transcription co-repressor in vertebrates,12,15,16 e(r) may be transcriptionally regulating the activity of the Notch signaling pathway, possibly through dampening the activity of a negative regulator of the Notch signaling pathway, such as H.
Two ligands for the N receptor have been identified in Drosophila, Dl and Ser. Each binds to N to activate the Notch signaling pathway. Genetic data presented in this paper suggest that e(r) may be interacting with Dl-N signaling but not with Ser-N signaling and present the possibility that e(r) is interacting with the Notch signaling pathway through regulating Dl expression. Interestingly, the expression pattern of e(r) in embryos is very similar to that of Dl.28,29 It is possible that e(r) is regulating Dl expression directly or by controlling the expression of a regulator of Dl activity. Two genes, neuralized26 and mind bomb27 encode ubiquitin ligases that have been shown to activate Dl activity post translationally. ER could be interacting with the Notch pathway through regulating the expression of one of these genes. Alternatively, ER could be interacting with the Notch signaling pathway further downstream in cells that interact with the Dl ligand but not in cells that interact with the Ser ligand.
Materials and Methods
Drosophila strains and crosses.
Tr[e(r)+]SS is a transgene, cloned into pCasper-4 and inserted into chromosome 3. It contains a 6.1-kb SalI fragment that has wild-type e(r) activity and a white, w+, allele.1 The e(r) null, e(r)27-1, is an 1,888-bp deletion that removes the promoter, start of transcription and 43% of the coding region.17 The e(r) null, e(r)37-6, is a 1,406-bp deletion that removes the promoter, start of transcription, but leaves the coding region intact.17 Dp(1;3)DC109 is a 93-kb genomic fragment that contains the entire N gene inserted into the third chromosome. It does not carry any other active genes. Dp(1;3) DC157 is a 107-kb genomic fragment. We have verified that it rescues a dx mutation and thus contains the wild-type dx gene. All other Drosophila mutations and stocks, other than those generated in this study, are described in FlyBase.18 Stocks that were not generated in this study were obtained from the Bloomington Drosophila Stock Center at Indiana University. All stocks were grown on yeast-glucose medium19 and all crosses were performed at 25°.
Drosophila melanogaster alleles, chromosomes and transformation constructs used in this study:
C(1)DX: FBab0000080
Df(1)N81k1: FBab0000580
Dl3: FBal0002460
Dl6B: FBal0002465
Dp(1;3)DC109: FBab0046325
Dp(1;3)DC15: FBab0046359
dx: FBal0003261
e(r)27-1: FBal0217524
e(r)37-6: FBal0217532
e(r)p2: FBti0101498
FM7c y31d sc8 wa snX2 vOf g4 B: FBba0000009
H/In(3R)C, sprd e: FBst0000515
N8: FBab0000579
N55e11: FBal0012701
Nco: FBal0012759
Nfa-g: FBal0012868
Nfa-swb: FBal0012874
Nnd-1: FBal0012890
Nnd-3: FBal0012892
Nnd-p: FBal0141642
Nspl-1: FBal0012900
pak7: FBal0098341
Ser1: FBal0015427
TM3, ryRK Sb1 Ser1 P{Δ2–3}99B/Df(3R)C7, ry506: FBst0305182
TM3: FBba0000047
TM6C, Sb: FBba0000071
Tr[e(r)+]SS: FBtp0003720
w: FBal0018074
y2: FBal0018612
Isolation of Nnd-p.
The screen was set up to isolate hybrid-dysgenesis-induced, X-linked lethals that were rescued by an e(r)+ transgene (Fig. 1). The starting X chromosome contained an e(r) allele, e(r)p2, which contained the only P element in the stock. In cross 1, The P element Δ2–3 was introduced in the stock to mobilize the P element in e(r)p2. In cross 2, Tr[e(r)+]SS was introduced into the stock to cover any X-linked lethals that were able to be rescued by e(r)+. Males containing this transgene and a mutagenized X chromosome were crossed to C(1)DX, y f females. Any male containing an X-linked lethal that was rescued by Tr[e(r)+]SS produced male progeny, all of which carried Tr[e(r)+]SS, and no males that lacked the transgene. These stocks were identified as those lacking white-eye males.
DNA sequencing of Nnd-p and its revertants.
PCR analyses identified a P element in the 5′ end of the N gene. The fragment containing this P element, or lacking it in the case of the two revertants, was amplified. The amplified fragment was isolated using a QIAquick PCR Purification Kit (QIAGEN, Inc.,) and partially sequenced using BigDye Terminator Cycle Sequencing (Applied Biosystems). The sequencing determined the exact location of the P element and the nature of the excision of the P element in the two revertants.
Isolation of revertants Nnd-p.
Given that Nnd-p is associated with a P-element insertion, it was important to show that the P element was responsible for both the phenotype of Nnd-p and its proposed lethal interaction with e(r) mutations. The P element in Nnd-p was mobilized by crossing in an autonomous P element, Δ2–3. In the first screen a revertant of Nnd-p was isolated by reverting the lethal interaction with e(r)27-1. This revertant was isolated as a viable male derived from a Nnd-p e(r)27-1 X chromosome. The second revertant was isolated taking advantage of the fact that N8/Nnd-p heterozygous females are lethal. The revertant was isolated as a viable heterozygous female.
Viability measurements.
Since the mutations being studied are on the X chromosome, viability was measured in hemizygous males and compared to the viability of their heterozygous sisters, who served as a control. Heterozygous females carrying the tester chromosome and the balancer FM7c were crossed to FM7c males. Among the progeny, the number of tester males and heterozygous tester females were counted. The two groups should be in roughly equal number if the viabilities are the same.
In the production of Dl e(r)27-1 double mutants, y w e(r)27-1/FM7c females were crossed to either ss1 Dl6B e1/TM6C, Sb1 or Dl3/In(3R)C, e1 males. The viability of e(r)27-1 males, that were either Dl/+ or +/+ was scored. As controls, e(r)27-1/+ females and FM7c males were scored. For the production of Ser e(r)27-1 double mutants, y w e(r)27-1/FM7c females were crossed to either pak7/TM3, Ser1. The viability of the flies was scored as in the Dl crosses. The wing phenotypes of the Dl and Ser flies were scored for any changes in the background of e(r)27-1.
Immunostaining drosophila larvae.
Third instar male larvae were obtained from the following cross:
e(r)27-1/FM7c females x FM7c males.
The male third instar larvae were collected and rinsed in PBS. Under a dissecting microscope, forceps were used to grab the mouth-piece to gently remove the head. The heads were immediately placed in the cap of a SNAPSTRIP PCR tube (Midwest Scientific, SST) containing approximately 30 ul of PEM-formaldehyde fix. The tissue was fixed for 30 min. followed by three 30 min. rinses in PBS/0.03% Triton X-100. The tissue was blocked in PBS/0.03% Triton X-100/10X BSA for 30 min. followed by a single 30 min rinse in PBS. Staining was performed by an overnight incubation at 4° with rabbit anti-ER and mouse anti-E(spl) at 1:1,000 in PBS/0.03% Triton X-100/10X BSA. Anti-E(spl) was obtained from Sarah Bray. The primary stain was removed by three 30 min. rinses in PBS/0.03% Triton X-100. The primary antibodies were detected by a FITC conjugated goat anti-rabbit antibody (Sigma, F-6005) and a Cy3 conjugated donkey anti-mouse antibody (Vector Labs and Invitrogen). The tissue was further dissected under a dissecting microscope, mounted in VectaSheild with DAPI and viewed as described previously. Since e(r)27-1 is an e(r) protein-null, the e(r)27-1 tissue was identified by lack of ER staining, while the FM7c e(r)+ larvae were identified by positive ER staining. Staining was visualized using a Leica DM4000 B fluorescent microscope attached to a QICAM FAST 1394 camera and Openlab 3.5 imaging software.
Immunostaining Drosophila whole-mount embryos.
Adult flies were allowed to lay eggs on molasses-agar collection plates (5 g sugar, 5 g agar, 5 g yeast and 50 ml molasses per liter) for 0–12 hours. The embryos were rinsed from the plate onto a Spectra/Mesh Nylon Filter (Spectrum, 146-502). On the mesh sieve, the embryos were rinsed well with water containing 0.05% Triton X-100. The embryos were flooded with 50% bleach diluted with distilled sterile water for 3–4 min. to remove the chorion. After bleaching, the embryos were fixed for 30 min. at 22° temperature in 50% n-Heptane and 50% PEM-formaldehyde fix (0.1 M PIPES, 2 mM EGTA, 1 mM MgSO4, 4% formaldehyde). An equal volume of 100% methanol was added to the embryos, and the tube was shaken vigorously by hand for 1 min. to remove the vitelline membrane. The de-vitellinized embryos were rinsed with 100% methanol. Before staining, the embryos were re-hydrated in 1 ml PBS for 30 min. The PBS was removed and replaced with 1 ml PBS/0.03% Triton X-100 for 30 min. This solution was removed and the embryos were blocked for 30 min in 1 ml PBS/0.03% Triton X-100/10X BSA, with gentle swirling. After blocking the embryos, the block was removed, and the embryos were rinsed for 10 min in PBS. The embryos were stained with anti-ER antibodies at 1:1,000 overnight with swirling at 4°. The embryos were rinsed with 1 ml PBS/0.03% Triton X-100 for 10 minutes, followed by a 20 min. rinse in PBS/0.03% Triton X-100, and a final 20 min rinse in PBS. The embryos were re-suspended in 1 ml PBS/0.03% Triton X-100/10X BSA and incubated with a donkey anti-rabbit Cy3 antibody (1:1,000) overnight with swirling at 4°. Whole mount embryos were visualized using a Leica DM4000 B fluorescent microscope attached to a QICAM FAST 1394 camera and Openlab 3.5 imaging software.
Immunostaining Drosophila Schneider 2 (S2) cells.
Drosophila Schneider 2 cells were grown in a Corning 24-well (16 mm) polystyrene tray at 28° (without CO2) for two days in Schneider's medium supplemented with 10% heat-inactivated fetal calf serum and penicillin/streptomycin at a final concentration of 50 units penicillin G and 50 µg streptomycin sulfate per milliliter of medium. The cells were fixed with 4% formaldehyde in PBS for 30 min, and stained with the rabbit anti-ER antibody and counterstained with a mouse anti-rabbit Cy3 conjugated secondary antibody. S2 cells immunostained with rabbit pre-bleed serum and the mouse anti-rabbit Cy3 conjugated secondary antibody failed to show any staining within the cell, demonstrating specificity of the signal (data not shown).
Acknowledgments
This work was supported by a grant from the National Institutes of Health (R15 GM64364) and by funds from Saint Louis University and The College at Brockport.
Abbreviations
- e(r)
enhancer of rudimentary
- ER
enhancer of rudimentary protein
- N
Notch
- N
Notch protein
- dx
deltex
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Wojcik E, Murphy AM, Fares H, Dang-Vu K, Tsubota SI. Enhancer of rudimentaryp1, e(r)p1, a highly conserved enhancer of the rudimentary gene. Genetics. 1994;138:1163–1170. doi: 10.1093/genetics/138.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gelsthorpe M, Pulumati M, McCallum C, Dang-Vu K, Tsubota SI. The putative cell cycle gene, enhancer of rudimentary, encodes a highly conserved protein found in plants and animals. Gene. 1997;186:189–195. doi: 10.1016/s0378-1119(96)00701-9. [DOI] [PubMed] [Google Scholar]
- 3.Gelsthorpe ME, Tan Z, Phillips A, Eissenberg JC, Miller A, Wallace J, Tsubota SI. Regulation of the Drosophila melanogaster protein, Enhancer of Rudimentary, by casein kinase II. Genetics. 2006;174:265–270. doi: 10.1534/genetics.106.061465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarry B, Falk D. Functional diversity within the rudimentary locus of Drosophila melanogaster. Mol Gen Gene. 1974;135:113–122. doi: 10.1007/BF00264779. [DOI] [PubMed] [Google Scholar]
- 5.Rawls JM, Fristrom J. A complex genetic locus that controls the first three steps of pyrimidine biosynthesis in Drosophila. Nature. 1975;255:738–740. doi: 10.1038/255738a0. [DOI] [PubMed] [Google Scholar]
- 6.Brothers V, Tsubota S, Germeraad S, Fristrom J. The rudimentary locus of Drosophila melanogaster: partial purification of a carbamyl phosphate synthetase-aspartate transcarbamylase-dihydroorotase complex. Biochem Genet. 1978;16:321–332. doi: 10.1007/BF00484088. [DOI] [PubMed] [Google Scholar]
- 7.Aoki T, Weber G. Carbamoyl phosphate synthetase (glutamine hydrolyzing): increased activity in cancer cells. Science. 1981;212:463–465. doi: 10.1126/science.7209543. [DOI] [PubMed] [Google Scholar]
- 8.Aoki T, Morris H, Weber G. Regulatory properties and behavior of activity of carbamoyl phosphate synthetase (glutamine hydrolyzing) in normal and proliferating tissues. J Biol Chem. 1982;257:432–438. [PubMed] [Google Scholar]
- 9.Rao G, Davidson J. CAD gene expression in serum-starved and serum-stimulated hamster cell. DNA. 1988;7:423–432. doi: 10.1089/dna.1.1988.7.423. [DOI] [PubMed] [Google Scholar]
- 10.Zafrakas M, Losen I, Knüchel R, Dahl E. Enhancer of the rudimentary gene homologue (ERH) expression pattern in sporadic human breast cancer and normal breast tissue. BMC Cancer. 2008;8:145. doi: 10.1186/1471-2407-8-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smyk A, Szuminska M, Uniewicz KA, Graves LM, Kozlowski P. Human enhancer of rudimentary is a molecular partner of PDIP46/SKAR, a protein interacting with DNA polymerase δ and S6K1 and regulating cell growth. FEBS J. 2006;273:4728–4741. doi: 10.1111/j.1742-4658.2006.05477.x. [DOI] [PubMed] [Google Scholar]
- 12.Pogge von Strandmann E, Senkel S, Ryffel GU. ERH (enhancer of rudimentary homologue), a conserved factor identical between frog and human, is a transcriptional repressor. Biol Chem. 2001;382:1379–1385. doi: 10.1515/BC.2001.170. [DOI] [PubMed] [Google Scholar]
- 13.Lukasik A, Uniewicz KA, Kulis M, Koslowski P. Ciz1, a p21Cip1/Waf1-interacting zinc finger protein and DNA replication factor, is a novel molecular partner for human enhancer of rudimentary homolog. FEBS J. 2008;275:332–340. doi: 10.1111/j.1742-4658.2007.06203.x. [DOI] [PubMed] [Google Scholar]
- 14.Coverley D, Marr J, Ainscough J. Ciz1 promotes mammalian DNA replication. J Cell Sci. 2005;118:101–112. doi: 10.1242/jcs.01599. [DOI] [PubMed] [Google Scholar]
- 15.Kwak YT, Guo J, Prajapati S, Park KJ, Surabhi RM, Miller B, et al. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol Cell. 2003;11:1055–1066. doi: 10.1016/s1097-2765(03)00101-1. [DOI] [PubMed] [Google Scholar]
- 16.Amente S, Napolitano G, Licciardo P, Monti M, Pucci P, Lania L, Majello B. Identification of proteins interacting with the RNAPII FCP1 phosphatase: FCP1 forms a complex with arginine methyltransferase PRMT5 and it is a substrate for PRMT5-mediated methylation. FEBS Lett. 2005;579:683–689. doi: 10.1016/j.febslet.2004.12.045. [DOI] [PubMed] [Google Scholar]
- 17.Roykhman M, Ibach S, Harding D, Wolff B, Vowels M, Gelsthorpe ME, et al. The generation and analysis of deficiencies within a small genomic region on the X chromosome of Drosophila melanogaster containing two genes, enhancer of rudimentary and CG15352. Fly. 2007;1:245–250. doi: 10.4161/fly.5117. [DOI] [PubMed] [Google Scholar]
- 18.Drysdale R, Crosby M. FlyBase Consortium. FlyBase: genes and gene models. Nuc Acids Res. 2005;33:390–395. doi: 10.1093/nar/gki046. http://flybase.bio.indiana.edu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts DB, editor. Drosophila a Practical Approach. Oxford: IRL Press; 1986. [Google Scholar]
- 20.Close D, Tsubota SI. A hot spot for P-element insertion in the enhancer of rudimentary gene. Dros Inf Serv. 2004;87:24. [Google Scholar]
- 21.Kelley MR, Kidd S, Berg RL, Young MW. Restriction of P-element insertions at the Notch locus of Drosophila melanogaster. Molec Cell Biol. 1987;7:1545–1548. doi: 10.1128/mcb.7.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vassin H, Vielmetter J, Campos-Ortega JA. Genetic interactions in early neurogenesis of Drosophila melanogaster. J Neurogenet. 1985;2:291–308. doi: 10.3109/01677068509102325. [DOI] [PubMed] [Google Scholar]
- 23.Brou C, Logeat F, Lecourtois M, Vandekerckhove J, Kourilsky P, Schweisguth F, Israäl A. Inhibition of the DNA-binding activity of Drosophila Suppressor of Hairless and of its human homolog, kbf2/rbp-j kappa, by direct protein-protein interaction with Drosophila Hairless. Genes Dev. 1994;8:2491–2503. doi: 10.1101/gad.8.20.2491. [DOI] [PubMed] [Google Scholar]
- 24.Matsuno K, Diederich RJ, Go MJ, Blaumueller CM, Artavanis-Tsakonas S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development. 1995;121:2633–2644. doi: 10.1242/dev.121.8.2633. [DOI] [PubMed] [Google Scholar]
- 25.Jennings B, Preiss A, Delidakis C, Bray S. The Notch signaling pathway is required for Enhancer of Split bHLH protein expression during neurogenesis in the Drosophila embryo. Development. 1994;120:3537–3548. doi: 10.1242/dev.120.12.3537. [DOI] [PubMed] [Google Scholar]
- 26.Boulianne GL, de la Concha A, Campos-Ortega JA, Jan LY, Jan YN. The Drosophila neurogenic gene neuralized encodes a novel protein and is expressed in precursors of larval and adult neurons. EMBO J. 1991;10:2975–2983. doi: 10.1002/j.1460-2075.1991.tb07848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai EC, Roegiers F, Qin X, Jan YN, Rubin GM. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development. 2005;132:2319–2332. doi: 10.1242/dev.01825. [DOI] [PubMed] [Google Scholar]
- 28.Vaessin H, Bremer KA, Knust E, Campos-Ortega JA. The neurogenic Gene Delta of Drosophila melanogaster is expressed in neurogenic territories and encodes a putative transmembrane protein with EGF-like repeats. EMBO J. 1987;6:3431–3440. doi: 10.1002/j.1460-2075.1987.tb02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haenlin M, Kramatschek B, Campos-Ortega JA. The pattern of transcription of the neurogenic gene Delta of Drosophila melanogaster. Development. 1990;110:905–914. doi: 10.1242/dev.110.3.905. [DOI] [PubMed] [Google Scholar]