Abstract
Disulfide bonds between Cys residues in adjacent strands of parallel β-sheet are rare among proteins, which suggests that parallel β-sheet structure is not stabilized by such disulfide crosslinks. We report experimental results that show, surprisingly, that an inter-strand disulfide bond can stabilize parallel β-sheet formed by an autonomously folding peptide in aqueous solution. NMR analysis reveals that parallel β-sheet structure is terminated beyond the disulfide bond, which causes deviation from the extended backbone conformation at one of the Cys residues.
Keywords: Disulfide Bonds, Parallel Beta-sheet, Secondary Structure, Covalent Cross-links, Protein Design
Helices and sheets are dominant structural motifs within proteins, but these secondary structures are generally not very stable in isolation, particularly for linear peptides of ≤ 20 residues. Helical conformations (α, 310 and π) and β-sheets can be stabilized via crosslinking, i.e., macrocycle formation, involving side chains and/or the backbone.1,2 This structural fortification strategy is observed among biological proteins and peptides, with cyclization most commonly achieved via disulfide formation between cysteine side chains.3 Other crosslinking modes are found among natural polypeptides as well, and an even wider variety has been explored in synthetic systems.4 Polypeptides generated via ribosomal biosynthesis and not modified post-translationally, however, are limited to disulfide crosslinks, which can form spontaneously under mildly oxidizing conditions.
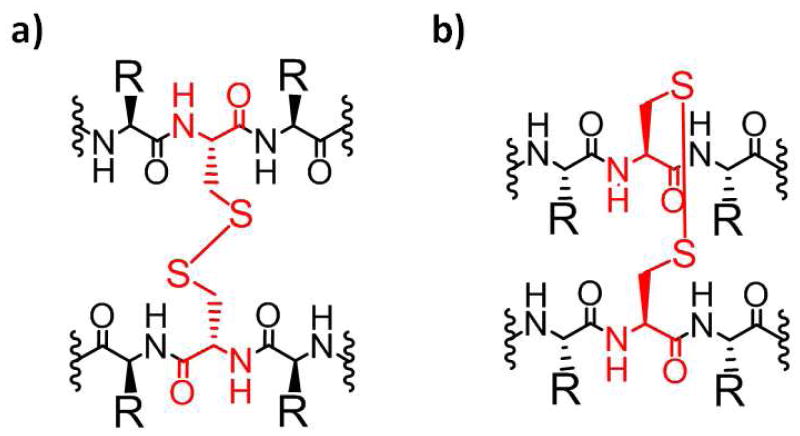
Bioinformatics analysis of antiparallel β-sheets reveals that pairs of disulfide-linked Cys residues often occur at non-hydrogen bonded positions that are aligned on adjacent strands5 (Figure 1). This observation and geometric considerations suggest that such inter-strand crosslinks can stabilize antiparallel β-sheet secondary structure, a hypothesis that has been supported by numerous studies with designed β-hairpins that fold in aqueous solution.6 In contrast, disulfide crosslinks between adjacent strands are very rare within parallel β-sheets,5f–h,7 which suggests that disulfide-based macrocyclization should destabilize autonomously folding parallel β-sheets. We report the first test of this hypothesis. Our experiments lead to the surprising discovery that an interstrand disulfide can stabilize parallel β-sheet secondary structure to one side along the strand direction; however, the parallel sheet cannot propagate beyond the disulfide position.
Figure 1.
Disulfide bonds between Cys in adjacent strands of (a) anti-parallel β-sheet, in non-hydrogen bond positions, and (b) parallel β-sheet.
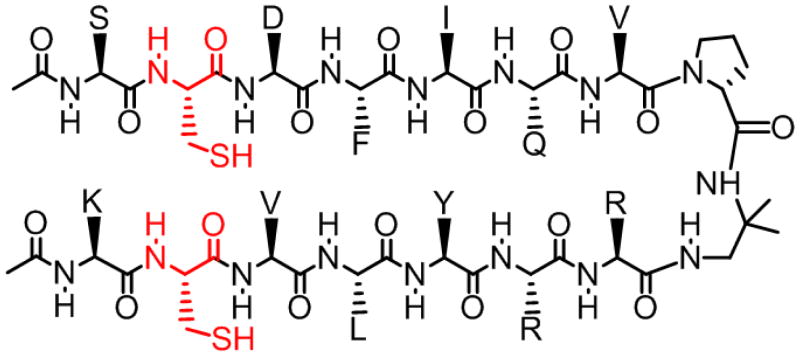
Our experimental design builds upon guidelines we have previously developed for creating short peptides that can form two-stranded parallel β-sheets in aqueous solution.8 Use of an autonomously folding β-sheet rather than a full-fledged protein is intended to avoid the influence of a specific tertiary context; therefore, our findings should reflect the intrinsic conformational behavior of parallel β-sheet secondary structure. The diamine segment D-prolyl-1,1-dimethyl-1,2-diaminoethane (D-Pro-DADME) promotes but does not enforce β-sheet interactions between peptide segments linked in parallel. Peptide 1-SH (Figure 2) is related to a molecule previously used to assess the thermodynamics of parallel β-sheet formation in aqueous solution.8c The two Cys residues, near the N-terminus of each strand, enable oxidative cyclization to generate 1-SS. 2D NMR analysis reveals multiple NOEs between protons on residues that are not adjacent in sequence for both forms of 1, and in each case all NOEs are consistent with the expected parallel β-sheet folding pattern.9 NOE-restrained molecular dynamics calculations for 1-SS (based only on NOEs and the CNS force field; no coupling-constant or H-bond constraintswere used) suggest canonical parallel β-sheet interactions between the segments bounded by the D-Pro-DADME unit and the Cys residues, i.e., between DFIQV on the upper strand (as drawn) and VLYRR on the lower strand (Figure 3). The β-sheet structure appears to be present throughout the hairpin, up to the location of the disulfide. Only the Cys residue of the lower strand, however, is part of the β-sheet; a kink is observed at the other Cys, which indicates that β-sheet secondary structure is terminated at this point.
Figure 2.
Chemical structure of peptide 1-SH. Cysteine residues are shown in red.
Figure 3.
NMR-based structural analysis of 1-SS based on data obtained for 2.5 mM peptide in 100 mM deuterioacetate, pH=3.8, 4°C: (a, b) two views of the overlay of the 10 structures with lowest calculated energies from NOE-restrained molecular dynamics simulations; (c, d) two views of the average minimized structure. RMSD among backbone atoms is 1.1 ± 0.4 Å.
Fraying at the open ends of autonomously folding hairpins has previously been noted,8a,10 but the NMR data suggest that the deviation from local β-sheet structure we observe at the disulfide position in 1-SS is not attributable to fraying. Comparisons among the 10 lowest-energy structures from the NOE-restrained dynamics calculations for 1-SS indicate an overall RMSD of 1.1 ± 0.4 Å. Similar RMSD values are obtained for comparisons focused on just a single Cys residue in either strand (1.0 ± 0.3 and 1.1 ± 0.5 Å, respectively, for the top and bottom Cys residues as drawn in Figure 2). If these Cys residues were frayed relative to the core of the β-sheet, then the Cys RMSD values should be larger than the overall RMSD.
For a residue that participates in β-sheet secondary structure, the α-proton chemical shift (δCαH) is generally downfield of the position expected for that residue in an unfolded (“random coil”) state.11 We have shown that δCαH data can be used to assess parallel β-sheet population for molecules such as 1-SH,8c–e which are anticipated to equilibrate rapidly between folded and unfolded states on the NMR time scale. This analysis requires two reference compounds, one to provide δCαH values for the fully unfolded state and another to provide δCαH values for the fully folded state. The former goal is achieved by replacing D-Pro in the diamine linker with L-Pro,8a and the latter goal is achieved by backbone cyclization with a diacid linking segment.8c,d,9
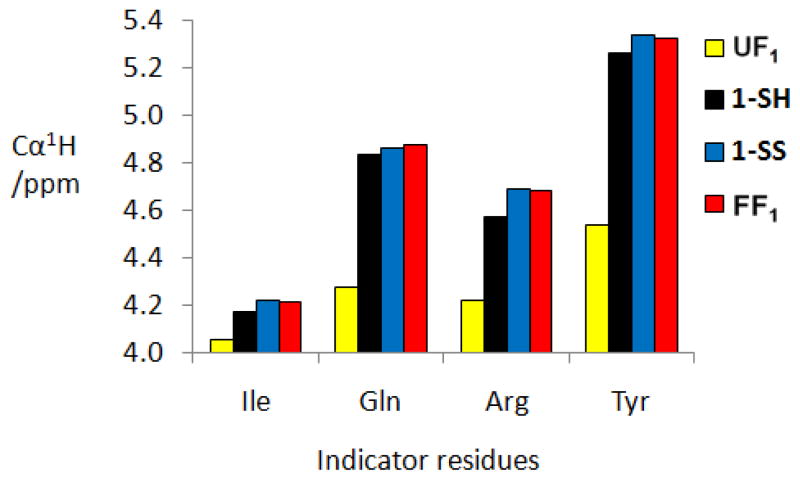
We used this approach to estimate the extent of parallel β-sheet folding in 1-SH and 1-SS by focusing on four ‘indicator residues’, Ile and Gln in the upper strand and Tyr and the more N-terminal Arg in the lower strand (Figure 2). These residues provide four independent measurements of parallel β-sheet population, and each is sufficiently isolated from the variable C-terminal portion of its strand to be free of chemical shift influences induced by covalent changes. For 1-SH, δCαH values for all four indicator residues are significantly down-field of the δCαH values in the unfolded reference peptide (Figure 4), which is consistent with the NOE data in indicating substantial parallel β-sheet formation. However, the indicator δCαH values for 1-SH do not differ significantly from the corresponding values for 1-SS or the fully folded reference peptide, which makes it impossible to determine whether interstrand disulfide formation enhances parallel β-sheet stability.
Figure 4.
1H NMR chemical shift data for protons attached to Cα (δCαH) of indicator residues in the unfolded reference peptide (UF1), 1-SH, 1-SS and the folded reference peptide (FF1).
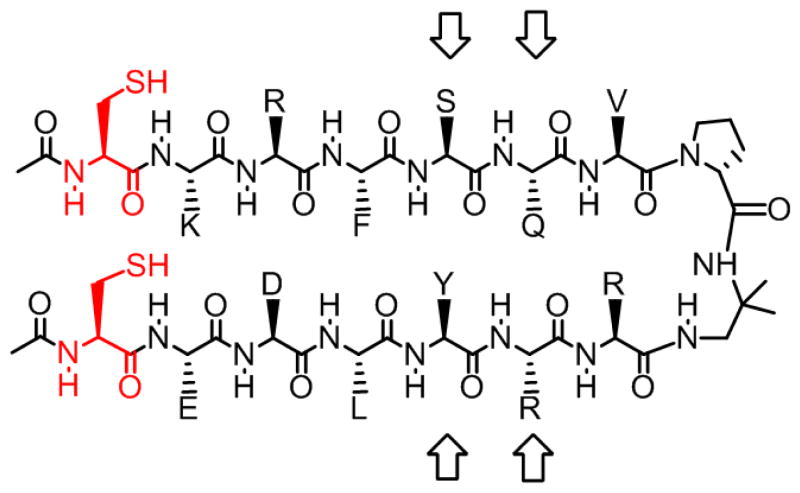
We examined a second design that was intended to manifest a lower folding propensity in order to assess the thermodynamic impact of an inter-strand disulfide on parallel β-sheet stability. The sequence of 2-SH/2-SS (Figure 5) differs at several points from that of 1-SH/1-SS. Two changes are particularly noteworthy: (1) the Cys residues occur at the N-termini of the strands in 2, rather than adjacent to the N-termini in 1, and (2) a cross-strand Ile-Tyr pairing that was intended to provide a hydrophobic driving force for folding of 1 has been replaced by a Ser-Tyr pairing, which should be less conducive to parallel β-sheet formation. 2D NMR analysis suggests qualitatively that 2-SH and 2-SS display smaller extents of parallel β-sheet folding relative to 1-SH and 1-SS, because the number of inter-strand NOEs is smaller for both forms of 2 than for either form of 1.9 Nevertheless, the 2D NMR data for 2-SH and 2-SS indicate adoption of the expected parallel β-sheet conformation, in the population of molecules that is folded, because all NOEs involving protons from sequentially non-adjacent protons are consistent with this conformation.9
Figure 5.
Chemical structure of peptide 2-SH. Cysteine residues are shown in red. Indicator residues are denoted by arrows.
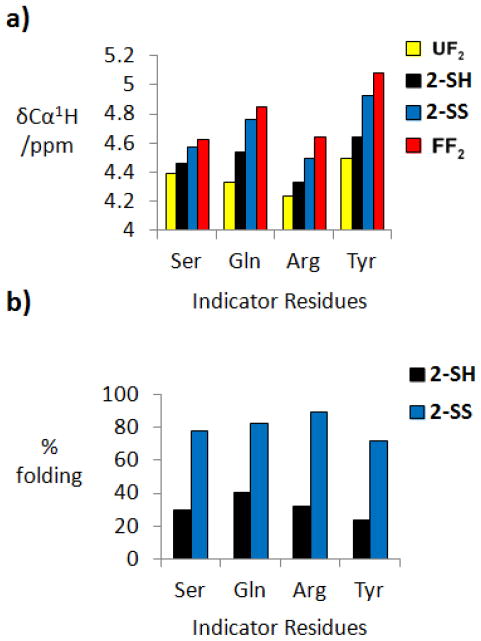
The extent of parallel β-sheet folding for 2-SH and 2-SS in aqueous solution was estimated based on δCαH data, via comparisons with appropriate fully-folded and fully-unfolded reference peptides.8e,9 Following the approach used for series 1, we focused on δCαH data from four indicator residues, Ser and Gln in the upper strand and Tyr and the more N-terminal Arg in the lower strand. At each position, δCαH values show a steady downfield movement in the order: unfolded reference, 2-SH, 2-SS, folded reference (Figure 6a). This consistent trend qualitatively suggests that neither version of 2 is fully folded, and, more important, that disulfide formation causes an increase in the extent of parallel β-sheet formation. δCαH-based population analysis9 provides a reasonably consistent conclusion across the four indicator positions, suggesting that 2-SH is ~20% folded and that 2-SS is ~70% folded in 9:1 H2O:D2O, pH 3.8, 100 mM sodium deuterioacetate buffer (buffer pH was not corrected for isotope effects) at 4°C (Figure 6b). These folded state population values can be converted to Gibbs free energy of folding (ΔGF) based on a two-state conformational model, random coil vs. parallel β-sheet conformation.8c–e Data from the four indicator residues indicate a ΔΔGF value of −1.1 ± 0.1 kcal/mol, which shows that the interstrand disulfide in 2-SS provides significant stabilization to the parallel β-sheet conformation.
Figure 6.
(a) 1H NMR chemical shift data for protons attached to Cα (δCαH) of indicator residues in the unfolded reference peptide (UF2), 2-SH, 2-SS and the folded reference peptide (FF2). (b) Percent folding calculated for 2-SH and 2-SS based on data obtained for 2.5 mM peptide in 100 mM deuterioacetate, pH 3.8, 4°C.
Previous bioinformatics analysis suggested that interstrand disulfide crosslinks are not well accommodated in parallel β-sheet secondary structure,5f–h,7 and this conclusion is supported by our experimental findings with autonomously folding peptides, since the backbone kinks at the disulfide. However, our studies reveal an unexpected insight: an interstrand disulfide can stabilize parallel β-sheet secondary structure that forms in the C-terminal direction relative to the Cys residues. Covalent bonding between Cys side chains appears to require that the backbone deviate from the extended conformation, at least in one strand, which prevents the sheet from propagating through the disulfide position in our peptides. Overall these results suggest that interstrand cystine crosslinks can both stabilize and define the extent of parallel β-sheet secondary structure in designed peptides and proteins.
Supplementary Material
Acknowledgments
This research was supported by the NIH (GM061238). R. L. was supported in part by fellowship from the UW Department of Chemistry and by NSF grant CHE-0848847. NMR spectrometers were purchased with partial support from NIH (Grant # 1 S10 RR13866-01) and NSF.
Footnotes
Author Contributions
The manuscript was written through contributions of all authors./All authors have given approval to the final version of the manuscript.
Supporting Information Available: Experimental details including compound characterization, NMR data, and thermodynamic analysis is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Henchey LK, Jochim AL, Arora PS. J Am Chem Soc. 2008;12:692–697. doi: 10.1016/j.cbpa.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Phelan JC, Skelton NJ, Braisted AC, McDowell RS. J Am Chem Soc. 1997;119:455–460. [Google Scholar]; (c) Haubner R, Schmitt W, Holzemann G, Goodman SL, Jonczyk A, Kessler H. J Am Chem Soc. 1996;118:7881–7891. [Google Scholar]; (d) Lutgring R, Chmielewski J. J Am Chem Soc. 1994;116:6451–6452. [Google Scholar]; (e) Betz SF. Protein Sci. 1993;2:1551–1558. doi: 10.1002/pro.5560021002. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Harrison PM, Sternberg MJE. J Mol Biol. 1994;244:448–463. doi: 10.1006/jmbi.1994.1742. [DOI] [PubMed] [Google Scholar]
- 2.Selected examples: Felix AM, Heimer EP, Wang CT, Lambros TJ, Fournier A, Mowles TF, Maines S, Campbell RM, Wegrzynski BB, Toome V, Fry D, Madison VS. Int J Pept Protein Res. 1988;32:441–454. doi: 10.1111/j.1399-3011.1988.tb01375.x.Osapay G, Taylor JW. J Am Chem Soc. 1992;114:6966–6973.Leduc AM, Trent JO, Wittlif JL, Bramlett KS, Briggs SL, Chirgadze NY, Wang Y, Burris TP, Spatola AF. Proc Natl Acad Sci USA. 2003;100:11273–11278. doi: 10.1073/pnas.1934759100.Zhou HXX, Hull LA, Kallenbach NR, Mayne L, Bai YW, Englander SW. J Am Chem Soc. 1994;116:6482–6483.Jackson D, King D, Chmielewski J, Singh S, Schultz P. J Am Chem Soc. 1991;113:9391–9392.
- 3.(a) McDonald NQ, Hendrickson WA. Cell. 1993:421–424. doi: 10.1016/0092-8674(93)90127-c. [DOI] [PubMed] [Google Scholar]; (b) Willey JM, van der Donk WA. Annu Rev Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]; (c) Ganz T. Nat Rev Immun. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]; (d) Cheek S, Krishna SS, Grishin NV. J Mol Biol. 2006;359:215–237. doi: 10.1016/j.jmb.2006.03.017. [DOI] [PubMed] [Google Scholar]; (e) Lin SL, Nussinov R. Nature Struct Biol. 1995;2:835–837. doi: 10.1038/nsb1095-835. [DOI] [PubMed] [Google Scholar]; (f) Gause GF, Brazhnikova MG. Nature. 1944;154:703. [Google Scholar]
- 4.Selected examples: Ghadiri MR, Choi C. J Am Chem Soc. 1990;112:1630–1632.Ruan FQ, Chen YQ, Hopkins PB. J Am Chem Soc. 1990;112:9403–9404.Tatko CD, Waters ML. J Am Chem Soc. 2002;124:9372–9373. doi: 10.1021/ja0262481.Kelso MJ, Beyer RL, Hoangt HN, Lakdawala AS, Snyder JP, Oliver WV, Robertson TA, Appleton TG, Fairlie DP. J Am Chem Soc. 2004;126:4828–4842. doi: 10.1021/ja037980i.Blackwell HE, Grubbs RH. Angew Chem Int Ed Engl. 1998;37:3281–3284. doi: 10.1002/(SICI)1521-3773(19981217)37:23<3281::AID-ANIE3281>3.0.CO;2-V.Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. J Am Chem Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191.Nowick JS. Acc Chem Res. 2008;41:1319–1330. doi: 10.1021/ar800064f.Fujimoto K, Kajino M, Inouye M. Chem Eur J. 2008;14:857–863. doi: 10.1002/chem.200700843.Liu J, Wang D, Zheng Q, Lu M, Arora PS. J Am Chem Soc. 2008;130:4334–4337. doi: 10.1021/ja077704u.Wang D, Chen K, Kulp JL, III, Arora PS. J Am Chem Soc. 2006;128:9248–9256. doi: 10.1021/ja062710w.Haney CM, Loch MT, Horne WS. Chem Commun. 2011 doi: 10.1039/C1CC12010G.Davies JS. J Pept Sci. 2003;9:471–501. doi: 10.1002/psc.491.Miller SJ, Blackwell HE, Grubbs RH. J Am Chem Soc. 1996;118:9606–9614.Schafmeister CE, Po J, Verdine GL. J Am Chem Soc. 2000;122:5891–5892.Brunel FM, Dawson PE. Chem Commun. 2005:2552–2554. doi: 10.1039/b419015g.Judice JK, Tom JY, Huang W, Wrin T, Vennari J, Petropoulos CJ, McDowell RS. Proc Natl Acad Sci USA. 1997;94:13426–13430. doi: 10.1073/pnas.94.25.13426.Sia SK, Carr PA, Cochran AG, Malashkevich VN, Kim PS. Proc Natl Acad Sci USA. 2002;99:14664–14669. doi: 10.1073/pnas.232566599.Balaram H, Uma K, Balaram P. Int J Peptide Protein Res. 1990;35:495–500. doi: 10.1111/j.1399-3011.1990.tb00253.x.Holland-Nell K, Meldal M. Angew Chem. 2011;123:5310–5312. doi: 10.1002/anie.201005846.Liu C, Sawaya MR, Cheng PN, Zheng J, Nowick JS, Eisenberg D. J Am Chem Soc. 2011;133:6736–6744. doi: 10.1021/ja200222n.
- 5.(a) Hutchinson EG, Sessions RB, Thornton JM, Woolfson DN. Protein Sci. 1998;7:2287–2300. doi: 10.1002/pro.5560071106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wouters MA, Curmi PMG. Proteins: Struct Funct Genet. 22:119–131. doi: 10.1002/prot.340220205. [DOI] [PubMed] [Google Scholar]; (c) Lifson S, Sander C. J Mol Biol. 1980;139:627–639. doi: 10.1016/0022-2836(80)90052-2. [DOI] [PubMed] [Google Scholar]; (d) Richardson JS. Advanc Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]; (e) Harrison PM, Sternberg MJ. J Mol Biol. 1996;264:603–623. doi: 10.1006/jmbi.1996.0664. [DOI] [PubMed] [Google Scholar]; (f) Srinivisan N, Sowdhamini R, Ramakrishnan C, Balaram P. Int J Pept Protein Res. 1990;36:147–155. doi: 10.1111/j.1399-3011.1990.tb00958.x. [DOI] [PubMed] [Google Scholar]; (g) Mao B. J Am Chem Soc. 1988;111:6132–6136. [Google Scholar]; (h) Chou PY, Fasman GD. Biochemistry. 1974;13:221–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]; (i) Levitt M. Biochemistry. 1978;17:4277–4285. doi: 10.1021/bi00613a026. [DOI] [PubMed] [Google Scholar]
- 6.Selected examples: Cochran AG, Tong RT, Starovasnik MA, Park EJ, McDowell RS, Theaker JE, Skelton NJ. J Am Chem Soc. 2001;123:625–632. doi: 10.1021/ja003369x.Gunasekaran K, Ramakrishnan C, Balaram P. Protein Eng. 1997;10:1131–1141. doi: 10.1093/protein/10.10.1131.Santiveri CM, Leon E, Rico M, Jimenez MA. Chemistry. 2008;14(2):488–499. doi: 10.1002/chem.200700845.Martinek TA, Fulop F. Eur J Biochem. 2003;270:3657–3666. doi: 10.1046/j.1432-1033.2003.03756.x.Mirassou Y, Santiveri CM, Perez de Vega MJ, Gonzalez-Muniz R, Jiminez MA. ChemBio- Chem. 2009;10:902–910. doi: 10.1002/cbic.200800834.
- 7.Fooks HM, Martin AC, Woolfson DN, Sessions RB, Hutchinson EG. J Mol Biol. 2006;356:32–44. doi: 10.1016/j.jmb.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 8.(a) Fisk JD, Gellman SH. J Am Chem Soc. 2001;123:343–344. doi: 10.1021/ja002493d. [DOI] [PubMed] [Google Scholar]; (b) Fisk JD, Powell DR, Gellman SH. J Am Chem Soc. 2000;122:5443–5447. [Google Scholar]; (c) Fisk JD, Schmitt MA, Gellman SH. J Am Chem Soc. 2006;128:7148–7149. doi: 10.1021/ja060942p. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Freire F, Gellman SH. J Am Chem Soc. 2009;131:7970–7972. doi: 10.1021/ja902210f. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Freire F, Almeida AM, Fisk JD, Steinkruger JD, Gellman SH. Angewandte Chemie. 2011;50:8735–8738. doi: 10.1002/anie.201102986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.See the Supporting Information for details.
- 10.Selected examples: Baldwin RL. Biophys Chem. 1995;55:127–135. doi: 10.1016/0301-4622(94)00146-b.de Alba E, Jimenez MA, Rico M. J Am Chem Soc. 1997;119:175–183.Syud FA, Stanger HE, Gellman SH. J Am Chem Soc. 2001;123:8667–8677. doi: 10.1021/ja0109803.Ciani B, Jourdan M, Searle MS. J Am Chem Soc. 2003;125:9038–9047. doi: 10.1021/ja030074l.Rai R, Raghothama S, Balaram P. J Am Chem Soc. 2006;128:2675–2681. doi: 10.1021/ja056861v.Rieman AL, Waters ML. J Am Chem Soc. 2009;131:14081–14087. doi: 10.1021/ja9047575.Kier BL, Shu I, Eidenschink LA, Andersen NH. Proc Nat Acad Sci. 2010;107:10466–10471. doi: 10.1073/pnas.0913534107.Eidenschink L, Crabbe E, Andersen NH. Biopolymers. 2009;91:557–564. doi: 10.1002/bip.21177.
- 11.Wishart DS, Sykes BD, Richards FM. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.