Abstract
In vertebrate somatic cells, the centrosome functions as the major microtubule-organizing center (MTOC), which splits and separates to form the poles of the mitotic spindle. However, the role of the centriole-containing centrosome in the formation of bipolar mitotic spindles continues to be controversial. Cells normally containing centrosomes are still able to build bipolar spindles after their centrioles have been removed or ablated. In naturally occurring cellular systems that lack centrioles, such as plant cells and many oocytes, bipolar spindles form in the complete absence of canonical centrosomes. These observations have led to the notion that centrosomes play no role during mitosis. However, recent work has re-examined spindle assembly in the absence of centrosomes, both in cells that naturally lack them and those that have had them experimentally removed. The results of these studies suggest that an appreciation of microtubule network organization, both before and after nuclear envelope breakdown (NEB), is the key to understanding the mechanisms that regulate spindle assembly and the generation of bipolarity.
Key words: centrosome, centriole, mitosis, spindle, cell cycle, meiosis, plant cell, microsurgery
Introduction
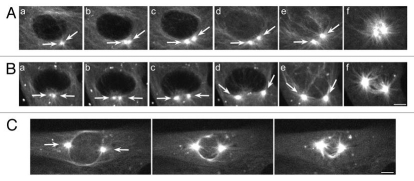
An ongoing question in cell biology is the role played by the centriole-containing centrosome in establishing the polarity of the mitotic spindle.1–3 In dividing vertebrate somatic cells, the major microtubule-organizing center (MTOC) is the centrosome, an organelle consisting of a pair of centrioles surrounded by a matrix of pericentriolar material (PCM).4 The MTOC generates a polarized array of microtubules (MTs) with their slow-growing minus ends concentrated at the cell center and their dynamic plus ends facing the cell periphery.5 Centrosomes undergo a duplication process during interphase, where the centrioles replicate in a semi-conservative fashion, resulting in two closely associated centriole pairs (called diplosomes) surrounded by a common PCM.4,6–8 The two pairs of diplosomes then disjoin, split apart and are separated prior to the onset of mitosis.6 As these daughter centrosomes move apart, they drive the separation of the single interphase MT focus into a pair of distinct MT arrays called asters (“stars”) in preparation for mitotic spindle assembly.3,9,10 As a unit, the pair of separated astral MT arrays has been referred to as the amphiaster (a “star on both sides”),11,12 though this term has also been linked to a distinct stage in the cell cycle of marine invertebrates, most notably, sea urchin zygotes.13 The extent of aster separation prior to nuclear envelope breakdown (NEB) varies (Fig. 1), but even modest aster separation does not affect the ability of cells to form a bipolar spindle.14
Figure 1.
Amphiastral spindle assembly in control BSC-1 cells. (A) The separation of the centrosomes is limited prior to NEB. The duplicated interphase centrosomes (part a, arrows) split apart but do not separate extensively (b–d, arrows). As the nuclear envelope breaks down (e and f), the bipolar spindle assembles. (B) The duplicated interphase centrosomes (part a, arrows) split and separate further apart (b–d, arrows). As the nuclear envelope breaks down (e and f), the bipolar spindle assembles. (C) The duplicated centrosomes (arrows) have migrated to opposite poles prior to nuclear envelope. GFP-α tubulin, fluorescence optics. Bar = 5 µm.
Coordinate with NEB, the asters use dynein to recruit microtubule-cross-linking proteins, such as NuMA released from the nucleus, to the focused MT minus ends at the spindle poles.15 Meanwhile the dynamic plus ends of the MTs interact with and cross-link interpolar MTs emanating from the opposite pole.16–18 As the asters separate, the forces of opposing motor proteins dynein and kinesin 5 drive the spindle poles apart and stabilize the anti-parallel MTs as the bipolar mitotic spindle begins to form.19–24 Forces intrinsic to each aster can also drive the poles apart through interactions between astral MTs and the actin, myosin II and dynein at the cell cortex.25–27 After NEB, MTs are also nucleated on or around mitotic chromosomes, producing extra-centrosomal polymers that must be captured and organized into the developing bipolar spindle.10,28–30 Finally, kinetochore MTs may also serve to drive apart the centrosomes and help finalize the establishment of spindle bipolarity.31
While the centrosome in vertebrate somatic cells has historically been viewed as necessary for both the organization of the interphase MT array and for the assembly of the bipolar mitotic spindle,32 it has been shown experimentally to be dispensable for both of these processes. First, microsurgery can generate centrosome- free cell fragments or “microcells,” which reorganize their random MTs into a central radial array with conventional ± MT polarity.33,34 It is important to note that this acentrosomal organization of interphase MTs may be mediated by the trans-Golgi network, which serves to organize non-centrosomal MTs, even in centrosome-containing cells.35–37 Other factors implicated in aMTOC organization include cytoplasmic dynein, dynactin and pericentrin.38
Second, laser ablation of centrosomes in mammalian somatic cells just prior to the onset of mitosis results in the release of the MTs into the mitotic cytoplasm.39 Initially, these free MTs are randomly oriented, but they rapidly become bundled and polarized by interactions with the mitotic chromosomes, forming a functional bipolar mitotic spindle.
Acentrosomal spindle formation in vertebrate cells takes place via a mechanism that is consistent with that used during spindle assembly in centrosome-free Xenopus egg cytoplasmic extracts.40,41 In both cases, chromosome-mediated organization of random MTs occurs via the Ran-GTP pathway,42–45 which activates spindle assembly factors, like TPX2, which bundle free MTs in the vicinity of the chromosomes; these become sorted into a bipolar spindle.46,47 The chromatinmediated spindles form essentially inside out, from the center to the poles. The acentrosomal poles lack extensive astral arrays but otherwise assemble properly into a bipolar configuration and with astonishingly high fidelity.39,40
Acentrosomal spindles have also been shown to form in experimentally manipulated crane fly spermatocytes,48 Drosophila embryos,49 cultured fly cells,50 and artificially activated sea urchin zygotes.51 A most notable case for experimentally-derived acentrosomal mitosis is found in mutant Drosophilia that completely lack centrioles or distinct centrosomes, yet can develop into morphologically normal adult animals, albeit ones lacking the cilia and flagella that are dependant on centrioles for their basal bodies.52,53
Further evidence supporting the case that centrosomes are not required for spindle assembly comes from observations in cell types that naturally lack canonical centrosomes, such as certain oocytes and all of the cells in higher plants.54–56 The assumption has been that these cells assemble bipolar spindles via the chromatin-mediated pathway, and that this mechanism is masked in centriolecontaining cells. The chromatin-mediated pathway appears to be able to compensate for the loss of the centrosome.
This then begs the question: “Are centrosomes actually necessary for bipolar mitotic spindle assembly at all?” The aforementioned experiments would conclude that they are not. Fully functional spindles can form in their absence, although there can be other mitotic and cytokinetic defects in spindles lacking functional centrosomes.57–59 Taken together, the results of experimental manipulation, which rid cells of their resident centrosomes, as well as observations in cells that never had centrosomes in the first place has supported the notion that centrosomes do not actually play a role in building a normal mitotic spindle. Rather, a contemporary view suggests these organelles are merely passengers “brought along for the ride,” with their major role being to template the formation of cilia and flagella; the centrioles that reside at the core of the centrosome also serve as the basal body, which nucleates the microtubules of the axoneme.60,61 Therefore, it is reasonable to conclude that the reproduction, splitting and segregation of diplosomes during the cell cycle is simply a mechanism to ensure the proper distribution of ciliary basal bodies to the daughter cells at each division (discussed in refs. 1 and 2).
Microtubule Reorganization during Spindle Assembly
Before the centrosome can be excluded from its potential role in spindle assembly, a major issue must first be addressed, namely, the mechanics of MT reorganization during the transition from interphase into mitosis and how this reorganization effects the establishment of spindle polarity. This is a key point, because the organization of the MT network in vertebrate cells remains intact throughout the cell cycle, and though it can be extensively remodeled, the basic polarity of MT plus ends at the centers and MT minus ends at the periphery persists.
It is important to remember that in vertebrate somatic cells, the establishment of spindle polarity begins well before the onset of NEB. The splitting of the MTOC into a pair of astral arrays takes place prior to NEB, though the extent to which this splitting occurs varies between cell types, and also within a population of cells.3,14,26 While it has been traditionally held that this splitting requires the disjunction and separation of the duplicated centrosomes,32 it is not clear that the presence of centrosomes is absolutely required for the formation of the amphiaster. What is clear is that, in these cells, the polarized organization of the MT network is maintained throughout the transition from interphase into mitosis. The MT minus ends remain at each MTOC, and the MT plus ends extend out toward the MTs from the opposite aster (Fig. 1). The MT network itself becomes highly reorganized during this transition. There is a dramatic increase in the MT-nucleating capacity of the MTOCs, coordinate with recruitment of κ-tubulin ring complexe.62 The long interphase MTs are severed by katanin,63 and the MT plus ends themselves increase their dynamic behavior, until the MT tips are stabilized by interactions with the kinetochores, the cell cortex, or they form into anti-parallel arrays.10,17 Any MTs released free into the mitotic cytoplasm are rapidly drawn into the nascent spindle poles through the action of cytoplasmic dynein.64
The observations that experimentally derived acentrosomal bipolar spindles form after NEB, as free MTs are bundled and sorted by activated SAFs in the region near chromosomes, has been extrapolated to suggest that centrosomes in vertebrate somatic cells do no real work at all during the early stages of mitosis. What is clear is that spindle assembly, at least in vertebrate somatic cells, does not involve the reorganization of randomly oriented, free cytoplasmic MTs. Instead, the process is dependant on maintaining the order and orientation of the preexisting MT network and reordering this network through a combination of MT severing, increased MT nucleation/dynamics, and the splitting of the MTOC into the amphiaster.
Both the splitting/separation of the MTOC and the increase in MT-nucleating capacity of the asters has been ascribed to the presence of the centrosome, but what about naturally occurring systems that do not have centrosomes in the first place? Do these acentrosomal cells organize and sort their MT arrays prior to NEB, or do they enter mitosis/meiosis with randomly oriented MTs that are then bundled, sorted and polarized by the chromatin-mediated pathway? An examination of the mechanisms used by traditionally “acentrosomal” cells may provide the answer.
Microtubule Organization in Plants
Flowering plants do not have canonical centrosomes, and, instead, create highly organized MT arrays in the absence of a centralized MTOC.55,65 In contrast to animal cells, plant cells completely disassemble their existing interphase MT network and assemble mitotic arrays de novo on either side of the nucleus at or just prior to NEB.65,66 Higher plants also lack obvious dynein and NuMA orthologs and do not focus their MT minus ends into compact spindle poles.66 However, while cells in higher plants lack centrosomes, emerging evidence suggests that they do generate distinct centers of MT organization coordinate with mitotic onset.65 The most widely recognized mitotic MT structure in plants is the preprophase band (PPB). The PPB is formed from cortical MTs67 that usually encircle the nuclear region and predict the division plane.65 In addition to the PPB, in many plant cells, there is another class of mitotic MTs that form prior to NEB. These are the polar cap microtubules (PCMTs), and they arise from the nuclear surface at opposite poles of the cell. Some PCMTs extend from the nuclear envelope to the cell cortex, and others enclose the nuclear surface.65 That these PCMTs form prior to NEB further suggests that bipolarization without interactions with mitotic chromatin is a mechanism conserved between animals and plants. Though plants re-order their MT network in a fashion distinct from vertebrate somatic cells, they do not appear to use chromatin-mediated MT nucleation/bundling as a primary source of spindle bipolarity, at least in somatic cells.65 Plant cells are still constrained by the requirement of building a spindle from polarized MTs, albeit those polymerized de novo.
Mouse Oocytes Meiotic Spindles
Mouse oocytes do not use a pair of centrosomes to sort their MT array28,54 and are a classical example of acentrosomal spindle assembly. However, recent work has shown that they do assemble multiple acentriolar MTOCs prior to NEB.68 On average, ∼80 MTOCs or asters form in each oocytes; these form both out in the cytoplasm and along the nuclear surface. The cytoplasmic asters were observed to migrate toward the nuclear region, and both cytoplasmic and nuclear asters contribute to spindle assembly. These multiple asters first assemble into a flexible multipolar spindle and then are sorted into a bi-astral array by the chromosomes in a RAN-GTP-dependent manner.68 This is reminiscent of spindle formation in Taxol-treated somatic cells, where multiple asters bundle into spindle poles, and the centrosomes have no connection to the asters.69 Again, this model for spindle assembly depends on organizing and sorting the MT array well before the nuclear envelope breaks down. Mouse oocytes must also sort their MT network during the transition into mitosis, but they use an alternative mechanism that does not rely on centrosomes.
Centrosomes in Drosophilia: To Be or Not to Be
Drosophilia meiosis also occurs in the absence of centrosomes,56 and, indeed, bipolarity is established after NEB, which along with C. elegans oocytes70 are examples of true chromatin-mediated spindle assembly found in nature.71,72 However, like most other animals flies use the paternally-inherited centrosome to organize the poles of the mitotic spindle during development and in the adult animal. Surprisingly, certain Drosophilia mutants can be generated that completely lack centrosomes, yet these can still develop,49 even forming morphologically normal adults, though ones lacking the cilia and flagella dependant on the presence of basal bodies.52 These studies provide the strongest evidence yet that a cell type that normally contains centrosomes can form bipolar spindles in their absence, suggesting that centrosomes are not necessary to establish spindle polarity.
However, recent studies have examined the organization of the MT network in fly cells as they transition into mitosis and have revealed that flies do not make extensive use of their centrosomes to organize the interphase MTs.53 Instead, Drosophilia cells normally lack a centralized focus of MT minus ends,73 and at least some of their MTs are organized by acentriolar MTOCs (aMTOCs74). In the absence of radial MT organization during the interphase-mitotic transition, persistent monopolar spindles would not be expected to form, and indeed, these are not observed.50 In fact, Drosophilia urchin mutants can form a bipolar spindle with one astral pole and one pole that is anastral.75 This suggests that the centrosome has failed to split and forms the astral array, and free MTs in the cytoplasm bundle and sort via the chromatin-mediated pathway to build the second, anastral pole. Without the need to manage the splitting of the MT network, the chromatin-mediated spindle assembly pathway appears to be able to function freely during mutant Drosophila development, resulting in morphologically normal adult flies, albeit those that lack centriole-dependant structures, such as cilia and flagella.1
Sea Urchin Zygotes
In sea urchin zygotes the mitotic spindle poles are established using the paternally inherited centrosome.76 Following fertilization the male and female pronuclei fuse together, and the centrosome provided by the sperm basal body duplicates and then splits apart to organize the mitotic spindle poles.13,76 The female centrosome used during oogenesis is degraded prior to fertilization.77
Interestingly, artificially activated sea urchin zygotes, which lack centrioles or centrosomes, assemble acentrosomal bipolar spindles, presumably via the chromosome-mediated spindle assembly pathway.51 These spindles are anastral and capable of undergoing anaphase. Thus, parthenogenic sea urchin zygotes are fully capable of chromatin-mediated spindle assembly when centrosomes are lacking.
However, the results of an interesting study in sea urchins suggest that the interrelationship between centrosomes, chromosomes and MTs is more complex. Experimental separation of the pronuclei before syngamy revealed that only the male chromosomes (with their associated centrosomes) could form a spindle. The female chromosomes, which lack a centrosome, could not bundle MTs nor form a spindle.78 The recent work of Henson and colleagues reveals that sea urchin zygotic chromosomes are fully capable of supporting RAN-induced MT bundling and sorting necessary to generate a function, anastral spindle. Then why do isolated female chromosomes fail to do so, while sharing common mitotic cytoplasm with the male chromosomes/centrosomes? The answer may be that the male-derived centrosomes nucleate and organize essentially all of the MTs within this common cytoplasm, depriving the female chromosomes of any free MTs needed to build an anastral spindle. Thus, it is not merely sufficient for chromosomes to have MT nucleating and/or bundling activity. They must also have access to free MTs.
Mammalian Cultured Cells
Unlike higher plants or meiotic systems, vertebrate somatic cells have an organized MT arrangement with a single MTOC and a polarized, radial array of MTs emanating from it. Much like the aforementioned experiments in sea urchins, it is not clear how vertebrate somatic cells lacking centrosomes but retaining a polarized MT array emanating from a single MTOC would behave as they progressed through the cell cycle into mitosis.
To address this, we recently published the results of a study in which the centrosome was removed from monkey kidney (BSC-1) cells using microsurgery.38 These acentrosomal cells (termed Karyoplasts) were generated during early interphase, and they had sufficient time to re-organize an acentrosomal MTOC (aMTOC) before entering into mitosis, much like that observed in enucleate cell fragments.33,34 By using cells constitutively expressing α-tubulin-GFP, we were able to use live-cell imaging to examine the mode of spindle assembly in these cells. We found that in a vast majority of karyoplasts (∼65%), the aMTOC underwent splitting and separation before NEB, forming an amphiaster. These cells built bipolar spindles that almost exclusively underwent mitosis and cytokinesis with relatively normal timings. This revealed that the centrosomes are not absolutely required to organize an MTOC, nor to bias its splitting into an amphiaster, thus supporting the case that centrosomes are not required to form bipolar mitotic spindles.
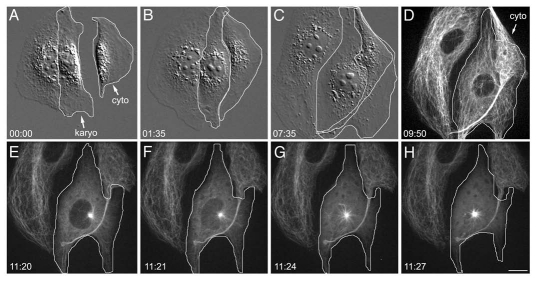
However, the fate of the other karyoplasts reveals an important function for centrosomes. In ∼35% of karyoplasts, the aMTOC never splits, and a monopolar spindle forms that persists throughout mitosis (Fig. 2). In these cells, the MTs remain organized, and thus, there are no free MTs capable of interacting with the chromatin and forming a second pole. So while vertebrate cultured cells may have the means to form chromatin-mediated spindles, if the MTs remain organized by a single MTOC, then there are no free MTs available to assemble this pole. Interestingly, these monopolar cells eventually exit mitosis and undergo cytokinesis with multiple cleavage furrows, which eventually retract, yielding a single tetraploid daughter cell.
Figure 2.
Behavior of acentrosomal cell undergoing monopolar division. (A) MT organization following microsurgery. (A–C) Microsurgery and flattening of the karyoplast-cytoplast pair (surrounded by white outline). (D) Confocal fluorescence image of interphase karyoplast (white outline). There are MTs throughout the karyoplast. (E–H) Monopolar spindle formation. The single MT focus (E) is directly adjacent to the nuclear envelope. As the interphase MT network disassembles, cytoplasmic MTs are being drawn into the monopole. The nuclear envelope breaks down (F) and a monopolar spindle forms (H). GFP-α tubulin, fluorescence optics. Bar = 10 µm.
This study clearly reveals the importance of centrosomes for bipolar spindle assembly. They ensure that the MT array is properly split into two. The consequence for centrosomal loss is a dramatic increase in the number of persistent monopolar spindles.
Equally important is the idea that cells that normally contain centrosomes can eventually form bipolar spindles, even if they enter mitosis with a monopole built on partially separated centrosomes. Such is the case for monopolar spindles assembled in the presence of the Eg5 (kinesin 5) inhibitor monastrol. These cells form monopoles that will persist until cytokinesis.79,80 If the inhibitor is washed out, then these cells eventually bipolarize.22,79 However, in karyoplasts, the monopoles persist, and bipolarization after NEB was extremely rare (∼1% of cases examined38). This suggests that centrosomes play a key role in ensuring spindle bipolarity, even after the onset of mitosis, by acting as a focal point for MT nucleation. Because there are two such focal points (duplicated centrosomes), correcting polarity errors is simply the process of driving these focal points apart.
What then is the role of the centrosome during mitotic spindle assembly? We hypothesize that it serves to bias the splitting and separation of the MT network at the time of mitotic onset (Fig. 3). In the absence of a centrosome, the reformed MT can still become separated into two distinct asters. However, this process is inefficient and error prone. There is also no opportunity for the spindle to bipolarize once the cell has progressed in mitosis, as the monopole is dominant unlike the case of Drosophilia, where an anastral pole can form along with an astral pole.75
Figure 3.
Model for spindle pole separation in centriole-containing vs. acentrosomal cells. (A) Centriole-containing cells. The duplicated centrosome contains a connected pair of diplosomes (red) surrounded by a common PCM (green). The pairs of centrioles split and separate along the nuclear envelope; each organizes a separate MT array that will become the poles of the spindle. (B) Bipolar spindle assembly in an acentriolar karyoplast. The PCM is focused but lacks centrioles. The focus splits and separates as above, and a pair of spindle poles forms. (C) Monopolar spindle assembly in an acentriolar karyoplast. The single focus of PCM fails to split and separate, and the cell enters mitosis with a single MT array; this array cannot bipolarize, because all of the MT minus ends are held in place by dynein and NuMA.
The results of our study in karyoplasts have also left several unanswered questions. First, what are the differences between the aMTOCs that bipolarize and those that don't? Is it merely a case of inefficiency, with an aMTOC struggling to split and separate the MT network in the absence of a centrosome? Or is the answer more complicated? One intriguing possibility is that the extent to which the Golgi reforms could influence the ability of the aMTOC to form, split and separate. In our experiments, we could not control for removal of the Golgi apparatus; our observations post-microsurgery revealed that varying amounts of Golgi remained in the karyoplasts. These Golgi remnants would reform to morphologically “normal” Golgi stacks, but it is not clear what “normal” means in this context. Perhaps there is some lower limit of Golgi organization that influences the size, composition or function of the aMTOC. In normal (i.e., untreated) cells, the centrosome may contribute to the morphology of the Golgi and vice versa. The amount of Golgi considered normal ensures that there is a robust organization of the MT network, and this organization provides for the accurate splitting of the centrosomes at the G2/M transition. In experimentally acentrosomal cells (karyoplasts), the Golgi reforms but may be altered in size, shape or composition. If this, in turn, influences the extent of aMTOC formation, then the MT network may not be able to reform to the degree necessary to support efficient splitting or separation at the onset of mitosis, particularly in the absence of centrosomes, which themselves provide a strong bipolarizing cue.32
Summary
In cells that do organize a focused interphase MT array, we find that centrosomes are necessary in order to bias the separation of the MT network during spindle assembly, in addition to forming cilia and flagella.1 Whereas additional centrosomes increase the likelihood of multipolar mitotic spindles,6 it appears that the loss of the centrosome transiently disrupts interphase MT organization but ultimately increases the risk of monopolar spindles, as the acentrosomal MTOC reforms in vertebrate somatic cells, but fails to split and separate at mitotic onset. Given our findings, it is not surprising that in cells without a centralized interphase MT network, such as flowering plants, flies and mouse oocytes, the centrosome is either completely lacking or dispensable for building spindle poles. These cells have simply evolved mechanisms that allow them to do without.
Acknowledgments
The author would like to thank Dr. Sidney Shaw for insightful comments on mitotic spindle assembly in plants, Dr. Kevin Vaughan for assistance with all things dynein and Mr. Kul Karanjeet for expert technical assistance. The work in the author's lab is supported by a grant from the NIH (GM072754).
References
- 1.Marshall WF. What is the function of centrioles? J Cell Biochem. 2007;100:916–922. doi: 10.1002/jcb.21117. [DOI] [PubMed] [Google Scholar]
- 2.Debec A, Sullivan W, Bettencourt-Dias M. Centrioles: active players or passengers during mitosis? Cell Mol Life Sci. 2010;67:2173–2194. doi: 10.1007/s00018-010-0323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanenbaum ME, Medema RH. Mechanisms of centrosome separation and bipolar spindle assembly. Dev Cell. 2010;19:797–806. doi: 10.1016/j.devcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/S0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- 5.Li R, Gundersen GG. Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat Rev Mol Cell Biol. 2008;9:860–873. doi: 10.1038/nrm2522. [DOI] [PubMed] [Google Scholar]
- 6.Hinchcliffe EH, Sluder G. “It takes two to tango”: Understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 2001;15:1167–1181. doi: 10.1101/gad.894001. [DOI] [PubMed] [Google Scholar]
- 7.Rodrigues-Martins A, Riparbelli M, Callani G, Glover DM, Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007;316:1046–1050. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- 8.Holland AJ, Lan W, Cleveland DW. Centriole duplication: A lesson in self-control. Cell Cycle. 2010;9:2731–2736. doi: 10.4161/cc.9.14.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wadsworth P, Khodjakov A. E pluribus unum: towards a universal mechanism for spindle assembly. Trends Cell Biol. 2004;14:413–419. doi: 10.1016/j.tcb.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. Int Rev Cytol. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- 11.Wilson EB. The cell in development and inheritance. New York NY: Macmillan; 1896. [Google Scholar]
- 12.Taylor EW. Dynamics of spindle formation and its inhibition by chemicals. J Biophys Biochem Cytol. 1959;6:193–196. doi: 10.1083/jcb.6.2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris P, Osborn M, Weber K. Distribution of tubulin-containing structures in the egg of the sea urchin Strongylocentrotus purpuratus from fertilization through first cleavage. J Cell Biol. 1980;84:668–679. doi: 10.1083/jcb.84.3.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rieder CL. Formation of the astral mitotic spindle: ultrastructural basis for the centrosome-kinetochore interaction. Electron Microsc Rev. 1990;3:269–300. doi: 10.1016/0892-0354(90)90005-D. [DOI] [PubMed] [Google Scholar]
- 15.Manning AL, Compton DA. Structural and regulatory roles of nonmotor spindle proteins. Curr Opin Cell Biol. 2008;20:101–106. doi: 10.1016/j.ceb.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khmelinskii A, Schiebel E. Assembling the spindle midzone in the right place at the right time. Cell Cycle. 2008;7:283–286. doi: 10.4161/cc.7.3.5349. [DOI] [PubMed] [Google Scholar]
- 17.Hornick JE, Karanjeet KB, Collins ES, Hinchcliffe EH. Kinesins to the core: The role of microtubule-based motor proteins in building the mitotic spindle midzone. Semin Cell Dev Biol. 2010;21:290–299. doi: 10.1016/j.semcdb.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pal D, Wu D, Haruta A, Matsumura F, Wei Q. Role of a novel coiled-coil domain-containing protein CCDC69 in regulating central spindle assembly. Cell Cycle. 2010;9:4117–4129. doi: 10.4161/cc.9.20.13387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaisberg EA, Koonce MP, McIntosh JR. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walczak CE, Vernos I, Mitchison TJ, Karsenti E, Heald R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr Biol. 1998;8:903–913. doi: 10.1016/S0960-9822(07)00370-3. [DOI] [PubMed] [Google Scholar]
- 21.Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- 22.Ferenz NP, Paul R, Fagerstrom C, Mogilner A, Wadsworth P. Dynein antagonizes Eg5 by cross-linking and sliding antiparallel microtubules. Curr Biol. 2009;19:1833–1838. doi: 10.1016/j.cub.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanneste D, Takagi M, Imamoto N, Vernos I. The Role of Hklp2 in the stabilization and maintenance of spindle bipolarity. Curr Biol. 2009;19:1712–1717. doi: 10.1016/j.cub.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 24.Gatlin JC, Matov A, Groen AC, Needleman DJ, Maresca TJ, Danuser G, et al. Spindle fusion requires dynein-mediated sliding of oppositely oriented microtubules. Curr Biol. 2009;19:287–296. doi: 10.1016/j.cub.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waters JC, Cole RW, Rieder CL. The force-producing mechanism for centrosome separation during spindle formation in vertebrates is intrinsic to each aster. J Cell Biol. 1993;122:361–372. doi: 10.1083/jcb.122.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenblatt J. Spindle assembly: asters part their separate ways. Nat Cell Biol. 2005;7:219–222. doi: 10.1038/ncb0305-219. [DOI] [PubMed] [Google Scholar]
- 27.Moore JK, Cooper JA. Coordinating mitosis with cell polarity: Molecular motors at the cell cortex. Semin Cell Dev Biol. 2010;21:283–289. doi: 10.1016/j.semcdb.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karsenti E, Vernos I. The mitotic spindle: A self-made machine. Science. 2001;294:543–547. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- 29.Tulu US, Rusan NM, Wadsworth P. Peripheral, non-centrosome-associated microtubules contribute to spindle formation in centrosome-containing cells. Curr Biol. 2003;13:1894–1899. doi: 10.1016/j.cub.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Gadde S, Heald R. Mechanisms and molecules of the mitotic spindle. Curr Biol. 2004;14:797–805. doi: 10.1016/j.cub.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 31.Toso A, Winter JR, Garrod AJ, Amaro AC, Meraldi P, McAinsh AD. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J Cell Biol. 2009;184:365–372. doi: 10.1083/jcb.200809055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazia D. The chromosome cycle and the centrosome cycle in the mitotic cycle. Int Rev Cytol. 1987;100:49–92. doi: 10.1016/S0074-7696(08)61698-8. [DOI] [PubMed] [Google Scholar]
- 33.McNiven MA, Porter KR. Organization of microtubules in centrosome-free cytoplasm. J Cell Biol. 1988;106:1593–1605. doi: 10.1083/jcb.106.5.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodionov VI, Borisy GG. Self-centering activity of cytoplasm. Nature. 1997;386:170–173. doi: 10.1038/386170a0. [DOI] [PubMed] [Google Scholar]
- 35.Rıos RM, Sanchís A, Tassin AM, Fedriani C, Bornens M. GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell. 2004;118:323–335. doi: 10.1016/j.cell.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 36.Efimov A, Kharitonov AA, Efimova N, Lonarek J, Miller PM, Andreyeva N, et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-golgi network. Dev Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sütterlin C, Colanzi A. The Golgi and the centrosome: building a functional partnership. J Cell Biol. 2010;188:621–628. doi: 10.1083/jcb.200910001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hornick JE, Mader CC, Tribble EK, Bagne CC, Vaughan KT, Shaw SL, et al. Amphiastral mitotic spindle assembly in vertebrate cells lacking centrosomes. Curr Biol. 2011;21:598–605. doi: 10.1016/j.cub.2011.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/S0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- 40.Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, et al. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 41.Heald R, Tournebize R, Habermann A, Karsenti E, Hyman A. Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J Cell Biol. 1997;138:615–628. doi: 10.1083/jcb.138.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- 43.Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- 44.Kaláb P, Pu RT, Dasso M. The ran GTPase regulates mitotic spindle assembly. Curr Biol. 1999;9:481–484. doi: 10.1016/S0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- 45.Kaláb P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 46.Gruss OJ, Wittman M, Yokoyama H, Pepperkok R, Kufer T, Sillje H, et al. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat Cell Biol. 2002;4:871–879. doi: 10.1038/ncb870. [DOI] [PubMed] [Google Scholar]
- 47.Loughlin R, Heald R, Nedelec F. A computational model predicts Xenopus meiotic spindle organization. J Cell Biol. 2010;191:1239–1249. doi: 10.1083/jcb.201006076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steffen W, Fuge H, Dietz R, Bastmeyer M, Muller G. Asterfree spindle poles in insect spermatocytes: Evidence for chromosome induced spindle formation. J Cell Biol. 1986;102:1679–1687. doi: 10.1083/jcb.102.5.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Megraw TL, Kao LR, Kaufman TC. Zygotic development without functional mitotic centrosomes. Curr Biol. 2001;11:116–120. doi: 10.1016/S0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- 50.Mahoney NM, Goshima G, Douglass A, Vale RD. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr Biol. 2006;16:564–569. doi: 10.1016/j.cub.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 51.Henson JH, Fried CA, McClellan MK, Ader J, Davis JE, Oldenbourg R, et al. Bipolar, anastral spindle development in artificially activated sea urchin eggs. Dev Dyn. 2008;237:1348–1358. doi: 10.1002/dvdy.21533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, et al. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 53.Gogendeau D, Basto R. Centrioles in flies: The exception to the rule? Semin Cell Dev Biol. 2010;21:163–173. doi: 10.1016/j.semcdb.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 54.Szollosi D, Calarco P, Donahue RP. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J Cell Sci. 1972;11:521–541. doi: 10.1242/jcs.11.2.521. [DOI] [PubMed] [Google Scholar]
- 55.Wolniak SM. Patterns of regulation during mitosis. In: Lloyd CW, editor. In “The cytoskeletal basis of plant growth and form”. London: Academic Press; 1991. pp. 209–226. [Google Scholar]
- 56.Theurkauf WE, Hawley RS. Meiotic spindle assembly in Drosophila females: Behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J Cell Biol. 1992;116:1167–1180. doi: 10.1083/jcb.116.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khodjakov A, Rieder CL. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J Cell Biol. 2001;153:237–242. doi: 10.1083/jcb.153.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hinchcliffe EH, Miller FJ, Cham M, Khodjakov A, Sluder G. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 2001;291:1547–1550. doi: 10.1126/science.1056866. [DOI] [PubMed] [Google Scholar]
- 59.Zamora I, Marshall WF. A mutation in the centriole associated protein centrin causes genomic instability via increased chromosome loss. BMC Biol. 2005;3:15. doi: 10.1186/1741-7007-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- 61.Rosales JL, Rattner JB, Lee KY. Cdk5 in the centriolar appendages mediates cenexin1 localization and primary cilia formation. Cell Cycle. 2010;9:2037–2039. doi: 10.4161/cc.9.10.11600. [DOI] [PubMed] [Google Scholar]
- 62.Khodjakov A, Rieder CL. The sudden recruitment of gamma-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J Cell Biol. 1999;146:585–596. doi: 10.1083/jcb.146.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roll-Mecak A, McNally FJ. Microtubule-severing enzymes. Curr Opin Cell Biol. 2010;22:96–103. doi: 10.1016/j.ceb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rusan NM, Tulu US, Fagerstrom C, Wadsworth P. Reorganization of the microtubule array in prophase/prometaphase requires cytoplasmic dynein-dependent microtubule transport. J Cell Biol. 2002;158:997–1003. doi: 10.1083/jcb.200204109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lloyd C, Chan J. Not so divided: the common basis of plant and animal cell division. Nat Rev Mol Cell Biol. 2006;7:147–152. doi: 10.1038/nrm1831. [DOI] [PubMed] [Google Scholar]
- 66.Bannigan A, Lizotte-Waniewski M, Riley M, Baskin TI. Emerging molecular mechanisms that power and regulate the anastral mitotic spindle of flowering plants. Cell Motil Cytoskeleton. 2008;65:1–11. doi: 10.1002/cm.20247. [DOI] [PubMed] [Google Scholar]
- 67.Shaw SL, Kamyar R, Ehrhardt DW. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science. 2003;300:1715–1718. doi: 10.1126/science.1083529. [DOI] [PubMed] [Google Scholar]
- 68.Schuh M, Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 2007;130:484–498. doi: 10.1016/j.cell.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 69.Hornick JE, Bader JR, Trimble K, Tribble E, Breunig JS, Halpin E, et al. Live-cell analysis of mitotic spindle formation in Taxol-treated cells. Cell Motil Cytoskeleton. 2008;65:595–613. doi: 10.1002/cm.20283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wignall SM, Villeneuve AM. Lateral microtubule bundles promote chromosome alignment during acentrosomal oocytes meiosis. Nat Cell Biol. 2009;11:839–844. doi: 10.1038/ncb1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matthies HJ, McDonald HB, Goldstein LS, Theurkauf WE. Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J Cell Biol. 1996;134:455–464. doi: 10.1083/jcb.134.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sköld HN, Komma DJ, Endow SA. Assembly pathway of the anastral Drosophila oocyte meiosis I spindle. J Cell Sci. 2005;118:1745–1755. doi: 10.1242/jcs.02304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rogers GC, Rusan NM, Peifer M, Rogers SL. A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol Biol Cell. 2008;19:3163–3178. doi: 10.1091/mbc.E07-10-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moutinho-Pereira S, Debec A, Maiato H. Microtubule cytoskeleton remodeling by acentriolar microtubule-organizing centers at the entry and exit from mitosis in Drosophila somatic cells. Mol Biol Cell. 2009;20:2796–2808. doi: 10.1091/mbc.E09-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilson PG, Fuller MT, Borisy GG. Monastral bipolar spindles: implications for dynamic centrosome organization. J Cell Sci. 1997;110:451–464. doi: 10.1242/jcs.110.4.451. [DOI] [PubMed] [Google Scholar]
- 76.Halpin ES, Hinchcliffe EH. Birth of the Cool: Using sea urchin zygotes to study centrosome duplication, cell division and cytokinesis. Signal Transduct. 2007;7:154–163. doi: 10.1002/sita.200600124. [DOI] [Google Scholar]
- 77.Sluder G, Miller FJ, Rieder CL. Reproductive capacity of sea urchin centrosomes without centrioles. Cell Motil Cytoskeleton. 1989;13:264–273. doi: 10.1002/cm.970130405. [DOI] [PubMed] [Google Scholar]
- 78.Sluder G, Reider CL. Experimental separation of pronuclei in fertilized sea urchin eggs: chromosomes do not organize a spindle in the absence of centrosomes. J Cell Biol. 1985;100:897–903. doi: 10.1083/jcb.100.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hu CK, Coughlin M, Field CM, Mitchison TJ. Cell polarization during monopolar cytokinesis. J Cell Biol. 2008;181:195–202. doi: 10.1083/jcb.200711105. [DOI] [PMC free article] [PubMed] [Google Scholar]