Abstract
Lipid-modified transcription factors (TFs) are biomolecular oddities, since their reduced mobility and membrane attachment appear to contradict nuclear import required for their gene-regulatory function. NFAT5 isoform a (selected from an in silico screen for predicted lipid-modified TFs) is shown to contribute about half of all endogenous expression of human NFAT5 isoforms in the isotonic state. Wild-type NFAT5a protein is, indeed, myristoylated and palmitoylated on its transport to the plasmalemma via the endoplasmic reticulum and the Golgi. In contrast, its lipid anchor-deficient mutants as well as isoforms NFAT5b/c are diffusely localized in the cytoplasm without preference to vesicular structures. Quantitative/live microscopy shows the plasma membrane-bound fraction of NFAT5a moving into the nucleus upon osmotic stress despite the lipid anchoring. The mobilization mechanism is not based on proteolytic processing of the lipid-anchored N terminus but appears to involve reversible palmitoylation. Thus, NFAT5a is an example of TFs immobilized with lipid anchors at cyotoplasmic membranes in the resting state and that, nevertheless, can translocate into the nucleus upon signal induction.
Key word: lipid-modified transcription factor, NFAT5, nuclear import, myristoylation, palmitoylation, osmotic stress, hypertonic stress, reversible palmitoylation
Introduction
Regulated shuttling between cytoplasm and the nucleus controls the availability of transcription factors (TFs) for gene expression induction. To enter the nucleus, the TFs are supposed to be soluble and mobile, yet TFs tied to cytoplasmic membranes with transmembrane helices do exist and are taxonomically widely spread.1 This paradox has been resolved with TF release via regulated intra-membrane proteolysis.2 Surprisingly, in silico screens for targets with potential lipid posttranslational modifications3–6 hit numerous proteins with known nucleic acid binding domains, among them many TFs. Are they all just computational artifacts, or do they have only cytoplasmic functions? A single example, the long, myristoylated isoform of OCA-B (accession NP_006226.2) has been experimentally studied. It was found to remain chained to the cytoplasmic compartment and to non-transcriptionally regulate SYK.7,8 To date, there is no biomolecular mechanism known that would import a lipid-modified TF into the nucleus in a regulated manner.
We wanted to understand whether nuclear import is possible for some of these potentially lipid modified TFs. For this purpose, the list of a few thousand sequences from the protein database having both a globular domain annotated as nucleic acid binding and a predicted lipid anchor site were reduced with plausibility criteria to a single candidate for experimental validation. We unselected all potentially incomplete sequences and limited ourselves to proteins from well-studied model organisms with evolutionary conserved predicted lipid anchor sites and an experimentally verified TF domain. Finally, we picked NFAT5a fromHomo sapiens (nuclear factor of activated T-cells 5, isoform a) with a predicted N-terminal myristoylation signal and cysteines close to the N-terminal glycine as potential palmitoylation sites.
NFAT5 [also called TonEBP (tonicity enhancer-binding protein) or OREBP (osmotic response element-binding protein)] is the only known osmo-sensitive transcription factor in mammalian cells with additional roles in immunity, infection, cancer, development and cell migration.9 Upon hyperosmotic stress, NFAT5 induces the transcription of osmo-protective genes, such as aldose reductase (AR, reduces glucose to sorbitol),10 betaine/γ-aminobutyric acid transporter (BGT1),11 taurine transporter (TauT),12 Na+-dependent myoinositol transporter (SMIT)13 andHSP70.14
Although the existence of various NFAT5 isoforms has become clear early, their potentially differential role in various biological processes has never been a target of investigation. In this work, we show that NFAT5a mobilization for nuclear import is mechanistically different from that of other NFAT5 isoforms. NFAT5a is myristoylated (at Gly2) and palmitoylated (at Cys5) in vitro and in vivo as predicted in silico. The lipid anchors affect subcellular localization since the wild-type (wt) form is transported to the plasma membrane (PM) via the endoplasmic reticulum (ER) and the Golgi, whereas the isoforms NFAT5b/c are diffusely spread within the cytoplasm without preference for intracellular vesicular systems. We show that the ability for myristoylation and palmitoylation is critical for NFAT5a accumulation at the PM. High salt stress initiates translocation of wt NFAT5a from the plasma membrane into the nucleus despite the lipid anchoring. Nuclear translocation of NFAT5a is not accompanied with proteolytical processing of the N-terminal sequence. We suggest a mechanism based on reversible palmitoylation for the regulated mobilization of NFAT5a for nuclear import.
Results
Isoforms of NFAT5 have an almost identical sequence architecture, but only isoform a can be N-terminally N-myristoylated.
The protein NFAT5a has been identified by an in silico screen for DNA-binding proteins with lipid anchors as described inSupplemental Material A. Knowing NFAT5 as the osmotic stress response TF provided us with initial hope that testing isoform a for nuclear import upon salt stress might be a functional assay.10,15
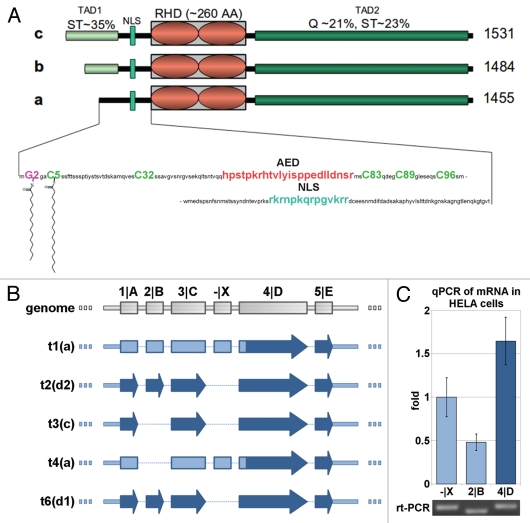
Human NFAT5 has first been identified via its isoform a.16–19 In the present protein database, five variants of the NFAT5 protein are described: (1) [NP_619728.2, 1,455 amino acids (AAs)], (2) (AAD48441, 1,484 AAs), (3) (O94916, 1,531 AAs), (4) (called “d1” in this work; NP_001106649, 1,548 AAs) and (5) another variant d2 differing from isoform d by an inserted alanine after A568 (NP_619727, 1,549 AAs). Except for this polymorphism, the C-terminal 1,455 AAs (i.e., the sequence of NFAT5a) are identical among all variants. All isoforms but isoform a have the same sequence architecture20–22 (Fig. 1A) including (1) a N-terminal compositionally biased region rich in serine/threonine (35% in first 76 AAs of NFAT5c) and proline [12% in first 76 AAs of NFAT5c; a most likely non-globular region of variable length TAD1 (transactivation domain 1)], (2) an AED (auxiliary export domain) and a NLS (nuclear localization signal),15 (3) a RHD (Rel homology domain, PFAM PF00554 and PF01833)23 mediating both protein-DNA interactions and NFAT5 dimerization17,24 and (4) a C-terminal low-complexity region (glutamine 21%, serine/threonine 23%) as a TAD2.25
Figure 1.
Protein isoforms, transcripts and expression of NFAT5. (A) Schematic alignment of NFAT5 isoforms a, b and c. The 3 main protein domains are two transactivation domains (TAD1/2) and a Rel homology domain (RHD). Tad1 contains a hypothetical auxiliary export domain (AED) followed by a nuclear localization sequence (NLS). The three isoforms differ in their N-terminal sequence. Only NFAT5a has the N-terminal glycine for myristoylation (purple). Possible cysteines for palmitoylation (green) are indicated.Supplemental Material A contains a Table of NFAT5 orthologs and their alignment. (B) Schematic representation of the mRNA architecture for all known NFAT5 transcripts. The respective protein isoforms are indicated next to the transcript number by their letter codes. (C) Endogenous NFAT5a mRNA expression in HeLa cells was tested with qPCR. All three mRNA segments targeted, namely, -|X (occurring in transcripts 1 and 4, seeTable 1 andSup. Material A for the aligment of the transcripts), exon 2|B (occurring in transcripts 1, 2 and 6) and exon 4|D (occurring in transcripts 1–4 and 6), were found to be present in the cells. The PCR products were analyzed on an agarose gel, and the bands were sequenced to confirm the identity of the respective mRNA segments.
Isoform a, the shortest among them, lacks the N-terminal TAD1 region and, instead, has a confidently predicted N-terminal N-myristoylation motif (a stretch of 17 amino acids with a N-terminal glycine, NMT/MyrPS score = 1.846 and p = 0.0005).3,26,27 Close to Gly2, there is a cysteine at position 5, and further downstream, there are a few more cysteines. Cys5 especially is a prime candidate for side chain acylation with a palmitoyl anchor given the vicinity of the myristoyl anchor.4,28
So far, the exact homolog of human NFAT5a is annotated in mouse (NP_598718), rat (EDL92475) and chicken (BAG70407). NFAT5a-containing conceptual translations are reported for many more proteomes in Mammalia, Aves and Amphibia (Sup. Material A). For all these sequences, the segments containing the myristoylation signal with Gly2 and the Cys5 palmitoylation site are strongly conserved.
Several NFAT5 isoforms including isoform a are concurrently expressed in the isotonic state, and isoform a contributes about half of all NFAT5 expression.
Despite their similar sequences, the mRNAs of the human NFAT5 isoforms have distinct differences.29,30Figure 1B shows a schematic view andTable 1 provides accession numbers and genome positional details of the five known transcripts and their intron and exon arrangements (see alignment inSup. Material A). We attempted to determine the relative expression of NFAT5a-related transcripts compared with other NFAT5 transcripts by quantitative real-time PCR (RT-PCR). For this purpose, we selected three RNA sequence segments for expression profiling.
Table 1.
Transcripts of NFAT5
| Location on chromosome 16 (NC_000016.9) | Transcripts: | |||||||
| Exon | Alignment indication | Start | Stop | Transcript 1 NM_138714.2 Isoform a | Transcript 2 NM_138713.2 Isoform d2 | Transcript 3 NM_006599.2 Isoform c | Transcript 4 NM_173214.1 Isoform a | Transcript 6 NM_001113178.1 soform d1 |
| 1 | A | 69600205 | 69600277 | + | + | + | + | + |
| 2 | B | 69602398 | 69602451 | + | + | absent | absent | + |
| 3 | C | 69660306 | 69660431 | + | + | + | + | + |
| X | 69680420 | 68680481 | + | absent | absent | + | absent | |
| 4 | D | 69680931 | 69681489 | + | + | + | + | + |
| 5 | E | 69687139 | 69687331 | + | + | + | + | + |
| 6 | F | 69689512 | 69689702 | + | + | + | + | + |
| 7 | G | 69693630 | 69693802 | + | + | + | + | + |
| 8 | H | 69703880 | 69704014 | + | + | + | + | + |
| 9 | I | 69704139 | 69704191 | + | + | + | + | + |
| 10 | K | 69711106 | 69711238 | + | + | + | + | + |
| 11 | L | 69718790 | 69718873 | + | + | + | + | shortened (start at 69718793) |
| 12 | M | 69724843 | 69724991 | + | + | + | + | + |
| 13 | N | 69725652 | 69728142 | + | + | + | + | + |
| 14 | P | 69729039 | 69729274 | + | + | + | + | + |
The data in this table complements the scheme in Figure 1B with genomic positional data of all five known transcripts of NFAT5.
Both transcripts 1 and 4 can only be translated into the protein isoform NFAT5a. They contain a unique sequence segment designated -|X in the 5′-untranslated region. The second segment selected is exon 2|B, which is part of the transcripts 1 (for NFAT5a), 2 (for NFAT5d2) and 6 (for NFAT5d1). All five transcripts (1–4 and 6), including the mRNA for NFAT5c (transcript 3) contain the third selected segment 4|D. It should be noted that all five transcripts contain the correct order of segments for translation into NFAT5a but not necessarily for the longer, N-terminally extended NFAT5 isoforms.
RNA was extracted from HeLa cells in isotonic medium and cDNA was synthesized. After digestion of genomic DNA, qPCR with primers designed for target segments -|X, 2|B and 4|D was used to quantify expression of NFAT5-related mRNAs (Fig. 1C). We also analyzed the PCR products using electrophoresis (Fig. 1C) and sequenced all three bands to ensure the correct products. The qPCR results indicate that mRNAs containing segment -|X are somewhat more frequent than those with segment 2|B. Taking into account that the expression of segment 4|D is within a factor below two of segment -|X or a factor of about three of segment 2|B, we conclude that NFAT5a-related transcripts make up a sizeable portion of all NFAT5 transcripts (about half of all NFAT5 expression) and are present in similar order of magnitude compared with other NFAT5 mRNAs. This implies that various isoforms including NFAT5a are endogenously expressed concurrently at the isotonic, resting stage.
NFAT5a is myristoylated and palmitoylated in vitro and in vivo.
To test the lipid anchoring of NFAT5a, we expressed C-terminally tagged NFAT5a fusion proteins in the presence of radioactively labeled anchor precursors under various conditions. We scanned for radioactivity using a thin layer chromatography (TLC) linear analyzer and monitored protein expression on the same protein gel blot membrane.31,32 Besides achieving higher experimental accuracy by testing protein extraction levels and radioactive anchor incorporation in one and the same experiment, this technique has also the advantage of higher sensitivity and saving time (readouts are completed in ∼20 min per sample). This allows the variation of experimental parameters, which is helpful when dealing with a protein that might be notoriously difficult to express, such as the long NFAT5a.
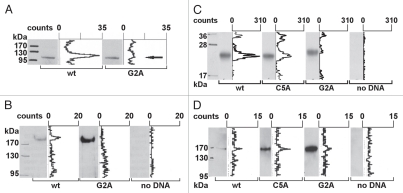
Figure 2A shows the results for the first 541 N-terminal AAs of NFAT5a and its G2A mutant expressed as a C-terminally GST-tagged fusion protein by coupled in vitro transcription/translation (TNT) in the presence of 3H-labeled myristate (TNT expression of full-length C-terminally tagged NFAT5a did not yield substantial amounts of protein to be detected in a protein gel blot). Similar efficiency of expression of both protein forms was monitored by anti-GST protein gel blot detection. TLC scanning for radioactivity of the same western membrane demonstrated a large peak corresponding to the wt protein band, thus, indicating incorporation of radioactive myristate. For the G2A mutant, no radioactive signal was detected.
Figure 2.
Transcription factor NFAT5a is N-terminal myristoylated and palmitoylated. (A) In vitro transcription/translation (TNT) of NFAT5a(AA1–541)-GST (wt) and its G2A mutant was performed in the presence of3H-labeled myristate. Expression was monitored by protein gel blot detection.3H incorporation was measured with a TLC linear analyzer. The scan shows that only NFAT5a-GST with an intact glycine at position 2 incorporates the myristoyl anchor. (B) HeLa cells were transiently transfected with NFAT5a-HA (wt) and NFAT5a(G2A)-HA. Proteins were purified from cells after in vivo labeling with3H-myristate, detected by immunoblotting and scanned for3H incorporation. Although the western signal is significantly lower for wt NFAT5a-HA in comparison to the mutant, only wt gives rise to a radioactive signal. As a control, non-transfected cells were used, and no signal could be detected. (C) In vitro TNT of NFAT5a(AA1–123)-HA (wt) and its mutants C5A and G2A was performed in the presence of activated3H-palmitate-CoA. Incorporation of radioactive palmitate was monitored by TLC scanning. Immunoblotting was used to monitor protein expression. The western bands are comparable in strength. The TLC signal for the wt is about double the size of the C5A mutant's signal. There is no TLC signal for the G2A mutant. No plasmid was used for the control reaction. (D) HeLa cells were transiently transfected with NFAT5a-HA (wt) and its mutants C5A and G2A. After transfection, cells were subjected to in vivo labeling with3H-palmitate. Proteins were immunoprecipitated, quantified by immunoblotting and scanned for labeled palmitate attachment. Although the expression of the wt is significantly weaker than the mutant ones, the TLC signal strength is comparable. Non-transfected cells were used as a background control.
NFAT5a is also myristoylated in vivo (Fig. 2B). We transiently transfected HeLa cells to express full-length NFAT5a-HA in the presence of radioactive myristate. IP of the wt protein resulted in a faint band, a relative small amount of protein as monitored by anti-HA immunoblot detection; nevertheless, a clear radioactive signal is associated with the band. With the G2A mutant, we could purify a much higher yield of protein, but no detectable radioactive signal was observed.
Palmitoylation of cysteines near the N terminus often correlates with N-terminal myristoylation.4,28 Testing palmitoylation directly with an in vitro TNT assay complemented with radioactive [9,10-3H]-palmitic acid is not possible. Therefore, it is necessary to activate the palmitic acid by enzymatically labeling it with Coenzyme A (CoA) to facilitate spontaneous S-acylation of the cysteine thiol.33,34 NFAT5a-HA (N-terminal 123 AAs) and the mutants C5A and G2A were TNT-expressed in the presence of palmitoyl-CoA and purified in nearly equal amounts (Fig. 2C). The radioactive signal for the wild type is around 300 counts. The signal for the C5A mutant is reduced by half. The G2A mutant generated no radioactive signal. This in vitro experiment shows that NFAT5a can be palmitoylated when it is directed to a vesicular membrane by the myristoyl anchor, and the most likely position for the palmitoylation is Cys5.
We also monitored NFAT5a-HA for palmitoylation in vivo. The wt fusion protein as well as the G2A and C5A mutated versions were expressed in transiently transfected HeLa cells in the presence of3H-labeled palmitate and purified using IP (Fig. 2D). Purification of the wt fusion protein and of the C5A and G2A variants resulted in dramatically different protein amounts. In the protein gel blot strips, the band corresponding to wt NFAT5a is almost invisible. It is a reproducible phenomenon that even overexpressed wt NFAT5a is hard to purify from cell culture. The respective bands are comparatively strong for the C5A mutant and much stronger for the G2A mutant. Nevertheless, the intensities of the recorded radioactive signals are of comparable and relatively low strength for all three conditions indicating anchor incorporation. The amount of protein in the protein gel blot is reversely indicative for the degree of protein palmitoylation. Thus, these results confirm the incorporation of palmitoyl anchors into NFAT5a in vivo and support the following conclusions: (1) Initial myristoylation of NFAT5a is a prerequisite for efficient palmitoylation in agreement with other cases in the literature.28 (2) Cys5 is the prime anchor attachment site, since the C5A mutation severely impairs palmitoylation, but it might not be the only palmitoylation site.
Subcellular localization of NFAT5a is influenced by the two lipid anchors in the resting state.
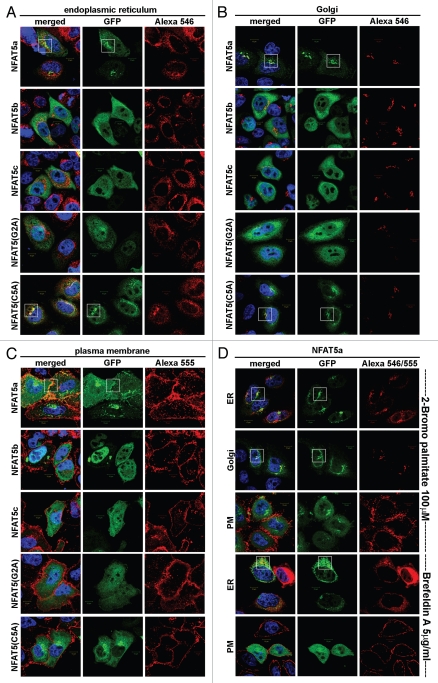
To compare the cellular localization of doubly lipid-modified NFAT5a-GFP, its mutants and the non-lipid anchored isoforms b and c under resting (isotonic) conditions, we conducted a series of co-localization experiments35,36 with markers for the ER, the Golgi and the PM in fixed cells (Fig. 3).
Figure 3.
Localization studies of NFAT5 isoforms and their mutants. HeLa cells were transfected with C-terminal GFP-tagged NFAT5 isoforms a, b and c and the mutants G2A and C5A (green). Squares indicate areas of interest. Full-size images are available athttp://mendel.bii.a-star.edu.sg/SEQUENCES/NFAT5_2011/. (A) The ER was stained with anti-PDI antibodies (red). NFAT5a and the C5A mutant co-localize with the ER. The isoforms b and c and the G2A mutant do not co-localize. (B) Golgi was stained with Giantin (red). NFAT5a and NFAT5a(C5A) co-localize with the Golgi. Isoforms b and c and the G2A mutant do not show a specific Golgi localization pattern. (C) The plasma membrane (PM) was stained with wheat germ agglutinin Alexa 555 (red). NFAT5a-GFP co-localizes with the PM. NFAT5b/c and the mutants G2A and C5A do not co-localize. (D) This part shows NFAT5a transfected cells treated with two inhibitors. With treatment of 2-Bromo palmitate, which inhibits palmitoylation, NFAT5a accumulates in the ER and the Golgi but not at the PM. Brefeldin A disrupts the Golgi. Localization to the ER can still be observed, but co-localization with the PM is inhibited (Scale bars 10 µm).
NFAT5a-GFP and the C5A mutant spread as discrete spherical vesicles (through the cell's cytoplasm and adjacent to the nucleus) co-localizing with the ER and the Golgi. Co-localization with the PM can only be claimed for wt NFAT5a. Therefore, we conclude that palmitoylation on Cys5 appears decisive for PM localization. The isoforms NFAT5b/c as well as the myristoylation-deficient mutant G2A of NFAT5a are retained in the cytoplasmic region, show a diffuse staining and do not co-localize with the organelles examined or the PM (Fig. 3A–C). Thus, the difference in the N termini of NFAT5 isoforms has a biological importance. It affects their specific subcellular localization, although all NFAT5 isoforms are retained to considerable extent cytoplasmically under isotonic conditions.
Treatment of HeLa cells with 2-Bromo palmitate (2-BP) inhibits palmitoylation, yet it does not affect the trend of NFAT5a to localize to ER and Golgi (Fig. 3D). Whereas the palmitoylated NFAT5a traffics to the PM along the secretory pathway via the Golgi complex (Fig. 3C), NFAT5a disappears at the PM after 2-BP treatment. Treatment with Brefeldin A (BFA) disrupts the Golgi network. It does not affect NFAT5a co-localization with the ER, yet NFAT5a at the PM is seemingly absent (Fig. 3D). Both treatments dramatically reduce NFAT5a trafficking to the PM whereas the co-localization with the ER/Golgi system is retained. These results also provide independent, indirect support for NFAT5a palmitoylation.
Quantitative differences in the translocation of NFAT5a, its mutants and other NFAT5 isoforms into nucleus under osmotic stress conditions.
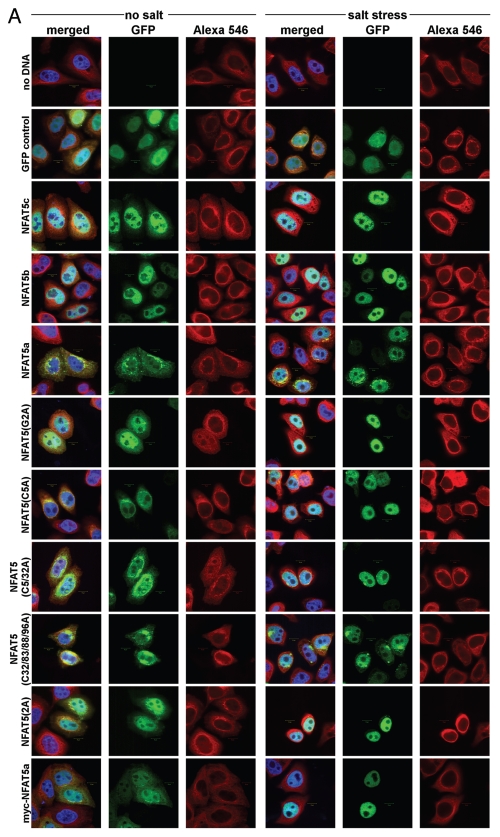
As a next step, we explored the effect of osmotic stress on the subcellular localization of NFAT5 isoforms and mutants. For this purpose, C-terminally GFP-tagged constructs of wt NFAT5c/b/a as well as the NFAT5a single mutants G2A, C5A, the double mutant C5/32A and the multiple mutant C32/83/88/96A (to test the physiological role of more distant N-terminal cysteines) were used to transfect HeLa cells. We also studied the two myristoylation-deficient constructs myc-NFAT5a-GFP37 and 2A (an alanine inserted before Gly2). Each culture was incubated for one hour either in isotonic medium or in a medium with 350 mOsmol NaCl (osmotic stress), and cells were studied by microscopy after fixation (Fig. 4A). For each construct and condition, we accumulated images with 25–30 cells in total for subsequent quantification of the nuclear fraction of the NFAT5 protein with image-analytic procedures.
Figure 4A.
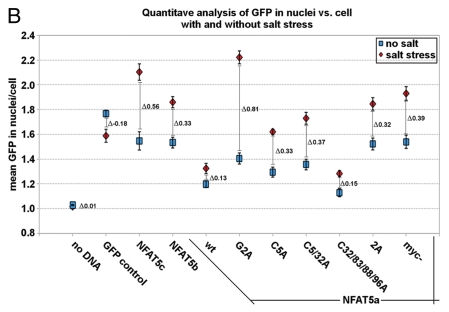
Quantification of NFAT5 translocation from the cytoplasm to the nucleus upon salt stress. Full-size images are available athttp://mendel.bii.a-star.edu.sg/SEQUENCES/NFAT5_2011/. Methodical detail of the image analysis proceduresis provided inSupplemental Material B. (A) Representative confocal images of HeLa cells transfected with the following GFP-tagged constructs: no DNA (negative control), GFP control (empty pEGFP vector), NFAT5c, NFAT5b, NFAT5a and its mutants G2A, C5A, C5/32A, C32/83/88/96A, 2A and myc-NFAT5a. Images were taken for all constructs without or with salt stress (350 mOsmol NaCl). The GFP channel shows the expression of the different constructs. Beta-tubulin was used for staining the cytoplasm (Alexa 546). DAP I (blue) stains the DNA in the nucleus (Scale bars-10 µm). (B) Representative graph showing the quantitative analysis of all chosen cells for each construct under each condition. 25–30 cells were used per construct. The graph shows the constructs on the x-axis. The y-axis shows the mean GFP in the nucleus vs. the GFP in the cell. Blue squares represent the values before salt stress and red diamonds after salt stress. The standard errors as well as the changes (Δ values) between the two conditions “no salt” and “salt stress” are shown.
A dramatic shift of the NFAT5 protein versions from the cytoplasm to the nucleus is evident in all cases studied (Fig. 4A). In contrast, GFP as control seems to move out of the nucleus to some extent under osmotic stress. The GFP control also shows that cells do not change their morphology under osmotic stress (except for some shrinking), and that the GFP tag does not influence the stress-induced nuclear translocation of NFAT5 versions. There are two principal cases to be distinguished: overexpressed NFAT5b/c already have a considerable nuclear fraction at isotonic conditions, and essentially all protein molecules appear imported into the nucleus upon stress. The N-terminally myristoylation- and/or palmitoylation impaired mutants G2A, 2A, C5A, C5/32A and myc-NFAT5a-GFP seem to behave essentially as NFAT5b/c. In contrast, NFAT5a with intact capability for the double lipid anchoring and its C32/83/88/96A variant appear almost exclusively outside of the nucleus under isotonic conditions; nevertheless, substantial amounts are imported into the nucleus upon stress.
The observed trends require quantification of the GFP-tagged protein distribution with regard to the subcellular compartments (nucleus and cytoplasm). Because the co-localization of NFAT5a with the cytoplasmic vesicular system in the vicinity of the nucleus makes the nuclear/cytoplasm fractionation procedure37 unreliable, we decided to carry out the quantification within an automated image analysis approach (Sup. Material B). In brief, the relative brightness of the GFP channel (the total intensity over a cellular compartment averaged by its area) is essentially a measure of protein concentration. After cellular morphology segmentation and background intensity correction, we determined the relative brightness values and corresponding to the nuclear compartment and to the whole cell separately. Since the absolute expression of NFAT5 constructs may differ among cells, the ratio γ of these two relative brightness values as inequation 1
| [1] |
normalizes for expression differences and is used to quantify the nuclear localization of the NFAT5 protein variant for a given condition (isotonic or osmotic stress). For biochemical analysis of protein expression using protein gel blot and cell sorting, seeSupplemental Material B.
The results for the various constructs are provided inFigure 4B. The blue squares represent the data for constructs without salt stress, and the red diamonds correspond to the salt stress condition. The difference Δ between those two γ values is indicative of the relative amount of GFP-tagged protein that is transferred from the cytoplasm into the nucleus within one hour of 350 mOsmol salt stress. E.g., γ = 1 implies equal relative brightness both of the nucleus and averaged over the whole cell (no DNA is transfected). Salt stress does not influence the value in this case (Δ ∼ 0). The GFP control cells show a small net shift of GFP into the cytoplasm from the nucleus accompanying the shrinkage of the cell (Δ = −0.18). Despite of the shrinkage, we could see a significant translocation into the nucleus for all NFAT5 constructs. This indicates active nuclear import upon osmotic stress. The relative distribution of overexpressed wt NFAT5b/c, the 2A and the myc mutants of NFAT5a at resting conditions is about the same, and the shift toward the nucleus at osmotic stress is dramatic (Δ between 0.32 and 0.56). The G2A, C5A and C5/32A mutants of NFAT5a behave similarly with a large shift in Δ (between 0.35 and 0.81) and a slightly lower nuclear concentration at resting conditions. NFAT5a and the lipid-anchoring competent mutant form C32/83/88/96A start with clearly reduced relative nuclear concentration compared with the other forms and show a smaller shift (Δ ∼ 0.15).
The quantitative data inFigure 4B supports the visual impression that impaired lipid anchoring of the G2A and the C5A mutants leads to their enhanced (apparently constitutive) nuclear localization in the resting state. The nuclear brightness caused by their GFP constructs is similar to the level of wt NFAT5a only under osmotic stress. Since overexpression of a construct after transfection is not properly synchronized with the other genes of the host cells, it cannot be excluded that comparatively large amounts of NFAT5-derived protein exhaust some retention and/or transport mechanisms of the cell. Possibly, overexpression of soluble NFAT5b/c as well as of NFAT5a mutants with impaired lipid-anchoring goes beyond the cytoplasmic retention capacity, and this might explain premature nuclear entry of some fraction of the protein molecules in the resting state. In contrast, wt NFAT5a as well as the NFAT5a mutant C32/83/88/96A that leaves the sites Gly2 and Cys5 untouched seem well-retained at membranes in isotonic conditions by the lipid anchors despite overexpression (Fig. 4A).
Only the plasma membrane-bound fraction of NFAT5a is sensitive to osmotic stress and is translocated to the nucleus.
The relative nuclear concentrations under osmotic stress are highest for the non-myristoylated isoforms NFAT5b/c and the NFAT5a mutants G2A, 2A and myc (Fig. 4B). The concentrations are less for the myristoylated but palmitoylation-deficient group of NFAT5a mutants (C5A and C5/33A) and even less for the potentially doubly lipid-anchored third group (wt NFAT5a and the mutant C32/83/88/96A;Fig. 4B). In parallel, the microscopy images (Fig. 4A) show even, and essentially complete, depletion of NFAT5-GFP constructs of the first group from all cytoplasm upon hypertonic stress, but, in contrast, some GFP-containing granules remained in the cytoplasm for the other constructs, especially for wt NFAT5a and the mutant C32/83/88/96A. It appears that the fraction of NFAT5a at intracellular, most likely ER/Golgi membranes [granular structures in row 5 (NFAT5a wt) and in row 9 (C32/83/88/96A) inFig. 4A] that did not yet reach the PM appears insensitive to osmotic stress and does not shuttle to the nucleus in the time frame studied.
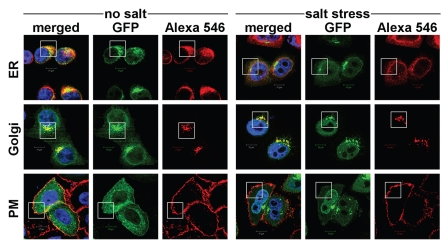
We tested the subcellular localization of NFAT5a without/with osmotic stress (350 Osmol NaCl for 1 h) in greater detail (Fig. 5). As observed previously, NFAT5a co-localizes with the ER, Golgi and the PM in the isotonic state. Upon salt stress, the co-localization pattern of NFAT5a to the ER and Golgi appears unchanged. On the other hand, there are dramatic changes at the PM. Essentially, NFAT5a does not co-localize to the PM under the hypertonic conditions studied. This shows that only NFAT5a previously attached to the PM transfers to the nucleus, whereas NFAT5a that is still processed in the ER/Golgi cannot shuttle.
Figure 5.
Localization studies of NFAT5a under isotonic and hypertonic conditions. HeLa cells were transfected with C-terminal GFP-tagged NFAT5a and were subjected to 1 h salt stress (350 mOsmol NaCl). Squares indicate areas of interest. The ER was stained with anti-PDI antibodies, the Golgi was stained with Giantin, and the plasma membrane (PM) was stained with wheat germ agglutinin Alexa 555 (red). NFAT5a co-localizes with the ER and the Golgi under isotonic- (no salt) and hypertonic-conditions (salt stress) to a similar extent. Co-localization with the PM is dramatically diminished under salt stress (Scale bars10 µm). Full-size images are available athttp://mendel.bii.a-star.edu.sg/SEQUENCES/NFAT5_2011/.
Osmotic stress directs isoforms a, b and c steadily into the nucleus.
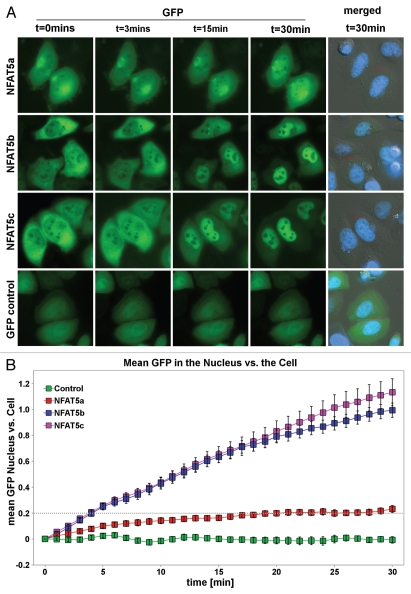
To estimate the time dependence of the import, we performed a “race” between the isoforms a, b and c of NFAT5 using live cell imaging. Cells were transfected with NFAT5a/b/c-GFP, exposed to osmotic stress, and images were taken every minute for 30 min directly after onset of stress. This specific setup did not allow us to stain the nucleus or the cell cytoplasm during the time course; we only applied a nuclear dye after completing the time course to avoid additional cell stress and death. Images were captured using intrinsic GFP fluorescence and phase contrast.Figure 6A shows representative images from the live cell imaging at 3, 15 and 30 min after onset of salt stress (images at 0 min show the resting state). All videos are available from the websitehttp://mendel.bii.a-star.edu.sg/SEQUENCES/NFAT5_2011/.
Figure 6.
Live cell imaging of the nuclear import of NFAT5 isoforms under salt stress. (A) Representative live cell images from each construct at time points 0, 3, 15 and 30 min after salt stress (350 mOsmol NaCl). HeLa cells were transfected with NFAT5a/b/c-GFP and pEGFP-N3 (GFP control). The images show that NFAT5b and c shuttle into the nucleus nearly completely in 30 min. For NFAT5a, a significant part did remain in the cytoplasm. GFP control does not change the localization pattern. The cell nuclei (blue) were stained after the time course was completed (Scale bars-10 µm). Full-size images and movies are available athttp://mendel.bii.a-star.edu.sg/SEQUENCES/NFAT5_2011/. Methodical detail of the image analysis proceduresis provided inSupplemental Material B. (B) The graph shows the statistical analysis of the mean GFP values in the nucleus vs. the cell of the in vivo imaging experiment. The slope for NFAT5b and c is steeper than the one for NFAT5a.
Phase contrast images were used to draw the boundary of the cell cytoplasm and the nuclei for cell morphology segmentation (seeSup. Material B for image analysis procedures). In brief, we calculated the relative brightness in the nucleus and in the total cell for each time point and we determined their ratio γ(t) as a function of timet (eq. 1). Since the starting values of γ(t1) (t1 is the video frame directly after the onset of osmotic stress) may be different for various constructs, we analyze the change γ(t) − γ(t1) relative tot1 after the onset of osmotic stress. The graph inFigure 6B shows that the nuclear relative brightness of GFP control (green) remains about constant over the video frames corresponding to 0–30 min aftert1. The two isoforms NFAT5b (blue) and NFAT5c (pink) first translocate into the nucleus at about the same rate. Toward the end of the 30 min, the rate of protein import decreases, and it seems to saturate thereafter.
The curve for NFAT5a (red) shows a visible incline after the onset of salt stress in comparison to the control (green) but progresses much more slowly compared with isoforms b and c. It reaches a similar nuclear localization levels after about 30 min of stress onset that NFAT5b or NFAT5c reach after about 5 min.
Thus, all three isoforms as well as the lipid-modified form NFAT5a, are mobilized in a regulated manner and move steadily into the nucleus upon salt stress. The relative nuclear concentration grows linearly with time. Whereas NFAT5b/c is evenly depleted from the cytoplasm during the time course, NFAT5a at some granules (intracellular membranes shown to be the ER/Golgi system,Fig. 5) does not participate in the translocation to the nucleus in the time frame studied.
It appears that the import of NFAT5a is delayed compared with non-lipid-anchored isoforms (Fig. 6B). Given that only the PM-bound fraction of NFAT5a can shuttle to the nucleus, but the rate of nuclear import is normalized with the total amount of NFAT5a in the cell, the nuclear import rate might be more similar to that of NFAT5b/c if normalization were performed with only the translocatable, PM-bound fraction. Yet, the latter is difficult to determine.
NFAT5a tied to the PM with a transmembrane segment is insensitive to osmotic stress and does not translocate to the nucleus.
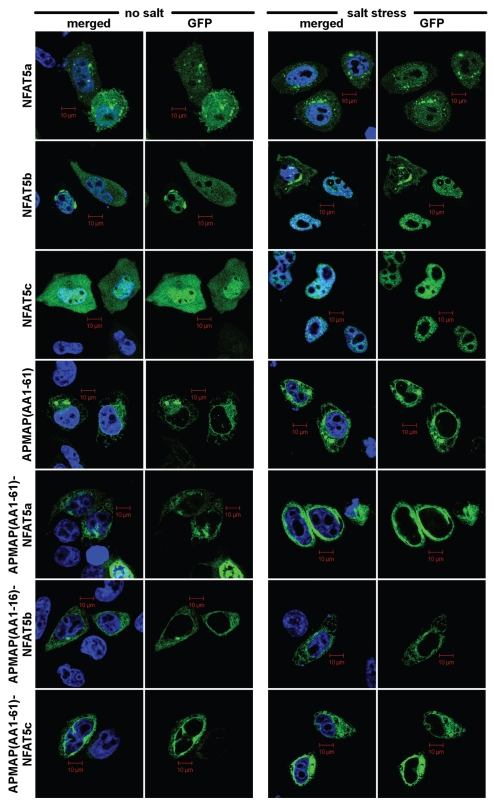
It is difficult to imagine mechanistically how a doubly lipid-modified TF can get mobilized for nuclear import. Proteolytic cleavage of a few N-terminal AAs is a possibility to solubilize NFAT5a. Since this region is non-globular, one might expect that this part of the sequence should readily be accessible to the active site of a potentially membrane-resident protease. To test this hypothesis, GPF-tagged NFAT5a/b/c were N-terminally linked with the transmembrane domain-containing segment AA1–61 of the constitutively PM localized protein APMAP.38 These constructs were expressed in HeLa cells that were either exposed or not exposed to one hour of 350 mOsmol salt stress.Figure 7 shows that APMAP(AA1–61) is sufficient to localize GFP to the PM as well as the NFAT5 isoforms. Upon osmotic stress, the APMAP(AA1–61)-fused proteins of NFAT5a/b/c all remain membrane-localized and cannot transfer to the nucleus. The outcome of this experiment does not support proteolytic cleavage of the N terminus as a mobilization mechanism for nuclear import.
Figure 7.
Anchoring of NFAT5 isoforms to the PM with the AP MAP N terminus. Selected subset of confocal images of HeLa cells transiently transfected with the following GFP-tagged (green) constructs: isoforms NFAT5a/b/c, AP MAP (AA1–61) (control), AP MAP (AA1–61)-NFAT5a/b/c. Salt stress (350 mM NaCl) was applied for 1 h. Nuclei were stained with DAP I (blue). Full-size images are available onhttp://mendel.bii.a-star.edu.sg/SEQUENCES/NFAT5_2011/. APMAP (AA1–61) represent the transmembrane region of APMAP for anchoring at the PM. All NFAT5 isoforms shuttle into the nucleus upon salt stress. No shuttling is observed after anchoring them to the PM (Scale bars 10 µm).
Discussion
In reply to the introductory question of this article, whether all transcription factors with lipid anchors have only cytoplasmic functions, we can confidently answer that this is not the case. With the example of NFAT5a, we demonstrate that lipid-modified TFs can be subject to regulated translocation into the nucleus. Cytoplasmic retention of NFAT5a in the resting state is the result of lipid anchoring. For efficient retention of NFAT5a outside of the nucleus in its non-induced state, both myristoylation as well as palmitoylation are necessary. Myristoylation targets NFAT5a to the ER/Golgi system, where it is palmitoylated (most likely in the Golgi35,39) and transferred to the PM. Upon a stress signal, wt NFAT5a localized at the PM is mobilized for translocation into the nucleus despite its two lipid anchors. Thus, there is a novel mechanism of regulated nuclear import of NFAT5a as part of a signal cascade that launches the cellular reaction on osmotic stress.
Subsequently, we will touch the following questions: (1) What is the likely mechanism of NFAT5a release from cytosolic membranes upon salt stress induction? (2) What is the physiological role of NFAT5a compared with that of the longer isoforms? (3) To what extent is the nuclear import and the involvement of lipid-modified transcription factors in transcription regulation a more general phenomenon?
Mechanism of release of NFAT5a from the membrane.
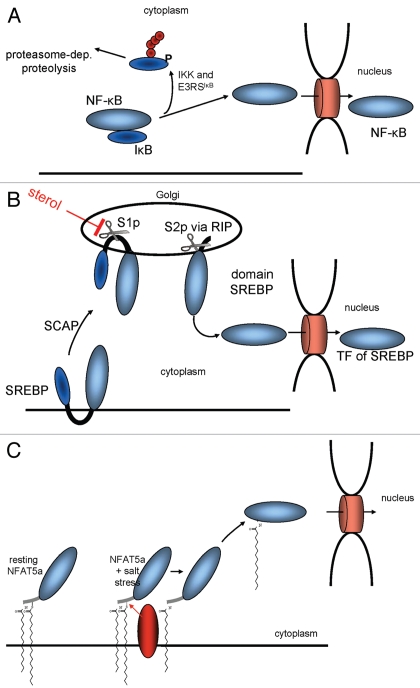
Currently, two major mechanisms for cytoplasmic retention are known to prevent TFs from entering the nucleus (Fig. 8). NFκB exemplifies the one that involves soluble TFs as part of protein complexes. It is retained in the cytoplasm as long as its nuclear localization signal (NLS) is masked by its inhibitor IκB.40 Upon stimulation, IκB dissociates, NFκB's NLS is exposed and, thereby, can guide the protein into the nucleus (Fig. 8A). A second mechanism involves TFs with transmembrane helices such as the sterol regulatory element binding protein (SREBP). In its non-activated state, it is attached to the ER via two transmembrane regions.41 During activation, two proteolytic cleavage events occur, allowing the TF domain to detach from the membrane and to accumulate in the nucleus2 (Fig. 8B).
Figure 8.
Mechanisms for the mobilization of TFs for nuclear import. (A) IκB prevents NFκB from entering the nucleus by masking its NLS. (B) The sterol regulatory element binding protein (SREBP) is retained at the ER via two transmembrane domains until its transcriptional active part gets cleaved off via two proteolytical cleavages. (C) A TF such as NFAT5a is attached to the plasma membrane via two lipid anchors. A stimulatory signal that causes its depalmitoylation allows its dissociation from the membrane and nuclear import.
Mechanisms for nuclear import of lipid-modified TFs have not been described yet. The hydrophobicity of two lipid anchors ties a TF such as NFAT5a tightly to cytoplasmic membranes. The hydrophobicities of farnesyl, myristoyl and palmitoyl anchors measured as membrane affinity are 100 µM, 80 µM and 5 µM; i.e., the palmitoyl anchor is ∼16 times more hydrophobic than myristoyl and 20 times more hydrophobic than farnesyl anchors.42–44 Whereas a protein with one myristoyl anchor can be solubilized to some extent, palmitoylated, prenylated or multiply lipid-anchored proteins are expected to be tightly bound to membranes.
Analogies with known biomolecular mechanisms suggest several explanations on how NFAT5a can be released from cytosolic membranes upon salt stress induction. First, NFAT5a might lose both lipid anchors as a result of proteolytic cleavage of the lipid anchored N terminus. Yet, we did not find any detectable change of migration speed of NFAT5a-HA on SDS gels and no additionally appearing band upon salt stress induction, indicating no major change of its molecular weight (data not shown). To note, several factors make this specific testing difficult. For NFAT5a getting rid of its lipid-anchor-laden N terminus, one cannot exclude a molecular weight change as low as ∼800 Da (for the minimal peptide GmyrGACpalm), which is difficult to detect for a ∼1,500 AA protein (around 160 kDa). C-terminally truncated versions of NFAT5a smaller than 450 AA do not transfer to the nucleus upon salt stress (data not shown). Further, lipid anchors themselves affect the electrophoretic migration speed, and this makes it even harder to distinguish between those two effects.45,46 As another argument, the construct APMAP(1–61)-NFAT5a is localized to the PM in a similar manner as wt, yet it does not translocate to the nucleus upon salt stress (Fig. 6). Thus, a proteolytic mechanism appears unlikely.
Second, NFAT5a might be mobilized for nuclear import via reversible depalmitoylation by a yet to determine thioesterase following a biomolecular mechanism known in the context of G proteins47,48 (Fig. 8C). Several experimental observations support this model. NFAT5a is myristoylated, palmitoylated and sorted to the PM via the ER and the Golgi (Figs. 2 and3). Quantified and live microscopic analysis confirms that NFAT5a moves into the nucleus upon salt stress (Figs. 4 and 6) but only the fraction that was previously palmitoylated and completed the translocation to the PM (Fig. 5). When overexpressed, the palmitoylationdeficient NFAT5a mutants (with G2A or C5A mutations) have high constitutive levels of nuclear concentration in the isotonic state similar to that of wt NFAT5a upon osmotic stress. Thus, NFAT5a after depalmitoylation is transported to the nucleus.
Physiological role of NFAT5 isoforms.
Although about 120 publications listed in PubMed describe various aspects of NFAT5 biology, the differential role of isoforms has so far not received proper attention. We find that the mechanisms of involving NFAT5a or NFAT5b/c in osmotic stress response are markedly different. First, NFAT5a localizes to the membranes of intracellular vesicular systems and is sorted to the PM via the ER and the Golgi in the resting state. In contrast, NFAT5b and NFAT5c do not show preference for the membrane systems and are diffusely spread in the cytoplasm. Second, salt stress leads to accumulation in the nucleus of all isoforms linearly growing with time. In the case of NFAT5a, only the PM-localized fraction is imported. In contrast, NFAT5b/c is evenly depleted from the cytoplasm upon hypertonic stress. It is necessary to study the tissue- and development-specific expression of the various NFAT5 isoforms and to test whether the sets of genes under control of NFAT5 or the onset and duration of their induction vary.
About other TFs with predicted lipid anchors.
Finally, the NFAT5a example provides a new paradigm for understanding how proteins can be retained from entering and mobilized for import into the nucleus and, therefore, it adds another facet to understanding transcriptional control. The most interesting question for general biology is whether lipid-modified TFs that are imported into the nucleus in a regulated manner are a more general phenomenon and whether reversible palmitoylation as a mobilization mechanism might be found in contexts other than NFAT5a.
It appears impossible to get an exact overview with regard to lipid modified TFs from a renewed in silico screen. Despite more than a decade of availability of “complete genomes,” new releases of genome assemblies and their reference proteomes remain accompanied with addition/losses of thousands of proteins, not to speak about isoforms. As potential examples for further study, we propose the following transcription regulators with conserved N-terminal myristoylation and palmitoylation sites: (1) human BTBD7 (Q9P203) (2) BAC20790 fromOryza sativa, (3) CAD60697 (Podospora anserine), (4) human LZTS1/FEZ1 (Q9Y250, isoform 1) and (5) LZTS2/LAPSER1 (isoform AAK31577; seeSup. Material A for detail).
From the methodological point of view, the case of NFAT5a shows that the analysis of biomolecular oddities, of proteins that unify seemingly contradictive elements in their molecular architecture, can guide scientific search toward the discovery of new biological mechanisms.
Experimental Procedures
For more detailed information refer toSupplemental Material C or tohttp://mendel.bii.a-star.edu.sg/SEQUENCES/NFAT5_2011/.
Cloning and constructs.
All NFAT5 constructs are based on a plasmid generously provided by Joan Ferraris containing NFAT5c (c: NM_006599, b: NM_138713, a: NM_173215). The APMAP (NM_020531) DNA was obtained via total cDNA from HeLa cells using RT-PCR [iScript cDNA Synthesis Kit (BioRad)]. Plasmids had to be shortened for several experiments. The following plasmids were used in this paper. Italian font stands for possible mutations of the plasmid. Plasmids were checked via sequencing:
T7_NFAT5a(AA1–541)(G2A)-GST,
NFAT5a(G2A)(C5A)/b/c-HA,
T7-NFAT5a(AA1–123)(G2A)(C5A)-HA,
NFAT5a(G2A)(C5A)(C5/32A)(C32/83/88/96A) (2A)/b/c-GFP,
myc-NFAT5a-GFP, APMAP(AA1–61)-GFP and
APMAP(AA1–61)-NFAT5a/b/c-GFP.
Cell culture.
HeLa and HEK293 cells were used as standard cell lines. For introduction of plasmids into cells Lipofectamine Plus™ Reagent or Lipofectamine 2000 (Invitrogen) were used according to the manual.
qPCR and rtPCR.
Total RNA was purified from HeLa cells with Trizol Reagent (Invitrogen). RNA concentration was determined using NanoDrop. cDNAs were synthesized using Maxima First strand cDNA synthesis kit for RT-qPCR (Fermentas). Realtime PCR was run using Maxima SYBR Green qPCR Master mix (Fermentas) on Rotor-Gene Q PCR cycler (Qiagen). PCR condition were 95°C for 10 min, 40 cycles of 95°C for 10 sec, 55°C for 15 sec, 72°C for 20 sec. A serial dilution test of the primers was done to determine the threshold use for Ct calculation. Triplicates of each sample were analyzed by DDCt method. RT-PCR was done using the cDNA protocol from above and iDNA 5x Mastermix for PCR. PCR products were separated by electrophoresis on 2% agarose gels with SYBR-Safe (Invitrogen) and sequenced using BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems). The following primers were used: GAPDH fwd 5′-GAG TCA ACG GAT TTG GTC GT-3′, rev: 5′-TTG ATT TTG GAG GGA TCT CG-3′, segment -|X fwd 5′-GAT TTG CCT CTG AAG CAG GGA G-3′, rev 5′-CCT TGC TGT CGG TGA CTG AGG TAG-3′, exon 2|B fwd 5′-ATTCTCTGAAGTTACACCCATC 3′, rev 5′-CTC ACC ACG GCT TGT CTG ACT C-3′, exon 4|D fwd 5′-ACC ACC TCT TCC AGC CCT ACC A-3′, rev 5′-TGT GCC TCT TCG GTG TTG ATG-3′.
Protein gel blot procedure and antibodies.
Cells were seeded, transfected and grown in 6-well plates (Nunc). Cells were scraped in 200 µl 1x Sample-Buffer. Samples were cooked at 95°C for 5 min. Ten µl lysate was loaded on self made 6% SDS-Page gels. The buffer was optimized for achieving the best resolution: Tris-Glycine-SDS (0.025 M Tris, 0.192 M Glycine, 0.1% SDS). For western transfer, either wet or semi-dry blotting was used with a Tris-Glycine Buffer used as transfer buffer. Antibody dilution and incubation was performed according to the manufacturer's protocols. Primary antibodies for western detection: Anti-GS-THRP Conjugate (Amersham) and mouse anti-HA BEAM (ETC). As secondary antibody Goat anti-mouse-HRP conjugated (Santa Cruz) was used. For protein gel blot detection ECL Plus™ protein gel blotting Detection Reagent (Amersham) was used.
Myristoylation and palmitoylation.
The in vitro assay for protein N-myristoylation is a modified version of the prenylation assay published previously in reference31 and32. All tags were C-terminal and used for immunodetection. The purification step of the fusion protein with glutathion beads was omitted. In vivo labeling assays were performed as described involving an immuno-purification step based on the HA tag. For labeling [9,10-3H]-myristic acid (NET-830) and [9,10-3H]-palmitic acid (NET-043) were used (both Perkin Elmer). [9,10-3H]-palmityl-CoA was generated following the protocol described earlier (Berthiaume et al. 1995).
Microscopy.
Mathematical procedures for processing image data are described inSupplemental Material B.
Confocal images. Cells were seeded on ethanol-flamed coverslips. The inhibitors 100 µM 2-Bromo-palmitate (Sigma) and 5 µg/ml BrefeldinA (Calbiochem) were incubated with the cells for around 12 h. For salt stress experiments NaCl was added to a final concentration of 350 mOsmol per well and incubated for 1 h. Cells were washed and immuno-staining was preformed. For localization studies the following primary AB were used: mouse anti-PDI (ER) (Abcam) and mouse anti-Giantin (Golgi) (Abcam), wheat germ agglutinin Alexa 555 (PM) (Invitrogen), anti-HA antibody BEAM (ETC) and rabbit anti β-tubulin (Abcam). Second AB: anti-mouse IgG Alexa 546 and anti-rabbit IgG Alexa 546 (Invitrogen). Coverslips were inverted onto glass slides with Vectashield, Hard SET Mounting Medium with DAPI (Vector Laboratories). Images were taken with upright LSM 510 META and LSM 5 LIFE duo scan (Carl Zeiss).
Live cell imaging. Transfected HeLa cells were transfered to coverslip chambers before mounting onto the Nikon TI-E inverted microscope platform. The Nikon TI-E inverted microscope with a heated stage, humidified CO2 chamber and motorized X, Y and Z stage used for these experiments were courtesy of SBIC-Nikon Centre, Singapore. Images were acquired every 1 min for 30 min. DMEM medium was gently withdrawn salt solution (350 mOsmol) was added. Cells were stained with Hoescht 33,342 dye at the end of the 30 min image capturing and a separate image was taken including the nucleii.
Acknowledgments
We are grateful to Joan D. Ferraris for her gift of NFAT5c cDNA and to Jose Aramburu for providing us with a myc-NFAT5a construct. Several authors (B.E., M.S., W.B., M.K., F.E.) were affiliated with the Institute of Molecular Pathology in Vienna (Austria) until July 2007, where this research was initiated in 2004. W.B. was affiliated with the Institute of Biochemistry of Vienna University until April 2009. Early financial support was provided by Boehringer Ingelheim and a grant to FE within Gen-AU BIN II. Since August 2007, support by the Biomedical Research Council of the Agency for Science, Technology and Research (A*STAR) is gratefully acknowledged. The authors thank Sun Tian for technical participation in the in silico screen and Fritz Pittner for laboratory support.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Zupicich J, Brenner SE, Skarnes WC. Computational prediction of membrane-tethered transcription factors. Genome Biol. 2001;2:50. doi: 10.1186/gb-2001-2-12-research0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ebinu JO, Yankner BA. A RIP tide in neuronal signal transduction. Neuron. 2002;34:499–502. doi: 10.1016/S0896-6273(02)00704-3. [DOI] [PubMed] [Google Scholar]
- 3.Maurer-Stroh S, Eisenhaber B, Eisenhaber F. N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J Mol Biol. 2002;317:541–557. doi: 10.1006/jmbi.2002.5426. [DOI] [PubMed] [Google Scholar]
- 4.Maurer-Stroh S, Gouda M, Novatchkova M, Schleiffer A, Schneider G, Sirota FL, et al. MYRbase: analysis of genome-wide glycine myristoylation enlarges the functional spectrum of eukaryotic myristoylated proteins. Genome Biol. 2004;5:21. doi: 10.1186/gb-2004-5-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maurer-Stroh S, Eisenhaber F. Refinement and prediction of protein prenylation motifs. Genome Biol. 2005;6:55. doi: 10.1186/gb-2005-6-6-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maurer-Stroh S, Koranda M, Benetka W, Schneider G, Sirota FL, Eisenhaber F. Towards complete sets of farnesylated and geranylgeranylated proteins. PLOS Comput Biol. 2007;3:66. doi: 10.1371/journal.pcbi.0030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegel R, Kim U, Patke A, Yu X, Ren X, Tarakhovsky A, et al. Nontranscriptional regulation of SYK by the coactivator OCA-B is required at multiple stages of B cell development. Cell. 2006;125:761–774. doi: 10.1016/j.cell.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 8.Yu X, Wang L, Luo Y, Roeder RG. Identification and characterization of a novel OCA-B isoform. implications for a role in B cell signaling pathways. Immunity. 2001;14:157–167. [PubMed] [Google Scholar]
- 9.Aramburu J, Drews-Elger K, Estrada-Gelonch A, Minguillon J, Morancho B, Santiago V, et al. Regulation of the hypertonic stress response and other cellular functions by the Rel-like transcription factor NFAT5. Biochem Pharmacol. 2006;72:1597–1604. doi: 10.1016/j.bcp.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Ko BC, Ruepp B, Bohren KM, Gabbay KH, Chung SS. Identification and characterization of multiple osmotic response sequences in the human aldose reductase gene. J Biol Chem. 1997;272:16431–16437. doi: 10.1074/jbc.272.26.16431. [DOI] [PubMed] [Google Scholar]
- 11.Takenaka M, Preston AS, Kwon HM, Handler JS. The tonicity-sensitive element that mediates increased transcription of the betaine transporter gene in response to hypertonic stress. J Biol Chem. 1994;269:29379–29381. [PubMed] [Google Scholar]
- 12.Ito T, Fujio Y, Hirata M, Takatani T, Matsuda T, Muraoka S, et al. Expression of taurine transporter is regulated through the TonE (tonicity-responsive element)/TonEBP (TonE-binding protein) pathway and contributes to cytoprotection in HepG2 cells. Biochem J. 2004;382:177–182. doi: 10.1042/BJ20031838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rim JS, Atta MG, Dahl SC, Berry GT, Handler JS, Kwon HM. Transcription of the sodium/myo-inositol cotransporter gene is regulated by multiple tonicity-responsive enhancers spread over 50 kilobase pairs in the 5′-flanking region. J Biol Chem. 1998;273:20615–20621. doi: 10.1074/jbc.273.32.20615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woo SK, Lee SD, Na KY, Park WK, Kwon HM. TonEBP/NFAT5 stimulates transcription of HSP70 in response to hypertonicity. Mol Cell Biol. 2002;22:5753–5760. doi: 10.1128/MCB.22.16.5753-60.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong EH, Guo JJ, Huang AL, Liu H, Hu CD, Chung SS, et al. Regulation of nucleocytoplasmic trafficking of transcription factor OREBP/TonEBP/NFAT5. J Biol Chem. 2006;281:23870–23879. doi: 10.1074/jbc.M602556200. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Rodríguez C, Aramburu J, Rakeman AS, Rao A. NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc Natl Acad Sci USA. 1999;96:7214–7219. doi: 10.1073/pnas.96.13.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyakawa H, Woo SK, Dahl SC, Handler JS, Kwon HM. Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity. Proc Natl Acad Sci USA. 1999;96:2538–2542. doi: 10.1073/pnas.96.5.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan S, Tsuruta R, Masuda ES, Imamura R, Bazan F, Arai K, et al. NFATz: a novel rel similarity domain containing protein. Biochem Biophys Res Commun. 2000;272:765–776. doi: 10.1006/bbrc.2000.2831. [DOI] [PubMed] [Google Scholar]
- 19.Trama J, Lu Q, Hawley RG, Ho SN. The NFAT-related protein NFATL1 (TonEBP/NFAT5) is induced upon T cell activation in a calcineurin-dependent manner. J Immunol. 2000;165:4884–4894. doi: 10.4049/jimmunol.165.9.4884. [DOI] [PubMed] [Google Scholar]
- 20.Ooi HS, Kwo CY, Wildpaner M, Sirota FL, Eisenhaber B, Maurer-Stroh S, et al. ANNIE: integrated de novo protein sequence annotation. Nucleic Acids Res. 2009;37:435–440. doi: 10.1093/nar/gkp254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider G, Wildpaner M, Sirota FL, Maurer-Stroh S, Eisenhaber B, Eisenhaber F. Integrated tools for biomolecular sequence-based function prediction as exemplified by the ANNOTATOR software environment. Methods Mol Biol. 2010;609:257–267. doi: 10.1007/978-1-60327-241-4_15. [DOI] [PubMed] [Google Scholar]
- 22.Eisenhaber F. Prediction of Protein Function: Two Basic Concepts and One Practical Recipe. In: Eisenhaber F, editor. Discovering Biomolecular Mechanisms with Computational Biology. Georgetown and New York: Landes Biosciences and Springer; 2006. [Google Scholar]
- 23.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stroud JC, Lopez-Rodriguez C, Rao A, Chen L. Structure of a TonEBP-DNA complex reveals DNA encircled by a transcription factor. Nat Struct Biol. 2002;9:90–94. doi: 10.1038/nsb749. [DOI] [PubMed] [Google Scholar]
- 25.Ferraris JD, Williams CK, Persaud P, Zhang Z, Chen Y, Burg MB. Activity of the TonEBP/OREBP transactivation domain varies directly with extracellular NaCl concentration. Proc Natl Acad Sci USA. 2002;99:739–744. doi: 10.1073/pnas.241637298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maurer-Stroh S, Eisenhaber B, Eisenhaber F. N-terminal N-myristoylation of proteins: refinement of the sequence motif and its taxon-specific differences. J Mol Biol. 2002;317:523–540. doi: 10.1006/jmbi.2002.5425. [DOI] [PubMed] [Google Scholar]
- 27.Eisenhaber F, Eisenhaber B, Kubina W, Maurer-Stroh S, Neuberger G, Schneider G, et al. Prediction of lipid posttranslational modifications and localization signals from protein sequences: big-Pi, NMT and PTS1. Nucleic Acids Res. 2003;31:3631–3634. doi: 10.1093/nar/gkg537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarro-Lérida I, varez-Barrientos A, Gavilanes F, Rodriguez-Crespo I. Distance-dependent cellular palmitoylation of de-novo-designed sequences and their translocation to plasma membrane subdomains. J Cell Sci. 2002;115:3119–3130. doi: 10.1242/jcs.115.15.3119. [DOI] [PubMed] [Google Scholar]
- 29.Dalski A, Wagner HJ, Schwinger E, Zuhlke C. Quantitative PCR analysis of different splice forms of NFAT5 revealed specific gene expression in fetal and adult brain. Brain Res Mol Brain Res. 2000;83:125–127. doi: 10.1016/S0169-328X(00)00202-3. [DOI] [PubMed] [Google Scholar]
- 30.Dalski A, Schwinger E, Zuhlke C. Genomic organization of the human NFAT5 gene: exon-intron structure of the 14-kb transcript and CpG-island analysis of the promoter region. Cytogenet Cell Genet. 2001;93:239–241. doi: 10.1159/000056990. [DOI] [PubMed] [Google Scholar]
- 31.Benetka W, Koranda M, Maurer-Stroh S, Pittner F, Eisenhaber F. Farnesylation or geranylgeranylation? Efficient assays for testing protein prenylation in vitro and in vivo. BMC Biochem. 2006;7:6. doi: 10.1186/1471-2091-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benetka W, Mehlmer N, Maurer-Stroh S, Sammer M, Koranda M, Neumuller R, et al. Experimental testing of predicted myristoylation targets involved in asymmetric cell division and calcium-dependent signalling. Cell Cycle. 2008;7:3709–3719. doi: 10.4161/cc.7.23.7176. [DOI] [PubMed] [Google Scholar]
- 33.Berthiaume L, Peseckis SM, Resh MD. Synthesis and use of iodo-fatty acid analogs. Methods Enzymol. 1995;250:454–466. doi: 10.1016/0076-6879(95)50090-1. [DOI] [PubMed] [Google Scholar]
- 34.Linder ME, Deschenes RJ. New insights into the mechanisms of protein palmitoylation. Biochemistry. 2003;42:4311–4320. doi: 10.1021/bi034159a. [DOI] [PubMed] [Google Scholar]
- 35.Draper JM, Xia Z, Smith CD. Cellular palmitoylation and trafficking of lipidated peptides. J Lipid Res. 2007;48:1873–1884. doi: 10.1194/jlr.M700179-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCabe JB, Berthiaume LG. Functional roles for fatty acylated amino-terminal domains in subcellular localization. Mol Biol Cell. 1999;10:3771–3786. doi: 10.1091/mbc.10.11.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Estrada-Gelonch A, Aramburu J, Lopez-Rodriguez C. Exclusion of NFAT5 from mitotic chromatin resets its nucleo-cytoplasmic distribution in interphase. PLoS ONE. 2009;4:7036. doi: 10.1371/journal.pone.0007036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bogner-Strauss JG, Prokesch A, Sanchez-Cabo F, Rieder D, Hackl H, Duszka K, et al. Reconstruction of gene association network reveals a transmembrane protein required for adipogenesis and targeted by PPARgamma. Cell Mol Life Sci. 2010;67:4049–4064. doi: 10.1007/s00018-010-0424-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, et al. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell. 2010;141:458–471. doi: 10.1016/j.cell.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 40.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 41.Osborne TF. Cholesterol homeostasis: clipping out a slippery regulator. Curr Biol. 1997;7:172–174. doi: 10.1016/S0960-9822(97)70081-2. [DOI] [PubMed] [Google Scholar]
- 42.Dunphy JT, Greentree WK, Manahan CL, Linder ME. G-protein palmitoyltransferase activity is enriched in plasma membranes. J Biol Chem. 1996;271:7154–7159. doi: 10.1074/jbc.271.12.7154. [DOI] [PubMed] [Google Scholar]
- 43.McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem Sci. 1995;20:272–276. doi: 10.1016/S0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- 44.Thompson GA, Jr, Okuyama H. Lipid-linked proteins of plants. Prog Lipid Res. 2000;39:19–39. doi: 10.1016/S0163-7827(99)00014-4. [DOI] [PubMed] [Google Scholar]
- 45.Breuer S, Gerlach H, Kolaric B, Urbanke C, Opitz N, Geyer M. Biochemical indication for myristoylation-dependent conformational changes in HIV-1 Nef. Biochemistry. 2006;45:2339–2349. doi: 10.1021/bi052052c. [DOI] [PubMed] [Google Scholar]
- 46.Franco M, Chardin P, Chabre M, Paris S. Myristoylation of ADP-ribosylation factor 1 facilitates nucleotide exchange at physiological Mg2+ levels. J Biol Chem. 1995;270:1337–1341. doi: 10.1074/jbc.270.3.1337. [DOI] [PubMed] [Google Scholar]
- 47.Chen CA, Manning DR. Regulation of G proteins by covalent modification. Oncogene. 2001;20:1643–1652. doi: 10.1038/sj.onc.1204185. [DOI] [PubMed] [Google Scholar]
- 48.Wedegaertner PB, Bourne HR. Activation and depalmitoylation of Gsalpha. Cell. 1994;77:1063–1070. doi: 10.1016/0092-8674(94)90445?6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.