Abstract
Introduction
Residing within human dental pulp are cells of an ectomesenchymal origin that have the potential to differentiate into odontoblast-like cells. These cells have a limited growth potential owing to the effects of cell senescence. This study examines the effects of immortalizing odontoblast-like cells on cell proliferation and mineralization by comparing transformed dental pulp stem cells (tDPSCs) and non-transformed dental pulp stem cells (nDPSCs).
Results
With the exogenous expression of hTERT, tDPSCs maintained a continued expression of odontogenic markers for cell proliferation and mineralization (ALP, COL-1, DMP-1, DSPP, OCN and OPN), as did nDPSCs. Oncoprotein expression was seen in both groups except for a noted absence of p16 in the tDPSCs. nDPSCs also showed lower levels of total ALP and DNA activity in comparison to tDPSCs when assayed, as well as low telomerase activity readings.
Methods
Using a retroviral vector, exogenous human telomerase reverse transcriptase (hTERT) was expressed in tDPSCs. Both cell groups were cultured, and their telomerase activities were determined using a telomerase quantification assay. Also examined, were the expression of genes involved in proliferation and mineralization, such as human alkaline phosphatase (ALP), β-actin, collagen I (col-1), core binding factor (cbfa)-1, dentin matrix protein (DMP-1), dentin sialophosphoprotein (DSPP), GAPDH, hTERT, osteocalcin (OCN), osteopontin (OPN) as well as oncoproteins involved in senescence (p16, p21 and p53) using RT-PCR. DNA and alkaline phosphate activity was also assayed in both cell groups.
Conclusion
These results indicate maintenance of odontoblast-like differentiation characteristics after retroviral transformation with hTERT and suggest a possible link with a reduced p16 expression.
Key words: dental pulp stem cells, odontoblasts, immortalization, senescence, oncoproteins, cell proliferation, cell mineralization
Introduction
Human pulp tissue is populated by a group of multipotent cells of mesenchymal origin known as dental pulp stem cells (DPSCs). These cells are heterogeneous in nature and have a potential for pluripotency.1–3 During the Bell stage of tooth development, the pulp is formed by the differentiation of the dental papilla.4 Separating the pulp from tooth dentine are mature odontoblasts that form the outer layer of the dental pulp and are responsible for the secretion of the collagenous and non-collagenous proteins5 needed to produce reparative (secondary) dentine.6–8 These odontoblasts have been referred to as the regenerative unit of the dentine-pulp complex.2,9 Lying beneath this layer of odontoblasts is a mixed population of undifferentiated cells that are able to differentiate under appropriate stimuli to yield different cell types.10 The process of dentinogenesis is influenced by the presence of an enabling environment controlled by several growth factors and signaling pathways.4,11 In appropriate culture media, isolated DPSCs have the potential to differentiate into adipocytes, neural-like cells and odontoblasts.1,2,12,13 The subject of a considerable amount of research interest are the odontoblasts and primary cell cultures of these cells will provide better understanding of their roles in dentin matrix formation. As is common with most human cell cultures, odontoblasts in vitro exhibit limited proliferation and senescence within several generations owing to a loss of telomeres.14
The in vitro study of primary cultures of human cells is dependent either on a constant supply of fresh tissue or the ability to maintain the cells in a steady proliferative state. Many cell/tissue types are only rarely available, depending on the frequency of biopsy. This difficulty is not associated with teeth, which are routinely extracted during many dental procedures, providing a ready supply of tissue. However, extraction of the pulp and expansion of the pulp cells is time-consuming, and cultures are often contaminated by bacteria from infected teeth, so the availability of immortalized pulp cells would be advantageous. Attempts to obtain cultures of human cells have been successful in the main, although often they only have limited proliferative ability.15 The death of these cells is mostly due to oncogenic stress as a result of an increased amount of cell divisions.14–16 As cell division progresses, there is a shortening of telomeres, which causes cellular instability and activates an arrest of cell division. This cell cycle arrest is mediated by growth suppressing proteins p16 and p53.16,17 The protein p53 controls the senescence of damaged or mutated cells, and its activity is dependent on the presence of p1618 and p21,19 which are both damage-response proteins.
The maintenance of telomere length of cells in culture is dependent on the expression of human telomerase reverse transcriptase (hTERT).20,21 This gene encodes the catalytic subunit of a protein complex known as telomerase that permits the addition of telomeric DNA repeat sequences during cell division.22 Uninhibited cell division has been demonstrated by research groups who have used a combination of viral oncogenes that allow for the ectopic expression of the telomerase enzyme to successfully establish human cell lines with increased periods of cell growth. Protein products of Simian Virus 40 (SV40) or retroviral vectors have been used to transduce the hTERT gene into human cell lines.23–25 This deactivates the p53 pathway and immortalizes the cells.26,27 A disadvantage in some human cell lines is the possibility of development of tumor cells.28 There is a suggestion that the expression of p16 may still prevent further cell division as its function is not dependent on telomerase expression.29
To further understand the effect of ectopic telomerase expression and senescence, we have used a retroviral vector to transfect human DPSCs with odontoblastic differentiation. While this method of transduction has been reported in references 24 and 30, this study examines the effects of p16, p21 and p53 levels on the proliferative and mineralising activities of odontoblast-like cells following ectopic telomerase expression.
Results
Cell morphology, phase contrast microscopy.
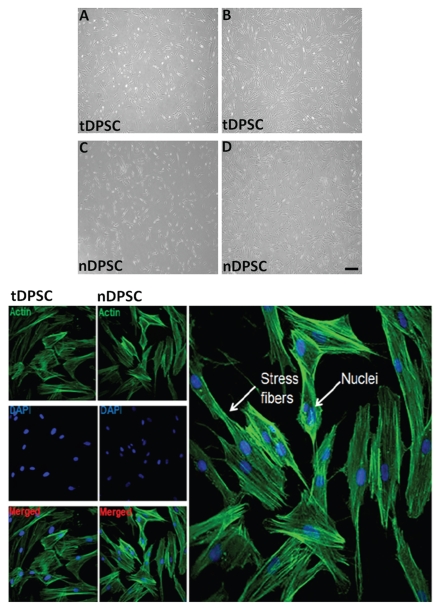
The morphology of tDPSCs and nDPSCs was analyzed microscopically at different time periods (Fig. 1). Both cells, as depicted by the optical phase contrast microphotograph reported in Figure 1A, spread along the surface of the culture plates, showing a rapid phase of growth within the first day. By day 7, both tDPSCs and nDPSCs had assumed a heterogeneous morphology typified by the appearance of spindle-like shaped cells with extensive cellular processes, and this phenotype was seen as the cells (tDPSCs and nDPSCs) grew to confluence (Fig. 1B).
Figure 1.
Optical phase contrast microphotographs showing the morphology tDPSCs and nDPSCs under phase contrast microscopy (x400) during the growth phase, day 3 (A and C) and at confluence, day 7 (B and D). Also shown is some immunopositive staining of tDPSCs and nDPSCs.
The tDPSCs maintained this spindle-like phenotype beyond day 7 through further culture passages, in keeping with the normal growth morphology observed by other groups.3,11,30,31
Cytoskeletal structure analysis by fluorescence microscopy.
Using a fluorescent-labeled phalloidin, the actin filaments of tDPSCs and nDPSCs were observed. In both cell groups, there was an identical pattern of parallel fiber orientation (Fig. 1). This was mainly in the cytoplasmic areas of the visualized cells, transversing the nuclei of the spindle-like cells.
TRAP (telomere repeat amplification protocol) assay.
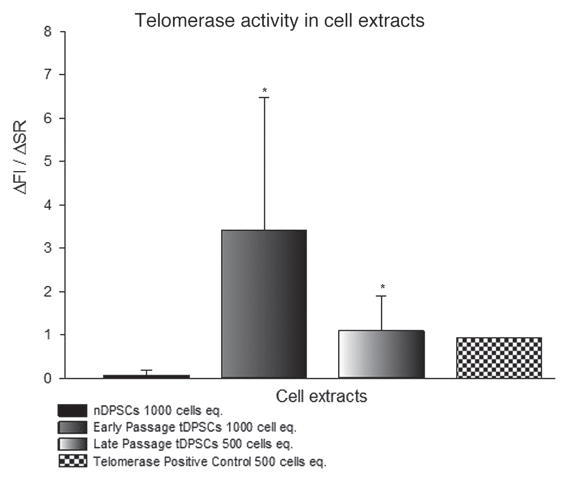
Cell extracts of tDPSCs, nDPSCs, telomerase-positive and telomerase-negative samples were made using equivalents of 500 and 1,000 cells (Fig. 2). The fluorescence was measured and showed a significant change in the ΔFl/ΔR values for tDPSCs at 500 and 1,000 cell equivalents. The telomerase-positive extracts showed ΔFl/ΔR values indicative of telomerase activity at 500 cell equivalents. ΔFl/ΔR values for nDPSCs at 500 and 1,000 cell equivalents were zero (data not shown) and less than 1 % of the telomerase positive cells, respectively, indicating a low level of telomerase activity. This would suggest background levels of telomerase activity in nDPSCs, in keeping with the faint expression bands detected on mRNA levels by PCR (Fig. 4). Telomerase-negative samples showed a zero ΔFl/ΔR at 500 cell equivalents (data not shown).
Figure 2.
Relative fluorescent readings used to assess telomerase activity in tDPSCs and nDPSCs after the introduction of exogenous HTERT. The results are expressed as the means ± SD (n = 4, *p < 0.001).
Figure 4.
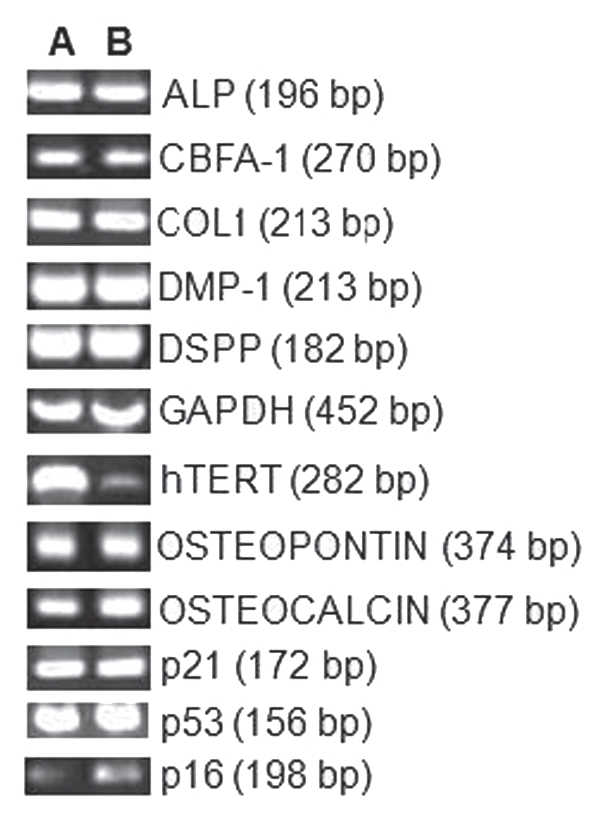
RT-PCR results for odontoblastic differentiation genes (ALP, cbfa-1, Col-1, DMP-1, DSPP, Osteopontin and Osteocalcin) and growth regulating factors p16, p21 and p53. Lane A, tDPSCs and Lane B, nDPSCs. GAPDH was run as an internal control and water as the negative control for both groups.
β-galactosidase staining.
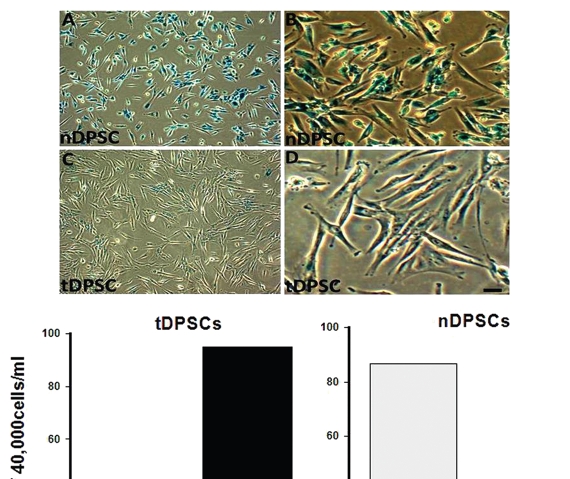
DPSCs undergoing senescence showed a blue peri-nuclear stain indicating positivity for β-galactosidase (at a pH of 6.5) activity in these cells (Fig. 3). The early passage tDPSCs (passage 6–9) did not stain for β-galactosidase. A few late passage tDPSCs (passage 30) and a significant number of nDPSCs (passage 9) showed β-galactosidase activity. The cells were re-plated after the experiment, and there was a failure of the positively stained nDPSCs to proliferate.
Figure 3.
β-galactosidase staining in cultured human dental pulp stem cells. Cells were stained and photographed at 100x (A and C) or 200x (B and D) magnification. (A and B) Early passage nDPS C showing > 80% positivity and (C and D) Late passage tDPSCs showing < 10% positivity for β-galactosidase staining both depicted in the graph above. Positive staining disappeared after re-plating tDPSCs.
RT-PCR analysis.
Initially, the presence of hTERT mRNA was confirmed using RT-PCR. There was expression in both nDPSCs and tDPSCs, with a fainter hTERT band appearing in nDPSCs (Fig. 4). The expression of mRNA for the growth regulating factors p16, p21 and p53 was also investigated in both cell groups. There was no evidence of downregulation of p16, p21 and p53. However, the p16 bands were fainter in both cell groups, even more so in the nDPSCs (Fig. 4). To confirm the presence of odontoblastic differentiation markers, we examined the presence of cbfa-1, DMP-1, DSPP and osteopontin in both cell groups. Also performed was the detection of mRNA expression of markers of mineralization such as ALP, COL1 and osteocalcin (Fig. 4).
Colorimetric assays. DNA assay.
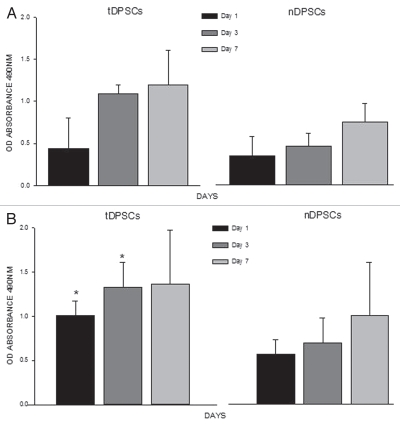
To analyze the proliferative activities of tDPSCs and nDPSCs, both cell groups (40,000 cells/ml) were cultured in DMEM/S growth medium for a week. Lysates were taken from both groups, and a DNA assay was performed. The optical absorbance values were obtained and represented graphically (Fig. 5A). There was a steady increase in the total proliferative activity in the tDPSCs compared with the nDPSCs over the 7-d culture period.
Figure 5.
Analyses of cell proliferation using a DNA assay (A) and cell mineralization by way of an ALP assay (B) in tDPS C and nDPS C cell groups. The experiments were performed in triplicate. The values are means of the absorbance readings at 490 nm ± SD (n = 4, *p < 0.001).
ALP assay. The mineralizing activity of both tDPSCs and nDPSCs was evaluated using lysates from cells from each group (40,000 cells/ml) cultured in DMEM/S growth media over a 7 d period. The data obtained showed a steady-state of increase in total ALP activity in the tDPSCs compared with the nDPSCs, rising to significant levels (p ≤ 0.001) from day 1 to day 7 (Fig. 5B).
Discussion
The immortalization of human dental cells is not entirely novel. A literature review demonstrated that several authors have successfully transformed and immortalized these cells, having obtained them from a variety of oral sources. Kamata and colleagues in 2004 performed a series of transformation studies using cells from human dental papilla, dental pulp, periodontal ligament and gingival fibroblasts.24 In the same year, Kitagawa and coworkers also successfully immortalized dental pulp cells.30 In 2005 Fujita and colleagues immortalized periodontal ligament cells.14 Previous studies have demonstrated that transfected cells are able to avoid the normal senescence program and hence extend the lifespan of the cells.32
In this study, we used the HIV retroviral packaging system intending to explore the possibility of establishing a hDPSC cell line that would circumvent normal programmed senescence and maintain its usefulness as a tool for further study of cellular activity. The detection of hTERT expression at mRNA levels following induction in a variety of cell lines has been widely reported in references 24, 30 and 33. The presence of hTERT mRNA was successfully demonstrated in this study following transfection of the tDPSCs with the hTERT gene. Also, the telomerase activity in tDPSCs was three times greater than in nDPSCs (Fig. 2). We have demonstrated perpetual cell growth over 30 population doublings while maintaining characteristics of differentiation and phenotypic cellular identity, in line with literature-defining parameters for a successful immortalization of dental pulp stem cells. The evidence to support continued odontoblastic differentiation is demonstrated by the expression of mRNA for cbfa-1, DMP-1, DSPP and osteopontin (Fig. 4). This corroborates the typical features of odontoblasts in culture.1–3,30 The genetic expression of ALP, COL1 and osteocalcin,1–3 in addition to the mineralization activities of the tDPSCs is further evidence to show a lack of change in cellular characteristics as demonstrated in the morphology and cytoskeletal organization of the cell (see Fig. 1).
In addition, the non transduced DPSCs (nDPSCs), demonstrated a very high degree of β-galactosidase-associated senescence activity (Fig. 3), a feature absent from a vast majority of the transduced DPSCs (tDPSCs) owing to the introduction of hTERT into these cells. With the prolongation of the lifespan of the DPSCs, we considered the possible effects of transfection on the silencing of senescence associated factors (oncoproteins). hTERT has long been associated with oncoproteins such as p16, p21 and p53 owing to the link between these proteins and telomerase activity.18,34–36 To the knowledge of the authors, this is the first study in dental pulp stem cells showing a possible link between the introduction of exogenous hTERT and mRNA expression of p16. The presence of p16 has been attributed to unfavorable culture conditions.37 Also noteworthy is its inverse relationship with the aging and proliferation of some cell types.38,39 It is uncertain whether the alteration of the pathways controlled by any of the three oncoproteins examined is solely dependent on the functional presence of hTERT.29 It is our opinion that this “downregulation” of p16 activity maybe promoted by the arresting of senescence of DPSCs owing to the introduction of exogenous hTERT, which promotes p53 activity in certain human cell types.18,28,29,34,40,41 The results from this study suggest an ability of DPSCs to maintain phenotypic characteristics of odontoblasts and circumvent senescence in the presence of ectopic hTERT, which may alter p16 expression.
Materials and Methods
Cell culture.
The dental pulp cells were prepared from human third molars extracted from four adult patients at the Department of Oral Surgery, Dental Institute, King's College London [with ethical approval (KCH REC ref. 08/H0808/104) and patient consent]. After collection, the teeth were washed with 70% ethanol and then with Hank's balanced saline solution (HBSS) (Invitrogen), pH 7.4. The crowns and roots of the teeth were separated aseptically by making horizontal indentations along the cervical margin using a low speed circular diamond saw (Agar Scientific Ltd.) and the pulp tissue retrieved from the pulp chamber using sterile forceps. After pooling the pulp cell samples, disaggregation was effected through enzymatic digestion in a solution of 3 mg/ml collagenase type I and 4 mg/ml dispase (Sigma-Aldrich®) for 30 min at 37°C after initially mincing the pulp tissue into small pieces measuring 1–2 mm by 3–4 mm with a scalpel blade under sterile conditions.
The cells were cultured following modification of the method described by Gronthos and coworkers (Gronthos et al. 2000). Following digestion, the pulpal tissue explants were grown in 25 cm3 culture flasks, T25 (Cell Star®), using Dulbecco's modified Eagle's medium DMEM (Pharmakine®) supplemented with 10% fetal bovine serum, FBS, 2.5 × 1,000 U/ml penicillin and 2.5 mg/ml streptomycin. The cultures were incubated at 37°C in an atmosphere of 95% O2 and 5% CO2. The media in the culture flasks was changed every 4 d to remove non adherent cells and cellular debris. After 10–14 d, outgrowths from the minced pulp tissue explants were observed. At confluence, the cells were trypsinized and passaged in T25 flasks. These cells were designated Passage 2. The cells were maintained for three passages before differentiation was induced by using a supplemented media. This supplementation was effected by conditioning the culture media with 50 µg/ml ascorbic acid, 10 mM β-glycerol phosphate and 10−7 M dexamethasone. A batch of the “non-differentiated” cells was harvested, frozen and stored in liquid nitrogen. The culture media was changed to a high glucose DMEM supplemented with 10% FCS, 5 × 1,000 U/ml Penicillin, 5 mg/ml Streptomycin and 0.11 g/L Sodium pyruvate (Sigma Aldrich®), DMEM/S. All other culture conditions were kept constant. Periodic monitoring of the cells was continued and sub culturing at confluence was done at a ratio of 1:4 to new culture 25 cm3 flasks.
Transduction of DPSCs with hTERT.
The retroviral vector containing the cloned hTERT gene, pBABE-puro-hTERT (Fig. 1),42 was a kind gift from Dr. J.M. Funes, University College London Cancer Institute. 1 × 104 DPSCs were plated in each well of a 24-well plate. This was followed by exposure to the hTERT retrovirus that was added to DMEM/S growth media, for 24 h. After 24 h, the virus was removed and replaced with DMEM/S growth media for a further 72–90 h period at 37°C in a humidified incubator. This was followed by selection using 1 µg/ml puromycin (Sigma) for between 7–14 d. The cells were then expanded. The DPSCs that survived were used for the rest of this study and are referred to as tDPSCs.
Cell extract TRAP (telomere repeat amplification protocol) assay.
Telomerase activity was determined using a TRAPeze®XL Telomerase Detection Kit (Millipore™). The cells were pelleted, washed in PBS and dissolved in 200 µL of 1x CHAPS lysis buffer. After a 30 min incubation period on ice, the cell samples were centrifuged at 12,000 g for 20 min at 4°C. 160 ul from each of the sample supernatants were collected for protein content analysis using a standard by Bicinchoninic acid protein assay. Two µL of the cell-extract (an equivalent of 2.5 µg protein/sample supernatant) was added to a 48 µL reaction solution consisting of 5x TRAPeze®XL reaction mix, a cloned and unmodified Taq polymerase (Sigma-Aldrich®) and distilled water. A 1,000 cell equivalents of telomerase positive cell extract (provided with the kit) was used as a positive control. Heat inactivated samples, a minus Taq polymerase and a minus telomerase sample were used as negative controls. The tubes containing the mixture were incubated at 30°C for 30 min in thermocycler block. This was followed by a four-step PCR at 94°C for 30 sec, 59°C for 30 sec, 72°C for 1 min (36 cycles), a 72°C for 3 min extension step, 55°C for 25 min and a 4°C incubation phase. The 50 µl PCR reaction product for each sample was transferred into a black-walled 96-well plate containing 150 µl of 10 nM TRIS-HCl/0.15 M NaCl/2 mM MgCl2 buffer (pH 7.4). Using a fluorescent plate reader, fluorescence was measured for each sample using excitation/emission parameters (in line with the manufacturers' protocol) for fluorescein, F (485 nm/534 nm) and sulforhodamine, R (584 nm/610 nm). The net change in fluorescence (ΔFl/ΔR) was determined for each sample.
β-galactosidase staining.
The expression of β-galactosidase by the nDPSCs and tDPSCs was assessed using a Senescence Cells Histochemical Kit (Sigma-Aldrich®). The cells were washed in PBS, fixed for 6–7 min (room temperature) in a 1 × 20% formaldehyde/2% glutaraldehyde buffer. After three washes, the cells were incubated at 37°C in a non-CO2 oven with a β-galactosidase senescence associated stain solution. The staining of cells was evident (under a microscope) after 4 h and was at a maximum at 12 hrs.
RT-PCR analysis.
Using an RNA extraction kit (Qiagen), the total cellular RNA was isolated from the cells as per the manufacturer's instructions. First-strand cDNA was synthesized from 1 µg of total RNA using a High Capacity RNA-to-cDNA Master Mix (Applied Biosystems). Products were amplified by PCR using primers specific for human alkaline phosphatase (ALP), β-actin, collagen I (col-1), core binding factor (cbfa)-1, dentin matrix protein (DMP-1), dentin sialophosphoprotein (DSPP), GAPDH, hTERT, osteocalcin (OCN), osteopontin (OPN), p16, p21 and p53 (see Table 1). PCR products were separated by electrophoresis (1.5% agarose gel) and detected with ethidium bromide under UV light.
Table 1.
Primer sequences and cycling conditions for the genes analyzed
| Primer | Sequence Product | Length (bp) |
| ALP | F: 5′-CCA CGT CTT CAC ATT TGG TG-3′ R: 5′-AGA CTG CGC CTG GTA GTT GT-3′ |
196 bp |
| β-ACTIN | F: 5′-ATG AGG ATG CTC ACG GAG CGC GGC TAC AGC-3′ R: 5′-ACA CCA CTG TGT TGG CGT ACA GGT CTT TGC-3′ |
331 bp |
| CBFA-1 | F: 5′-TGA GAG CCG CTT CTC CAA CC-3′ R: 5′-GCG GAA GCA TTC TGG AAG GA-3′ |
270 bp |
| COL-1 | F: 5′-CCA AAT CTG TCT CCC CAG A-3′ R: 5′-TCA AAA ACG AAG GGG AGA T-3′ |
213 bp |
| DSPP | F: 5′-TCA CAA GGG AGA AGG GAA TG-3′ R: 5′-TGC CAT TTG CTG TGA TGT TT-3′ |
182 bp |
| DMP-1 | F: 5′-CAG GAG CAC AGG AAA AGG AG-3′ R: 5′-CTG GTG GTA TCT TGG GCA CT-3′ |
213 bp |
| GAPDH | F: 5′-ACC ACA GTC CAT GCC ATC AC-3′ R: 5′-TCC ACC ACC CTG TTG CTG TA-3′ |
452 bp |
| hTERT | F: 5′-CCT CTG TGC TGG GCC TGG ACG ATA-3′ R: 5′-ACG GCT GGA GGT CTG TCA AGG TAG-3′ |
282 bp |
| OCN | F: 5′-ATG AGA GCC CTC ACA CTC CT-3′ R: 5′-CAA GGG GAA GAG GAA AGA AG-3′ |
377bp |
| OPN | F: 5′-CAG TGA CCA GTT CAT CAG ATT CAT C-3′ R: 5′-CTA GGC ATC ACC TGT GCC ATA CC-3′ |
374bp |
| p16 | F: 5′-GAC ATC CCC GAT TGA AAG AA-3′ R: 5′-TTT ACG GTA GTG GGG GAA GG-3′ |
198 bp |
| p21 | F: 5′-GAC ACC ACT GGA GGG TGA CT-3′ R: 5′-CAG GTC CAC ATG GTC TTC CT-3′ |
172 bp |
| p53 | F: 5′-GGC CCA CTT CAC CGT ACT AA-3′ R: 5′-GTG GTT TCA AGG CCA GAT GT-3′ |
156 bp |
Colorimetric assays.
Alkaline phosphatase (ALP) assay. tDPSCs and nDPSCs were seeded in 24-well plates at a density of 1 × 104 cells/well. ALP activity was measured after 1, 3 and 7 d cultures in DMEM/S using a modified assay that employs p-nitro phenyl phosphate as a substrate for measuring the enzyme protein present.43 The alkaline phosphatase enzyme cleaves the phosphate group from p-nitro phenyl phosphate to yield p-nitro phenol, which is yellow at an alkaline pH (10.3). The absorbance was then measured at 405 nm with a spectrophotometer (Opsys MR Microplate reader).
DNA assay. Following cell seeding at 1 × 104 cells/well in 24-well plates, the DNA content of tDPSCs and nDPSCs was examined using the method of Rao and colleagues:44 three cycles of freezing, thawing at −80°C and 37°C, respectively; 100 µl aliquots of each cell group was then transferred to a 96-well plate; 100 µl Hoechst 33,258 (Sigma®) fluorimetric dye was added to each well; florescence was measured at an excitation of 355 nm and emission wavelength of 450 nm on a plate reader (Chameleon™, Hidex, Finland). DNA standards (Sigma®) were prepared, and the DNA assay was performed in parallel for standardization. The cell lysates used for the experiment were taken at days 1, 3 and 7 of the cell culture and the experiment was performed using triplicate samples.
Immunocytochemistry.
To analyze the actin microfilament cytoskeletal protein, visualization was performed using FITC-conjugated phalloidin (1/1,000). The cells were cultured to 70% confluence in 6-well plates and fixed in 4% paraformaldehyde supplemented with 1% (w/v) sucrose at 37°C for 15 min. After washing in PBS, the cells were permeabilized at room temperature with 0.1% Triton-X 100 for 5 min and incubated with 1% bovine serum albumin at 37°C for 10 min to saturate non specific binding sites. This was followed by incubation with phalloidin at 37°C for 1 h. Control wells were incubated without phalloidin. The nuclei were counterstained with DAPI for 10 min. The cells were washed before viewing. The plates were visualized using a Leica DMRE upright confocal microscope (Leica Microsystems Heidelberg GmbH).
Statistical analyses.
The analyses of the data were performed using independent t-test with Sigma Plot 11.0 software (©Systat Software, Inc.). Statistical significance of the data was determined at p ≤ 0.001 and the values were expressed as mean ± standard deviation (SD).
Conclusions
We have isolated hTERT immortalized DPSCs that exhibit and maintain phenotypic characteristics of odontoblastic differentiation. The “downregulation” of p16 activity is an important feature in the characterization of these cells, as it may have an important role in the understanding of the mechanisms that control cellular senescence. This may provide a greater understanding of cellular mechanisms with respect to tissue regeneration. As a more readily available source of stem cells, involving a less invasive procedure compared with bone marrow aspiration, for example, it may provide a viable method for cell-seeded scaffold in situ and lead to the development of new clinical treatment modalities.
Acknowledgments
The authors acknowledge financial support from the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy's & St Thomas' NHS Foundation Trust in partnership with King's College London and King's College Hospital NHS Foundation Trust. The authors would also like to thank Professor Chris Boshoff from the University College London Cancer Institute for his expert advice.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Gronthos S, Brahim J, Li W, Fisher LW, Cherman N, Boyde A, et al. Stem Cell Properties of Human Dental Pulp Stem Cells. J Dent Res. 2002;81:531–535. doi: 10.1177/154405910208100806. [DOI] [PubMed] [Google Scholar]
- 2.Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and invivo. Proc Natl Acad Sci USA. 2000;97:13625–13630. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karaöz E, Do an B, Aksoy A, Gacar G, Akyüz S, Ayhan S, et al. Isolation and in vitro characterisation of dental pulp stem cells from natal teeth. Histochem Cell Biol. 2010;133:95–112. doi: 10.1007/s00418-009-0646-5. [DOI] [PubMed] [Google Scholar]
- 4.Thesleff I, Aberg T. Molecular regulation of tooth development. Bone. 1999;25:123–125. doi: 10.1016/S8756-3282(99)00119-2. [DOI] [PubMed] [Google Scholar]
- 5.Chen S, Gluhak-Heinrich J, Wang YH, Wu YM, Chuang HH, Chen L, et al. Runx2, Osx and Dspp in Tooth Development. J Dent Res. 2009;88:904–909. doi: 10.1177/0022034509342873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tziafas D, Kodonas K. Differentiation Potential of Dental Papilla, Dental Pulp and Apical Papilla Progenitor Cells. J Endod. 2010;36:781–789. doi: 10.1016/j.joen.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Tziafas D, Smith AJ, Lesot H. Designing new treatment strategies in vital pulp therapy. J Dent. 2000;28:77–92. doi: 10.1016/S0300-5712(99)00047-0. [DOI] [PubMed] [Google Scholar]
- 8.Yamamura T. Differentiation of pulpal cells and inductive influences of various matrices with reference to pulpal wound-healing. J Dent Res. 1985;64:530–540. doi: 10.1177/002203458506400406. [DOI] [PubMed] [Google Scholar]
- 9.Bender IB. Pulpal Pain Diagnosis—A Review. J Endod. 2000;26:175–179. doi: 10.1097/00004770-200003000-00012. [DOI] [PubMed] [Google Scholar]
- 10.Jo YY, Lee HJ, Kook SY, Choung HW, Park JY, Chung JH, et al. Isolation and characterization of postnatal stem cells from human dental tissues. Tissue Eng. 2007;13:767–773. doi: 10.1089/ten.2006.0192. [DOI] [PubMed] [Google Scholar]
- 11.Mesgouez C, Oboeuf M, Mauro N, Colon P, MacDougall M, Machtou P, et al. Ultrastructural and immunocytochemical characterization of immortalized odontoblast MO6-G3. Int Endod J. 2006;39:453–463. doi: 10.1111/j.1365-2591.2006.01089.x. [DOI] [PubMed] [Google Scholar]
- 12.Huang AHC, Chen YK, Lin LM, Shieh TY, Chan AWS. Isolation and characterization of dental pulp stem cells from a supernumerary tooth. J Oral Pathol Med. 2008;37:571–574. doi: 10.1111/j.1600-0714.2008.00654.x. [DOI] [PubMed] [Google Scholar]
- 13.Huang AHC, Chen YK, Chan AWS, Shieh TY, Lin LM. Isolation and Characterization of Human Dental Pulp Stem/Stromal Cells From Nonextracted Crown-fractured Teeth Requiring Root Canal Therapy. J Endod. 2009;35:673–681. doi: 10.1016/j.joen.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 14.Fujita T, Otsuka-Tanaka Y, Tahara H, Ide T, Abiko Y, Mega Ji. Establishment of immortalized clonal cells derived from periodontal ligament cells by induction of the hTERT gene. J Oral Sci. 2005;47:177–184. doi: 10.2334/josnusd.47.177. [DOI] [PubMed] [Google Scholar]
- 15.Dirac AMG, Bernards R. Reversal of Senescence in Mouse Fibroblasts through Lentiviral Suppression of p53. J Biol Chem. 2003;278:11731–11734. doi: 10.1074/jbc.C300023200. [DOI] [PubMed] [Google Scholar]
- 16.Ohtani N, Yamakoshi K, Takahashi A, Hara E. Realtime in vivo imaging of p16Ink4a gene expression: a new approach to study senescence stress signaling in living animals. Cell Div. 2010;5:1. doi: 10.1186/1747-1028-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 18.Beauséjour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P, et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 2003;22:4212–4222. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman EJ, Hurst CD, Pitt E, Chambers P, Aveyard JS, Knowles MA. Expression of hTERT immortalises normal human urothelial cells without inactivation of the p16/Rb pathway. Oncogene. 2006;25:5037–5045. doi: 10.1038/sj.onc.1209513. [DOI] [PubMed] [Google Scholar]
- 20.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 21.Carney SA, Tahara H, Swartz CD, Risinger JI, He H, Moore AB, et al. Immortalization of Human Uterine Leiomyoma and Myometrial Cell Lines After Induction of Telomerase Activity: Molecular and Phenotypic Characteristics. Lab Invest. 2002;82:719–728. doi: 10.1097/01.lab.0000017499.51216.3e. [DOI] [PubMed] [Google Scholar]
- 22.Greider CW. Telomerase RNA levels limit the telomere length equilibrium. Cold Spring Harb Symp Quant Biol. 2006;71:225–229. doi: 10.1101/sqb.2006.71.063. [DOI] [PubMed] [Google Scholar]
- 23.Galler KM, Schweikl H, Thonemann B, D'Souza RN, Schmalz G. Human pulp-derived cells immortalized with Simian Virus 40 T-antigen. Eur J Oral Sci. 2006;114:138–146. doi: 10.1111/j.1600-0722.2006.00327.x. [DOI] [PubMed] [Google Scholar]
- 24.Kamata N, Fujimoto R, Tomonari M, Taki M, Nagayama M, Yasumoto S. Immortalization of human dental papilla, dental pulp, periodontal ligament cells and gingival fibroblasts by telomerase reverse transcriptase. J Oral Pathol Med. 2004;33:417–423. doi: 10.1111/j.1600-0714.2004.00228.x. [DOI] [PubMed] [Google Scholar]
- 25.Böker W, Yin Z, Drosse I, Haasters F, Rossmann O, Wierer M, et al. Introducing a single-cell-derived human mesenchymal stem cell line expressing hTERT after lentiviral gene transfer. J Cell Mol Med. 2008;12:1347–1359. doi: 10.1111/j.1582-4934.2008.00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shay JW, Pereira-Smith OM, Wright WE. A role for both RB and p53 in the regulation of human cellular senescence. Exp Cell Res. 1991;196:33–39. doi: 10.1016/0014-4827(91)90453?2. [DOI] [PubMed] [Google Scholar]
- 27.Harvey DM, Levine AJ. p53 alteration is a common event in the spontaneous immortalization of primary balb/c murine embryo fibroblasts. Genes Dev. 1991;5:2375–2385. doi: 10.1101/gad.5.12b.2375. [DOI] [PubMed] [Google Scholar]
- 28.Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, et al. Human Keratinocytes That Express hTERT and Also Bypass a p16INK4a-Enforced Mechanism That Limits Lifespan Become Immortal yet Retain Normal Growth and Differentiation Characteristics. Mol Cell Biol. 2000;20:1436–1447. doi: 10.1128/MCB.20.4.1436-47.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 30.Kitagawa M, Ueda H, Iizuka S, Sakamoto K, Oka H, Kudo Y, et al. Immortalization and characterization of human dental pulp cells with odontoblastic differentiation. Arch Oral Biol. 2007;52:727–731. doi: 10.1016/j.archoralbio.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Huang GTJ, Sonoyama W, Chen J, Park SH. In vitro characterization of human dental pulp cells: various isolation methods and culturing environments. Cell Tissue Res. 2006;324:225–236. doi: 10.1007/s00441-005-0117-9. [DOI] [PubMed] [Google Scholar]
- 32.Counter CM, Hahn WC, Wei W, Caddle SD, Beijersbergen RL, Lansdorp PM, et al. Dissociation among in vitro telomerase activity, telomere maintenance and cellular immortalization. Proc Natl Acad Sci USA. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang MK, Kameta A, Shin KH, Baluda MA, Park N-H. Senescence occurs with hTERT repression and limited telomere shortening in human oral keratinocytes cultured with feeder cells. J Cell Physiol. 2004;199:364–370. doi: 10.1002/jcp.10410. [DOI] [PubMed] [Google Scholar]
- 34.Tao Q, Lv B, Qiao B, Zheng Cq, Chen Zf. Immortalization of ameloblastoma cells via reactivation of telomerase function: Phenotypic and molecular characteristics. Oral Oncol. 2009;45:239–244. doi: 10.1016/j.oraloncology.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Dakic A, Chen RX, Disbrow GL, Zhang YY, Dai YH, et al. Cell-Restricted Immortalization by Human Papillomavirus Correlates with Telomerase Activation and Engagement of the hTERT Promoter by Myc. J Virol. 2008;82:11568–11576. doi: 10.1128/JVI.01318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haferkamp S, Tran SL, Becker TM, Scurr LL, Kefford RF, Rizos H. The relative contributions of the p53 and pRb pathways in oncogene induced melanocyte senescence. Aging (Albany, NY) 2009;1:542–556. doi: 10.18632/aging.100051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright WE, Shay JW. Telomere dynamics in cancer progression and prevention: fundamental differences in human and mouse telomere biology. Nat Med. 2000;6:849–851. doi: 10.1038/78592. [DOI] [PubMed] [Google Scholar]
- 38.Chimenti C, Kajstura J, Torella D, Urbanek K, Heleniak H, Colussi C, et al. Senescence and death of primitive cells and myocytes lead to premature cardiac aging and heart failure. Circ Res. 2003;93:604–613. doi: 10.1161/01.RES.0000093985.76901.AF. [DOI] [PubMed] [Google Scholar]
- 39.Torella D, Rota M, Nurzynska D, Musso E, Monsen A, Shiraishi I, et al. Cardiac stem cell and myocyte aging, heart failure and insulin-like growth factor-1 overexpression. Circ Res. 2004;94:514–524. doi: 10.1161/01.RES.0000117306.10142.50. [DOI] [PubMed] [Google Scholar]
- 40.Migliaccio M, Amacker M, Just T, Reichenbach P, Valmori D, Cerottini JC, et al. Ectopic Human Telomerase Catalytic Subunit Expression Maintains Telomere Length But Is Not Sufficient for CD8+ T Lymphocyte Immortalization. J Immunol. 2000;165:4978–4984. doi: 10.4049/jimmunol.165.9.4978. [DOI] [PubMed] [Google Scholar]
- 41.de Jonge HJM, Woolthuis CM, de Bont E, Huls G. Paradoxical downregulation of p16(INK4a) mRNA with advancing age in Acute Myeloid Leukemia. Aging (Albany, NY) 2009;1:949–953. doi: 10.18632/aging.100096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Counter CM, Hahn WC, Wei WY, Caddle SD, Beijersbergen RL, Lansdorp PM, et al. Dissociation among in vitro telomerase activity, telomere maintenance and cellular immortalization. Proc Natl Acad Sci USA. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oreffo ROC, Driessens FCM, Planell JA, Triffitt JT. Growth and differentiation of human bone marrow osteoprogenitors on novel calcium phosphate cements. Biomaterials. 1998;19:1845–1854. doi: 10.1016/S0142-9612(98)00084-2. [DOI] [PubMed] [Google Scholar]
- 44.Rao J, Otto WR. Fluorimetric DNA assay for cell growth estimation. Anal Biochem. 1992;207:186–192. doi: 10.1016/0003-2697(92)90521?8. [DOI] [PubMed] [Google Scholar]