Abstract
Most bona fide centrosome proteins, including centrins, small calcium-binding proteins, participate in spindle function during mitosis and play a role in cilia assembly in non-cycling cells. Although the basic cellular functions of centrins have been studied in lower eukaryotes and vertebrate cells in culture, phenotypes associated with centrin depletion in vertebrates in vivo has not been directly addressed. To test this, we depleted centrin2 in zebrafish and found that it leads to ciliopathy phenotypes, including enlarged pronephric tubules and pronephric cysts. Consistent with the ciliopathy phenotypes, cilia defects were observed in differentiated epithelial cells of ciliated organs, such as the olfactory bulb and pronephric duct. The organ phenotypes were also accompanied by cell cycle deregulation, namely, mitotic delay resulting from mitotic defects. Overall, this work demonstrates that centrin2 depletion causes cilia-related disorders in zebrafish. Moreover, given the presence of both cilia and mitotic defects in the affected organs, it suggests that cilia disorders may arise from a combination of these defects.
Key words: centrosome, cilia, centrin, mitosis, cystogenesis, ciliopathies, zebrafish
Introduction
Ciliopathies and cystogenesis have long been associated exclusively with cilia dysfunction.1 Recent results suggest that cilia proteins, well-characterized for their function in cilia in non-cycling cells, also contribute to cell cycle progression2 and spindle function in dividing cells.3,4 This suggests that multiple pathways may participate in the ciliopathy-related phenotypes. However, the contribution of cell cycle deregulation and mitotic dysfunction in cilia disorders is unclear.
Centrosomes are microtubule organizing centers that function at multiple cell cycle stages and are tightly linked to cell cycle progression.5,6 In mitotic cells, centrosomes contribute to spindle organization and orientation.7 In non-cycling cells, they are required for the formation of primary as well as motile cilia.8 In most cases, the mother centriole serves as the template for the assembly of these cilia. The exception is motile cilia of multi-ciliated epithelial cells, which assemble from centrioles produced de novo.9,10 Several protein components of the centrosome, including the small calcium-binding protein centrin, participate in assembly and maintenance of primary cilia in human cells.11–14
Centrin is a widely conserved core component of the centrosome, as it can be found in the proteomes of centrioles and basal bodies from various organisms.15–18 Up to four centrin isoforms have been described in mammals.19 Centrins associate with basal bodies in ciliated cells and with spindle poles in mitotic cells.20–25 Centrin1 and −4 appear to be expressed in terminally differentiated ciliated cells, whereas centrin2 and −3 are ubiquitously expressed in all somatic cells.26 Centrin2 is subject to extensive posttranslational modifications that regulate its function.22,26–29 It is required for centrosome organization and duplication and thus, plays a role in spindle function and cilia assembly and function.24,30–36 Non-centrosomal roles of centrins have also been described that include nucleotide excision repair,37 mRNA and protein export38,39 and proteasome activities.40 Although the basic cellular functions of centrins have been extensively studied over the past two decades, their biological roles in vivo in vertebrate organisms remain to be directly tested. In fact, the fundamental role of centrins has thwarted all known attempts to construct centrin-knockout mice, as they die early in development.35 To overcome this problem and directly test for phenotypes associated with centrin2 disruption in vertebrates, we targeted the centrin2 gene using morpholino oligonucleotides in zebrafish and monitored defects in embryos.
Results
Centrin2-depleted zebrafish embryos exhibit gross anatomical features of ciliopathies.
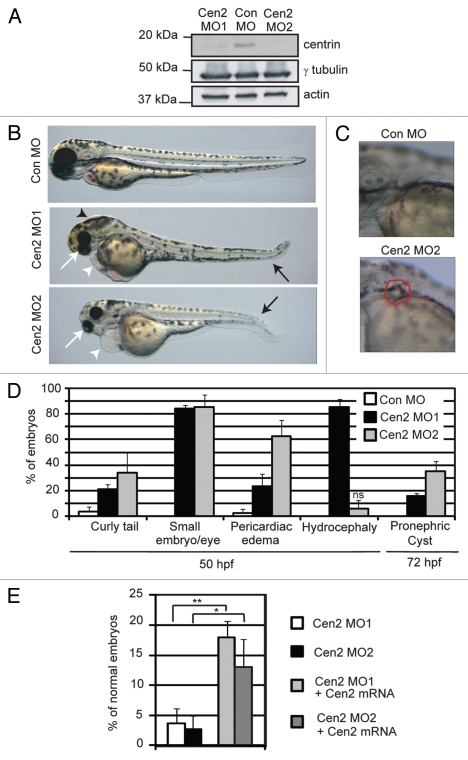
Centrin2 was depleted in zebrafish embryos, as it contributes to centrosome functions including cilia and mitotic defects in cultured cells,11,32 and its mRNA is localized to ciliated tissues in zebrafish, such as the olfactory organ, pronephric duct and spinal cord (http://zfin.org). Depletion of centrin2 revealed gross anatomical phenotypes characteristic of genes involved in ciliopathies (Fig. 1A–D).41,42 These included smaller embryos with smaller eyes, increased tail curvature, pericardiac edema and hydrocephaly (Fig. 1B and D). Some embryos also presented with cysts in the proximal region of the duct, one of the most common phenotypes associated with ciliopathies (Fig. 1C and D).41 Similar phenotypes were observed with two independent morpholinos demonstrating specificity for the centrin target gene. Differences in phenotypes (i.e., hydrocephaly) were most likely due to the use of ATG- vs. splice-site-blocking morpholinos, the former inhibiting translation of the maternal protein leading to earlier/stronger defects. Importantly, all global defects were partially complemented by re-expression of zebrafish centrin2 protein from injected mRNA (Fig. 1E).
Figure 1.
Centrin2-depleted zebrafish embryos exhibit gross anatomical features of ciliopathies. (A) Immunoblot from lysates of control (Con) or centrin2 (Cen2) morphants probed for centrin, γ tubulin and actin antibodies shows loss of centrin2 protein in centrin2 morphants (Cen2 MO1 and MO2). Actin and γ tubulin, loading controls. (B) Control and centrin2 morphants 48 h post fertilization (48 hpf). Defects in centrin2 morphants include smaller eyes (white arrows), curly tails (black arrows), pericardiac edema (white arrowheads) and hydrocephaly (black arrowhead). (C) Cyst in the proximal region of the pronephric duct (red line around cyst) in 72 hpf centrin2 morphant (lower part) compared with control (upper part). (D) Quantification of defects in centrin2 morphants compared with control. p < 0.01 unless otherwise stated. ns, p-value not significant. (E) Quantification of centrin morphants (Morpholino, MO1 and MO2) with defects after complementation with zebrafish centrin2 mRNA. Error bars, mean of three independent experiments ± SD (n > 50 embryo). *p < 0.05 and **p < 0.01.
Pronephric ducts of centrin2-depleted embryos are enlarged and disorganized.
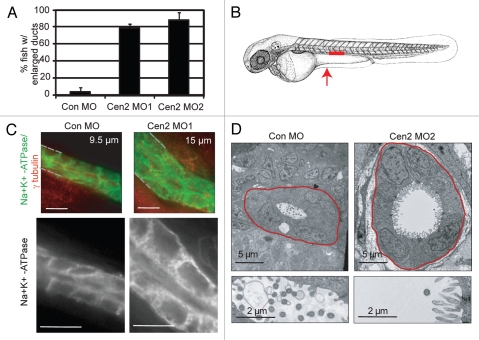
The appearance of cysts in the proximal region of the duct in zebrafish has been shown to result from defects in pronephric duct formation leading to obstruction in the distal region. Indeed, as previously shown for cilia proteins that lead to cyst formation in zebrafish,41 closer inspection of pronephric ducts of centrin2-depleted embryos revealed that, in over 80% of embryos, ducts were enlarged and had lost their linear organization at 48 h post-fertilization (Fig. 2A). This result was observed by immunohistochemistry (not shown), immunofluorescence and electron microscopy (Fig. 2A–D) and was consistent with the appearance of cysts in the proximal region of the duct in about 30% of the embryos later in development (Fig. 1C and D). Epithelial cells in centrin-depleted embryos showed loss of the tight organization typical of control embryos (Fig. 2C, lower parts). These results confirm that centrin depletion perturbs the overall organization of the pronephric ducts, subsequently leading to cyst formation in the proximal region of the duct.
Figure 2.
Centrin2 depletion in zebrafish embryos leads to enlarged pronephric ducts. (A) Quantification of 48 hpf embryos with enlarged pronephric ducts. Error bars represent an average of three independent experiments ± SD (n > 25 fish); p < 0.001. (B) Schematic showing region of the duct imaged in (C) (red line) and position of cross-sections used in (D) (red arrow). (C) Whole mount embryos stained by immunofluorescence for Na+ K+ ATPase (pronephric duct marker, green) and γ tubulin (centrosome, red); shown are maximum projections from stacks of confocal images. Dotted line indicates duct border. Diameter of the duct is shown in upper right corner. Scale bar, 10 µm. Insets (below), single plane images and enlargements showing cell disorganization and diffuse Na+ K+ ATPase marker in centrin2 morphants compared with control. (D) Electron micrographs showing crosssections of proximal pronephric duct regions in control (upper left) and centrin2 morphants (upper right). Enlargements (lower parts) show decreased number of cilia (cross-section) in the pronephric tubule lumen of centrin2 morphant (right) compared with control (left). Note: similar magnifications; red line indicates duct border.
Cilia are disrupted in centrin2-depleted zebrafish embryos.
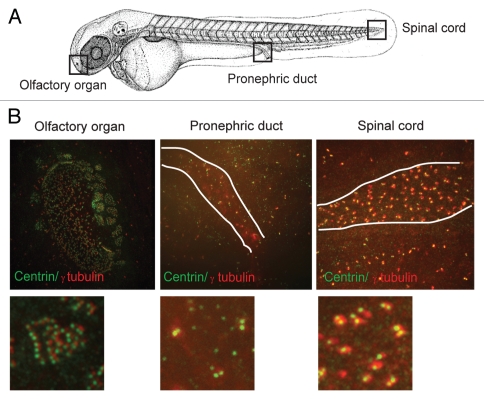
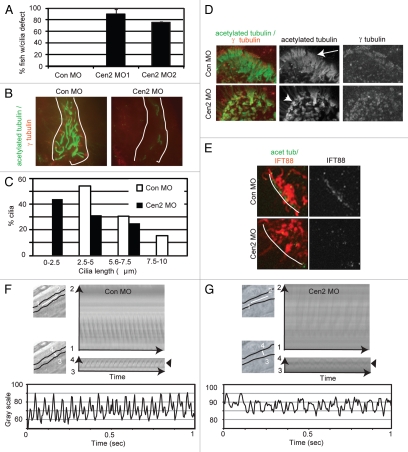
The ciliopathy phenotypes described above were previously observed in the context of cilia gene disruption and were attributed to cilia dysfunction.41,42 This suggested that centrin2 played a role in cilia formation in zebrafish. In fact, centrin2 localized to basal bodies at the base of cilia in ciliated tissues in zebrafish (Fig. 3), and 80% of whole-mount centrin2-depleted embryos exhibited cilia defects (Fig. 4A), including fewer (Fig. 4B) and shorter cilia in the pronephric duct (Fig. 4C). The defects seen by immunofluorescence microscopy using markers for stabilized microtubules (acetylation, polyglutamylation) or total microtubule polymer (α tubulin) (Fig. 4A–C) were also confirmed by electron microscopy on pronephric ducts cross-sections (Fig. 2D). Cilia defects were also observed in multiciliated cells of the olfactory organ (Fig. 4D). As expected, centrin2 was dramatically reduced at centrioles at the base of olfactory cilia, confirming loss of the targeted protein (data not shown). IFT88, a well-characterized cilia protein was also lost from centrosomes in centrin2-depleted embryos (Fig. 4E) suggesting that cilia defects resulted from centrosome disruption through the loss of proteins required for cilia assembly from this site. We next assessed the motility of the remaining motile cilia in the pronephric duct and olfactory organ of living embryos using time-lapse differential interference microscopy (Movies S1–3; Fig. 4F and G). In the pronephric duct of control embryos, cilia located at intervals along the duct beat coordinately producing a metachronous wave (Movie S1, control; Fig. 4F). In contrast, the sparse cilia in centrin2-depleted embryos showed uncoordinated (Movie S2, centrin2; Fig. 4G) or undetectable beating (Movie S3, centrin2). Similarly, cilia in the olfactory organ of control embryos showed strong beating along most of the edge of the olfactory pit, whereas little to no ciliary beating was observed in similar regions of centrin2-depleted embryos (data not shown). These results demonstrate cilia disruption in centrin2-depleted embryos and indicate that cilia defects could contribute to the ciliopathy phenotypes induced by centrin2 depletion in zebrafish.
Figure 3.
Centrin2 localization in ciliated tissues. (A) Schematic showing the olfactory organ, distal pronephric duct and spinal cord. (B) Centrin and γ tubulin localization in these tissues. White line, pronephric duct lumen and central canal of spinal cord.
Figure 4.
Centrin2 depletion in zebrafish leads to cilia defects. (A) Quantification of centrin2 morphants with cilia defects (stained as in B). Error bars, average of three independent experiments ± SD (n = 25 fish). p < 0.001. (B) Immunofluorescence images of the distal pronephric duct (cloaca region) of whole mount 48 hpf zebrafish embryos, stained for cilia (acetylated tubulin, green) and centrosomes (γ tubulin, red); white lines, lumen border. Fewer cilia are observed in centrin2 morphants. Scale bar, 10 µm. (C) Quantification of cilia length in the distal region of the pronephric duct (n = 30 individual cilia measured from at least three different embryos; stained as in D). Immunofluorescence stainning: α tubulin, cilia marker. (D) Immunofluorescence images of the olfactory organ of whole mount control and centrin2 zebrafish embryos stained for cilia and centrosomes as in (B) showing cilia defects in the olfactory organ of centrin2 morphant compared with control. Arrow, cilia; arrowhead, shorter cilia. (E) Immunofluorescence images of whole mount embryos showing cilia (acetylated tubulin, red) and IFT88 (green) in the olfactory organ. Basal body-associated IFT88 is reduced in centrin2 morphant compared with control. White line, edge of the olfactory pit. (F) Still frames from videos (left, upper and lower), kymograph (right, upper and lower) and corresponding plot (bottom part) of cilia movement in the proximal pronephric duct of control embryo. The position of the line used to create the kymograph along the tubule (upper left) and across the tubule (lower left) is indicated by the white line on the corresponding still frame. Arrowhead indicates the part of the kymograph used for the corresponding plot; the dotted black line outlines lumen of the duct. (G) Same as (F). for centrin2 morphant. All cilia analysis were done in 48 hpf embryos.
Centrin2 depletion in zebrafish embryos induces a delay in mitosis due to mitotic defects.
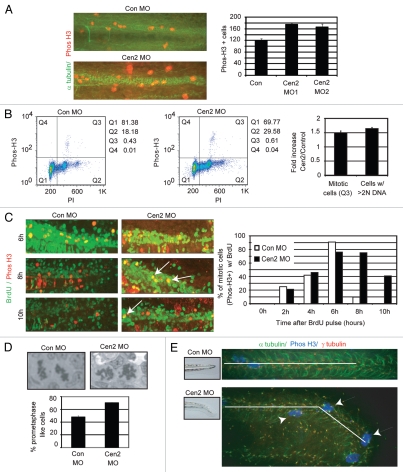
In addition to its role in cilia formation, centrin2 is required for proper cell division in human cultured cells.32 To determine if mitotic dysfunction contributes to global ciliopathy phenotypes observed in centrin-depleted embryos, we tested if centin depletion induced cell division defects in vivo in zebrafish. We first observed a 1.5-fold increase in the mitotic index as determined by manual counting of phospho-Histone H3-positive (mitotic) cells in the trunk, tail and pronephric duct of whole-mount embryo preparations (Fig. 5A). This was confirmed by flow cytometric analysis of cells dissociated from whole zebrafish embryos (Fig. 5B, mitotic cells, Q3 and summary graph fold increase). To determine whether the increased mitotic index reflected a mitotic delay due to mitotic defects or to increased cell proliferation, we assessed cell cycle progression in living zebrafish embryos by pulse labeling with BrdU followed by fixation at different chase times then staining for phos-H3 to identify mitotic cells. BrdU/phos-H3 double-positive cells (S-phase cells entering mitosis) were observed 2 h after the pulse in both control and centrin2-depleted embryos, suggesting that progression from S to M phase was similar under both conditions (Fig. 5C). Double-positive control cells peaked (∼80% of cells) ∼6 h post-pulse, decreased dramatically by 8 h (exit from mitosis) and were gone by 10 h. Double-positive cells in centrin2-depleted embryos peaked at 6–8 h, but in contrast to controls, a significant fraction (∼40%) remained in mitosis at least 10 h after pulse labeling (Fig. 5C). These data demonstrate that accumulation of mitotic cells is, at least in part, due to a delay in mitotic progression and most likely a consequence of mitotic defects. To directly test for mitotic defects in vivo, we analyzed histological sections of whole embryos. Centrin2-depleted embryos presented with an increase in bipolar prometaphase-like cells with misaligned chromosomes (∼70% vs. ∼50% in control, Fig. 5D) in the trunk, tail and pronephric duct. These mitotic defects most likely resulted in the observed cell cycle delay, the increase in cells with enlarged nuclei (Fig. 2D) and the 1.5-fold increase in aneuploid cells (> 2N DNA, Q2 + Q3 and summary graph fold increase; Fig. 5B). The accumulation of mitotic cells in zebrafish organs exhibiting ciliopathy phenotypes, namely, the pronephric duct and the tail (Fig. 5E), suggested that mitotic dysfunction may contribute cilia disorders in addition to cilia defects.
Figure 5.
Centrin2 depletion in zebrafish leads to mitotic defects and a delay in mitosis. (A) Immunofluorescence images of phospho-histone H3-positive cells (red) in the tail of control and centrin2 morphants. α tubulin, green. Quantification of phos-H3 positive cells in control and centrin2 morphants (right). p < 0.01. 48 hpf embryos. (B) Dot plots showing cell cycle analysis by flow cytometry analysis of cells from 48 hpf dissociated embryos stained for propidium iodide (PI) and phospho-histone H3 (phos-H3). Graph (right): fold increase in mitotic cells (phos-H3-positive and PI positive; Q3) and cells with DNA content > 2N in centrin2 morphants compared with control; average of three independent experiments ± SD, n = 30 dissociated embryos. n > 20,000 cells. (C) Immunofluoresence images of the tail of control and centrin2 morphants stained with anti-BrdU (green) and anti-phos-H3 (red) at selected time points (T, h) after BrdU incorporation. Quantification (right), time course of mitotic cell progressing from S phase into and out of G2/M as shown by BrdU incorporation followed by double phos-H3/BrdU staining at varying time points post-incorporation. Control shows peak colocalization (entry in mitosis) 6 h after BrdU pulse. Centrin2 MO shows peak colocalization 6 h after pulse, but, in contrast to control colocalization, remains even 10 h after BrdU pulse indicating a delay in exiting mitosis. Brdu incorporation done on 32 hpf embryos. (D) Toluidine blue staining of longitudinal histological sections of embryos showing mitotic cells with condensed DNA. Well-congressed chromosomes on metaphase plates in control embryos compared with prometaphase-like chromosome configurations in centrin2 morphants. Graph, prevalence of prometaphase-like cells in histological sections of 48 hpf embryos. n > 100 mitotic cells/fish. Average of three independent experiments ± SD p < 0.01. (E) Immunofluorescence images of the the spinal cord (tail bud region, inset) of whole mount embryos showing α tubulin (green), γ tubulin (red) and phos-H3 (blue) of control or centrin2 morphants. Mitotic cells (arrows) accumulate in the defective organs in centrin2-depleted embryos (48 hpf). White line, straight tail of control or downward curving tail of centrin2 morphant.
Discussion
This work leads to three major conclusions. First, we demonstrate a role for centrin2 in cilia disorders and cyst formation in a vertebrate organism. Given the fundamental role of centrosome proteins during the cell cycle, all known attempts to deplete centrosomal proteins, such as centrin in mice, have led to difficulties due to early embryonic lethality. Using an alternative strategy in zebrafish, we demonstrate a key role for centrin2 in vertebrate development. Second, taking advantage of zebrafish embryos as a platform for cell biology in vivo our data define a cellular role for centrin2 in both cilia formation and mitosis in vivo in a vertebrate organism. Finally, given that both cilia and mitotic defects are observed in defective zebrafish organs, this work suggests that like cilia, mitotic dysfunction may contribute to global ciliopathy phenotypes, including cyst formation. Along these lines, our data highlight the potential of zebrafish as a model organism to study cellular mechanisms contributing to developmental abnormalities, particularly embryonic defects.
Centrin2 is well-characterized for its function at the basal body11,24,34,35 and in dividing cells.32,43 In this context, our work confirms previously described roles of centrin in cilia formation and mitosis in vivo in a vertebrate and suggests that both defects can contribute to the disease phenotype. Further work will be required to address wether noncentrosomal roles of centrins37–40 can also contribute to the disease phenotype in vivo in vertebrates.
The phenotype of centrin2-depleted zebrafish embryos described here is essentially identical to that of the extensively characterized cilia protein depletion model of ciliopathy, from the gross anatomical features (e.g., cysts, curly tail or trunk) to the cell biological details (e.g., cilia and mitotic defects).3,41,42 In this context, our work suggests a centrosome-mediated model for ciliopathies involving both cilia and mitotic dysfunctions. In non-cycling cells, loss of centrosome integrity would lead to defects in cilia formation. In mitosis, centrosome/spindle pole defects would lead to mitotic dysfunction. In agreement with this model, recent studies suggest mitotic roles for well-characterized cilia proteins,3,4 although the mechanism of function remains to be fully addressed. Moreover, the fact that cell division is required for cyst formation supports a role for mitotic defects in cystogenesis/ciliopathy. Indeed, the fact that high proliferation rates during early development44–46 and during tissue regeneration following injury47 exacerbate cyst formation indicates that cell division is an important parameter in the induction of cysts and suggests that defects occurring during mitosis could contribute to ciliopathy-related phenotypes.
Depletion of a diversity of centrosome proteins disrupts primary cilia formation in cell culture.11–13 Centrosome proteins such as pericentrin, when mutated, affect cilia organization in vivo.48,49 It will be interesting to determine if centrin2 is in a subclass of centrosome proteins involved in ciliopathies or if centrosome proteins of diverse mitotic functions and locations within the centrosome (e.g., centrioles, pericentriolar material, centriole linkers, subdistal and distal appendages) contribute to these disorders in vertebrates. This work and future studies will provide insight into the precise contributions of centrosome proteins to cilia related disorders.
Finally, it is important to note that the term “ciliopathy,” accurately describes the disorganization/dysfunction of cilia commonly observed in these disorders. However, this study, together with other work showing that processes other than cilia formation and function are disrupted when cilia proteins are depleted (i.e., spindle function, cell division, cell polarity),3–5 suggests that cilia dysfunction may explain only part of the mechanistic underpinnings of these disorders. Further work will be required to clearly define the primary cause(s) of these disorders.
Methods
Zebrafish lines, morpholino injection, phenotype analysis and rescue experiment.
Wild-type zebrafish were raised according to standard protocols.51 1-Phenyl-2-thiourea (Sigma, PTU) was used to suppress pigmentation when needed according to standard protocols.51 Embryos were staged according to hours post-fertilization (hpf). Two types of morpholino antisense oligonucleotides (MO) were designed to target either the translation of the mRNA (AUG MO) or an exon splice donor site (exon 3) causing splicing defects of the mRNA (SP MO), both leading to a protein knockdown phenotype (Fig. 1A). MO were obtained from GENE TOOLS: centrin2 MO1-ATG, TGC TTT TCC TGA AGC CGG ACG CCA T; centrin2 MO2-SP GTT TGC TCA CCA TTT TCT GTG TCA T and standard control MO. 10 ng of control and centrin2 ATG MOs and 7.5 ng of centrin2 SP MO were used for injection in 1 cell stage embryo as previously described. Centrin2 splice MO efficacy was determined by RT-PCR. RNA was isolated using TRIzol reagent (Invitrogen) at 24 hpf, reverse-transcribed and subjected to PCR spanning targeted exon junctions (Cetn2ex3Forward TCG GGC GTA ATC GGC TTC AGC G; Cetn2ex4Reverse TCA TCG TCA AAC AGC CGG AAA GC; Cetn2ex5Reverse TCT CCC CGT CAC CGT CTC TGT CG). MO efficacy was also assessed by immunoblotting (see below). Global defects and cyst formation were observed in 48 hpf and 72 hpf embryos, respectively, with a MZFLIII dissection microscope (Zeiss). Rescue experiments were performed by co-injection of capped zebrafish centrin2 mRNA together with a MO. Capped mRNA was made using mMESSAGE mMACHINE (Ambion) and 15 pg was injected together with MO. Low amounts of mRNA were used as injection of higher levels of mRNA alone induced defects in embryos. Zebrafish mutant (ATG region) centrin2 cDNA was cloned using Gateway system (Invitrogen).
Histology and electron microscopy in zebrafish.
48 hpf embryos were fixed in 2% formaldehyde/2% glutaraldehyde, post-fixed in osmium tetroxide and processed for embedding in SPI-pon/Araldite. For light microscopic examination, 1-micron longitudinal serial sections were stained with toluidine blue. For electron microscopy, 80 nm sections were studied. Conventional protocols for electron microscopy were employed (Basic Techniques for Transmission Electron Microscopy. M.A. Hayat. Academic Press).
BrdU assay in zebrafish and flow cytometry.
BrdU assay in zebrafish were done as previously described in reference 52. For flow cytometry, embryos were grown in egg water to 48 hpf stage and were dechorionated by pronase treatment.51 Embryos were rinsed for 15 min in calcium free Ringer and passed several times through a 200 µL pipet tip to remove their yolk. Embryos were transferred into a 35 mm culture dish with 2 mL phosphate buffered saline (PBS, pH 8) containing 0.25% trypsin and 1 mM EDTA and incubated for 30 to 60 min at 28.5°C. The digest was stopped by adding CaCl2 to a final concentration of 1 mM and fetal calf serum to 10%. Cells were centrifuged for 3 min at 3,000 rpm, rinsed once with PBS and fixed and processed for flow cytometry. Phos-H3 staining (Ser 10 Phospho-Histone 3, alexa fluor 488 conjugate) was performed according to manufacturers' instructions (Cell Signaling).
Antibodies.
We are indebted to the following investigators for providing antibodies: 20H5 Centrin2 (J. Salisbury); IFT88 (G. Pazour); polyglutamylated tubulin (GT335) antibody (P. Denoulet, C. Janke). Anti-γ-tubulin polyclonal peptide antibody53 was also used (HM2569, S. Doxsey). Commercially available antibodies were also used: α-tubulin, FITC conjugate α-tubulin, BrdU and acetylated tubulin (Sigma); Ser10 Phos-H3 (Cell Signaling); Na+K+ ATPase (Developmental Studies Hybridoma Bank, University of Iowa).
Immunoblotting.
Lysates were obtained from 24 hpf zebrafish embryos. Lysis buffer included 50 mM Hepes (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% IGEPAL CA-630 and protease inhibitors (Mini tablets, Roche Diagnostics). Proteins were separated by SDS-PAGE and analyzed by protein gel blotting.
Immunofluorescence, microscopes and imaging software.
48 hpf whole embryos were fixed for immunofluorescence in Dent's Fix (80% methanol/20% DMSO) at 4°C overnight. Before antibody labeling, the fixed specimens were rehydrated, washed with PBS containing 0.5% Tween 20 (PBST), and blocked in 1X PBS-DBT (1% DMSO/1% BSA/0.5% Tween20) at room temperature for 2 h. Primary and then secondary antibody incubation were performed in 1x PBS-DBT at 4°C overnight and 1 h at room temperature, respectively, using 1x PBS-DBT washes between incubations. After rinsing in 1x PBS, the embryos were mounted and examined using a Perkin Elmer Ultraview spinning disk confocal microscope: Zeiss Axiovert 200 M, 100x Plan-APOCROMAT NA1.4 Oil or 63x Plan-APOCROMAT NA1.4 Oil and Hamamatsu ORCA-ER camera. Z stacks were acquired and used for creation of maximum projections. Images were processed on a MetaMorph workstation (Molecular Devices). MetaMorph software was used to measure distances required to mesure cilia length in the distal region of duct using confocal z-series projections. All immunofluorescence images are presented with anterior axis of the embryo to the left.
High-speed videomicroscopy in zebrafish.
Embryos were prepared as previously described in reference 42. Live embryos were observed with differential interference contrast microscopy using an inverted microscope (IX71; Olympus) equipped with a 60x oil immersion objective (NA 1.42). Images of ciliary activity were recorded at 200 frames per second with a high-speed, progressive scan charge-coupled device camera (TM-6740; Pulnix) and image acquisition software (Video Savant; IO Industries) as described previously in reference 54. Videos were displayed at 25 frames per second. Kymographs were prepared by extracting a row of pixels from each image of a series and placing them sequentially in time to create a single image using Scion Image (Scion Corporation).
Statistical analysis.
The statistical analysis and number of embryo or cells counted are indicated in figure legends. Error bars, average of at least at least three independent experiments ± SD. Statistical analysis was performed using Student t-test. p < 0.05 were considered as statistically significant.
Acknowledgments
We thank C. Powers for performing electron microscopy and histology experiments, M. Sanderson and P. Delmotte for assistance in filming and analyzing cilia movement. This work was supported by funding from the National Institutes of Health (GM51994) to S.J.D., the Polycystic Kidney Disease Foundation to B.D.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Hildebrandt F, Otto E. Cilia and centrosomes: a unifying pathogenic concept for cystic kidney disease? Nat Rev Genet. 2005;6:928–940. doi: 10.1038/nrg1727. [DOI] [PubMed] [Google Scholar]
- 2.Robert A, Margall-Ducos G, Guidotti JE, Bregerie O, Celati C, Brechot C, et al. The intraflagellar transport component IFT88/polaris is a centrosomal protein regulating G1-S transition in non-ciliated cells. J Cell Sci. 2007;120:628–637. doi: 10.1242/jcs.03366. [DOI] [PubMed] [Google Scholar]
- 3.Delaval B, Bright A, Lawson ND, Doxsey S. The cilia protein IFT88 is required for spindle orientation in mitosis. Nat Cell Biol. 2011;13:461–468. doi: 10.1038/ncb2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Wu M, Wang S, Shah JV, Wilson PD, Zhou J. Polycystic kidney disease protein fibrocystin localizes to the mitotic spindle and regulates spindle bipolarity. Hum Mol Genet. 2010;19:3306–3319. doi: 10.1093/hmg/ddq233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krämer A, Lukas J, Bartek J. Checking out the centrosome. Cell Cycle. 2004;3:1390–1393. doi: 10.4161/cc.3.11.1252. [DOI] [PubMed] [Google Scholar]
- 6.Holland AJ, Lan W, Cleveland DW. Centriole duplication: A lesson in self-control. Cell Cycle. 2010;9:2731–2736. doi: 10.4161/cc.9.14.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lüders J, Stearns T. Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol. 2007;8:161–167. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- 8.Rieder CL, Faruki S, Khodjakov A. The centrosome in vertebrates: more than a microtubule-organizing center. Trends Cell Biol. 2001;11:413–419. doi: 10.1016/S0962-8924(01)02085-2. [DOI] [PubMed] [Google Scholar]
- 9.Dawe HR, Farr H, Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J Cell Sci. 2007;120:7–15. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- 10.Mizukami I, Gall J. Centriole replication. II. Sperm formation in the fern, Marsilea, and the cycad, Zamia. J Cell Biol. 1966;29:97–111. doi: 10.1083/jcb.29.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikule K, Delaval B, Kaldis P, Jurcyzk A, Hergert P, Doxsey S. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat Cell Biol. 2007;9:160–170. doi: 10.1038/ncb1529. [DOI] [PubMed] [Google Scholar]
- 12.Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, et al. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jurczyk A, Gromley A, Redick S, San Agustin J, Witman G, Pazour GJ, et al. Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly. J Cell Biol. 2004;166:637–643. doi: 10.1083/jcb.200405023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosales JL, Rattner JB, Lee KY. Cdk5 in the centriolar appendages mediates cenexin1 localization and primary cilia formation. Cell Cycle. 2010;9:2037–2039. doi: 10.4161/cc.9.10.11600. [DOI] [PubMed] [Google Scholar]
- 15.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 16.Keller LC, Romijn EP, Zamora I, Yates JR, 3rd, Marshall WF. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 17.Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Jr, Culver BP, et al. New Tetrahymena basal body protein components identify basal body domain structure. J Cell Biol. 2007;178:905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Q, Tan G, Levenkova N, Li T, Pugh EN, Jr, Rux JJ, et al. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6:1299–1317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedberg F. Centrin isoforms in mammals. Relation to calmodulin. Mol Biol Rep. 2006;33:243–252. doi: 10.1007/s11033-006-9004-z. [DOI] [PubMed] [Google Scholar]
- 20.Spang A, Courtney I, Fackler U, Matzner M, Schiebel E. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J Cell Biol. 1993;123:405–416. doi: 10.1083/jcb.123.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Errabolu R, Sanders MA, Salisbury JL. Cloning of a cDNA encoding human centrin, an EF-hand protein of centrosomes and mitotic spindle poles. J Cell Sci. 1994;107:9–16. doi: 10.1242/jcs.107.1.9. [DOI] [PubMed] [Google Scholar]
- 22.Paoletti A, Moudjou M, Paintrand M, Salisbury JL, Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J Cell Sci. 1996;109:3089–3102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- 23.Baron AT, Greenwood TM, Bazinet CW, Salisbury JL. Centrin is a component of the pericentriolar lattice. Biol Cell. 1992;76:383–388. doi: 10.1016/0248-4900(92)90442-4. [DOI] [PubMed] [Google Scholar]
- 24.Ruiz F, Garreau de Loubresse N, Klotz C, Beisson J, Koll F. Centrin deficiency in Paramecium affects the geometry of basal-body duplication. Curr Biol. 2005;15:2097–2106. doi: 10.1016/j.cub.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 25.Laoukili J, Perret E, Middendorp S, Houcine O, Guennou C, Marano F, et al. Differential expression and cellular distribution of centrin isoforms during human ciliated cell differentiation in vitro. J Cell Sci. 2000;113:1355–1364. doi: 10.1242/jcs.113.8.1355. [DOI] [PubMed] [Google Scholar]
- 26.Salisbury JL. A mechanistic view on the evolutionary origin for centrin-based control of centriole duplication. J Cell Physiol. 2007;213:420–428. doi: 10.1002/jcp.21226. [DOI] [PubMed] [Google Scholar]
- 27.Yang CH, Kasbek C, Majumder S, Yusof AM, Fisk HA. Mps1 phosphorylation sites regulate the function of centrin 2 in centriole assembly. Mol Biol Cell. 2010;21:4361–4372. doi: 10.1091/mbc.E10-04-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein UR, Nigg EA. SUMO-dependent regulation of centrin-2. J Cell Sci. 2009;122:3312–3321. doi: 10.1242/jcs.050245. [DOI] [PubMed] [Google Scholar]
- 29.Lutz W, Lingle WL, McCormick D, Greenwood TM, Salisbury JL. Phosphorylation of centrin during the cell cycle and its role in centriole separation preceding centrosome duplication. J Biol Chem. 2001;276:20774–20780. doi: 10.1074/jbc.M101324200. [DOI] [PubMed] [Google Scholar]
- 30.Stemm-Wolf AJ, Morgan G, Giddings TH, Jr, White EA, Marchione R, McDonald HB, et al. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol Biol Cell. 2005;16:3606–3619. doi: 10.1091/mbc.E04-10-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giessl A, Pulvermuller A, Trojan P, Park JH, Choe HW, Ernst OP, et al. Differential expression and interaction with the visual G-protein transducin of centrin isoforms in mammalian photoreceptor cells. J Biol Chem. 2004;279:51472–51481. doi: 10.1074/jbc.M406770200. [DOI] [PubMed] [Google Scholar]
- 32.Salisbury JL, Suino KM, Busby R, Springett M. Centrin-2 is required for centriole duplication in mammalian cells. Curr Biol. 2002;12:1287–1292. doi: 10.1016/S0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- 33.Mana-Capelli S, Graf R, Larochelle DA. Dictyostelium discoideum CenB is a bona fide centrin essential for nuclear architecture and centrosome stability. Eukaryot Cell. 2009;8:1106–1117. doi: 10.1128/EC.00025-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vonderfecht T, Stemm-Wolf AJ, Hendershott M, Giddings TH, Jr, Meehl JB, Winey M. The two domains of centrin have distinct basal body functions in Tetrahymena. Mol Biol Cell. 2011:2221–2234. doi: 10.1091/mbc.E11-02-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trojan P, Krauss N, Choe HW, Giessl A, Pulvermuller A, Wolfrum U. Centrins in retinal photoreceptor cells: regulators in the connecting cilium. Prog Retin Eye Res. 2008;27:237–259. doi: 10.1016/j.preteyeres.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Schiebel E, Bornens M. In search of a function for centrins. Trends Cell Biol. 1995;5:197–201. doi: 10.1016/S0962-8924(00)88999-0. [DOI] [PubMed] [Google Scholar]
- 37.Dantas TJ, Wang Y, Lalor P, Dockery P, Morrison CG. Defective nucleotide excision repair with normal centrosome structures and functions in the absence of all vertebrate centrins. J Cell Biol. 2011;193:307–318. doi: 10.1083/jcb.201012093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischer T, Rodriguez-Navarro S, Pereira G, Racz A, Schiebel E, Hurt E. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat Cell Biol. 2004;6:840–848. doi: 10.1038/ncb1163. [DOI] [PubMed] [Google Scholar]
- 39.Resendes KK, Rasala BA, Forbes DJ. Centrin 2 localizes to the vertebrate nuclear pore and plays a role in mRNA and protein export. Mol Cell Biol. 2008;28:1755–1769. doi: 10.1128/MCB.01697-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen L, Madura K. Centrin/Cdc31 is a novel regulator of protein degradation. Mol Cell Biol. 2008;28:1829–1840. doi: 10.1128/MCB.01256-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun Z, Amsterdam A, Pazour GJ, Cole DG, Miller MS, Hopkins N. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 2004;131:4085–4093. doi: 10.1242/dev.01240. [DOI] [PubMed] [Google Scholar]
- 42.Kramer-Zucker AG, Olale F, Haycraft CJ, Yoder BK, Schier AF, Drummond IA. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer's vesicle is required for normal organogenesis. Development. 2005;132:1907–1921. doi: 10.1242/dev.01772. [DOI] [PubMed] [Google Scholar]
- 43.Tsang WY, Spektor A, Luciano DJ, Indjeian VB, Chen Z, Salisbury JL, et al. CP110 cooperates with two calcium-binding proteins to regulate cytokinesis and genome stability. Mol Biol Cell. 2006;17:3423–3434. doi: 10.1091/mbc.E06-04-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piontek K, Menezes LF, Garcia-Gonzalez MA, Huso DL, Germino GG. A critical developmental switch defines the kinetics of kidney cyst formation after loss of Pkd1. Nat Med. 2007;13:1490–1495. doi: 10.1038/nm1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davenport JR, Watts AJ, Roper VC, Croyle MJ, van Groen T, Wyss JM, et al. Disruption of intraflagellar transport in adult mice leads to obesity and slow-onset cystic kidney disease. Curr Biol. 2007;17:1586–1594. doi: 10.1016/j.cub.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lantinga-van Leeuwen IS, Leonhard WN, van der Wal A, Breuning MH, de Heer E, Peters DJ. Kidney-specific inactivation of the Pkd1 gene induces rapid cyst formation in developing kidneys and a slow onset of disease in adult mice. Hum Mol Genet. 2007;16:3188–3196. doi: 10.1093/hmg/ddm299. [DOI] [PubMed] [Google Scholar]
- 47.Patel V, Li L, Cobo-Stark P, Shao X, Somlo S, Lin F, et al. Acute kidney injury and aberrant planar cell polarity induce cyst formation in mice lacking renal cilia. Hum Mol Genet. 2008;17:1578–1590. doi: 10.1093/hmg/ddn045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Campos M, Basto R, Baker J, Kernan M, Raff JW. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J Cell Biol. 2004;165:673–683. doi: 10.1083/jcb.200402130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyoshi K, Kasahara K, Miyazaki I, Shimizu S, Taniguchi M, Matsuzaki S, et al. Pericentrin, a centrosomal protein related to microcephalic primordial dwarfism, is required for olfactory cilia assembly in mice. FASEB J. 2009;23:3289–3297. doi: 10.1096/fj.08-124420. [DOI] [PubMed] [Google Scholar]
- 50.Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, Otto EA, et al. Mapping the NPHP-JBTS-MKS Protein Network Reveals Ciliopathy Disease Genes and Pathways. Cell. 2011;145:513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westerfield M. The Zebrafish book: a guide for the laboratory use of zebrafish (Brachydanio rerio) Eugene, OR: University of Oregon Press; 1993. [Google Scholar]
- 52.Shepard JL, Amatruda JF, Stern HM, Subramanian A, Finkelstein D, Ziai J, et al. A zebrafish bmyb mutation causes genome instability and increased cancer susceptibility. Proc Natl Acad Sci USA. 2005;102:13194–13199. doi: 10.1073/pnas.0506583102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin OC, Gunawardane RN, Iwamatsu A, Zheng Y. Xgrip109: a gamma tubulin-associated protein with an essential role in gamma tubulin ring complex (gamma-TuRC) assembly and centrosome function. J Cell Biol. 1998;141:675–687. doi: 10.1083/jcb.141.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L, Sanderson MJ. Oscillations in ciliary beat frequency and intracellular calcium concentration in rabbit tracheal epithelial cells induced by ATP. J Physiol. 2003;546:733–749. doi: 10.1113/jphysiol.2002.028704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.