Abstract
Background. The dynamics of raltegravir-resistant variants and their impact on virologic response in 23 HIV-1–infected patients, who started a salvage raltegravir-containing regimen, were investigated.
Methods. Integrase population sequencing and Ultra-Deep-454 Pyrosequencing (UDPS) were performed on plasma samples at baseline and at raltegravir failure. All integrase mutations detected at a frequency ≥1% were considered to be reliable for the UDPS analyses. Phylogenetic and phenotypic resistance analyses were also performed.
Results. At baseline, primary resistance mutations were not detected by both population and UDPS genotypic assays; few secondary mutations (T97A-V151I-G163R) were rarely detected and did not show any statistically association either with virologic response at 24-weeks or with the development of resistant variants at failure. At UDPS, not all resistant variants appearing early during treatment evolved as major populations during failure; only specific resistance pathways (Y143R-Q148H/R-N155H) associated with an increased rate of fitness and phenotypic resistance were selected.
Conclusions. Resistance to raltegravir in integrase strand transfer inhibitor–naive patients remains today a rare event, which might be changed by future extensive use of such drugs. In our study, pathways of resistance at failure were not predicted by baseline mutations, suggesting that evolution plus stochastic selection plays a major role in the appearance of integrase-resistance mutations, whereas fitness and resistance are dominant factors acting for the late selection of resistant quasispecies.
The extreme variability and the high evolution rate of human immunodeficiency virus (HIV ) type 1 support the hypothesis that some drug resistance mutations may already exist as minor viral populations before the initiation of any antiretroviral therapy [1–3]. Such presence can be attributable to the natural background of mutations in the HIV-1 quasispecies, or vice versa, as an indication of transmission of drug-resistant HIV-1 strains. Several studies demonstrated that minority drug-resistant HIV-1 species, undetectable by standard genotyping assays, might increase the risk of virologic failure for the first-line highly active antiretroviral therapy (HAART) regimens, especially for patients treated with nonnucleoside reverse-transcriptase inhibitors (NNRTIs) [4–9].
Raltegravir is the first integrase strand transfer inhibitor (InStI) approved for use in clinical practice against HIV-1 infection [10–12]. For this reason, current use of this drug has created a requirement for resistance monitoring on the HIV-1 integrase gene [13]. Although it is an extremely potent antiretroviral drug, raltegravir showed a low-intermediate genetic barrier for drug-resistance development.
The resistance patterns associated with raltegravir failure include 7 primary mutations (E92Q, Y143C/R, Q148H/K/R, and N155H) directly involved in the development of resistance, and often associated with secondary mutations (L74M, T97A, E138A/K, G140A/C/S, Y143H, S147G, V151I, N155S, E157Q, G163K/R, I203M, and S230N/R), mostly able to reestablish the impaired fitness of resistant viruses [14–16].
Recent studies performed using standard sequencing reported the complete absence of primary InStI resistance mutations and the extremely infrequent detection of secondary resistance mutations in InStI-naive patients [15, 17–21]. Overall, a deep exploration of the natural resistance in InStI-naive patients needs further investigation, with special regard to whether natural polymorphisms and/or minority resistant variants may contribute to the efficacy and evolution of resistance under the InStI pressure. At present, the new technology of Ultra-Deep 454 Pyrosequencing (UDPS) is considered to be an ideal tool for investigating the presence of HIV-1 drug resistance mutations at frequency below the detection limit of standard genotyping assays [1, 22–26].
Therefore, on the basis of all these considerations, the goal of this study was to investigate and quantify, using UDPS, the presence of both primary and secondary resistance mutations and to evaluate their impact on the virologic response in a set of 23 HIV-1–infected InStI-naive patients who started a raltegravir-containing regimen.
MATERIALS AND METHODS
Patients
The study included 23 HIV-1–infected treatment-experienced patients followed up at 4 clinical centers in Italy who were mostly infected by subtype B strains and received raltegravir plus optimized background therapy. For patients with an available protease and reverse-transcriptase sequence at raltegravir therapy baseline, the genotypic sensitive score (GSS) according to the protease inhibitor (PI), NNRTI, or nucleoside reverse-transcriptase inhibitor (NRTI) coadministered with raltegravir was calculated using the Rega 8.02 algorithm (http://regaweb.med.kuleuven.be/sites/default/files/algorithms/Rega_HIV1_Rules_v8.0.2.pdf).
Integrase Population Sequencing
Integrase genotype analysis was performed at baseline and at raltegravir failure on plasma samples with use of a research-use protocol, as described elsewhere [18]. All integrase sequences were submitted to GenBank (accession No. JN544083-JN544122).
Massively Parallel Sequencing (UDPS)
Integrase UDPS was also performed at baseline and at raltegravir failure on plasma samples. Viral RNA was extracted from 1 mL of plasma (QIAamp Viral RNA kit; Qiagen). The same amount of viral RNA (21 μL), regardless of viral load, was reverse transcribed to cDNA, and the integrase region spanning amino acids 66–163 was amplified (HXB2 positions: forward primer, 4400→4423; reverse primer, 4743→4719). Amplicon primer pairs were tailed at their 5′end with the 454 specific sequencing primers, followed by a barcode. Addition of barcode sequences to the primers allowed the simultaneous processing of amplicons originating from multiple individuals in a single experiment [27]. To maximize the number of input templates and to minimize variation owing to polymerase chain reaction (PCR) drift, 7 parallel reverse-transcriptase PCR analyses were performed per patient sample and pooled [28–30]. Barcoded amplicons were equimolarly pooled and sequenced on the GS-FLX instrument, according to the manufacturer’s amplicon sequencing protocol (454 Life Sciences; Roche). Sequences were analyzed using Amplicon Variant Analyzer (AVA) software (454 Life Sciences; Roche), and haplotypes were extracted from the AVA alignments.
Mutations
Consensus B (http://hivdb.stanford.edu/) was used as a reference strain for the definition of mutations. Primary and secondary mutations associated (by in vitro or in vivo studies) with resistance to InStIs [12, 14, 16, 17, 19, 31–35], in addition to all other integrase mutations, were analyzed.
With use of UDPS, only mutations present in the integrase region (amino acids 66–163 ) were analyzed. In accordance with another study [30], mutations were accepted as real variants when present at a frequency of ≥1% among the total number of reads. In cases when the total number of reads was <5000, a fixed cutoff of 50 reads was used. The prevalence of each integrase mutation was calculated for each specific position according to all available sequences (range, 1189–16 300 sequences/residue) and to established reliability cutoffs. The linkage of resistance mutations was evaluated calculating their prevalence in unique overlapping sequences obtained by UDPS with a prevalence ≥1% (haplotypes spanning amino acids 90–163).
Phylogenetic Analyses
The phylogenetic analysis was performed for each patient on unique overlapping sequences obtained using UDPS (haplotypes spanning amino acids 90–163) and integrase population sequences. Phylogenetic trees were estimated using a maximum likelihood approach in PAUP, version 4.0 [36], using the transversion model (GTR + I + G) manually modified to optimize parameter settings. The statistical robustness and reliability of branching order were confirmed through a bootstrap analysis using 1000 replicates on a maximum likelihood tree produced by the PhyML 3.0 algorithm [37, 38] and through the Zero Branch Length Test.
Production of Recombinant Viruses and Replication Capacity Assays
Replication-competent recombinant viruses were generated by Amaxa nucleofection (Amaxa Biosystems) and tested for replication capacity in human T lymphocytic C8166 cells, as described elsewhere [18]. Integrase amplicons were cloned in an HXB2D-based integrase-deleted backbone with use of the In-Fusion Dry-Down PCR cloning technology (Clontech-Westburg), according to the manufacturer’s protocol [39, 40]. Cloning mixes were transformed into MAX Efficiency Stbl2 cells (Invitrogen) using the manufacturer’s procedure. Recombinant bacteria colonies populations from patients’ samples were washed and cultured to prepare DNA. Plasmid DNA was prepared using the QiaPrep Spin Miniprep system (Qiagen).
Drug Susceptibility Testing of Recombinant Viruses
Recombinant viruses were titrated and phenotypically tested in terms of drug susceptibility to raltegravir and elvitegravir, as described elsewhere [39, 40]. Biological cutoff values were 2.1 for raltegravir and 2.0 for elvitegravir [40]. Raltegravir and elvitegravir were obtained from Merck and Gilead Sciences, respectively.
Statistical Analyses
To assess the potential role of all integrase baseline mutations on raltegravir virologic response (in terms of frequency for interpatient comparisons and median percentage of variants for intrapatient comparisons), Fisher's exact test and median test were used to compare mutations among patients who achieved or did not achieve HIV-1 RNA levels <50 copies/mL at 24 weeks of raltegravir treatment. Wilcoxon matched-pairs signed rank test was used to compare the median number of mutations among patients detected using UDPS or population sequencing at baseline genotypes.
RESULTS
Patient Characteristics
Overall, 69.5% of the 23 study patients were male, with a mean age of 44 years; 91.3% were infected by HIV-1 B strains. All patients were heavily pretreated with antiretroviral therapy, including a median of 11 regimens containing NRTIs (interquartile range [IQR], 9–14), 2 with NNRTIs (IQR, 2–3), 10 with PIs (IQR, 5.5–11), and 1 with Enfuvirtide (T20) (IQR, 1–4). At baseline, among patients with available protease and reverse transcription sequences, the median GSS was 1 (IQR, 0–1.75). The median baseline CD4 cell count was 170 cell/mm3 (IQR, 21–276 cell/mm3), and the median HIV-1 RNA level was 5.1 log10 copies/mL (IQR, 4.7–5.4 log10 copies/mL). Fourteen of 23 patients did not achieve an HIV RNA level <50 copies/mL at 24 weeks of raltegravir treatment and were then considered as having experienced virologic failures (Table 1). No difference in demographic, viro-immunologic, GSS, and preexposure therapy data was observed among responding and nonresponding patients (data not shown). At raltegravir therapy baseline, both integrase population sequencing and integrase UDPS were performed for all patients. For 3 of 14 patients experiencing treatment failure, the HIV RNA level at failure was <500 copies/mL. For these patients, genotyping was not successful.
Table 1.
Patient Characteristics
| Genotypic Tests, No.a |
|||||||||||||
| Baseline |
Follow-up |
Overall |
|||||||||||
| Patient ID | Sex | CD4 Cell Count, Cells/mm3 | HIV RNA Level, log10 Copies/mL | Subtype | GSSb | Therapy With Raltegravir | HIV RNA <50 Copies/mL at 24 wk | UDPS | Popc | UDPS | Popc | UDPS | Popc |
| 12 | F | 24 | 5.1 | B | 0 | 3TC, DRV/r, T20 | No | 1 | 1 | 5 | 4 | 6 | 5 |
| 27 | M | 231 | 3.6 | B | 1.75 | DRV/r, TDF, ETR | No | 1 | 1 | 1 | 1 | 2 | 2 |
| 49 | F | 21 | 4.5 | B | 2.5 | DRV/r, FTC, TDF | No | 1 | 1 | 1 | 1 | ||
| 56 | M | 1 | 5.5 | B | 0 | 3TC, DDI, DRV/r, T20 | No | 1 | 1 | 1 | 1 | ||
| 69 | F | 328 | 4.4 | B | 0 | 3TC, DRV/r | No | 1 | 1 | 2 | 2 | 3 | 3 |
| 80 | M | 2 | 4.7 | F | ND | 3TC, AZT, DRV/r | No | 1 | 1 | 1 | 1 | 2 | 2 |
| 81 | M | 83 | 5.3 | B | ND | 3TC, ETR | No | 1 | 1 | 1 | 1 | 2 | 2 |
| 84 | M | 14 | 5.5 | B | 0.5 | 3TC, MVC, TDF | No | 1 | 1 | 7 | 4 | 8 | 5 |
| 141 | M | 276 | 4.9 | B | 1 | 3TC, AZT, DRV/r, ETR | No | 1 | 1 | 1 | 1 | 2 | 2 |
| 142 | M | 7 | 5.7 | B | ND | DRV/r, ETR, FTC, T20, TDF | No | 1 | 1 | 1 | 1 | ||
| 145 | M | 480 | 5 | B | 0 | 3TC, DRV/r | No | 1 | 1 | 1 | 1 | 2 | 2 |
| 156 | F | 320 | 5.4 | B | 1.5 | EFV, LPV/r | No | 1 | 1 | 1 | 2 | 1 | |
| 162 | M | 192 | 5.1 | B | 2.5 | ETR, SQV/r | No | 1 | 1 | 2 | 3 | 1 | |
| 229 | F | 8 | 4.7 | B | 1.5 | DRV/r, ETR, FTC, TDF | No | 1 | 1 | 3 | 2 | 4 | 3 |
| 5 | F | 315 | 5.3 | F | 2.5 | DRV/r, ETR | Yes | 1 | 1 | 1 | 1 | ||
| 15 | M | 178 | 5.2 | B | 0 | DRV/r, FTC, MVC, TDF | Yes | 1 | 1 | 1 | 1 | ||
| 16 | M | 132 | 4.6 | B | 0.5 | 3TC, DRV/r, TDF | Yes | 1 | 1 | 1 | 1 | ||
| 45 | F | 106 | 5.7 | B | ND | 3TC, DRV/r, EFV | Yes | 1 | 1 | 1 | 1 | ||
| 57 | M | 105 | 5.3 | B | ND | DRV/r, FTC, TDF | Yes | 1 | 1 | 1 | 1 | ||
| 58 | M | 563 | 4 | B | 0.75 | DRV/r, FTC, TDF | Yes | 1 | 1 | 1 | 1 | ||
| 63 | M | 170 | 5.6 | B | 1.75 | DRV/r, FTC, TDF | Yes | 1 | 1 | 1 | 1 | ||
| 151 | M | 207 | 4.7 | B | ND | DRV/r, FTC, TDF | Yes | 1 | 1 | 1 | 1 | ||
| 155 | M | 11 | 5.7 | B | 1 | ETR, T20 | Yes | 1 | 1 | 1 | 1 | ||
| Total (n = 23) | Median: 170 (IQR, 21–276) | Median: 5.1 (IQR, 4.7–5.4) | Median: 1.0 (IQR, 0–1.75) | 23 | 23 | 25 | 16 | 48 | 40 | ||||
Abbreviations: 3TC, lamivudine; AZT, zidovudine; DDI, didanosine; DRV/r, darunavir/ritonavir boosted; ETR, etravirine; FTC, emtricitabine; GSS, genotype sensitive score; HIV, human immunodeficiency virus; IQR, interquartile range; MVC, maraviroc; ND, not determined; Pop, population; SQV/r, saquinavir/ritonavir boosted; T20, enfuvirtide; TDF, tenofovir; .UDPS, Ultra-Deep-454 Pyrosequencing.
UDPS and population sequencing were not available for patients at failure ID49, ID56, ID142 (because with HIV-RNA levels < 500 copies/ml) and for all patients with HIV-RNA levels < 50 copies at 24 week.
The GSS to optimized background therapy was calculated for patients with an available protease and reverse-transcriptase sequence at raltegravir baseline, according to the coadministration of protease inhibitors, nonnucleoside reverse-transcriptase inhibitors, and nucleoside reverse-transcriptase inhibitors, obtained by using the Rega 8.02 algorithm.
Population sequencing.
Baseline Presence of Raltegravir Resistance Mutations and Virologic Response
No primary resistance mutations E92Q, Y143 C/R, Q148 H/K/R, and N155H were detected either by population sequencing or by UDPS. Secondary resistance mutations T97A, V151I, and G163R were detected in few patients as major and/or minor variants (Table 2). At baseline, as expected, UPDS was able to detect a higher number of integrase polymorphisms (median, 6; IQR, 5–11), compared with population sequencing (median, 3; IQR, 2–5; P < .001, by Wilcoxon test). However, no statistically significant difference was observed in the number of raltegravir resistance mutations between the 2 techniques used.
Table 2.
Baseline Ultra-Deep-454 Pyrosequencing (UDPS) Prevalence of Raltegravir Resistance Mutations and Virologic Response
| HIV RNA Level ≥50 Copies/mL at 24 wk (n = 14) |
HIV RNA Level <50 Copies/mL at 24 wk (n = 9) |
||||
| Mutationa | Frequency, No. (%) of Patients | Frequency Range, No. (%) of Variants | Frequency, No. (%) of Patients | Frequency Range, No. (%) of Variants | Overall Frequency, No. (%) of Patients (n = 23) |
| T97A | 1 (7.1) | 6491 (99.0) | 0 (0) | 1 (4.3) | |
| V151I | 2 (14.3) | 156–6106 (1.5–98.3) | 0 (0) | 2 (8.6) | |
| G163R | 0 (0) | 1 (11.1) | 500 (8.6) | 1 (4.3) | |
Abbreviation: HIV, human immunodeficiency virus.
Data are from http://hivdb.stanford.edu/cgi-bin/INIResiNote.cgi [16] and Johnson et al (2010) [14].
All primary raltegravir resistance mutations (E92Q, Y143C/R, Q148H/K/R, N155H) and the secondary mutations (L74M, F121Y, E138A/K, G140A/S, Y143H, S147G, N155S, E157Q) were absent.
None of the integrase mutations (found with UDPS or population sequencing) was statistically associated with virologic response or failure to raltegravir (median test, Fisher’s exact test; data not shown). However, the baseline secondary mutations T97A and V151I were found only in nonresponding patients. On the contrary, the G163R secondary resistance mutation was detected at baseline only in 1 responding patient (with a prevalence of 8.6%) (Table 2).
Dynamics of Raltegravir Resistance Mutations During Failure
The development of either primary or secondary resistance mutations at treatment failure did not statistically correlate with any baseline preexistent mutation. Among the 11 patients who experience treatment failure and were analyzed using UDPS at failure, the primary resistance mutations Y143R, Q148 H/R, and N155H, appeared in 1, 2, and 5 patients, respectively (Table 3). The other 3 patients experienced failure without any primary resistance mutation (data not shown).
Table 3.
Dynamics of Raltegravir Resistance Mutations Detected by Ultra-Deep-454 Pyrosequencing (UDPS)
| A: Patient Who Developed Primary Resistance Y143R Mutation | ||||||||||||
| Mutationsa |
Linked Mutations |
|||||||||||
| Patient and Time, mo | HIV RNA, log10 copies/mL | L74M | E92Q | T97A | T112Ab | Y143R | E157Q | Reads Analyzed, Mean ± SD | Mutationsc | Haplotypes, % | Haplotypes, No. | |
| 12 | 0 | 4.9 | 0 | 0 | 99.0 | 0 | 0 | 0 | 5266 ± 991 | 97A | 99.6 | 2746 |
| 1 | 5.0 | 0 | 1.9 | 99.8 | 0 | 95.0 | 0 | 9591 ± 3496 | 97A, 143R | 97.9 | 2986 | |
| 92Q, 97A | 1.8 | |||||||||||
| 3 | 5.2 | 0 | 0 | 99.6 | 0 | 99.8 | 0 | 10 809 ± 3912 | 97A, 143R | 100.0 | 3368 | |
| 7 | 5.1 | 1.7 | 0 | 99.4 | 31.0 | 99.7 | 0 | 12 805 ± 3083 | 97A, 143R | 100.0 | 427 | |
| 9 | 4.8 | 3.3 | 0 | 99.4 | 40.1 | 99.7 | 1.1 | 11 963 ± 3059 | 97A, 143R | 97.8 | 808 | |
| 97A, 112A, 143R | 2.2 | |||||||||||
| 12 | 4.8 | 9.0 | 0 | 99.7 | 66.9 | 99.6 | 6.9 | 12 154 ± 3199 | 97A, 143R | 65.7 | 487 | |
| 97A, 112A, 143R | 33.6 | |||||||||||
| B: Patients Who Developed Primary Resistance Q148H/R Mutations | ||||||||||||
| Mutationsa |
Linked Mutations |
|||||||||||
| Patient and Time, mo | HIV RNA, log10 copies/mL | G140A | G140S | Q148H | Q148R | Q140K | V151I | Reads Analyzed, Mean ± SD | Mutationsc | % | Haplotypes, No. | |
| 27 | 0 | 3.5 | 0 | 0 | 11 286 ± 2893 | Wild type | 100.0 | 4522 | ||||
| 10 | 3.6 | 99.4 | 99.4 | 10 604 ± 2618 | 140S, 148H | 100.0 | 5241 | |||||
| 84 | 0 | 5.7 | 0 | 0 | 0 | 0 | 6312 ± 2047 | Wild type | 100.0 | 1771 | ||
| 1 | 3.2 | 0 | 0 | 1.5 | 0 | 11 012 ± 2973 | Wild type | 100.0 | 5678 | |||
| 2 | 2.7 | 0 | 0 | 15.8 | 1.6 | 13 480 ± 3494 | 148R | 15.7 | 7488 | |||
| 3 | 4.7 | 4.3 | 34.0 | 63.3 | 0 | 6870 ± 2603 | 148R | 69.8 | 1443 | |||
| 140S, 148R | 12.0 | |||||||||||
| 4 | 5.4 | 1.0 | 66.7 | 70.7 | 0 | 7116 ± 2363 | 140S, 148R | 76.1 | 2362 | |||
| 5 | 5.1 | 3.7 | 68.1 | 76.3 | 0 | 5647 ± 2275 | 140S, 148R | 84.3 | 1198 | |||
| 3d | 5.5 | 0 | 1.6 | 1.8 | 0 | 6089 ± 1595 | 140S, 148R | 1.3 | 1810 | |||
| 9d | 5.8 | 0 | 0 | 1.2 | 0 | 10 909 ± 2952 | Wild type | 100.0 | 2908 | |||
| C: Patients Who Developed Primary Resistance N155H Mutation | |||||||||||||||
| Mutationsa |
Linked Mutations |
||||||||||||||
| Patient and Time, mo | HIV RNA, log10 copies/mL | L74M | E92Ab | E92Q | T97A | Y143C | V151I | N155H | E157Q | G163R | Reads Analyzed, Mean ± SD | Mutationsc | % | Haplotypes, No. | |
| 69 | 0 | 4.4 | 0 | 0 | 0 | 0 | 0 | 5014 ± 2215 | Wild type | 100.0 | 28 | ||||
| 155H | 51.3 | ||||||||||||||
| 5 | 3.8 | 30.6 | 4.1 | 6.5 | 98.1 | 2.2 | 4501 ± 768 | 92A, 155H | 38.6 | 2767 | |||||
| 151I, 155H | 6.4 | ||||||||||||||
| 92Q, 155H | 4.3 | ||||||||||||||
| 151I, 155H | 41.8 | ||||||||||||||
| 92Q, 155H | 22.1 | ||||||||||||||
| 7 | 3.6 | 8.7 | 21.2 | 41.6 | 97.4 | 4.3 | 5704 ± 1443 | 155H | 21.0 | 1766 | |||||
| 92A, 155H | 11.6 | ||||||||||||||
| 155H, 163R | 2.4 | ||||||||||||||
| 81 | 0 | 5.3 | 0 | 1190 ± 430 | Wild type | 100.0 | 856 | ||||||||
| 3 | 5.1 | 65.3 | 9861 ± 4011 | 155H | 64.5 | 2767 | |||||||||
| 141 | 0 | 4.9 | 98.3 | 0 | 0 | 0 | 9261 ± 2439 | 151I | 100 | 5832 | |||||
| 151I, 155H | 62.5 | ||||||||||||||
| 6 | 4.3 | 99.8 | 98.7 | 2.0 | 37.2 | 10 088 ± 2542 | 151I, 155H, 163R | 35.6 | 6433 | ||||||
| 151I, 155H, 157Q, 163R | 1.8 | ||||||||||||||
| 145 | 0e | 5.0 | 0 | 0 | 0 | 11 820 ± 3254 | Wild type | 100.0 | 6514 | ||||||
| 155H | 85.0 | ||||||||||||||
| 10e | 3.2 | 96.5 | 14.9 | 99.6 | 9195 ± 2522 | 151I, 155H | 15.0 | 5580 | |||||||
| 229 | 0e | 4.7 | 0 | 0 | 0 | 0 | 0 | 9300 ± 2361 | Wild type | 100 | 5164 | ||||
| 143C, 155H | 89.2 | ||||||||||||||
| 7e | 5.7 | 15.0 | 88.9 | 10.2 | 98.0 | 0 | 12 615 ± 3555 | 151I, 155H | 8.7 | 8922 | |||||
| 143C, 151I, 155H | 1.1 | ||||||||||||||
| 143C, 155H | 89.9 | ||||||||||||||
| 9e | 5.7 | 15.1 | 88.3 | 10.6 | 98.3 | 0 | 11 999 ± 3271 | 151I, 155H | 8.9 | 8201 | |||||
| 143C, 151I, 155H | 1.2 | ||||||||||||||
| 143C, 155H | 90.6 | ||||||||||||||
| 11e | 4.1 | 0 | 99.2 | 10.3 | 99.4 | 10.0 | 12 231 ± 3263 | 143C, 151I, 155H, 157Q | 9.4 | 8790 | |||||
Data include percentage of reads of raltegravir resistance mutations (as percentage ratio of number of reads mutated /total number of reads detected at each specific position) from integrase positions 66–163 (http://hivdb.stanford.edu/cgi-bin/INIResiNote.cgi [16]; Johnson et al, 2010 [14]) over time, relative mean ± standard deviation of reads detected at each specific position and HIV RNA level for patients with raltegravir failure. Linkage of resistance mutations is given as prevalence in unique overlapping sequences obtained with UDPS (haplotypes spanning amino acids 90–163) with the number of haplotypes reconstructed for each time point/patient.
Only raltegravir resistance mutations found in each patient are reported. Mutations F121Y-E138A/K-Y143H-S147G-Q148K were never detected. The first letter represents the wild type aminoacid (according to consensus B), followed by the integrase position and finally the mutated aminoacid.
Novel mutations.
Raltegravir-resistance and novel mutations found in haplotypes; “wild type” indicates haplotype without mutations.
Samples obtained after raltegravir interruption.
For these patients, only haplotypes spanning amino acid 130–163 integrase positions were available.
One patient immediately experienced treatment failure within the first month of raltegravir treatment, with the acquisition of primary Y143R mutation. At baseline, this patient harbored the secondary mutation T97A as the major variant (99% viral population) (Table 3). After 6 months, while still receiving raltegravir treatment despite failure, secondary mutations L74M, E157Q, and the novel T112A also gained their prevalence among other variants. At the latest time (M12), the mutations T97A, T112A, and Y143R were associated in 33.6% of haplotypes (Table 3)
Two patients developed the Q148H/R resistance pathway, including other secondary mutations (G140A/S, V151I) (Table 3). Patient ID84 showed a gradual increase in prevalence of Q148R, starting from month 1 (month 1, 1.5% of variants; month 2, 15.8%; month 3, 63.3%; month 4, 70.7%; month 5, 76.3%). Viruses harboring G140S+Q148R were detected at month 3, with an increase in prevalence of the G140S mutation during the failure (months 1 and 2, 0% of variants; month 3, 34.0%; month 4, 66.7%; month 5, 68.1%). Of interest, after raltegravir suspension, raltegravir-resistant viruses rapidly decreased over time: indeed, after 3 months of interruption, both G140S and Q148R mutations were still detectable but only by UDPS and at low frequency (<2%).
Of the 5 patients who developed the primary mutation N155H at raltegravir failure (Table 3), none had any secondary mutation at baseline, with the exception of a single patient who presented at baseline with the V151I secondary mutation (98.3% prevalence) (Table 2). The development of the N155H mutation was in nearly all patients, accompanied by the presence of other secondary mutations (L74M, E92Q, T97A, Y143C, V151I, E157Q, and/or G163R). In particular, the V151I mutation was selected in 3 of 5 N155H-failing patients, with a prevalence ranging from 6.5% to 41.6% of the total viral population. Of note, the patient already carrying the mutation V151I at baseline maintained it during raltegravir therapy failure.
Phylogenetic Analyses of Integrase Haplotypes During Raltegravir Treatment
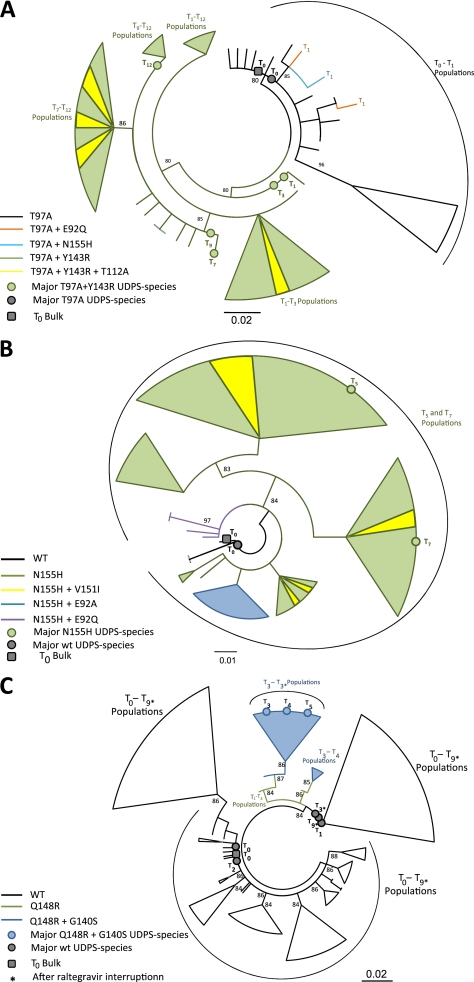
To better understand the evolution of integrase variants during raltegravir therapy failure, a phylogenetic analysis was performed for 3 representative patients who experienced treatment failure (ID12, ID69, ID84) on unique overlapping sequences obtained using UDPS and population sequencing of the integrase region spanning amino acids 90–163. Baseline sequences obtained by population sequencing were always found to be highly similar to the corresponding predominant viral strains, as determined by UDPS (Figure 1).
Figure 1.
Evolution of viral haplotypes during raltegravir failure. Maximum likelihood trees inferred for the haplotypes (90–163 integrase positions) from patients ID12 (A), ID69 (B), and ID84 (C). Dots indicate major variants (prevalence, >50%) detected by Ultra-Deep-454 Pyrosequencing, and squares indicate specimens by population sequencing. T0 indicates raltegravir baseline; T1−T12 , time (months) of sample collection during raltegravir treatment.
Patient ID12
The baseline (at T0) viral population consisted of different haplotype strains, all carrying T97A mutation. From the 1 with higher prevalence (53.4%), 3 different strains harboring specific resistance mutations (E92Q, N155H, or Y143R) developed after 1 month of failing raltegravir treatment. However, viral T1 strains expressing E92Q or N155H mutations were at low frequency and did not further evolve, because they were absent at later times. On the other hand, viral strains harboring the Y143R mutation rapidly took the advantage over all other T1 populations. Viral evolution since the third month was, thus, exclusively based on viral strains expressing T97A+Y143R. Several strains in T3, T7, T9, and T12 populations also developed the T112A mutation in addition to Y143R (Figure 1A).
Patient ID69
The baseline viral population consisted of different wild-type strains, from which subsequently, after 5 months of raltegravir treatment, different strains harboring the N155H resistance mutation developed. In N155H viral populations, mutations E92A, E92Q, and V151I were further developed at month 7, through different evolutionary pathways (prevalence, 11.6%, 22.1%, and 41.8%, respectively) (Figure 1B).
Patient ID84
In addition, in this case, the baseline viral population consisted of different wild-type strains. Major viral populations during the first 2 months were still mainly constituted by wild-type strains, even if some viral strains carrying Q148R were detected (at T1 and T2). Starting from month 3, a further acquisition of G140S mutation on Q148R resistant variants was observed. These resistant viruses were identified as the major viral populations starting from T3, T4, and T5 sampling times. Of interest, after raltegravir suspension, wild-type viruses dominated again in later time sampling, with resistant variants remaining detectable only at low frequency (haplotype Q148R+G140S after interruption at month 3, 1.3%) (Table 3; Figure 1C).
Phenotypic Resistance at Failure
Finally, to analyze the in vitro susceptibility to raltegravir in all patients who experienced treatment failure, phenotyping assays were performed on samples from both patients who developed and patients who did not develop the genotypic resistance. Phenotypic resistance was found only in samples with primary resistance mutations and it increased with further acquisition of other resistance mutations (Table 4). Indeed, the resistance associated with Y143R mutation, further increased by the presence of T112A and E157Q mutations during failure, was higher for raltegravir than for elvitegravir (fold change range, 28.2–205.5 for raltegravir and 4.6–14.2 for elvitegravir). On the contrary, the resistance associated with N155H alone was higher for elvitegravir than for raltegravir (fold change, 29.5 for elvitegravir and 4.5 for raltegravir). Of note, when N155H was associated with the novel mutation E92A, the resistance was ≥4-fold increased for both drugs (see patient 69 in Table 4). A very high phenotypic resistance to both drugs was observed in a single patient (ID 229) harboring N155H together with Y143C and S230R mutations (fold change, 1255.3 ± 297.1 for raltegravir and 625.3 ± 382.9 for elvitegravir). Finally, the Q148 pathway conferred high phenotypic resistance to both drugs (50–450-fold), especially for a combination of Q148H and G140S mutations (250–450-fold) (Table 4).
Table 4.
Phenotype Effect of Raltegravir Resistance Mutations on Raltegravir and Elvitegravir Susceptibility and Replication Capacity
| Fold Changeb |
||||
| Patient and Time, mo | Mutationsa | Raltegravir | Elvitegravir | Replication Capacityb |
| Patient 12 | ||||
| 0 | T97A | 1.2 ± 0.1 | 1.2 ± 0.4 | 100 ± 9.7 |
| 1 | T97A, Y143R | 28.0 ± 7.3 | 4.6 ± 0.9 | 17.3 ± 5.4 |
| 3 | T97A, Y143R | 33.5 ± 7.8 | 6.6 ± 2.9 | 13.3 ± 8.8 |
| 7 | T97A, Y143R, T112A/T | 59.4 | 4.8 | >100 |
| 9 | T97A, Y143R, T112A/T | 96.1 | 7.5 | >100 |
| 12 | T97A, Y143R, T112A, E157E/Q | 205.5 | 14.2 | >100 |
| Patient 27 | ||||
| 0 | No resistance mutations | 0.4 | 0.5 | 100 ± 2.8 |
| 6 | G140S, Q148H | 313.3 | >456.0 | 8.9 ± 2.4 |
| 9 | G140S, Q148H | 232.4 | >456.0 | 33.5 ± 2.3 |
| 12 | G140S, Q148H, S119S/T, T125T/A | 248.0 | >456.0 | 79.9 ± 7.0 |
| Patient 84 | ||||
| 0 | No resistance mutations | 0.9 ± 0.0 | 0.7 ± 0.1 | 100 |
| 4 | G140S, Q148R | 34.4 | 50.6 | ND |
| 5 | G140S, Q148Q/R | 1.4 ± 0.3 | 1.1 ± 0.2 | >100 |
| Patient 69 | ||||
| 0 | No resistance mutations | 1.3 | 0.8 | 100 ± 11.2 |
| 4 | E92E/A, N155Hc | 7.8 | 27.7 | 17.4 ± 5.5 |
| 5 | E92A, N155H | 31.6 ± 7.4 | 117.5 ± 34.4 | 41.3 ± 19.5 |
| 7 | V151I, N155H | 5.4 ± 1.9 | 7.5 ± 2.3 | 24.0 ± 7.8 |
| Patient 81 | ||||
| 0 | No resistance mutations | 0.77 | 1.0 | 100 ± 7.5 |
| 3 | N155H | 4.5 | 29.5 | 43.4 ± 15.7 |
| Patient 141 | ||||
| 0 | No resistance mutations | ND | ND | ND |
| 6 | V151I, N155H, G163G/R | 11.7 | 32.0 | ND |
| Patient 229 | ||||
| 0 | No resistance mutations | 1.2 | 0.8 | 100 ± 8.0 |
| 11 | Y143C, N155H, S230R | 1255.3 ± 297.1 | 625.3 ± 382.9 | 62.4 ± 19.3 |
Fold changes are mean values from 1 or 2 experiments, each performed with 4 replicate determinations in duplicate plates. Replication capacity experiments were performed in triplicate. Bold and underlined type indicates primary raltegravir resistance mutations; bold type, secondary raltegravir resistance mutations.
Mutations found in recombinant viruses.
Standard deviations were determined only when 2 experiments were performed. ND: not determined.
Mutations found in plasma sample.
To analyze the replication capacity of each recombinant virus that showed InStI resistance mutations, p24 gag antigen production in human C8166 T lymphocytes was also analyzed and compared with each baseline virus. After 5 days of infection, p24 production of recombinant viruses containing the primary mutations was, as expected, lower than the of the baseline virus. Of interest, in some cases, the replication capacity increased over time, according to the accumulation of mutations able to restore viral fitness, such as T112A and E157Q for the virus with T97A+Y143R (ID12) and S119T and T125A for the virus with Q148H+G140S (ID27).
DISCUSSION
In this study, with use of both population sequencing and UDPS, with a cutoff of reads of 1%, baseline primary resistance mutations for raltegravir (E92Q, Y143R/C, Q148H/K/R, and N155H) were absent in InStI-naive patients receiving raltegravir as a new drug in the context of a salvage regimen.
Few secondary resistance mutations, such as T97A, V151I, and G163R, were rarely detected at baseline, and their presence was not significantly associated with virologic response or failure to raltegravir at 24 weeks. In addition, the development of resistance at the time of failure did not statistically correlate with any baseline preexistent mutation but was associated only with the appearance of primary resistance mutations and further selection of specific pathways.
Nevertheless, mutations T97A and V151I were detected only in patients who experienced treatment failure, both at baseline (rarely) and at failure (more frequently). T97A is a compensatory InStI resistance mutation selected in vivo by raltegravir [31, 33, 34, 41–43], but its role in virologic failure is still unclear. Of interest, in both this and another recently published study [18], the T97A mutation, when found alone in the absence of primary mutations, does not change the raltegravir susceptibility in vitro. Similarly, V151I is a polymorphic mutation that has been selected in vitro by multiple Integrase Inhibitors, and it has no effect on raltegravir or elvitegravir susceptibility [18, 20, 34, 41, 44–46]. In this study, the V151I mutation was found at baseline in 2 patients, in both of whom the raltegravir-containing regimen failed, and at failure, it was often present with N155H as minor or major quasispecies.
Further investigation is needed to better assess the clinical impact of these (and all other) polymorphic mutations on raltegravir virologic response. For instance, in our study, phenotypic analyses confirmed the decreased susceptibility to raltegravir (and decreased replication capacity) only for viruses carrying primary resistance mutations (Y143C/R, Q148H/R, and N155H). The combination of primary mutations with other known mutations (L74M, E92Q, T97A, V151I, and E157Q) and the novel E92A and T112A was often found at failure and contributed to increasing the phenotypic resistance. Of interest, we found that the uncommon combination of N155H and Y143C mutations associated with a very high phenotypic resistance to both raltegravir (fold change, >1200) and elvitegravir (fold change, >600).
The evolution of resistance mutations over time was also inferred by phylogenetic analyses of the patients’ haplotypes generated by UDPS. By this approach, it was possible to analyze the intrapatient heterogeneity of wild-type HIV-1 integrase strains at baseline and their evolution under pharmacologic pressure during raltegravir treatment and after treatment interruption. In all 3 patients analyzed, we observed, at earlier times of failure, the presence of several haplotypes carrying different resistance mutations, with a different prevalence. However, not all of these variants evolved as major populations and/or accumulated additional mutations during failure. Indeed, at later times, only specific primary resistance mutations were selected, and specific pathways were generated and then maintained at low frequency after raltegravir interruption. This intrapatients UDPS analysis suggests that evolution plus stochastic selection plays a major role in the appearance of integrase resistance mutations, whereas fitness and resistance are dominant factors acting for the late selection of resistant quasispecies. Of interest, previous studies based on population sequencing similarly showed an evolution of resistance mutations during different phases of raltegravir failure [33, 34, 41, 43].
Overall, our results obtained by UDPS confirm other recent observations based on different ultrasensitive assays. Indeed, with use of a clonal approach, allele-specific PCR, or parallel allele-specific sequencing, primary resistance mutations were absent or very rare in InStI-naive patients [18, 47, 48], whereas secondary mutations were more frequently detected [47]. However, the frequency of nearly all detected resistance mutations was <1% of the viral population, and the frequencies of mutations between the raltegravir success and failure groups were similar [47–48], suggesting that these low-frequency resistance mutations do not contribute significantly to virologic failure. It is probable that additional studies with larger numbers of patients may improve the statistical power to better understand the clinical implications of these preexisting minority resistant mutations.
A potential limitation of our study is that resistant variants at frequencies <1% of the viral population were excluded and, thus, underestimated. For instance, and anecdotally, we found at baseline only in a single patient with multidrug-resistant virus (ID84) who was rapidly experiencing virologic failure to a raltegravir and maraviroc salvage treatment, 2 sequences (0.08% of the viral population, theoretically corresponding to 576 copies/mL of mutant virus in plasma) carrying the primary Q148R mutation below the UDPS reliability cutoff. By phylogenetic inference, when these 2 haplotypes potentially carrying Q148R were also exceptionally considered, 1 variant was identical to those found at failure. In a similar recently published clinical case report [49], a patient receiving raltegravir in the salvage regimen harbored at baseline the mutations N155H or Q148R at very low levels (4 sequences [0.12%] and 1 sequence [0.03%], respectively).
Therefore, according to these recent observations, the detection cutoff of minority resistance variants by ultrasensitive techniques should be defined and clinically validated. Virologic outcome is likely to be multifactorial and, thus, not simply dependent on the presence or absence of a baseline resistance mutation at low frequency [1–7]. Indeed, with use of allele-specific PCR, it was recently observed that, in treatment-naive patients, only the presence of K103N mutant virus in plasma at a level of >2000 copies/mL (eg, percentage of mutation multiplied by HIV-RNA level) correlated with an increased risk of virologic failure of the efavirenz-containing triple-drug regimens [6]. In this light, the mutational load of minority variants, considered as absolute number of viral variants with a specific mutation [6, 26], may have a different impact on virologic outcomes, especially for patients treated with drugs showing low-intermediate genetic barrier.
Taken together, our work suggests that primary resistance mutations detected at a frequency of <1% of the viral population should be considered with particular caution, especially in patients with high viral load and with many previous ARV-failures. In these patients with problematic cases, the risk of ineffectiveness of salvage regimens may favor the selection and the evolution of these potential preexisting minority resistant species.
In conclusion, natural resistance to raltegravir in InStI-naive patients remains a rare event that seems to not contribute significantly to virologic failure. In our study, pathways of resistance at failure were not predicted by baseline mutations present as minor or major quasispecies. Nevertheless, the current and future extensive use of InStI, increasing chances of InStI resistance transmission, could probably change these results, thus far, reported only in a single clinical case [50]. Therefore, the role of minority InStI resistance variants, together with the reliability and clinical cutoffs of ultrasensitive integrase genotyping, will deserve further investigation.
Notes
Acknowledgments.
We thank Michela Pollicita, Fabiola Di Santo, and Mario Santoro, for excellent technical assistance, and all the Resistance Group of the National Institute for Infectious Diseases ‘Lazzaro Spallanzani’: R. Acinapura, A. Ammassari, A. Antinori (cochair), G. Anzidei, F. Baldini, R. Bellagamba, A. Bertoli, E. Boumis, F. Ceccherini-Silberstein, S. Cerilli, R. D’Arrigo, P. De Longis, G. D’Offizi, F. Forbici, S. Galati, M. L. Giancola, E. Girardi, C. Gori, G. Liuzzi, P. Lorenzini, P. Marconi, S. Mosti, P. Narciso, V. Neri, E. Nicastri, C. F. Perno (cochair), M. M. Santoro, P. Sette, V Svicher, M. P. Trotta, V. Tozzi, U. Visco-Comandini, and M. Zaccarelli.
Financial support.
This work was supported by Merck Sharp & Dohme, the Italian National Institute of Health, the Ministry of University and Scientific Research, Current and Finalized Research of the Italian Ministry of Health, and the European Commission Framework 7 Program (Collaborative HIV and Anti-HIV Drug Resistance Network, Integrated Project 223131).
Potential conflicts of interest.
F. C. S. has received funds for attending symposia, speaking, and organizing educational activities, consultancy, and advisory board, grant research support, from Abbott, Merck Sharp & Dohme, Janssen Cilag, Gilead, and Virco. C. F. P. has received funds for attending symposia, speaking, organizing educational activities, grant research support, consultancy, and advisory board membership from Abbott, Boehringer Ingelheim, Bristol Myers Squibb, Gilead, Merck Sharp & Dohme, Janssen Cilag, Pfizer, Tibotec, Roche, and ViiV. L. S., I. V., H. V. M., L. V. W., and J. A. report being employees of Virco BVBA or Tibotec-Virco BVBA, Johnson & Johnson. All other authors: no reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Johnson JA, Geretti AM. Low-frequency HIV-1 drug resistance mutations can be clinically significant but must be interpreted with caution. J Antimicrob Chemother. 2010;65:1322–6. doi: 10.1093/jac/dkq139. [DOI] [PubMed] [Google Scholar]
- 2.Domingo E, Martin V, Perales C, Grande-Perez A, Garcia-Arriaza J, Arias A. Viruses as quasispecies: biological implications. Curr Top Microbiol Immunol. 2006;299:51–82. doi: 10.1007/3-540-26397-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–8. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paredes R, Lalama CM, Ribaudo HJ, et al. Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J Infect Dis. 2010;201:662–71. doi: 10.1086/650543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuritzkes DR, Lalama CM, Ribaudo HJ, et al. Preexisting resistance to nonnucleoside reverse-transcriptase inhibitors predicts virologic failure of an efavirenz-based regimen in treatment-naive HIV-1-infected subjects. J Infect Dis. 2008;197:867–70. doi: 10.1086/528802. [DOI] [PubMed] [Google Scholar]
- 6.Goodman DD, Zhou Y, Margot NA, et al. Low level of the K103N HIV-1 above a threshold is associated with virological failure in treatment-naive individuals undergoing efavirenz-containing therapy. AIDS. 2011;25:325–33. doi: 10.1097/QAD.0b013e3283427dcb. [DOI] [PubMed] [Google Scholar]
- 7.Buckton AJ, Harris RJ, Pillay D, Cane PA. HIV type-1 drug resistance in treatment-naive patients monitored using minority species assays: a systematic review and meta-analysis. Antivir Ther. 2011;16:9–16. doi: 10.3851/IMP1687. [DOI] [PubMed] [Google Scholar]
- 8.Metzner KJ, Giulieri SG, Knoepfel SA, et al. Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and -adherent patients. Clin Infect Dis. 2009;48:239–47. doi: 10.1086/595703. [DOI] [PubMed] [Google Scholar]
- 9.Geretti AM, Fox ZV, Booth CL, et al. Low-frequency K103N strengthens the impact of transmitted drug resistance on virologic responses to first-line efavirenz or nevirapine-based highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2009;52:569–73. doi: 10.1097/QAI.0b013e3181ba11e8. [DOI] [PubMed] [Google Scholar]
- 10.McColl DJ, Chen X. Strand transfer inhibitors of HIV-1 integrase: bringing IN a new era of antiretroviral therapy. Antiviral Res. 2010;85:101–18. doi: 10.1016/j.antiviral.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Bethesda, MD: Department of Health and Human Services; 2011. pp. 1–166. http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf. Accessed 1 February 2011. [Google Scholar]
- 12.Steigbigel RT, Cooper DA, Kumar PN, et al. Raltegravir with optimized background therapy for resistant HIV-1 infection. N Engl J Med. 2008;359:339–54. doi: 10.1056/NEJMoa0708975. [DOI] [PubMed] [Google Scholar]
- 13.Hirsch MS, Günthard HF, Schapiro JM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis. 2008;47:266–85. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 14.Johnson VA, Brun-Vezinet F, Clotet B, et al. Update of the drug resistance mutations in HIV-1: December 2010. Top HIV Med. 2010;18:156–63. [PubMed] [Google Scholar]
- 15.Varghese V, Liu TF, Rhee SY, et al. HIV-1 integrase sequence variability in antiretroviral naive patients and in triple-class experienced patients subsequently treated with raltgravir. AIDS Res Hum Retroviruses. 2010;26:1323–6. doi: 10.1089/aid.2010.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shafer RW. Rationale and Uses of a Public HIV Drug-Resistance Database. J Infect Dis. 2006;194 (Suppl 1):S51–8. doi: 10.1086/505356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sichtig N, Sierra S, Kaiser R, et al. Evolution of raltegravir resistance during therapy. J Antimicrob Chemother. 2009;64:25–32. doi: 10.1093/jac/dkp153. [DOI] [PubMed] [Google Scholar]
- 18.Ceccherini-Silberstein F, Baelen KV, Armenia D, et al. Secondary HIV-1 integrase resistance mutations, found as minority quasispecies in integrase therapy naive patients, have Little or no effect on susceptibility to integrase inhibitors. Antimicrob Agents Chemother. 2010;54:3938–48. doi: 10.1128/AAC.01720-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ceccherini-Silberstein F, Malet I, D'Arrigo R, Antinori A, Marcelin AG, Perno CF. Characterization and structural analysis of HIV-1 integrase conservation. AIDS Rev. 2009;11:17–29. [PubMed] [Google Scholar]
- 20.Low A, Prada N, Topper M, et al. Natural polymorphisms of human immunodeficiency virus type 1 integrase and inherent susceptibilities to a panel of integrase inhibitors. Antimicrob Agents Chemother. 2009;53:4275–82. doi: 10.1128/AAC.00397-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myers RE, Pillay D. Analysis of natural sequence variation and covariation in human immunodeficiency virus type 1 integrase. J Virol. 2008;82:9228–35. doi: 10.1128/JVI.01535-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simen BB, Simons JF, Hullsiek KH, et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis. 2009;199:693–701. doi: 10.1086/596736. [DOI] [PubMed] [Google Scholar]
- 23.Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res. 2007;17:1195–201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varghese V, Shahriar R, Rhee SY, et al. Minority variants associated with transmitted and acquired HIV-1 nonnucleoside reverse transcriptase inhibitor resistance: implications for the use of second-generation nonnucleoside reverse transcriptase inhibitors. J Acquir Immune Defic Syndr. 2009;52:309–15. doi: 10.1097/QAI.0b013e3181bca669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann C, Minkah N, Leipzig J, et al. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007;35:e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lataillade M, Chiarella J, Yang R, et al. Prevalence and clinical significance of HIV drug resistance mutations by ultra-deep sequencing in antiretroviral-naive subjects in the CASTLE study. PLoS One. 2010;5:e10952. doi: 10.1371/journal.pone.0010952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parameswaran P, Jalili R, Tao L, et al. A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res. 2007;35:e130. doi: 10.1093/nar/gkm760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polz MF. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–30. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandenbroucke I, Eygen VV, Rondelez E, Vermeiren H, Baelen KV, Stuyver LJ. Minor variant detection at different template concentrations in HIV-1 phenotypic and genotypic tropism testing. Open Virol J. 2008;2:8–14. doi: 10.2174/1874357900802010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vandenbroucke I, Van MH, Mostmans W, et al. HIV-1 V3 envelope deep sequencing for clinical plasma specimens failing in phenotypic tropism assays. AIDS Res Ther. 2010;7:4. doi: 10.1186/1742-6405-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fransen S, Gupta S, Danovich R, et al. Loss of raltegravir susceptibility by human immunodeficiency virus type 1 is conferred via multiple nonoverlapping genetic pathways. J Virol. 2009;83:11440–6. doi: 10.1128/JVI.01168-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hazuda D, Iwamoto M, Wenning L. Emerging pharmacology: inhibitors of human immunodeficiency virus integration. Annu Rev Pharmacol Toxicol. 2009;49:377–94. doi: 10.1146/annurev.pharmtox.011008.145553. [DOI] [PubMed] [Google Scholar]
- 33.Canducci F, Barda B, Ceresola E, et al. Evolution patterns of raltegravir resistance mutations after integrase inhibitor interruption. Clin Microbiol Infect. 2011;17:928–34. doi: 10.1111/j.1469-0691.2010.03375.x. [DOI] [PubMed] [Google Scholar]
- 34.Malet I, Delelis O, Valantin MA, et al. Mutations associated with failure of raltegravir treatment affect integrase sensitivity to the inhibitor in vitro. Antimicrob Agents Chemother. 2008;52:1351–8. doi: 10.1128/AAC.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baldanti F, Paolucci S, Gulminetti R, Brandolini M, Barbarini G, Maserati R. Early emergence of raltegravir resistance mutations in patients receiving HAART salvage regimens. J Med Virol. 2010;82:116–22. doi: 10.1002/jmv.21651. [DOI] [PubMed] [Google Scholar]
- 36.Wilgenbusch JC, Swofford D. Inferring evolutionary trees with PAUP*. Curr Protoc Bioinformatics. 2003 doi: 10.1002/0471250953.bi0604s00. Chapter 6:Unit 6.4. [DOI] [PubMed] [Google Scholar]
- 37.Guindon S, Delsuc F, Dufayard JF, Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol. 2009;537:113–37. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- 38.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 39.Baelen KV, Eygen VV, Rondelez E, Stuyver LJ. Clade-specific HIV-1 integrase polymorphisms do not reduce raltegravir and elvitegravir phenotypic susceptibility. AIDS. 2008;22:1877–80. doi: 10.1097/QAD.0b013e32830f9703. [DOI] [PubMed] [Google Scholar]
- 40.Baelen KV, Rondelez E, Eygen VV, et al. A combined genotypic and phenotypic human immunodeficiency virus type 1 recombinant virus assay for the reverse transcriptase and integrase genes. J Virol Methods. 2009;161:231–9. doi: 10.1016/j.jviromet.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 41.Canducci F, Sampaolo M, Marinozzi MC, et al. Dynamic patterns of human immunodeficiency virus type 1 integrase gene evolution in patients failing raltegravir-based salvage therapies. AIDS. 2009;23:455–60. doi: 10.1097/QAD.0b013e328323da60. [DOI] [PubMed] [Google Scholar]
- 42.Ceccherini-Silberstein F, Armenia D, D'Arrigo R, et al. Virological response and resistance in multi-experienced patients treated with raltegravir. Antivir Ther. 2008;13(A20Suppl 3) [Google Scholar]
- 43.Miller MD, Danovich RM, Yuxiong K, et al. Longitudinal analysis of resistance to the HIV-1 integrase inhibitor raltegravir: results from P005, a phase 2 study in treatment experienced patients. Antivir Ther. 2008;13A8(Suppl 3) [Google Scholar]
- 44.McColl DJ, Fransen S, Gupta S, et al. Resistance and crossresistance to first generation integrase inhibitors: insights from a phase 2 study of elvitegravir (GS-9137) Antivir Ther. 2007;12(A9Suppl 11) [Google Scholar]
- 45.Markowitz M, Nguyen BY, Gotuzzo E, et al. Rapid and durable antiretroviral effect of the HIV-1 Integrase inhibitor raltegravir as part of combination therapy in treatment-naive patients with HIV-1 infection: results of a 48-week controlled study. J Acquir Immune Defic Syndr. 2007;46:125–33. doi: 10.1097/QAI.0b013e318157131c. [DOI] [PubMed] [Google Scholar]
- 46.Rowley M. The discovery of raltegravir, an integrase inhibitor for the treatment of HIV infection. Prog Med Chem. 2008;46:1–28. doi: 10.1016/S0079-6468(07)00001-X. [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Miller MD, Danovich RM, et al. Analysis of low frequency mutations associated with drug-resistance to raltegravir before antiretroviral treatment. Antimicrob Agents Chemother. 2010;55:1114–9. doi: 10.1128/AAC.01492-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Charpentier C, Laureillard D, Piketty C, et al. High frequency of integrase Q148R minority variants in HIV-infected patients naive of integrase inhibitors. AIDS. 2010;24:867–73. doi: 10.1097/QAD.0b013e3283367796. [DOI] [PubMed] [Google Scholar]
- 49.Codoner FM, Pou C, Thielen A, et al. Dynamic escape of pre-existing raltegravir-resistant HIV-1 from raltegravir selection pressure. Antiviral Res. 2010;88:281–6. doi: 10.1016/j.antiviral.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 50.Young B, Fransen S, Greenberg K, et al. Program and abstracts of the 18th International AIDS Conference. Vienna: 2010. Transmission of integrase strand-transfer inhibitor multi-drug resistant HIV: case report and natural history of response to raltegravir-containing antiretroviral therapy. TUPE0163 http://pag.aids2010.org/Abstracts.aspx?AID=13018. [Google Scholar]