Abstract
The assembly of complex double-stranded DNA viruses includes a genome packaging step where viral DNA is translocated into the confines of a pre-formed procapsid shell. In most cases, the preferred packaging substrate is a linear concatemer of viral genomes linked head-to-tail. Viral terminase enzymes are responsible for both excision of an individual genome from the concatemer (DNA maturation) and translocation of the duplex into the capsid (DNA packaging). Bacteriophage λ terminase site-specifically nicks viral DNA at the cos site in a concatemer and then physically separates the nicked, annealed strands to mature the genome in preparation for packaging. Here we present biochemical studies on the so-called helicase activity of λ terminase. Previous studies reported that ATP is required for strand separation and it has been presumed that ATP hydrolysis is required to drive the reaction. We show that ADP and non-hydrolyzable ATP analogs also support strand separation at low (μM) concentrations. In addition, the Escherichia coli Integration Host Factor protein (IHF) strongly stimulates the reaction in a nucleotide-independent manner. Finally we show that elevated concentrations of nucleotide inhibit both ATP- and IHF-stimulated strand separation by λ terminase. We present a model where nucleotide and IHF interact with the large terminase subunit and viral DNA, respectively, to engender a site-specifically bound, catalytically-competent genome maturation complex. In contrast, binding of nucleotide to the low-affinity ATP binding site in the small terminase subunit mediates a conformational switch that down regulates maturation activities and activates the DNA packaging activity of the enzyme. This affords a motor complex that binds tightly, but non-specifically to DNA as it translocates the duplex into the capsid shell. These studies have yielded mechanistic insight into the assembly of the maturation complex on viral DNA and its transition to a mobile packaging motor that may be common to all of the complex double-stranded DNA viruses.
Keywords: Helicase, Viral Motor, Terminase, DNA Packaging, Virus Assembly
The assembly of a virus particle within an infected cell follows an ordered pathway that requires the coordinated activity of macromolecular complexes of both viral and host origins (1-3). These pathways are generally conserved within specific classes of viruses. For instance, the complex double stranded (ds) DNA viruses, such as the eukaryotic herpesvirus groups and many bacteriophages, require a DNA packaging step where the viral genome is inserted into a pre-assembled procapsid shell (4-6). This reaction is catalyzed by a terminase enzyme, fueled by the hydrolysis of ATP (5-7). The preferred packaging substrate is typically a linear concatemer of viral genomes linked head-to-tail and DNA packaging requires excision of a genome monomer from the concatemer (DNA maturation) and simultaneous translocation of the duplex into the confines of the capsid interior (DNA packaging). Terminase enzymes are responsible for both of these reactions.
All characterized terminase enzymes are hetero-oligomers composed of large (TerL, 49 to 72 kDa) and small (TerS, 18 to 21 kDa) subunits (5, 7). The TerL subunits possess all of the catalytic activities required for DNA maturation and packaging, while the TerS subunits are required for specific recognition of viral DNA. In most cases the subunit stoichiometry of the catalytically competent terminase motors remains ambiguous. The terminase enzyme from bacteriophage lambda (λ) has been intensively studied using genetic, biochemical, biophysical, and structural approaches and is ideally suited to mechanistic dissection of genome packaging reactions.
Infection of Escherichia coli by λ initiates with injection of its linear genome through the cell wall via the tail apparatus of the viral particle (8). The 48.5 kb duplex circularizes via 12 base, single-stranded “sticky” ends to form an intact cohesive end site, or cos (8, 9). During the latter stages of infection, the circular duplex is replicated by a rolling-circle mechanism that yields linear concatemers of the viral genome linked in a head-to-tail fashion (immature DNA) (10, 11); this is the preferred packaging substrate.
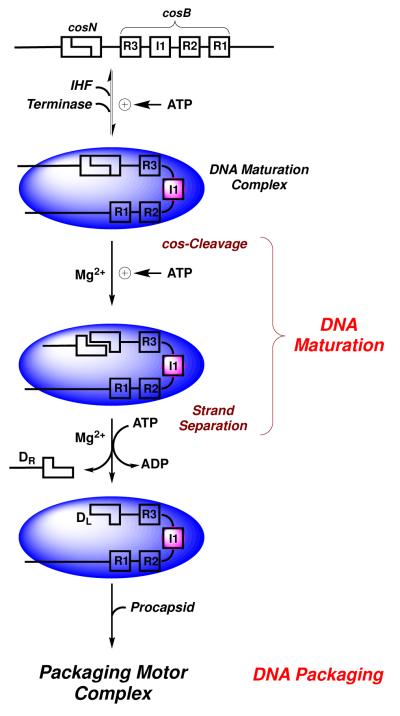
The λ terminase protomer is a stable, homogeneous heterotrimer composed of one TerL and two TerS subunits (TerL1•TerS2) (7, 12-15). The TerS subunit binds to the cosB (binding) subsite in λ DNA and is responsible for specific assembly of the packaging motor at cos (Figure 1) (13, 16). This positions TerL subunits at the cosN (nicking) subsite. A regulatory ATPase site in TerS modulates specific vs. non-specific DNA binding activity (17-19). Escherichia coli Integration Host Factor (IHF) promotes cooperative assembly of TerS, and thus terminase at cos (20-22). In all known cases, IHF binds to a consensus sequence, introducing a sharp bend in the duplex. This provides an architecture conducive to the assembly of additional proteins at that site (23). Consistently, we have demonstrated that IHF and TerS cooperatively bind and bend cosB-DNA (22). Based on a variety of biochemical, biophysical, and kinetic data, we have proposed that IHF and four terminase protomers cooperatively and specifically assemble at cos to engender the catalytically-competent DNA maturation/packaging complex, as depicted in Figure 1 (14, 15).
FIGURE 1. DNA Maturation and Packaging by λ Terminase.
E. coli IHF (pink circle) and λ terminase (blue oval) cooperatively assemble a DNA maturation complex at a cos site. Site-specific assembly is mediated by IHF and the terminase TerS subunit, which bind to the I1 element and to the three R-elements in cosB, respectively. Terminase is shown as a blue oval for simplicity. The TerL subunit sequentially nicks the duplex at cosN and then separates the strands to afford complex I (T1/2= 8 hours). TerL next binds to the portal ring of an empty procapsid, which triggers the transition to a powerful translocating motor complex that inserts DNA into the capsid interior.
Once assembled, the endonuclease activity of TerL introduces symmetric nicks into the cosN subsite (cos-cleavage reaction). The nicked, annealed duplex is G-C rich and strand separation requires the so-called “helicase” activity of TerL (24, 25). Both reactions, which in combination represent DNA maturation (Figure 1), require a dedicated maturation ATPase site in a C-terminal domain in gpA (26-28). DNA maturation affords complex I, a stable intermediate composed of terminase tightly and specifically bound to the mature left end of the λ genome (DL). Terminase next binds to the portal ring of a preassembled procapsid to afford the activated motor complex. The terminase motor translocates DNA into the capsid interior, powered by ATP hydrolysis at a dedicated packaging ATPase site in N-terminus of TerL (27, 29). Translocation continues until the packaging motor arrives at the next downstream cos site which signals the end of the packaged genome. The motor stops and the DNA maturation activities of terminase are reactivated; duplex nicking and strand separation yields the mature right genome end (DR) to complete the packaging of a unit-length genome. The DNA-filled capsid is further processed into an infectious viral particle with the addition of finishing proteins and a pre-assembled tail (7, 13).
We have previously described a kinetic interrogation of the cos-cleavage (21, 30) and strand separation (25) activities of λ terminase. Our initial studies demonstrated that ATP is required for strand separation. This was consistent with the presumption that the chemical energy of ATP hydrolysis is required to separate the annealed G-C rich duplex (13, 19, 25). More recently, however, we observed that ADP also supports the strand separation activity of the enzyme (31). Here we further investigate this surprising observation and provide evidence that separation of the nicked, annealed cosN duplex by terminase does not require the energy of ATP hydrolysis. We further interrogate the interaction of nucleotides and IHF in the sequential stimulation of cos-cleavage and strand separation activities of λ terminase. The results of these studies yield insight into a general mechanism for the nucleotide-regulated transition of a site-specifically bound genome maturation complex to a rapidly translocating packaging motor complex required for the assembly of double-stranded DNA viruses.
Experimental Procedures
Materials
Tryptone, yeast extract, agar, and ampicillin were purchased from Fisher Scientific. Terrific broth was purchased from Difco. Mature λ DNA (cI857ind 1 Sam 7) was purchased from Invitrogen. Restriction endonuclease AccI was purchased from New England BioLabs. All nucleoside triphosphates were purchased from Sigma-Aldrich. Thermo Scientific “Halt”® EDTA-free protease inhibitor cocktail (100X) was purchased from Thermo-Fisher. Chromatography media was purchased from GE Healthcare Life Sciences. All other materials were of the highest quality available. Unless otherwise stated, the pH of all buffers was adjusted at 4°C. Cell lysis utilized a Thermo Scientific IEC “French” laboratory press. All protein purifications utilized the Amersham Biosciences ÄKTApurifier™ core 10 System from GE Healthcare. UV-VIS absorbance spectra were recorded on a Hewlett-Packard HP8452A spectrophotometer.
Purification of Terminase Holoenzyme
Hexahistidine-tagged λ terminase was purified as described previously (32), with modification. Briefly, E. coli OR1265[pQH101] cells were grown to an OD600 of 0.6 - 0.8 AU at 32°C in one liter Terrific Broth containing 50 μg/ml ampicillin. Protein expression was then induced with the addition of an equal volume of fresh media at 65°C and the incubation was continued for 15 minutes at 45°C followed by 45 minutes at 42°C. The cells were then harvested by centrifugation, the cell pellet was taken into Buffer A (20 mM Tris, pH 8.6, containing 500 mM NaCl, 10% (v/v) glycerol, 7 mM β-ME, 1 mM EDTA, and 20 mM imidazole), and protease inhibitor cocktail (1X) was added to the mixture. The cells were lysed by 2 - 3 passages through the French Press (10,000 psi) and the lysates were clarified by centrifugation (6,000 × g × 40 minutes). The clarified lysate was loaded onto a HisTrap FF column (5 ml) and protein was eluted with a gradient to 500 mM imidazole in Buffer A. Eluting fractions were analyzed by SDS-PAGE and the terminase containing fractions (~150 mM imidazole) were pooled, diluted 5-fold with Buffer A, and then dialyzed against Buffer B (20 mM Na Phosphate, pH 6.8, containing 10% (v/v) glycerol, 1 mM EDTA, 7 mM β-ME, and 100 mM NaCl). The sample was loaded onto a HiTrap Q column (1 ml) and protein was eluted with a gradient to 1 M NaCl in Buffer B. Terminase containing fractions (~400 mM NaCl) were pooled and then stored at −80°C.
Purification of E. coli Integration Host Factor (IHF)
IHF was purified from HN880, a heat-inducible IHF overproducing strain (a kind gift of Howard Nash, National Institutes of Health, Bethesda, MD) using a protocol modified from Filutowicz, et al. (33). Briefly, the cells were grown to an OD600 of 0.6 at 32°C in 2 liters Terrific Broth containing 50 μg/ml ampicillin. Protein expression was induced with the addition of an equal volume of TB heated to 70°C and the cell culture was maintained at 46°C for 15 minutes followed by 42°C for 3 hours. The cells were harvested by centrifugation and the cell pellet was taken into Buffer C (50 mM Tris buffer, pH 7.0, containing 20 mM KCl, 1 mM EDTA, 7 mM β-mercaptoethanol, 10% glycerol v/v) with protease inhibitor cocktail. The cells were lysed by 2 passages through the French Press (10,000 psi), and the lysates were clarified by centrifugation (8,000 × g × 30 minutes). Solid ammonium sulfate was added to the clarified supernatant to 45% saturation and insoluble material was removed by centrifugation (8,000 × g × 30 minutes). Ammonium sulfate was then added to 85% saturation and the precipitated protein, which contained the majority of the IHF, was isolated by centrifugation (10,000 × g × 30 minutes). The pellet was taken into Buffer C and dialyzed against the same buffer overnight. The sample was loaded onto a HiTrap Q column (1 ml) equilibrated with Buffer C. The flow-through fraction, which contained IHF, was collected and then loaded onto a HiTrap Heparin HP (5 ml) column equilibrated with Buffer C. The protein was eluted with a linear gradient to 1.5 M KCl in Buffer C. IHF-containing fractions (~870 mM KCl) were pooled, dialyzed against Buffer C containing 20% glycerol (v/v), and stored at −20°C.
Preparation of the DNA Substrates
The DNA substrate used in the strand separation assay was prepared as previously described (25). Briefly, mature λ DNA was digested to completion with AccI. This yielded 10 fragments including 5.6 kb and 2.2 kb duplexes that contain the mature right (DR) and left (DL) genome ends, respectively. The digestion products were purified using the Wizard® SV Gel and PCR Clean-up System (Promega). The purified duplexes were incubated for 2 hours at 50°C in 50 mM Tris buffer, pH 8.0, containing 1 mM EDTA and 2.5 mM MgCl2 and then slowly cooled to room temperature. This afforded a 7.8 kb duplex that contains a nicked, annealed cos sequence in the background of 40.7 kb non-specific duplex DNA. The molar concentration of cos-DNA was determined spectrally as previously described (25).
The DNA substrate used in the cos cleavage assay, pCT-λ, is a 12.5 kb duplex that contains the intact cos sequence (30). The duplex was linearized with ScaI according to manufacturer’s recommendations, phenol-chloroform extracted, and precipitated with ethanol. The linearized DNA was resuspended with TE buffer and the final DNA concentration was determined by UV spectroscopy as previously described (30).
Strand Separation Activity Assay
Unless otherwise indicated, the reaction mixtures (10 μl) contained 5 nM cos-DNA, 10 μM ATP, and 50 nM IHF in 20 mM Tris buffer, pH 8, containing 6 mM MgCl2, 1 mM EDTA, and 7 mM β-ME. The reaction was initiated with the addition of 100 nM terminase and was allowed to proceed at 30°C for 15 minutes unless otherwise indicated. Aliquots were quenched at the indicated times with the addition of 4 μl helicase stop buffer (200 mM EDTA, 20% glycerol, 0.16% bromophenol blue, 0.16% xylene cyanol). The products were fractionated on a 0.8% agarose gel and visualized by staining with 1 μg/ml ethidium bromide. Quantitation of the strand separated cos-DNA products was performed by video densitometry as previously described (21, 25).
Kinetic Analysis
Strand separation time course data was analyzed according to a monophasic exponential time course,
| (1) |
where DNA products is the amount of DNA separated at time τ, A is the extent of the reaction at infinite time, and kobs is the observed rate of the reaction. The values of A and kobs were determined by nonlinear regression analysis of the experimental data using the IGOR® data analysis program (Wave Metrics, Portland, OR) (21, 34).
Calculation of the Nucleotide•Terminase Apparent Dissociation Constant (KD,app)
Nucleotides bind to the terminase enzyme and stimulate strand-separation. To characterize nucleotide binding, the strand separation assay was performed as described above with varying nucleotide concentration as indicated. The activity vs. concentration data were fit to a simple equilibrium binding model,
| (2) |
where DNA products is the amount of DNA separated in the presence of nucleotide at concentration [X], Min and Max represent the DNA products in the absence and presence of saturating nucleotide, respectively, and KD,app is the apparent equilibrium dissociation constant for nucleotide binding to terminase. The variables Max, Min, and KD,app were determined by nonlinear regression of the experimental data.
Calculation of the IHF•DNA Apparent Dissociation Constant (KD,app)
IHF binds to cos-DNA and promotes strand-separation by the terminase enzyme. The apparent affinity of IHF for cos-DNA was determined as described above except that nucleotides were omitted and that IHF was added to the reaction mixture at the indicated concentrations. The activity vs. IHF concentration data was fitted to equation 2 where [X] = [IHF]. The variables Max, Min, and KD,app were determined by nonlinear regression of the experimental data.
The IHF binding data were also analyzed according a multi-site, cooperative Hill model,
| (3) |
where DNA products is the amount of DNA separated in the presence of IHF at concentration [X], Min and Max represent the DNA products in the absence and presence of saturating IHF, respectively. KD,app is the apparent equilibrium dissociation constant, and n is the Hill coefficient. The variables Max, Min, and KD,app, and n were determined by nonlinear regression of the data.
cos-Cleavage and in situ Strand Separation Assay
The cos-cleavage reaction was performed as described previously (30). Briefly, the reaction mixtures (20 μl) contained 5 nM ScaI-linearized pCT-λ DNA in 20 mM Tris buffer, pH 8, containing 10 mM MgCl2, 23 mM NaCl, 7 mM β-ME, and 0.5% glycerol. Nucleotides and IHF were included as indicated in each individual experiment. The reaction was initiated with the addition of terminase to a final concentration of 100 nM and was allowed to proceed for 40 minutes at 37°C. Two μL of 500 mM EDTA was then added to stop the reaction and the extent of strand separation was determined by analysis of the products by 0.6% agarose gel, as described above. Alternatively, the extent of cos-cleavage (duplex nicking) was analyzed by heating the quenched reaction mixture at 75°C for 30 seconds prior to loading onto a 0.6% agarose gel.
Results
Requirements for Strand Separation
We previously reported that the strand separation activity of λ terminase is dependent on divalent metal and ATP (25). This is consistent with several published studies (24, 35) and it has been assumed that the energy of ATP hydrolysis is required to separate the 12 base pair, GC-rich duplex (Figure 1). More recently, we found that ADP also supports the strand separation activity of the enzyme (31), bringing into question the energetic requirement for the reaction. A major difference between our recent study and all prior studies is the inclusion of IHF in the reaction mixture. We previously postulated that IHF might drive a DNA conformation that is susceptible to duplex melting by terminase (31). Here, we directly test this hypothesis.
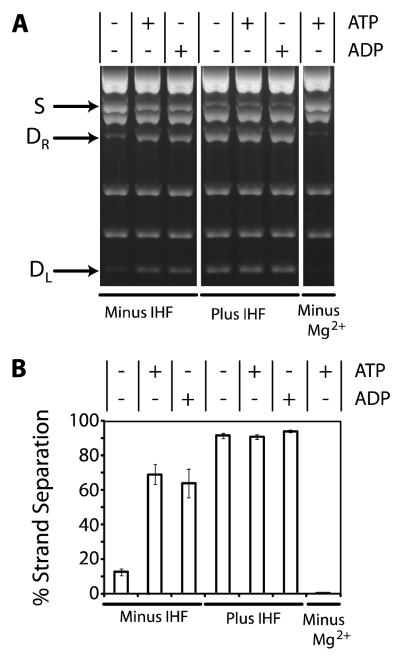
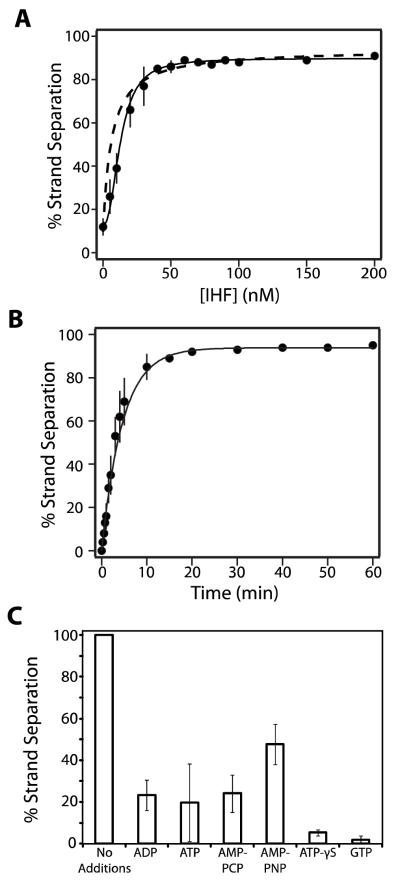
The data presented in Figure 2 reveals several important features of the terminase-mediated strand separation reaction. First, ATP strongly stimulates the reaction in an IHF-independent manner, as originally reported (25). Second, ATP hydrolysis is not required under these conditions, because ADP promotes strand separation with similar efficiency. Third, IHF strongly stimulates terminase mediated strand separation in a nucleotide-independent manner. By itself, however, IHF lacks any detectable activity (data not shown). Finally, magnesium is strictly required for terminase-mediated strand separation by both nucleotides and by IHF.
FIGURE 2. The Strand Separation Activity of λ Terminase.
Panel A The strand separation assay was performed as described in Experimental Procedures except that ATP (10 μM), ADP (10 μM), and/or IHF (50 nM) were included as indicated. The migration of the nicked, annealed substrate (S) and the strand separation products (DR and DL) in the agarose gel are indicated with arrows. Panel B. Quantitation of the data presented in Panel A. Each bar represents the average of at least three separate experiments with standard deviations indicated.
Nucleotide Requirement for Strand Separation
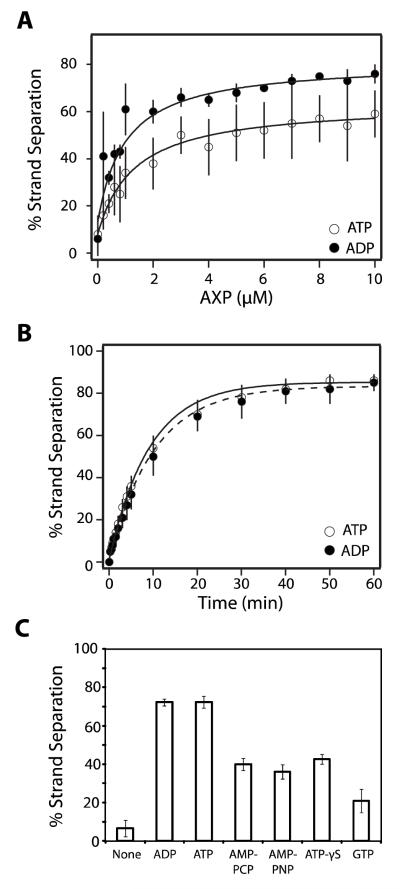
In the absence of IHF, the strand separation activity of terminase is strongly stimulated by both ATP and ADP in a concentration-dependent manner (Figure 3A). We presume that AXP1 binds to the maturation ATP binding site in TerL and that the binary terminase•AXP complex is catalytically active. Therefore, each data set was fit to a simple binary binding model (Equation 2), which indicates that both nucleotides bind to the maturation ATPase site with similar affinity (, ).
FIGURE 3. Nucleotides Stimulate Strand Separation Activity in the Absence of IHF.
Panel A. The strand separation assay was performed for 15 minutes as described except that IHF was omitted and the indicated concentration of nucleotide was included in the reaction mixture. Each data point represents the average of at least three separate experiments with standard deviation indicated with a bar. The solid lines represent the best fit of the data to equation 2. Panel B. The strand separation assay was performed in the absence of IHF and the presence of 10 μM nucleotide for the indicated time. Each data point represents the average of at least three separate experiments with standard deviation indicated. The solid lines represent the best fit of the data to equation 1. Panel C. The strand separation assay was performed as described except that IHF was omitted and ATP was replaced with the indicated nucleotide at a concentration of 10 μM AMP-PNP, β,γ-imidoadenosine 5′-triphosphate, AMP-PCP, β,γ-methylene-adenosine 5′-triphosphate. Each bar represents the average of at least three separate experiments with standard deviation indicated. We note that there is some day-to-day variability in the observed extent to which the strand separation reaction is stimulated by ATP (but not ADP); this is most evident in the data presented in panel A. Notwithstanding, the ensemble of data indicate that there is little difference between the two nucleotides in their capacity to stimulate strand separation (compare Figures 2, 3, 4, 5, and 6).
We next examined the time course for the reaction at saturating concentrations of nucleotide (10 μM). We note that strand separation affords complex I (Figure 1), which has a half-life of over 8 hours under these conditions (16, 21). Further, the reaction was performed in the presence of excess terminase and the time course represents a single catalytic turnover by the enzyme. Analysis of the data according to Equation 1 indicates that the rate of strand separation is virtually identical in the presence of either ATP or ADP ( and ; Figure 3B).
While it is clear that ADP efficiently stimulates the strand separation activity of terminase, the role of ATP is less clear. It is feasible that ATP binds to the maturation ATP binding site in TerL and directly stimulates the reaction. Alternatively, hydrolysis of ATP might be required to provide the ultimate stimulatory ligand, ADP. To further define the energetic requirement for the reaction, we examined a variety of nucleotides for their capacity to support strand separation activity. At a concentration of 10 μM, the poorly hydrolyzed ATP-γS, the non-hydrolyzable adenosine analogs AMP-PNP and AMP-PCP, and even GTP support the reaction, albeit at an attenuated extent (Figure 3C). The data indicate that hydrolysis of ATP to ADP at the maturation ATP binding site is not required and that occupancy of this site with any nucleotide is sufficient to stimulate terminase strand separation activity.
Elevated Nucleotide Concentrations Inhibit Strand Separation Activity
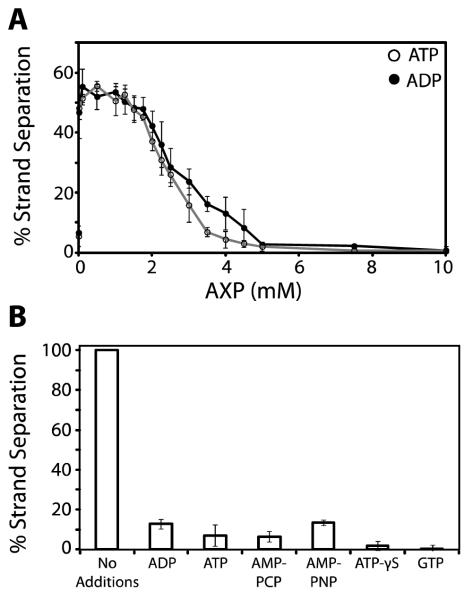
The data presented above indicate that the ATP analogs only partially support strand separation activity. We considered the possibility that this reflects weak binding to terminase and that that higher concentrations might be required to fully saturate the enzyme. We therefore repeated the experiment but increased the concentration of each nucleotide in the reaction mixture. Unexpectedly, while all of the nucleotides support the reaction at a concentration of 10 μM (Figure 3C), none support strand separation at a concentration of 5 mM (data not shown). This phenomenon was investigated further with ATP and ADP in a wider concentration range. As above, 10 μM AXP maximally stimulates the reaction which remains optimal up to ~ 2 mM nucleotide; however, strand separation activity drops to undetectable levels as concentration of AXP is further increased (Figure 4A).
FIGURE 4. Elevated Concentrations of Nucleotide Inhibit Terminase Strand Separation Activity.
Panel A. The strand separation assay was performed as described in Experimental Procedures except that IHF was omitted and the indicated concentration of nucleotide was included in the reaction mixture. Each data point represents the average of at least three separate experiments with standard deviation indicated with a bar. Panel B. The strand separation assay was performed in the absence of IHF and in the presence of 10 μM ADP. Additional nucleotides were added to a final concentration of 5 mM as indicated. Each bar represents the average of at least three separate experiments with standard deviation indicated.
We next examined the possibility that elevated nucleotide concentrations actively inhibit the strand separation activity of terminase. The reaction was performed using 10 μM ADP to drive strand separation and in the presence of 5 mM competing nucleotide. As shown in Figure 4B, all of the nucleotides examined directly inhibit the ADP-driven strand separation activity of terminase. These data suggest that two ATP binding sites are operational in this assay; (i) a high-affinity ATP binding site that saturates at 10 μM nucleotide (KD,app ~ 1 μM, Figure 3A) and that stimulates the strand separation reaction. Presumably, this represents the maturation ATPase binding site in TerL. (ii) a second very low affinity AXP binding site that becomes populated only at higher concentrations and that inhibits the reaction when nucleotide is bound. This is further discussed below.
IHF Stimulated Strand Separation Activity of Terminase
IHF strongly stimulates the strand separation activity of terminase in a nucleotide-independent manner (Figure 2). The data presented in Figure 5A demonstrates that IHF simulates the reaction in a concentration dependent and saturable manner. Direct interaction between terminase and IHF has not been reported. Rather, IHF binds to the I1 element in cos to provide a duplex architecture that promotes terminase assembly at that site (Figure 1) (21, 22). Presumably, the binary IHF-DNA complex is directly responsible for terminase assembly at cos and for stimulation of strand separation activity. Thus, the data were analyzed using a simple binary IHF•DNA binding model (Equation 2); this yields a modest fit to the data (Figure 5A, dashed line). Quantitative gel mobility shift studies from our lab have shown that IHF binding to cos-DNA is best described by a cooperative (Hill) binding model (22). We therefore analyzed the strand separation data assuming a cooperative binding model (equation 3), which yielded a significantly better fit (solid line). The latter analysis affords a and a Hill coefficient, n= 2.3 ± 0.3. These values are commensurate with those obtained in our published gel mobility shift studies (22). Finally, we examined the time course for the reaction at saturating concentrations of IHF (50 nM), which afforded a rate constant (Figure 5B).
FIGURE 5. IHF Stimulates Strand Separation Activity in the Absence of Nucleotides.
Panel A. The strand separation assay was performed as described except that nucleotide was omitted and the indicated concentration of IHF was included in the reaction mixture. Each data point represents the average of at least three separate experiments with standard deviation indicated with a bar. The dashed line represents the best fit of the data to a simple binary IHF-DNA binding model (equation 2). The solid line represents the best fit of the data to a cooperative (Hill) binding model (equation 3). Panel B. The strand separation assay was performed in the absence of nucleotide and the presence of 50 nM IHF for the indicated time. Each data point represents the average of at least three separate experiments with standard deviation indicated with a bar. The solid lines represent the best fit of the data to equation 1. Panel C. The strand separation assay was performed in the absence of nucleotide and in the presence of 50 nM IHF. Additional nucleotides were added to a final concentration of 5 mM as indicated. Each bar represents the average of at least three separate experiments with standard deviation indicated.
Elevated Nucleotide Concentrations Inhibit IHF-Stimulated Strand Separation Activity
The data presented in Figure 4 demonstrate that elevated concentrations of nucleotide inhibit the ADP-driven strand separation activity of terminase. We next asked whether elevated concentrations of nucleotide might also inhibit the IHF-stimulated reaction. Indeed, all of the nucleotides examined strongly inhibit the strand separation activity of terminase when included at 5 mM (Figure 5C). Importantly, nucleotides do not significantly affect IHF binding to cos-containing duplexes under similar conditions (Sanyal and Catalano, unpublished). In sum, the data indicate that binding of nucleotide to a low affinity ATP binding site in terminase abrogates assembly of the maturation complex at cos, the capacity of the enzyme to adopt a catalytically competent strand separation complex, or both.
Coupled cos-Cleavage Endonuclease and Strand Separation Activities of λ Terminase
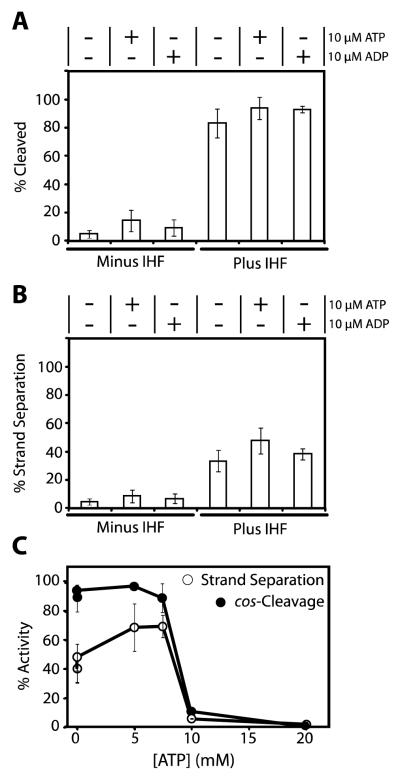
The studies described above utilized a pre-nicked DNA substrate to kinetically isolate the strand-separation activity of terminase. Maturation of an intact cos sequence requires duplex nicking by terminase followed by separation of the nicked, annealed strands in situ without release of the duplex from the enzyme (Figure 1). To examine the role of nucleotides and IHF on the sequential duplex nicking and strand separation activities of terminase, we utilized a 12 kb cos-containing duplex that is efficiently nicked by terminase. In the absence of enzyme catalyzed strand separation, the nicked duplex remains intact unless heated (75°C for 30 seconds). Thus, analysis of the heated and unheated reaction mixture allows quantitation of enzyme catalyzed duplex nicking and strand-separation, respectively. Duplex nicking and strand separation by the enzyme affords 8.7kb and 3.3 kb duplexes that can be quantified by gel assay.
In the absence of IHF, little to no stimulation of the cos-cleavage reaction is observed by 10 μM nucleotide (Figure 6A). Quantitation of the data under these conditions was highly error prone due to the limited extent of the reaction, so we did not pursue these studies in great detail. In contrast, IHF strongly stimulates cos-cleavage activity and further stimulation by 10 μM nucleotide is quite modest (6-7%; Figure 6A). Essentially identical results are observed for in situ strand separation by the enzyme (Figure 6B). Consistent with the strand separation studies above, elevated concentrations of ATP inhibit both the IHF-stimulated cos-cleavage and coupled in situ strand separation activities of terminase (Figures 6C).
FIGURE 6. Coupled cos-Cleavage Endonuclease and Strand Separation Activities of λ Terminase.
Panel A. The cos-cleavage assay was performed as described in Experimental Procedures in the absence or presence of 50 nM IHF and nucleotide, as indicated. The samples were heated prior to loading onto the gel to ensure physical separation of the nicked strands. Each bar represents the average of at least three separate experiments with standard deviation indicated. Panel B. The in situ strand separation assay was performed as described in Experimental Procedures in the absence or presence of 50 nM IHF and nucleotide, as indicated. The samples were not heated prior to loading onto the gel and only those strands separated by terminase are observed. Each bar represents the average of at least three separate experiments with standard deviation indicated. Panel C. The cos-cleavage and strand separation assays were performed as described except that nucleotide was added to the reaction mixture as indicated. Each data point represents the average of at least three separate experiments with standard deviation indicated with a bar.
Discussion
Replication of viral DNA within a cell must forestall damage by exonucleases that degrade exposed duplex ends and viruses have developed several strategies to overcome these anti-viral systems. This includes the synthesis of different forms of “endless” DNA such as (i) circular genomes (phages φX174 and PM2), (ii) linear monomers containing protective proteins at the duplex ends (adenovirus, phage φ29), and (iii) linear concatemers of the individual genomes (herpesviruses, phages λ and T4) (1, 2, 5, 7). The synthesis of linear concatemers is the most commonly used mechanism and in these cases a viral enzyme is required to specifically recognize the genome end within the context of the concatemer and to cut the duplex in preparation for packaging. Terminase enzymes provide this genome maturation activity and also serve as motors that catalyze duplex translocation into the capsid.
Mechanism of Strand Separation
λ terminase introduces site-specific nicks into the cosN subsite and then physically separates the nicked, annealed strands. We (19, 25) and others (24) have previously examined the kinetic features of this “helicase” reaction, reporting that ATP is required for strand-separation. The presumption has been that the energy of ATP hydrolysis is utilized to drive the reaction in analogy with the helicase enzymes (36, 37); however, the data presented here clearly demonstrate that ATP hydrolysis is not required for strand separation. Accordingly, our mechanistic pre-conceptions of terminase enzymology require modification. Within this context, we note that IHF alone in the absence of nucleotides fully activates the strand separation activity of terminase. These features are similarly observed in the coupled cos-cleavage/strand separation reactions and the ensemble of data suggests that both the architecture of the DNA and the conformation of the enzyme play a role in the maturation activity of terminase.
We previously proposed that strand separation is driven by a nucleotide-dependent terminase conformation that has a high affinity for the single stranded end that is generated in situ subsequent to duplex nicking (19, 25). This mechanism essentially represents a single-turnover of the active rolling strand separation helicase model proposed by Lohman and co-workers (25, 36, 38). The data presented here indicate that nucleotide or IHF are individually necessary and sufficient to assemble a catalytically-competent strand-separation complex on pre-nicked DNA. We suggest that bending of the DNA at the nicked cos site introduces torsional stress into the duplex and provides the “energy” required by terminase to separate the strands. This bend may be introduced by IHF in a nucleotide independent manner, or by a terminase conformation induced upon nucleotide binding to the maturation ATP binding site in TerL. Terminase utilizes this torsional stress to separate the strands and bind tightly to the single-stranded end of the DNA. This drives strand separation and yields the stable complex I packaging intermediate (Figure 1).
Maturation and Packaging by λ Terminase
Beyond the characterization of strand-separation by λ terminase, the data presented here provides mechanistic insight into the sequential assembly, maturation, and packaging activities of the enzyme. The terminase packaging motor alternates between a stable, site-specifically bound maturation complex and a powerful, mobile packaging motor. We have proposed that alternation between these two states is regulated by nucleotides and three ATPase sites have been identified in the λ terminase protomer: (i) a DNA-stimulated regulatory ATPase site located in TerS (; ). This site, which also hydrolyzes GTP (; (34)), modulates terminase binding to cos vs. non-specific DNA substrates (17-19); (ii) a packaging ATPase that is located in the N-terminus of TerL (). This site powers translocation of the terminase motor and active DNA packaging (39-41); and (iii) a DNA maturation ATP binding site that resides in a C-terminal domain of TerL. The latter ATP binding site is required for cos-cleavage and strand separation activities of terminase, though catalytic turnover has not been observed (26-28). The KD,app for nucleotide-stimulated strand separation activity measured here (~1 μM) represents AXP binding to the maturation ATP binding site in TerL.
How do these three nucleotide binding sites work in concert to assemble and regulate the terminase motor complexes? First we consider the observation that elevated concentrations of nucleotide strongly inhibit both the cos-cleavage and strand separation activities of terminase. The concentration range over which this occurs is commensurate with nucleotide binding to the low affinity regulatory ATPase site in TerS (vide supra) (17, 18). Next we note our published observation that nucleotide binding to the regulatory ATPase site in TerS stimulates ATP hydrolysis at the packaging ATPase site in TerL (34). Together, the data indicate that nucleotide binding to TerS (i) down-regulates the DNA maturation activities and (ii) up-regulates the packaging ATPase in the TerL subunit. We propose that this allosteric regulation is central to the alternation between a stationary maturation complex and a mobile packaging motor complex.
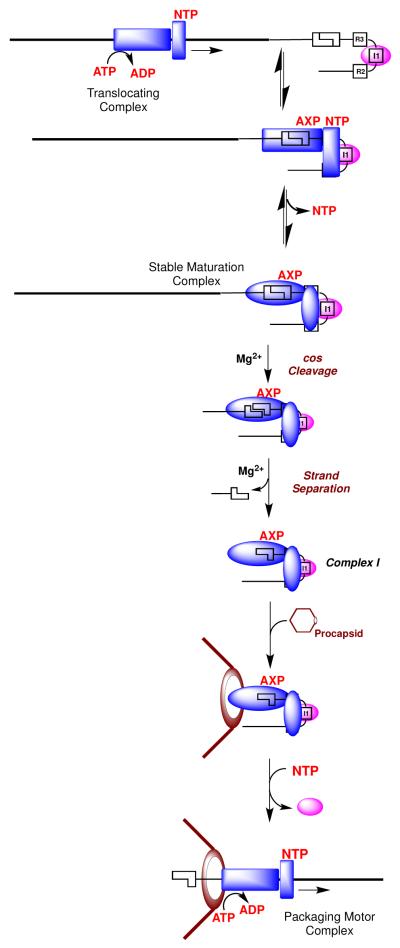
Model for DNA Maturation and Packaging in Lambda
Scheme 1 depicts a model for the sequential action of terminase in the maturation and subsequent packaging of viral DNA into the capsid. Initially, terminase binds non-specifically to DNA, which increases NTP binding to the regulatory ATP binding site in TerS. This is consistent with our demonstration that DNA decreases the KM for ATP hydrolysis at this site (17-19). Here we propose that nucleotide binding to this site down regulates the maturation activities and activates the packaging ATPase in TerL to afford a mobile complex (17-19). IHF binds to a cos-site and introduces a strong bend in the duplex, which promotes cooperative assembly of terminase at that site. This is consistent with our demonstration that IHF and gpNu1 cooperatively bind and bend a cos-containing DNA duplex (22). Once assembled specifically at the cos site, nucleotide is released from the TerS regulatory binding site, which down-regulates the packaging ATPase and engenders a stable, catalytically competent nuclease complex. The nuclease activity of TerL introduces nicks into the duplex, allowing a conformational change to an activated strand-separation complex. The activated complex is stabilized by nucleotide binding to the maturation ATP binding site in TerL and by the duplex architecture induced by IHF, both of which introduce strain into the DNA. We propose that this post-cleavage conformation binds tightly and specifically to single-stranded DNA, driving separation of the nicked, annealed strands to form complex I. This mechanism essentially represents a single-turnover of the active rolling strand separation helicase model proposed by Lohman and co-workers (25, 36, 38).
Scheme 1. Model for DNA Maturation and Packaging by λ Terminase.
In the next step, the TerL subunits in complex I bind to the portal ring of an empty procapsid, triggering a conformational change from a maturation complex to an activated motor complex (Scheme 1). This switch requires two fundamental alterations in terminase activity. First, terminase must switch from a specific DNA binding conformation that binds tightly to the single-stranded genome end, to a conformation that binds duplex DNA tightly but non-specifically during translocation by the packaging motor. Second, the packaging ATPase must be activated to power DNA translocation into the procapsid. As discussed above, nucleotide binding to the regulatory ATPase site in TerS modulates these changes. We propose that terminase binding to the portal decreases the at TerS, which promotes nucleotide binding and facilitates the transition to a mobile motor complex (Scheme 1). In sum, this model proposes that (i) the affinity of nucleotide for the maturation ATP binding site in TerS is modulated by DNA (cos- vs. non-specific DNA), by IHF, and by interaction with the procapsid, and that (ii) occupancy of this site in turn regulates the maturation and motor activities of the enzyme. Within this context, allosteric regulation of the TerS ATP binding site by DNA, nucleotides, and procapsids has been demonstrated in a concentration range commensurate with the in vivo concentration of nucleotides, as required for a biologically-relevant regulatory site (see above) (17-19, 33, 39).
A General Mechanism for Viral Genome Maturation
Lambda-like viruses package a unit-length genome with defined ends. Thus, lambda terminase must mature both ends of the genome in an identical manner to excise a single genome from the concatemer. The human herpes viruses (herpes simplex I, cytomegalovirus, varicella-zoster virus, etc.) similarly initiate and terminate packaging at specific “a” sequences in concatemeric DNA (42). A large group of related “head-full” viruses initiate packaging with an analogous DNA maturation complex that assembles at a specific “pac” site on concatemeric DNA (1, 5, 7, 43). While these initial complexes must also cut the duplex and subsequently transition to an active packaging motor complex, they do not terminate at a specific site and the motor continues to package until a “head-full” of DNA has been inserted into the capsid (102%-104% genome length) (5, 7). Notwithstanding, the essential functions required of terminase motors are recapitulated in virtually all of these viruses, both prokaryotic and eukaryotic. The basic architecture of the terminase motors is conserved among these viruses and we presume that structure follows function. Thus the catalytic model presented here may serve as a paradigm for understanding the essential mechanistic features of these complex biological motors and nucleotide-regulated motors throughout biology.
Acknowledgments
The authors further wish to thank Rishi Sanyal for providing the IHF used in these studies.
Funding. This work was supported by National Institutes of Health grants GM088186 (CEC) and F32GM-905652 (BTA).
Abbreviations
- cos
the cohesive end site of the bacteriophage lambda genome
- cos-cleavage reaction
the site-specific endonuclease activity of lambda terminase
- IHF
the Escherichia coli integration host factor protein
- immature DNA
concatemeric lambda DNA
- mature DNA
genome-length lambda DNA found within the viral capsid that contains the 12 base complementary single stranded ends
- terminase protomer
the lambda terminase complex composed of two TerS and one TerL subunits (TerS2•TerL1)
- TerL
the large terminase subunit
- TerS
the small terminase subunit
Footnotes
We use the term “AXP” to denote either ATP or ADP.
References
- 1.Calendar R, Abedon ST. The Bacteriophages. Oxford University Press; New York, N.Y: 2006. [Google Scholar]
- 2.Fields BN, Knipe DM, Howley PM. Fields Virology. Third ed. Lippincott-Raven; New York, NY: 1996. [Google Scholar]
- 3.Roizman B, Palese P. In: Multiplication of Viruses: An Overview, In Fields Virology. Third ed. Fields BN, Knipe DM, Howley PM, editors. Lippincott-Raven; New York, N.Y: 1996. pp. 101–111. [Google Scholar]
- 4.Roizman B, Knipe DM, Whitley RJ. In: Herpes Simplex Viruses, In Fields Virology. Fifth ed. Knipe DM, Howley PM, editors. Lippincott, Williams and Wilkins; Philidelphia, PA: 2007. pp. 2501–2602. [Google Scholar]
- 5.Catalano CE. In: Viral Genome Packaging Machines: An Overview, In Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Catalano CE, editor. Kluwer Academic/Plenum Publishers; New York, N.Y: 2005. pp. 1–4. [Google Scholar]
- 6.Jardine PJ, Anderson DL. In: DNA Packaging in Double-Stranded DNA Phages, In The Bacteriophages. 2nd ed. Calendar R, Abedon ST, editors. Oxford University Press; New York, N.Y: 2006. pp. 49–65. [Google Scholar]
- 7.Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat Rev Micro. 2011;9:647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- 8.Friedman D, Gottesman M. In: Lytic Mode of Lambda Development, In Lambda II. Hendrix RW, Roberts JW, Stahl FW, Weisberg RA, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor; New York: 1983. pp. 21–52. [Google Scholar]
- 9.Daniels D, Schroeder J, Szybalski W, Sanger F, Coulson A, Hong G, Hill D, Petersen G, Blattner F. In: Complete Annotated Lambda Sequence, In Lambda II. Hendrix RW, Roberts JW, Stahl FW, Weisberg RA, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor; NY: 1983. [Google Scholar]
- 10.Furth M, Wickner S. In: Lambda DNA Replication, In Lambda II. Hendrix RW, Roberts JW, Stahl FW, Weisberg RA, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor; NY: 1983. pp. 145–174. [Google Scholar]
- 11.Tomizawa J, Ogawa T. Replication of Phage Lambda DNA; Cold Spring Harbor Symposium on Quantitative Biology; 1968; pp. 533–551. [DOI] [PubMed] [Google Scholar]
- 12.Becker A, Murialdo H. Bacteriophage Lambda: the Beginning of the End. J. Bacteriol. 1990;172:2819–2824. doi: 10.1128/jb.172.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feiss M, Catalano CE. Bacteriophage Lambda Terminase and the Mechanism of Viral DNA Packaging. In: Catalano CE, editor. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Kluwer Academic/Plenum Publishers; New York, N.Y: 2005. pp. 5–39. [Google Scholar]
- 14.Maluf NK, Yang Q, Catalano CE. Self-association properties of the bacteriophage lambda terminase holoenzyme: implications for the DNA packaging motor. J Mol Biol. 2005;347:523–542. doi: 10.1016/j.jmb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 15.Maluf NK, Gaussier H, Bogner E, Feiss M, Catalano CE. Assembly of bacteriophage lambda terminase into a viral DNA maturation and packaging machine. Biochemistry. 2006;45:15259–15268. doi: 10.1021/bi0615036. [DOI] [PubMed] [Google Scholar]
- 16.Yang Q, Hanagan A, Catalano CE. Assembly of a nucleoprotein complex required for DNA packaging by bacteriophage lambda. Biochemistry. 1997;36:2744–2752. doi: 10.1021/bi9622682. [DOI] [PubMed] [Google Scholar]
- 17.Tomka MA, Catalano CE. Kinetic characterization of the ATPase activity of the DNA packaging enzyme from bacteriophage lambda. Biochemistry. 1993;32:11992–11997. doi: 10.1021/bi00096a008. [DOI] [PubMed] [Google Scholar]
- 18.Hwang Y, Catalano CE, Feiss M. Kinetic and mutational dissection of the two ATPase activities of terminase, the DNA packaging enzyme of bacteriophage Chi. Biochemistry. 1996;35:2796–2803. doi: 10.1021/bi952322z. [DOI] [PubMed] [Google Scholar]
- 19.Yang Q, Catalano CE. A minimal kinetic model for a viral DNA packaging machine. Biochemistry. 2004;43:289–299. doi: 10.1021/bi035532h. [DOI] [PubMed] [Google Scholar]
- 20.Miller G, Feiss M. The bacteriophage lambda cohesive end site: isolation of spacing/substitution mutations that result in dependence on Escherichia coli integration host factor. Mol Gen Genet. 1988;212:157–165. doi: 10.1007/BF00322459. [DOI] [PubMed] [Google Scholar]
- 21.Tomka MA, Catalano CE. Physical and kinetic characterization of the DNA packaging enzyme from bacteriophage lambda. J Biol Chem. 1993;268:3056–3065. [PubMed] [Google Scholar]
- 22.Ortega ME, Catalano CE. Bacteriophage lambda gpNu1 and Escherichia coli IHF proteins cooperatively bind and bend viral DNA: implications for the assembly of a genome-packaging motor. Biochemistry. 2006;45:5180–5189. doi: 10.1021/bi052284b. [DOI] [PubMed] [Google Scholar]
- 23.Swinger KK, Rice PA. IHF and HU: Flexible Architects of Bent DNA. Current Opinion in Structural Biology. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Parris W, Rubinchik S, Yang YC, Gold M. A new procedure for the purification of the bacteriophage lambda terminase enzyme and its subunits. Properties of gene product A, the large subunit. Journal of Biological Chemistry. 1994;269:13564–13574. [PubMed] [Google Scholar]
- 25.Yang Q, Catalano CE. Kinetic characterization of the strand separation (“helicase”) activity of the DNA packaging enzyme from bacteriophage lambda. Biochemistry. 1997;36:10638–10645. doi: 10.1021/bi970689t. [DOI] [PubMed] [Google Scholar]
- 26.Arens JS, Hang Q, Hwang Y, Tuma B, Max S, Feiss M. Mutations that extend the specificity of the endonuclease activity of lambda terminase. J Bacteriol. 1999;181:218–224. doi: 10.1128/jb.181.1.218-224.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duffy C, Feiss M. The large subunit of bacteriophage lambda’s terminase plays a role in DNA translocation and packaging termination. J Mol Biol. 2002;316:547–561. doi: 10.1006/jmbi.2001.5368. [DOI] [PubMed] [Google Scholar]
- 28.Ortega ME, Gaussier H, Catalano CE. The DNA maturation domain of gpA, the DNA packaging motor protein of bacteriophage lambda, contains an ATPase site associated with endonuclease activity. J Mol Biol. 2007;373:851–865. doi: 10.1016/j.jmb.2007.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hang JQ, Tack BF, Feiss M. ATPase center of bacteriophage lambda terminase involved in post-cleavage stages of DNA packaging: identification of ATP-interactive amino acids. J Mol Biol. 2000;302:777–795. doi: 10.1006/jmbi.2000.4086. [DOI] [PubMed] [Google Scholar]
- 30.Woods L, Terpening C, Catalano CE. Kinetic analysis of the endonuclease activity of phage lambda terminase: assembly of a catalytically competent nicking complex is rate-limiting. Biochemistry. 1997;36:5777–5785. doi: 10.1021/bi963044m. [DOI] [PubMed] [Google Scholar]
- 31.Yang Q, Catalano CE, Maluf NK. Kinetic Analysis of the Genome Packaging Reaction in Bacteriophage Lambda. Biochemistry. 2009;48:10705–10715. doi: 10.1021/bi901016n. [DOI] [PubMed] [Google Scholar]
- 32.Hang Q, Woods L, Feiss M, Catalano CE. Cloning, expression, and biochemical characterization of hexahistidine-tagged terminase proteins. J Biol Chem. 1999;274:15305–15314. doi: 10.1074/jbc.274.22.15305. [DOI] [PubMed] [Google Scholar]
- 33.Filutowicz M, Grimek H, Appekt K. Purification of the Escherichia coli Integration Host Factor (IHF) in One Chromatographic Step. Gene. 1996;147:149–150. doi: 10.1016/0378-1119(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 34.Woods L, Catalano CE. Kinetic characterization of the GTPase activity of phage lambda terminase: evidence for communication between the two “NTPase” catalytic sites of the enzyme. Biochemistry. 1999;38:14624–14630. doi: 10.1021/bi990866l. [DOI] [PubMed] [Google Scholar]
- 35.Higgins RR, Lucko HJ, Becker A. Mechanism of cos DNA Cleavage by Bacteriophage Lambda Terminase: Multiple Roles of ATP. Cell. 1988;54:765–775. doi: 10.1016/s0092-8674(88)91021-5. [DOI] [PubMed] [Google Scholar]
- 36.Lohman TM, Tomko EJ, Wu CG. Non-Hexameric DNA Helicases and Translocases: Mechanisms and Regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 37.Patel SS, Pandey M, Nandakumar D. Dynamic Coupling Between the Motors of DNA Replication: Hexameric Helicase, DNA Polymerase, and Primase. Current Opinion in Chemical Biology. 2011;15:595–605. doi: 10.1016/j.cbpa.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lohman TM, Bjornson KP. Mechanisms of Helicase-Catalyzed DNA Unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 39.Dhar A, Feiss M. Bacteriophage lambda terminase: alterations of the high-affinity ATPase affect viral DNA packaging. J Mol Biol. 2005;347:71–80. doi: 10.1016/j.jmb.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 40.Yang Q, Catalano CE. Biochemical characterization of bacteriophage lambda genome packaging in vitro. Virology. 2003;305:276–287. doi: 10.1006/viro.2002.1602. [DOI] [PubMed] [Google Scholar]
- 41.Gaussier H, Yang Q, Catalano CE. Building a virus from scratch: assembly of an infectious virus using purified components in a rigorously defined biochemical assay system. J Mol Biol. 2006;357:1154–1166. doi: 10.1016/j.jmb.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 42.Baines JD, Weller SK. Cleavage and Packaging of Herpes Simplex Virus 1 DNA. In: Catalano CE, editor. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Kluwer Academic/Plenum Publishers; New York, N.Y: 2005. pp. 135–149. [Google Scholar]
- 43.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]