Abstract
The latest scientific findings in the field of cancer research are redefining our understanding of the molecular and cellular basis of the disease, moving the emphasis toward the study of the mechanisms underlying the alteration of the normal processes of cellular differentiation. The concepts best exemplifying this new vision are those of cancer stem cells and tumoral reprogramming. The study of the biology of acute lymphoblastic leukemias (ALLs) has provided seminal experimental evidence supporting these new points of view. Furthermore, in the case of B cells, it has been shown that all the stages of their normal development show a tremendous degree of plasticity, allowing them to be reprogrammed to other cellular types, either normal or leukemic. Here we revise the most recent discoveries in the fields of B-cell developmental plasticity and B-ALL research and discuss their interrelationships and their implications for our understanding of the biology of the disease.
Key words: leukemia, hematopoietic development, leukemic stem cells, lymphopoiesis, developmental plasticity, B cells, stem cells, cancer, B-ALL
Introduction
The different subtypes of acute lymphoblastic leukemias (ALLs) are grouped under this umbrella term, because, as a whole, they present many similarities from the point of view of the differentiation phenotypes of the tumoral cells. However, ALLs are a heterogeneous group of diseases, not only between children and adults, but also from the aspect of the molecular pathogenesis and from the point of view of their clinical evolution and response to treatment. Indeed, some subtypes are now highly responsive to the currently used chemotherapy regimens, and complete long-term remissions (>10–15 y) can be achieved, while other subtypes still present very poor prognoses. This aspect is highly correlated with the characteristics of the molecular, leukemia-initiating, genetic anomalies that are at the root of the disease. In this way, the 5-y event-free survival estimate for TEM-AML1+ patients is of 89%,1 while for those carrying the MLL-AF4 translocation it is only of 32%.2,3 Therefore, given the high degree of similarity on their B-cell differentiation surface or intracellular markers, it is clear that the natural evolution and prognosis of B-ALLs is closely dictated by the molecular nature of the underlying genetic anomalies.
The normal development of B lymphocytes is a highly ordered process in which a tightly regulated interplay between the extrinsic cytokine and B-cell receptor (BCR) signaling and the intrinsic transcriptional and epigenetic programming leads the cells all the way from hematopoietic stem cells (HSCs) to the mature B cells. This is a multistep process in which several “quality control” checkpoints verify the presence of the correct immunoglobulin genes rearrangements and give access to the progressively more differentiated stages. Any deregulation of this delicate equilibrium will lead, in most cases, to developmental arrest and cellular death. However, there are specific circumstances in which this differentiation program can be subverted and rewired toward new developmental fates. This reprogramming can be caused by experimental manipulations in the laboratory, as in the case of reprogramming to pluripotency or programmed dedifferentiation, but it can also happen in vivo in human leukemias (Fig. 1). In the majority of these cases, the molecular players leading to these aberrant fates are in close relationship with the ones participating in normal development. The field of ALL development is a clear example of the mutual beneficial feedback existing between the study of the normal developmental biology of a lineage and that of its tumoral counterpart. In this review, we describe the fundamental and latest findings in the field of B-cell commitment and early development, and we discuss them in the context of the most recently proposed paradigms for our understanding of B-ALL development and biology.
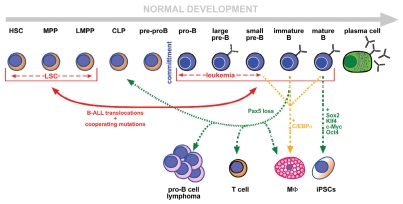
Figure 1.
Plasticity in B-cell development. B-cell differentiation is represented in a lineal configuration representing the stages from the HSC to the terminally differentiated plasma cells (see text for details). Committed B cells expressing Pax5 are shown with a blue cytoplasm. The red lines represent events occurring during the genesis and evolution of B-ALL. The red continuous double arrow reflects the fact that dedifferentiation can occur from committed B cells to earlier developmental stages by the action of the oncogenic alterations, but, also, the presence of the leukemia-inducing alterations in stem/progenitor cells programs these cells to acquire a B cell-like phenotype. The dashed red arrows reflect the fact that both LSCs and differentiated B-ALL cells can present with a variable and evolving surface phenotype, resembling different developmental stages. Green and orange arrows represent situations obtained experimentally in vivo or in vitro, that illustrate the high degree of plasticity of B cells that allows them to give rise to either normal T cells, macrophages, to progenitor B-cell lymphomas or to induced pluripotent stem cells.
Early B-Cell Development
The progression from hematopoietic stem cells to committed B cells.
The continuous technological and scientific advances in the fields of flow cytometry and mouse genetic engineering keep on adding new layers of complexity to our understanding of hematopoietic cell development. This is especially true for the early stages of differentiation, where progenitors' populations are formed by very low numbers of cells, and the use of novel combinations of phenotypic markers allows their subdivision into new subgroups. There are, however, several problems associated with this increased cytometric accuracy. First, the demonstration of the functional capabilities of these new populations in vivo is not always easy given their low numbers and the fact that the isolation procedure can condition and/or modify by itself the developmental potential of the purified population (on what could be the Heisenberg's uncertainty principle applied to development). Another important problem is the difficult inter-laboratory reproducibility of the experiments due to the advanced flow cytometric technologies required for the separation and the reduced numbers of progenitors obtained. This, together with the fact that different laboratories might have generated and studied their own genetically modified mice with specific developmental tracers, has caused quite some controversy in the field of early hematopoietic development and sometimes makes it difficult to unify the data coming from different research groups.
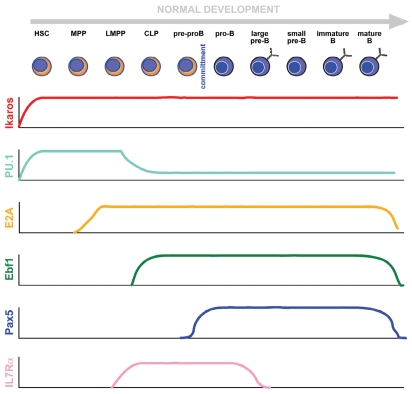
The maturation process from bone-marrow (BM) progenitors to mature B cells proceeds through a series of stages that can be identified by the expression of specific transcription factors and of cell surface markers as well as by the status of the immunoglobulin gene rearrangements and the pattern of expression of immunoglobulins (Fig. 2).
Figure 2.
Molecular control of B-cell development. The figure depicts the cellular windows of expression of the main molecular players participating in the building and stabilization of B-cell identity. It must be taken into account that the stem/progenitor populations are heterogeneous regarding their transcriptomes (see main text), and that some of the depicted factors possess several isoforms whose functions and individual patterns of expression have not yet been elucidated.
The first cell in the hematopoietic hierarchy is the hematopoietic stem cell (HSC). By definition, according to their stem cell nature, these cells have the capacity of extensive self-renewal and differentiation (i.e., they can originate all the cellular types within the hematopoietic system). From a functional point of view, this is defined as their capacity to maintain blood formation for at least 16 weeks after their transplantation into a lethally irradiated host.4 As we have just mentioned, the procedures for isolating hematopoietic stem cells are not standardized or 100% reproducible among different laboratories. For a detailed description of the combinations of markers used to define the subpopulations of early lymphoid progenitors, please see reference 5. Possibly the most common universal (but nowadays somewhat insufficient for advanced studies) definition is that of lineage (CD3, CD4, CD8, CD11c, CD19, DX5 or NK1.1, B220, TER119, GR1 and MAC1)-negative (Lin−) Sca1+ c-KitHIGH cells from the bone marrow (BM), containing all short- and long-term repopulating HSCs (LT-HSCs) and representing approximately 0.1% of the total cell pool in the BM of mice of the C57BL/6 strain. It is possible to further enrich LT-HSCs by excluding short-term HSCs and progenitors, distinguishable on the basis of their higher retention of the rhodamine 123 dye6 or their expression of markers like Mac-1,7 CD34,8,9 Flk2/Flt3 10–12 or CD49b/a2 integrin.13 But even within these highly purified populations, functional variability exists. For example, it has been recently shown that many of the cells that form part of highly enriched HSCs suspensions are, in fact, intermediate term HSCs that do not have the capacity to sustain long-term erythropoiesis.14 There are many accumulating evidences of the functional heterogeneity of HSCs. For example, the heritable properties of individually analyzed HSC clones indicated that a minority of HSCs have an unbalance regarding myeloid-vs.-lymphoid fates.15 This property was speculated to result from epigenetic events occurring early in life. However, this “fate instability” that seems intrinsic to the progenitor compartment can also arise as a logical consequence of the equilibrium between regulatory genes “pushing” in different directions. Indeed, using the well-established stem cell antigen Sca1 as a phenotypic marker, it has been demonstrated that its levels of expression follow a Gauss curve, even in a clonal population of progenitor cells.16 However, the cells at both ends of this curve are not static but, rather, can regenerate the whole population with all its range of expression levels. Furthermore, when analyzed at the molecular level, these cellular subcompartments with defined Sca1 expression levels present different transcriptomes, and they confer them specific developmental capacities toward given lineages.16 These results indicate that each individual cell at the progenitor stage is an almost unique transitional stage in a gamut of fluctuating transcriptomes. This equilibrium makes the population very resilient while, at the same time, making it strongly responsive to external or internal factors, like, for example, cytokines or pharmacological compounds or epigenetic regulators. Therefore, the heterogeneous expression of surface markers is a reflection of what happens inside the cells at the much more relevant level of the transcription factors and epigenetic regulators. In fact, progenitors at the population level have been shown to present a particular phenotypic heterogeneity in the form of the promiscuous activation of lineage-associated genes.17,18 In summary, we can see that, in the progenitors' population, multipotency is a dynamic state of heterogeneity. The cells that are in a more stable state, at the center of the attractor “valley” have less tendency to differentiate than the ones lying at the periphery of the distribution. These ones are partially poised to differentiate, suggesting that commitment toward a lineage is a spontaneous but infrequent phenomenon, unless it is brought forth by external signals.19,20 In this way, the equilibrium between plasticity and heterogeneity in multipotent populations is explained, and this also helps understanding how the balance between instructed and stochastic cell fate decisions takes place. In the context of leukemias (and cancer in general) from this point of view, malignant development could be considered as a particular natural state of cell regulation that normally is inaccessible, locked beneath many layers of molecular regulatory networks.21 The role of the oncogenic modifications would therefore be to tip the balance to make these hidden fates possible22 (see below).
Even though, as we have outlined, the precise way in which it happens is still not known, it is clear that there is a progressive loss of developmental potential as we move away from the LT-HSCs. CD34+Flt3+ cells present already a greatly reduced capacity to contribute to the megakaryocytic or erythroid lineages, and within this compartment, together with other progenitors, we can already find the lymphoid-primed multipotent progenitor cells (LMPPs).23 A subset within LMPPs is the one formed by the early lymphoid progenitors (ELPs), identified by using a GFP knock-in strategy within the RAG1 locus.24 ELPs are very effective in restoring lymphocytes in transplantation assays but have reduced potential for generating myeloid cells, therefore representing the earliest stages of lymphoid-lineage specification. Their progenitor nature is, however, evidenced by the fact that, like HSCs, they are not homogeneous, since many are quiescent, and individual cells express different combinations of lymphoid genes.25 The next stage toward B cells is formed by the common lymphoid progenitors (CLPs), first described by the Weissman laboratory26 as Lin-IL-7Rα+c−KitLoSca1+ BM cells. They are the main differentiation intermediate between ELPs and B lymphocytes or NK cells, and, although they were originally described as capable of generating T cells, this is currently the subject of intense discussion. Indeed, much confusion surrounds the CLPs in the literature, partly because their definition is not standardized, and they are isolated in different ways in different laboratories.27,28 What is clear is that CLPs harbor lymphoid, natural killer (NK) and dendritic cell (DC) potential, and that they constitute the last hematopoietic progenitor stage before B-cell differentiation. The most primitive cells in the thymus are referred to as early T-lineage progenitors (ETPs), but the nature of the lymphoid progenitor that seeds the thymus and is responsible for T-cell development is still highly controversial and falls out of the scope of this review.
The subsequent B-cell differentiation steps are represented in several schemes that account for the sequential loss and gain of expression of differentiation markers and correlate with the molecular events that take place along with B-cell differentiation (namely, but not only, the immunoglobulin gene rearrangements). One of the most widely used is the scheme of Hardy and colleagues, which classifies B-cell progenitors into several, alphabetically ordered, fractions: A (pre-pro-B cells), B (pro-B cells) and C (large pre-B cells)29 (Fig. 2). Fraction A (pre-pro-B cells, B220+CD24low) are most probably immediately downstream of ELPs and CLPs and still include cells belonging to the NK and DC lineages.30 Fraction A includes cells with a latent myeloid potential, consistent with the fact that they still don't express the B-cell commitment factor Pax5 (see below).31 The original definition of Fraction A has been expanded to B220+CD24lowLy6C− AA4.1hiCD43lowKITlowIL-7Rα+ to exclude cells with DC, NK or myeloid potential.28 The developmental transition to Fraction B (pro-B cell) is marked by the expression of CD19. This event is totally contingent on the activation of the transcription factor Pax5 and dictates in vivo the irreversible commitment to the B-cell lineage.32 From this point onwards, in normal conditions, B cells proceed through an ordered series of steps that lead to the formation of immature B220+CD19+IgM+IgD− B cells in the bone marrow that would mature in the peripheral lymphoid organs into B220+CD19+IgM+IgD+ mature B cells.32
All the developmental intermediates described above have been identified and studied in mice. Although differences exist with humans, especially at the levels of the markers defining the different compartments, the main concepts can be extrapolated from one organism to another. CD34 is expressed by the majority of the early precursors, which lack lineage markers, and CD38 expression correlates with lineage commitment. The LIN−CD34+CD38− fraction of human BM bone marrow can be subdivided into three subpopulations: CD90+CD45RA−, CD90−CD45RA− and CD90−CD45RA+.33 The LIN−CD34+CD38−CD90+CD45RA− fraction is highly enriched in HSCs, and the LIN−CD34+CD38−CD90−CD45RA− has been proposed as the candidate cord blood human multipotent progenitor,33 and the LMMPs have been proposed to be contained in the CD38−CD45RA+ population.33–35 The human candidate CLPs are LIN−CD10+CD34+, and they are principally biased toward the B-cell fate, both in terms of developmental potential and gene expression profiling.36–38 So, although there is much less information available for normal lymphopoiesis in humans than in mice, the systems seem to develop in a very similarly organized manner.
Transcriptional regulation of B-cell development.
The progression of the cells from HSCs to the B-cell lineage is paralleled by the loss of multipotency and self-renewal and the activation of the B cell-specific gene program. As previously explained, the characteristic promiscuous lineage-priming gene expression profile is lost, leading to an increased expression of B-cell genes.18 The genes that are expressed during the multilineage priming phase are characteristic examples of genes marked by bivalent chromatin, presenting both activating and inhibiting histone and DNA marks.39,40 In HSCs, B cell-restricted promoters, like those of ebf and pax5, present a bivalent histone modification status41 that will resolve upon commitment leading to their open expression in B cells. One of the main regulators of the progression of HSCs into the lymphoid lineages is the Ikaros transcription factor (Fig. 2). Ikaros is required for the suppression of stem cell-associated genes and for the initiation of the lymphoid priming even at the stage of HSCs.42 Ikaros proteins regulate lambda 5, an essential component of the pre-B-cell receptor (pre-BCR) complex, in a stage-specific manner.43 Ikaros also regulates the expression of flt3 and rag genes and contributes to the remodeling of the chromatin at the immunoglobulin loci.44 An essential role of Ikaros is to antagonize the expression of the transcription factor PU.1 in multipotential progenitors, achieved through the induction of the expression of the B-cell fate-promoting gene Gfi1. The equilibrium between Gfi1 and PU.1 activities regulates the lymphoid vs. myeloid fate choice,45 and in this indirect way Ikaros promotes lymphoid development. PU.1 (purine-box factor 1) is an Ets-family transcription factor that plays essential roles in the regulation of the specification of lymphoid, myeloid and erythroid lineages. It regulates a large number of genes belonging to several lineages. Its expression in progenitors counteracts that of the myelo-erythroid-promoting gene GATA-1 at the level of multipotential progenitors in such a way that the reciprocal activation of GATA-1 and PU.1 organizes the lineage fate choice to form a branchpoint between myelo-erythroid and myelo-lymphoid progenitors.46 After this, the interaction with the Ikarosinduced Gfi1 establishes myeloid vs. B-lymphoid lineage choice, so that PU.1 is kept at low levels in B cells and high levels in myeloid cells.47,48 In the lymphoid lineage, Gfi-1 will also suppress the expression of Id2 (inhibitor of DNA binding factor 2), an antagonist of the function of the E2A transcription factor, whose function is essential for the specification of the B-cell fate.49
As mentioned, the early specification of the B-cell lineage requires the transcription factor E2A, which is encoded by the Tcfe2 gene.50 The transcripts from the Tcfe2a gene give rise, by alternative splicing, to the E2A isoforms E12 and E47, differing in the dimerization basic-helix-loop-helix (bHLH) domain, through which they dimerize and bind to the DNA E-box motif. E2A proteins are ubiquitously expressed and normally form heterodimers with other tissue-restricted bHLH factors. However, in the lymphoid compartment they exist mainly as homodimers. As mentioned above, their activity can be regulated by heterodimerization with Id proteins, leading to the formation of inactive complexes.50 The entrance of the CLPs into the B-cell lineage is contingent on E2A and, in its absence, B-cell differentiation is completely blocked at the pre-pro-B-cell stage.51–53 The E2A-deficient CLPs and pre-pro-B cells lack rag1 expression and, therefore, fail to undergo immunoglobulin rearrangements. They express Il7rα, Igb, transcribe low levels of Ebf1 and do not express Pax5 and, therefore, its direct targets Cd19, mb1 or λ5. This suggests that E2A acts upstream of Ebf1 and Pax5 in the initiation of the B-cell expression program.51,52 In fact, E2A binds to and activates the distal promoter of the Ebf1 gene,54,55 and forced retroviral expression of EBF1 restores pro-B-cell development of in vitro cultured E2A-deficient progenitors.56 Afterwards, E2A cooperates with Ebf1 to coordinately regulate B cell-specific genes.57,58 E2A is also an essential regulator of light chain immunoglobulin rearrangement to such an extent that its ectopic expression, in combination with that of the RAG proteins, is enough to induce Igκ VJ recombination in nonlymphoid cells.59 By conditional mutagenesis, it has recently been shown that E2A is essential for the development of pro-B, pre-B and immature B cells in the bone marrow but is dispensable for the generation of mature B cells and plasma cells in peripheral lymphoid organs.60 At the molecular level, this is due to the fact that E2A is required not only for initiating but also for maintaining the expression of Ebf1, Pax5 and the B-cell gene program in pro-B cells. Also at later stages, germinal center B-cell development is impaired in the absence of E2A.60 Ebf1 and Pax5 are the definitive factors participating in the commitment to the B-cell fate. Forced expression of EBF in multipotential progenitors activates B cell-specific genes like Pax5 or its downstream targets and represses genes associated with alternative fates, like the myeloid ones.61 Ebf1 is first expressed in the CLP and regulates many genes involved in B-cell development.57,62 Consequently, Ebf1-knockout mice have a CLP compartment, but the cells lack B-cell developmental potential, since they lack the expression of Pax5 and other B-cell genes.63 The transcription of Ebf1 is dependent on two promoters that are differentially regulated. The distal one is controlled by activated STAT5 downstream of IL7 signaling by E47 and by autoregulation. The proximal promoter is upregulated by Pax5 and PU.1, creating, in this way, an autoregulatory loop during B-cell commitment and posterior development.54,55
Pax5 is the perfect example of a lineage commitment factor, since it controls the irreversible entrance of lymphoid progenitors into the B-cell pathway by a dual mechanism:32,64,65 it represses B lineage-inappropriate genes66 and activates B cell-specific genes.67 According to this B-cell commitment function, Pax5 expression is restricted to the B lymphoid lineage, from committed pro-B cells to the mature B-cell stage, and it has to be repressed afterwards for plasma cell differentiation.68 The expression of Pax5 is continuously required throughout B-cell development, and its conditional inactivation induces the conversion of mature B cells into functional T cells via dedifferentiation to uncommitted progenitors.69 Pax5 is activated in a stepwise manner during B-cell development. Its expression is controlled by an enhancer in intron 5 and a promoter element in the 5′ region of the Pax5 gene.70 In the transition from the HSC to multipotential progenitors, the CpG motifs in the enhancer are demethylated. The enhancer is activated by PU.1, IRF-4 and IRF-8 and NFκB already in multipotent hematopoietic and lymphoid progenitors. Therefore, the Pax5 enhancer is kept silent by DNA methylation in early progenitors and non-B cells and is only demethylated before or at the onset of hematopoiesis and organized into accessible chromatin in Ebf1-null lymphoid progenitors and at subsequent B-cell developmental stages.70 However, the promoter is demethylated and repressed by Polycomb group proteins in embryonic stem cells and other non-B cells. This repression is overcome only at the onset of B-cell development by the expression of Ebf1. Thus, Ebf1 is crucial for initiating modifications that result in an active chromatin configuration at the Pax5 promoter but not at the enhancer. Once it is established, Pax5 induces active chromatin at activated target genes and eliminates it at repressed genes in committed pro-B cells. It does this by recruiting chromatin-remodeling, histone-modifying and basal transcription factor complexes to its target genes.71
We have described the main intrinsic transcriptional regulators that coordinate the entry of hematopoietic progenitors into the B-cell lineage. However, this is not a cell-autonomous process, but it is dependent on developmental signals that have instructive and survival signals acting on the progenitors. The development and homeostasis of lymphocytes are regulated by cytokines as the interleukins IL-2, IL-4, IL-7, IL-9, IL-15 and IL-21. Among them, IL-7 is an essential, nonredundant cytokine for T- and B-cell development.72 All mentioned interleukins signal through receptors that contain a common molecule, the common γ chain (γc, Il2rg).73 In contrast to the broad expression of the γc chain in various hematopoietic lineages, the IL-7Rα subunit of IL7 is preferentially expressed in the lymphoid system from CLPs to large pre-B and pre-T cells as well as in thymic and peripheral CD4+ and CD8+ single-positive (SP) T cells.26,74 B-cell development is blocked at the pre-pro-B-cell stage in the BM of adult Il7r-null mice.75–77 IL-7 signaling is transduced by three main downstream pathways: the Jak-STAT, the PI3K-Akt and the RAS-MAPK pathways, with STAT5 being the predominant STAT protein activated by IL-7.78 The most recent data demonstrate that STAT5 and IL-7R signaling control cell survival during early B lymphopoiesis, therefore mainly playing a permissive role in early B-cell development.79 Also, it puts order into immunoglobulin gene rearrangement by repressing Igκ recombination in pro-B cells.80 Subsequently, IL-7 signaling is also required for the pro-B to pre-B-cell transition, acting in synergy with pre-BCR signaling to promote the expansion of large pre-B cells with correctly rearranged Igh genes.81 In this way, IL7 signaling, especially through STAT5, integrates with the process of rearrangement and signaling form the immunoglobulin genes. The pre-BCR and BCR are not only markers of developmental stages, but also essential signaling molecules themselves that regulate the development of B cells by acting as “quality control” checkpoints along the differentiation process. The precise mechanisms of this control at the signaling, transcriptional and epigenetic levels are still incompletely understood and are the subject of numerous exhaustive reviews by themselves.82–85 In our context, it is enough to mention that they are tightly intertwined with the transcriptional and epigenetic machinery that we have described so far, in such a way that any changes in one of the elements has profound consequences on the others.
Once the linage commitment has been established, B-cell identity will be stably maintained until the interaction of the BCR with its cognate antigen triggers the germinal center response and the posterior terminal differentiation into plasma cells. This actually implies a loss of B-cell identity that is brought about by the direct repression of Pax5 by the master regulator of plasma cell differentiation, the transcription factor Blimp1.86
Molecular Players and Postulated Cellular Origins of B-ALL
In the previous section, we succinctly described the intrinsic molecular mechanisms that lead cells through the early stages of hematopoietic development, from the multipotential HSC to the commitment to the B-cell lineage. From this description, the mechanistic importance of the cooperativity between transcription factors during lineage priming is evident, and it is easy to imagine how this delicate equilibrium can be perturbed by any interference with this machinery. B-cell acute lymphoblastic leukemia (B-ALL) is a clonal malignant disease characterized by an accumulation of early B cells. This abnormal growth causes the suppression of normal hematopoiesis and the infiltration of vital organs. ALLs form one of the four major categories of human leukemias, and most of the ALLs are of the B-cell type.3,87,88 ALLs, like every cancer, are clonal and originate from a single cell and, traditionally, B-ALL cells have been considered malignant counterparts of normal B-cell precursors of similar phenotype. However, this assumption is being heavily questioned in light of the most recent data. In normal clinical practice, the types of B-ALL have traditionally been correlated with the different normal B-cell developmental stages by studying microscopic appearance and immunophenotype. Using these criteria, in most cases, a relationship can be established between every type of B-cell leukemia and a given normal stage of B-cell differentiation.89 However, these associations are not always straightforward, since expression of myeloid markers has been found in up to 30% of precursorB-ALLs and since lymphoid markers can be found in acute myeloid leukemias (AML). Moreover, even the presence of immunoglobulin gene rearrangements, which is usually considered evidence of the previous passage of the tumor cells through the B-cell lineage, cannot be considered decisive evidence for the B-cell origin of a B-ALL: indeed, immunoglobulin gene rearrangements have also been described outside of the B-cell compartment,90,91 thus further complicating the issue of the B-ALL cellular origins. Finally, the plasticity of the different differentiation stages of B-cell development makes it very difficult to ascertain the precise nature of the first normal cell in which a B-ALL-associated genetic lesion takes place (Fig. 1). In human B-ALL patients, it is almost impossible to monitor the natural evolution of the disease from a normal hematopoietic cell (an HSC, a progenitor or a differentiated cell). In fact, oncogenic events might have occurred at various possible stages of B-cell development, after which the now aberrant B cell could have differentiated further and been arrested at a later stage of differentiation (see below). Nowadays, there is undeniable evidence indicating that several of the chromosomal translocations commonly found in childhood B-ALL originate prenatally in utero in fetal hematopoiesis92 (see below). This prenatal origin has now been confirmed for MLL fusion genes in infant B-ALL and for TEL-AML1 fusions in childhood B-ALL.92 These observations strongly suggest that these chromosomal changes have taken place either in the HSCs or even in earlier stages in the fetal liver or bone marrow. Still, the precise cellular origin of the translocations is difficult to ascertain, especially since the functional consequences of the translocation (that is, the clonal expansion) can appear in the cellular types whose phenotypic markers would place them either downstream or upstream of the point of origin of the translocation (Fig. 1). Thus, cells at later stages of B-cell development cannot be formally excluded as B-ALL cells-of-origin. One essential aspect to be considered in this context is that of the leukemic stem cells. This concept has been extensively discussed in previous reviews in references 93–95, so it will only be briefly summarized here, in relationship with the nature of the leukemic cell of origin. In almost all cases analyzed to date, the capacity of maintaining and repopulating the leukemic populations in human B-ALL samples has been shown to be restricted to a small subset of the total tumoral population;93 these are the so-called leukemic stem cells (and sometimes leukemia-initiating cells, but this can be confusing, see next section). Here, when we speak of the leukemic cell of origin (LCO), we refer to the first, normal hematopoietic cell in which the first genetic lesion linked to the development of B-ALL takes place and not to the leukemic stem cell (LSC) than can maintain the leukemic clone once this has been established in full-blown leukemia. Determining the nature of the LCO is difficult, since, as we have seen, normal HSCs are not a static, homogeneous population, but rather a continuum with highly variable proliferation and self-renewal properties.96,97 Using samples obtained from human acute myelogenous leukemias, it has been shown that, at the root of the disease, there is a hierarchical organization of LSCs differing in self-renewal capacity,98 therefore showing that heterogeneity can also be found in the LSCs compartment, similarly to how it happens with the normal HSCs.34,35 This fact is supported by the results obtained from the analyses of relapsed human acute lymphoblastic leukemias, which show that the cells of the relapse clone were already present as subpopulations at diagnosis.99–102 The most recent revealing studies about the clonal nature of cancer heterogeneity have, in fact, been performed on human B-ALL, either TEL-AML1+99 or BCR-ABL+.103 These studies have demonstrated that LSCs are genetically diverse, and that samples of patient at diagnosis contain multiple genetically distinct LSC subclones. Reconstructing the genetic ancestry of these subclones proves that leukemogenesis occurs in a Darwinian model of branching, multi-clonal evolution, rather than in a linear succession.99,103,104 This indicates that the cells responsible for the relapse were positioned developmentally upstream of the leukemic cells that were found at diagnosis. Therefore, the leukemic clone evolves with time and so do the LSCs, making their precise identification even more complicated. This has been also demonstrated using xenografts: when human cord blood stem cells were transduced with MLL-associated translocations and injected into mice, it was found that, from primary to secondary recipients, an increase in immunoglobulin rearrangements occurs.105 Furthermore, the rearrangements found in different secondary recipients were all coming from the same primary donor, indicating once more a clonal evolution implying that the properties of the LSC at a certain time point might not have any relationship with those of the initial LCO.105
By analogy to the study of the molecular mechanisms regulating normal lymphoid development, as described above, a great deal of knowledge about the pathophysiology of B-ALLs has been obtained by the identification of the genetic abnormalities consistently present in B-cell blasts. The most prevalent findings are gene fusions created by chromosomal translocations: TEL-AML1, MLL rearrangements, BCR-ABL and E2A-PBX1 (see below). The frequency of these genetically defined leukemia-associated molecular aberrations is different from children to adults, suggesting that there is a relationship between the development of normal B cells (which is known to change with age, see below) and that of leukemias. In this way, the TEL-AML1 fusion gene is almost exclusively present in childhood leukemias (22%, vs. 2% in adult leukemias), while BCR-ABL is much more frequent in adults (25%) than in children (3%). The molecular characteristics on these chimeric proteins have been very well-characterized in the last decade; however, the precise mechanisms by which they induce the B-cell developmental arrest characteristic of B-ALLs are unknown, although there are several possible ways to explain it. One possibility would be that the chimeric proteins interfere with the networks controlling B-cell differentiation (see below). Also, the translocation-carrying B-cell blasts might lose their capacity to respond to the external signals that regulate normal B-cell development. Finally, it could happen that the B-ALL-inducing oncogenic events might activate pathways that specifically reprogram the cells to phenotypically “copy” a particular stage of normal B-cell differentiation.
It is in this context where the unusually high degree of plasticity of B cells has to be especially taken into account (Fig. 1). This plasticity has been demonstrated for mouse mature peripheral B cells in various experimental settings. On one side, it has been described that transduction of BM CD19+ cells with the myeloid transcription factor C/EBPα leads to the full transdifferentiation of B cells into macrophages in vitro.106 Also, reprogramming of mature B cells to induced pluripotent stem cells (iPSCs) can be achieved by using the four Yamanaka transcription factors, but only if Pax5 activity is previously abolished.107 This Pax5-dependent high degree of developmental plasticity of B cells is best exemplified in the experiments showing that mature B cells can dedifferentiate all the way to multipotential progenitors after the genetic inactivation of Pax5.69 Furthermore, the complete loss of Pax5 in mature B cells also gives rise to tumors of an early pro-B-cell phenotype.69 Such plasticity was first evidenced in animal models but has since been proven in human patients, where examples of transdifferentiation from follicular lymphomas to myeloid histiocytic/dendritic cell sarcomas or from chronic lymphocytic leukemias/small lymphocytic lymphomas to histiocytic and dendritic cell sarcomas have been recently described.108,109 In human Hodgkin lymphomas, the overexpression of specific antagonists leads to the inactivation of the B-cell factor E2A and the consequent loss of B-cell markers and expression of lineage-inappropriate genes that characterizes the Reed-Sternberg cells.110,111 In children's B-ALLs, the involvement of stem cells in disease initiation and progression is a well-known fact,104 and it has recently been described that LSC properties can be found in blasts at several different maturational stages, that apparently can also interconvert among them to a certain degree,112 thus making it even more difficult to ascertain the nature of the cancer cell of origin. This high degree of lymphoid plasticity has led researchers to propose that, in some cases, even the cellular origin of CALM-AF10+ acute myeloid leukemias could reside in a lymphoid progenitor.113 Finally, it has to be considered that the problem of the LCO might not be restricted to the hematopoietic compartment, since mesenchymal stem cells obtained from the BM of infants with MLL-AF4+ B-ALL harbor the MLL-AF4 fusion gene and express it.114
Regulation of B-ALL Development
B-ALL-initiating lesions.
At the genetic level, as we have mentioned, B-ALL is characterized by a relatively small series of frequent chromosomal alterations, from aneuploidy to chromosomal rearrangements. Although in most cases they are accompanied by secondary mutations (which, as we will see below, often involve the main B-cell transcriptional regulators), the chromosomal alterations are the essential determinants of leukemogenesis and, in all likelihood, they are the earliest, leukemia-initiating genetic lesions. Aneuploidy is an important prognostic factor in B-ALL, and high hyperdiploidy (>50 chromosomes) present in 30% of childhood ALLs is associated with a favorable outcome,115 while hypodiploidy (less than 45 chromosomes), present in 6% of cases, implies a poor prognosis.116 However, the role that aneuploidies play at the molecular level in causing ALL is still poorly understood. Much more specific determinants of the disease are the products of chromosomal translocations. These are defined chimeric proteins with newly generated aberrant functions that interfere in several ways with the mechanisms of lineage differentiation that we have previously outlined. TEL-AML1 is the product of the t(12;21)(p13;q22) translocation. It comprises the 5′ region of the TEL gene from chromosome 12 and almost the entire coding region of AML1 from chromosome 21. TEL/ETV6 (translocation-ETS-leukemia) encodes a nuclear phospho-protein belonging to the ETS family of transcription factors and is required for maintenance of definitive adult hematopoiesis.117 AML1 (acute myeloid leukemia) encodes the α subunit of the core-binding factor (CBFA). In normal development, the heterodimeric CBFA2/CBFB transcription factor complex binds to specific enhancer sequences of genes that are important for HSC biology and differentiation. AML1 is essential for the emergence of HSCs from the embryonic hemogenic endothelium, and its loss results in embryonic death and lack of definitive hematopoiesis.118–121 TEL-AML1 retains its capacity to bind to the AML1-binding sequences and to heterodimerize with CBFB but, unlike the normal AML1 protein, it recruits a transcriptional corepressor complex with histone deacetylase activity (HDAC),122 therefore leading to the repression of transcription. In this way, the fusion protein represses the usually activated AML1 target genes, and it can also inhibit ETS family proteins via binding through the TEL DNA-binding domain, therefore globally altering the self-renewal and differentiation capacities of HSCs.
Another frequent translocation, present in 70% of infant leukemias, and with a very poor prognosis, is the t(4;11) resulting in a chimeric protein consisting of the N-terminal portion of the MLL (mixed-lineage leukemia) protein, from chromosome 11 and the C-terminal portion of AF4 (“ALL1 fused gene from chromosome 4”). The MLL gene encodes a large methyltransferase protein belonging to the thrithorax group, whose main biological function is to methylate the lysine 4 of histone H3 (H3K4) to regulate patterns of gene expression, particularly those of the genes of the HOX family, which are essential regulators of morphogenesis and development.123 More than 40 different translocations have been identified in which MLL is involved, and the most frequent ones in B-ALL (80% of the cases in which the MLL gene is involved in B-ALLs) are those affecting the genes AF4, AF9, ENL, AF10 and AF6.124,125 In spite of intensive studies, the precise mechanisms by which MLL-containing fusion proteins mediate leukemogenesis are not known, although the HOX genes, tightly controlled by MLL and which control the expansion of the HSC pool,126–128 are the most plausible critical targets involved in the process, together with the potential capacity of interfering with the cell cycle.129
The translocations involving the E2A gene are found in both childhood and adult B-ALLs. We have already described the essential role of E2A in B-cell development. Two are the main genes fused with E2A in B-ALLs. The t(1;19)(q23;p13.3) translocation gives rise to the E2A-PBX1 (pre-B-cell leukemia homeobox1) fusion gene, mainly present in childhood leukemias (5–6% of ALLs and 23% of pediatric B-ALLs), but also in adults (1–3%).130 The prognosis for E2A-PBX1+ childhood B-ALLs is good, with an 80–90% survival at 5 y. On the contrary, adult E2A-PBX1+ B-ALLs have a much poorer outcome (20–40% survival at 3 y). The E2A-HLF (hepatic leukemia factor) fusion gene is the result of the t(17;19)(q22; p13.3) translocation.131 It presents with a very low frequency in both children and adults (0.5–1%), but its prognosis is highly unfavorable (10% disease-free survival at 4 y). PBX1 is an homeobox-containing protein that is normally not expressed in the lymphoid compartment. Pbx1-/- mice die by embryonic day 15 or 16 with severe hypoplasia or aplasia of multiple organs and widespread patterning defects of the axial and appendicular skeleton, indicating a global role for this gene in development and organogenesis.132 The chimeric E2A-PBX1 transcription factor contains the N-terminal transactivation of E2A and the DNA-binding domain of PBX1.133 The t(1;19)(q23;p13.3) translocation has two simultaneous effects. First, it disrupts one allele of both E2A and PBX1, and this already can have a tumor-promoting effect. Second, it generates the novel fusion gene E2A-PBX1 expressed under the control of E2A regulatory sequences. In contrast to PBX1, E2A-PBX1 is a transcriptional activator of genes containing PBX1-binding sites.134–136 E2A-PBX1 can induce leukemogenesis by interacting and titrating away normal partners of the PBX proteins, such as the HOX proteins, and also by sequestering E2A coactivators, leading to the repression of E2A target genes and to uncontrolled cell cycle progression.130 HLF is a member of the PAR-bZIP (proline and acidic amino acid-rich basic leucine zipper) transcription factor family that is normally not expressed in the lymphoid compartment, and its transcripts are detected in the liver, kidney, lung and adult nervous system. It binds DNA as a homodimer or as a heterodimer with other PAR factors,137 and it has been proposed to play a role in the function of differentiated neurons in the adult nervous system.138 Although it is clear that the chimeric E2A-HLF protein interferes with normal lymphocyte development and renders lymphocytes susceptible to malignant transformation, the precise molecular mechanisms of this effect have not been completely elucidated.131 One of its main effectors is the transcriptional activation of SLUG/SNAI2, a multifunctional zinc-finger transcriptional regulator139 implicated in many cancerous processes. In the case of E2A-HLF+ B-ALLs, it seems that the main consequence of SLUG activation is the promotion of cell survival by counteracting the proapoptotic function of the BH3 protein.140 In any case, both E2A-involving translocations, together with the TEL-AML1 and MLL examples, demonstrate that the direct interference with transcriptional and epigenetic regulators of normal HSCs and B cells is at the base of the origin of B-ALLs.
Other type of interference with normal developmental regulatory controls can be mediated by deregulated signaling. The most classical example of this mechanism in the context of B-ALL is the product of the t(9;22)(q34;q11) translocation, the chimeric BCR-ABL oncogene. This is a rare alteration in childhood B-ALLs (3–5%), but it occurs in 20–40% of adult B-ALLs, and it has the most unfavorable prognosis among all B-ALL subtypes (37% 5-y event-free from diagnosis).141 There are two main BCR-ABL isoforms, p190 and p210, with different involvement in acute lymphoid or chronic myeloid leukemias. Only 33% of adult or 10% of BCR-ABL+ childhood B-ALLs express the p210 isoform, the p190 form being the main one involved in this type of BCR-ABL+ leukemias.142 ABL is a tyrosine kinase that presents normally in an inactive conformation, and it participates in the transduction of signals from many different pathways (implicated in processes like cell differentiation, cell division, cell adhesion and stress response) as well as in B-cell development. Its lack in ABL-knockout mice leads to death 1 to 2 weeks after birth with a loss of specific B- and T-cell populations.143,144 The BCR-ABL fusion proteins are deregulated, constitutively active nonreceptor tyrosine kinases, which directly activate signal transduction pathways controlling cell proliferation, survival and activation and, therefore, contribute to leukemogenic programming of the targeted cells.145
Cooperative mutations in B-ALL.
We have mentioned the main examples of B-ALL-initiating lesions and their basic known molecular mechanisms of leukemogenesis. However, human leukemias always present with additional mutations that clearly contribute to disease evolution, progression and malignancy. Most of the essential regulators of B cell commitment that we have described (Fig. 2) are altered in human B-ALLs. For example, using genome-wide analysis of leukemia samples from more than 300 ALL patients, deletions of the gene encoding the IKAROS protein (IKZF1) have been found in > 80% of BCR-ABL+ ALL cases,146 and they are accompanied by a much shorter survival in humans147 and by an acceleration of the disease in BCR-AB+ leukemic transgenic mice.148 These deletions resulted either in haploinsufficiency, expression of a dominant-negative IKAROS isoform or the complete loss of IKAROS expression. It has been recently found that the pre-BCR acts as a tumor suppressor upstream of IKAROS by inducing cell cycle arrest in BCR-ABL+ ALL cells. Therefore, the absence of IKAROS eliminates this control and promotes uncontrolled proliferation at the pre-B cell stage.149
Also PAX5 has been directly involved in the molecular pathogenesis of B-ALLs.32 Approximately 30% of all pediatric B-ALL and BCR-ABL+ adult B-ALL cases studied by genome-wide analyses showed monoallelic losses or point mutations of the PAX5 gene, that result either in haploinsufficiency or in the generation of hypomorphic alleles of PAX5.150,151 PAX5 is also involved in specific chromosomal translocations that give rise to chimeric transcription factors by fusing the N-terminal DNA-binding domain of PAX5 with C-terminal regulatory sequences of a second transcription factor that can be one of a list of several different partner genes described up to date, like ETV6, FOXP1, EVI3, ELN, JAK2, PML, BRD1, POM121, HIPK1 or DACH.152–155 It is believed that these fusion proteins exert their leukemogenic effect by acting as constitutive repressors by interfering with the transactivation function of the wild-type PAX5 protein encoded by the unaltered PAX5 allele.32
There are also examples showing the involvement of EBF1 in B-ALL. In genome-wide association studies, eight cases of pediatric progenitor B-ALL cases carried mono-allelic deletions of EBF1.151 Since EBF1 has been shown to be monoallelicaly expressed,156 and heterozygous Ebf+/− mice have a 50% reduction in the number of mature B cells, these observations suggest that EBF1 haploinsufficiency may contribute to leukemogenesis. In fact, overexpression of proteins known to interact with EBF1 and block its function, like ZFP521, have been shown to collaborate with E2A-HLF in B-ALL and to promote lymphomagenesis in mouse models.157–160
Not only are transcriptional regulators altered and mutated in B-ALL, but also the molecules involved in signaling, like STAT5. BCR-ABL-deregulated kinase activity leads to constitutive activation of STAT5 and, thus, likely contributes to leukemogenesis by signaling via the JAK-STAT pathway.161 In fact, BCR-ABL kinase cannot induce leukemia in Stat5-/- mice, therefore indicating an important role for STAT5 in the genesis of B-ALL.162 Increased STAT5 expression has been shown in cells from ALL patients, and it correlates with poor prognosis. Furthermore, using mouse models, it has been demonstrated that haploinsufficiency of either Pax5 or Ebf1 synergized with constitutively active STAT5 forms to rapidly induce ALL in 100% of the mice,163 indicating that small perturbations in a self-reinforcing network of transcription factors critical for B-cell development cooperate in the initiation and progression of B-ALL.
Molecular alterations in many other genes have been described by studying human B-ALL samples, for example affecting FTL3 or the RB or Tp53 pathways.88 However, the precise way in which they contribute to the disease origin and evolution is unclear. Also, by using genome-wide association studies, a small group of gene variants have been found (IKZF1, ARIDB5, CEBPE and CDKN2A) that affect susceptibility to ALL, although their individual effects are modest.164–168 As exemplified by the case of IKZF1 (IKAROS), these gene variants are also related to pathways controlling B-cell development and differentiation. However, how these germline variations might contribute to the development of ALL is still unclear. Finally, while several genetic alterations have been proven to be associated with the risk of relapse, the biologic and cellular components determining treatment failure are poorly understood.
The Role of Age in B-ALL Development
We have discussed how the developmental position of a cell within the differentiation cascade leading from HSCs to B cells can have a deep influence on the final leukemic phenotype. We have also seen how the molecular nature of the initial and secondary genetic leukemogenic lesions is an essential determinant of leukemic malignancy and response to therapy. One final essential aspect to consider is that of the age at which the oncogenic initiating lesion takes place. This is especially relevant in the case of B-ALL. As we have already briefly mentioned, the age of the patients at diagnosis is, together with the molecular nature of the chromosomal translocation involved, essential in establishing B-ALL prognosis.3,88 For example, BCR-ABL+ B-ALL is generally associated with a poor prognosis in adolescents, but in children with a low leukocyte count at presentation, it has a relatively favorable outcome; again, adults with this type of B-ALL have a dismal prognosis. Infants with MLL-related B-ALL fare considerably worse than older children with this alteration.3,88 This may be related, as explained, to the developmental stage of the target cell undergoing malignant transformation and also to the pharmacogenetic or pharmacokinetic features of the patient.3,88 However, age itself greatly affects the properties and functionality of the hematopoietic system,169 affecting both the HSC170,171 and lymphoid172–175 compartments. Therefore, age of onset must clearly affect the outcome of the disease.
For some B-cell precursor childhood B-ALLs, it has been clearly proven that the translocation occurs prenatally and constitutes the initiating event. The best example is that of TEL-AML1.92,99,176,177 Screening of cord blood samples found that around 1% of newborns have a functional TEL-AML1 fusion gene. Since the frequency of development of B-ALL is much lower, this implies that the conversion of the preleukemic clone to overt disease is a rare phenomenon, and that development of childhood B-ALL is a multi-step process requiring, besides the prenatal initiating event, secondary prenatal and/or postnatal events that would be the rate-limiting ones. While the initiating in utero event is believed to be common, the subsequent events act as the rate-determining steps in the development of the disease. If this “two hit” model is correct, it would mean that, for every child diagnosed with ALL, there are at least 20 “healthy” children who carry a covert preleukemic clone generated in utero. It must be mentioned that it has recently been challenged, however, that the prevalence of the TEL-AML1 gene fusion is really as high as 1%,178 and that this topic is still the matter of intense debate. But, even if the frequency was not that high, the “two hit” model nicely explains the biology of the disease and forces us to further understand if there is any external cause for the appearance of the secondary genetic alterations that lead to the development of overt leukemia. Many explanations have been postulated (the effect of electromagnetic fields, infections, parental cigarette smoking, etc.), but the etiology of B-ALL is still unknown.2
But also, for B-ALLs of assumed adult origin, age plays an essential role in the malignancy of the disease. From this point of view, it has recently been shown, using a BCR-ABLp190-inducible transgene179 or by transfecting mouse cells of different ages,180 that cells from old mice develop leukemias at a much faster rate than those from young animals, therefore suggesting some kind of competitive advantage of aged B-cell progenitors that could explain the more aggressive nature of B-ALL in advanced ages.181,182
Concluding Remarks
Leukemia is the most common type of childhood cancer, accounting for 30% of all cancers diagnosed in children younger than 15 y, and its incidence is on the rise.183 However, little is known about its etiology. Apart from some rare genetic syndromes, only exposure to ionizing radiation and a child's high birth weight have been identified as risk factors.184 Although treatments have improved the rate of childhood B-ALL cures tremendously in the last decades, relapses are still a serious problem,185 and in adults, B-ALL is a disease with a generally poor prognosis. As we have seen, the heterogeneity within the leukemias at all the levels is very high, thus making it difficult to find a common ground to fight the disease. The knowledge gained from the study of the normal development of the hematopoietic system and how the normal physiological choices are made during lineage differentiation has helped us to shed new light on the molecular mechanisms used by B-ALL-associated genetic lesions to subvert development and to reprogram normal stem/progenitors into leukemic or pre-leukemic cells. It has also allowed us to realize that the developmental organization of the leukemias follows a similar scheme to that of the normal hematopoietic system. Experimental approaches based both in the study of human patient samples and in the generation of animal models of disease have allowed us to gain new insights into the nature of the disease. We hope, in the near future, that both approaches will help us to translate all this new knowledge into efficient therapies.
Acknowledgments
Research in C.C. lab was partially supported by FEDER, Fondo de Investigaciones Sanitarias (PI080164), CSIC P.I.E., 200920I055 and 201120E060, from the ARIMMORA project (FP7-ENV-2011, European Union Seventh Framework Program) and from an institutional grant from the “Fundación Ramón Areces.” Research in I.S.G. group was partially supported by FEDER and by MICINN (SAF2009-08803 to I.S.G.), by Junta de Castilla y León (REF. CSI007A11-2 and Proyecto Biomedicina 2009-2010), by MEC OncoBIO Consolider-Ingenio 2010 (Ref. CSD2007-0017), by NIH grant (R01 CA109335-04A1), by Sandra Ibarra Foundation, by Group of Excellence Grant (GR15) from Junta de Castilla y Leon, and the ARIMMORA project (FP7-ENV-2011, European Union Seventh Framework Program) and by Proyecto en Red de Investigación en Celulas Madre Tumorales en Cancer de Mama, supported by Obra Social Kutxa y Conserjería de Sanidad de la Junta de Castilla y Leon. All Spanish funding is co-sponsored by the European Union FEDER program. I.S.G. is an API lab of the EuroSyStem project. E.C.S. and M.B.D. are recipients of JAE-predoc fellowships from CSIC. I.R.C. is recipient of a FPU fellowship from MICINN. E.C.S. is recipient of a “Residencia de Estudiantes” fellowship.
References
- 1.Evans WE, Relling MV. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429:464–468. doi: 10.1038/nature02626. [DOI] [PubMed] [Google Scholar]
- 2.Greaves M. Infection, immune responses and the aetiology of childhood leukaemia. Nat Rev Cancer. 2006;6:193–203. doi: 10.1038/nrc1816. [DOI] [PubMed] [Google Scholar]
- 3.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 4.Purton LE, Scadden DT. Limiting factors in murine hematopoietic stem cell assays. Cell Stem Cell. 2007;1:263–270. doi: 10.1016/j.stem.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 5.Welner RS, Pelayo R, Kincade PW. Evolving views on the genealogy of B cells. Nat Rev Immunol. 2008;8:95–106. doi: 10.1038/nri2234. [DOI] [PubMed] [Google Scholar]
- 6.Li CL, Johnson GR. Murine hematopoietic stem and progenitor cells: I. Enrichment and biologic characterization. Blood. 1995;85:1472–1479. [PubMed] [Google Scholar]
- 7.Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-X. [DOI] [PubMed] [Google Scholar]
- 8.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Lin Y, Zhan Y, Yang G, Louie J, Harrison DE, et al. Murine hematopoietic stem cell characterization and its regulation in BM transplantation. Blood. 2000;96:3016–3022. [PubMed] [Google Scholar]
- 10.Adolfsson J, Borge OJ, Bryder D, Theilgaard-Monch K, Astrand-Grundstrom I, Sitnicka E, et al. Upregulation of Flt3 expression within the bone marrow Lin(-) Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15:659–669. doi: 10.1016/S1074-7613(01)00220-5. [DOI] [PubMed] [Google Scholar]
- 11.Christensen JL, Weissman IL. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc Natl Acad Sci USA. 2001;98:14541–14546. doi: 10.1073/pnas.261562798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forsberg EC, Serwold T, Kogan S, Weissman IL, Passegue E. New evidence supporting megakaryocyte-erythrocyte potential of flk2/flt3+ multipotent hematopoietic progenitors. Cell. 2006;126:415–426. doi: 10.1016/j.cell.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 13.Wagers AJ, Weissman IL. Differential expression of alpha2 integrin separates long-term and short-term reconstituting Lin-/loThy1.1(lo)c-kit+ Sca-1+ hematopoietic stem cells. Stem Cells. 2006;24:1087–1094. doi: 10.1634/stemcells.2005-0396. [DOI] [PubMed] [Google Scholar]
- 14.Benveniste P, Frelin C, Janmohamed S, Barbara M, Herrington R, Hyam D, et al. Intermediate-term hematopoietic stem cells with extended but time-limited reconstitution potential. Cell Stem Cell. 2010;6:48–58. doi: 10.1016/j.stem.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Müller-Sieburg CE, Cho RH, Thoman M, Adkins B, Sieburg HB. Deterministic regulation of hematopoietic stem cell self-renewal and differentiation. Blood. 2002;100:1302–1309. [PubMed] [Google Scholar]
- 16.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, et al. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- 18.Månsson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Enver T, Pera M, Peterson C, Andrews PW. Stem cell states, fates and the rules of attraction. Cell Stem Cell. 2009;4:387–397. doi: 10.1016/j.stem.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Huang S, Ernberg I, Kauffman S. Cancer attractors: a systems view of tumors from a gene network dynamics and developmental perspective. Semin Cell Dev Biol. 2009;20:869–876. doi: 10.1016/j.semcdb.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang S. On the intrinsic inevitability of cancer: From foetal to fatal attraction. Semin Cancer Biol. 2011 doi: 10.1016/j.semcancer.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Abollo-Jiménez F, Jimenez R, Cobaleda C. Physiological cellular reprogramming and cancer. Semin Cancer Biol. 2010;20:98–106. doi: 10.1016/j.semcancer.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Igarashi H, Gregory SC, Yokota T, Sakaguchi N, Kincade PW. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 2002;17:117–130. doi: 10.1016/S1074-7613(02)00366-7. [DOI] [PubMed] [Google Scholar]
- 25.Ichii M, Shimazu T, Welner RS, Garrett KP, Zhang Q, Esplin BL, et al. Functional diversity of stem and progenitor cells with B-lymphopoietic potential. Immunol Rev. 2010;237:10–21. doi: 10.1111/j.1600-065X.2010.00933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/S0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 27.Izon D, Rudd K, DeMuth W, Pear WS, Clendenin C, Lindsley RC, et al. A common pathway for dendritic cell and early B cell development. J Immunol. 2001;167:1387–1392. doi: 10.4049/jimmunol.167.3.1387. [DOI] [PubMed] [Google Scholar]
- 28.Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, Hardy RR. Lineage specification and plasticity in CD19− early B cell precursors. J Exp Med. 2006;203:675–687. doi: 10.1084/jem.2005244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardy RR, Kincade PW, Dorshkind K. The protean nature of cells in the B lymphocyte lineage. Immunity. 2007;26:703–714. doi: 10.1016/j.immuni.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 31.Balciunaite G, Ceredig R, Massa S, Rolink AGA. B220+ CD117+ CD19− hematopoietic progenitor with potent lymphoid and myeloid developmental potential. Eur J Immunol. 2005;35:2019–2030. doi: 10.1002/eji.200526318. [DOI] [PubMed] [Google Scholar]
- 32.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 33.Majeti R, Park CY, Weissman IL. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 2007;1:635–645. doi: 10.1016/j.stem.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goardon N, Marchi E, Atzberger A, Quek L, Schuh A, Soneji S, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011;19:138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 35.Majeti R, Weissman IL. Human acute myelogenous leukemia stem cells revisited: there's more than meets the eye. Cancer Cell. 2011;19:9–10. doi: 10.1016/j.ccr.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galy A, Travis M, Cen D, Chen B, Human TB. Natural killer and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity. 1995;3:459–473. doi: 10.1016/1074-7613(95)90175-2. [DOI] [PubMed] [Google Scholar]
- 37.Haddad R, Guardiola P, Izac B, Thibault C, Radich J, Delezoide AL, et al. Molecular characterization of early human T/NK and B-lymphoid progenitor cells in umbilical cord blood. Blood. 2004;104:3918–3926. doi: 10.1182/blood-2004-05-1845. [DOI] [PubMed] [Google Scholar]
- 38.Miller JS, McCullar V, Punzel M, Lemischka IR, Moore KA. Single adult human CD34(+)/Lin−/CD38(-) progenitors give rise to natural killer cells, B-lineage cells, dendritic cells and myeloid cells. Blood. 1999;93:96–106. [PubMed] [Google Scholar]
- 39.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 40.Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 41.Weishaupt H, Sigvardsson M, Attema JL. Epigenetic chromatin states uniquely define the developmental plasticity of murine hematopoietic stem cells. Blood. 2010;115:247–256. doi: 10.1182/blood-2009-07-235176. [DOI] [PubMed] [Google Scholar]
- 42.Ng SY, Yoshida T, Zhang J, Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 2009;30:493–507. doi: 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson EC, Cobb BS, Sabbattini P, Meixlsperger S, Parelho V, Liberg D, et al. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity. 2007;26:335–344. doi: 10.1016/j.immuni.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 44.Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, et al. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9:927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh H. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31:576–586. doi: 10.1016/j.immuni.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arinobu Y, Mizuno S, Chong Y, Shigematsu H, Iino T, Iwasaki H, et al. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 47.DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 48.Nutt SL, Metcalf D, D'Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med. 2005;201:221–231. doi: 10.1084/jem.20041535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Ji M, Klarmann KD, Keller JR. Repression of Id2 expression by Gfi-1 is required for B-cell and myeloid development. Blood. 2010;116:1060–1069. doi: 10.1182/blood-2009-11-255075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 51.Bain G, Maandag EC, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 52.Borghesi L, Aites J, Nelson S, Lefterov P, James P, Gerstein R. E47 is required for V(D)J recombinase activity in common lymphoid progenitors. J Exp Med. 2005;202:1669–1677. doi: 10.1084/jem.20051190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 54.Roessler S, Gyory I, Imhof S, Spivakov M, Williams RR, Busslinger M, et al. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol Cell Biol. 2007;27:579–594. doi: 10.1128/MCB.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith EM, Gisler R, Sigvardsson M. Cloning and characterization of a promoter flanking the early B cell factor (EBF) gene indicates roles for E-proteins and autoregulation in the control of EBF expression. J Immunol. 2002;169:261–270. doi: 10.4049/jimmunol.169.1.261. [DOI] [PubMed] [Google Scholar]
- 56.Seet CS, Brumbaugh RL, Kee BL. Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A. J Exp Med. 2004;199:1689–1700. doi: 10.1084/jem.20032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O'Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/S1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 58.Sigvardsson M, O'Riordan M, Grosschedl R. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity. 1997;7:25–36. doi: 10.1016/S1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- 59.Romanow WJ, Langerak AW, Goebel P, Wolvers-Tettero IL, van Dongen JJ, Feeney AJ, et al. E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Mol Cell. 2000;5:343–353. doi: 10.1016/S1097-2765(00)80429-3. [DOI] [PubMed] [Google Scholar]
- 60.Kwon K, Hutter C, Sun Q, Bilic I, Cobaleda C, Malin S, et al. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 2008;28:751–762. doi: 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 61.Pongubala JM, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E, et al. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol. 2008;9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- 62.Sigvardsson M, Clark DR, Fitzsimmons D, Doyle M, Akerblad P, Breslin T, et al. Early B-cell factor, E2A and Pax-5 cooperate to activate the early B cell-specific mb-1 promoter. Mol Cell Biol. 2002;22:8539–8551. doi: 10.1128/MCB.22.24.8539-51.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zandi S, Mansson R, Tsapogas P, Zetterblad J, Bryder D, Sigvardsson M. EBF1 is essential for B-lineage priming and establishment of a transcription factor network in common lymphoid progenitors. J Immunol. 2008;181:3364–3372. doi: 10.4049/jimmunol.181.5.3364. [DOI] [PubMed] [Google Scholar]
- 64.Carotta S, Holmes ML, Pridans C, Nutt SL. Pax5 maintains cellular identity by repressing gene expression throughout B cell differentiation. Cell Cycle. 2006;5:2452–2456. doi: 10.4161/cc.5.21.3396. [DOI] [PubMed] [Google Scholar]
- 65.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 66.Delogu A, Schebesta A, Sun Q, Aschenbrenner K, Perlot T, Busslinger M. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 2006;24:269–281. doi: 10.1016/j.immuni.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 67.Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration and immune function. Immunity. 2007;27:49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 68.Fuxa M, Busslinger M. Reporter gene insertions reveal a strictly B lymphoid-specific expression pattern of Pax5 in support of its B cell identity function. J Immunol. 2007;178:8222–8228. doi: 10.4049/jimmunol.178.12.8221-a. [DOI] [PubMed] [Google Scholar]
- 69.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 70.Decker T, Pasca di Magliano M, McManus S, Sun Q, Bonifer C, Tagoh H, et al. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 2009;30:508–520. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 71.McManus S, Ebert A, Salvagiotto G, Medvedovic J, Sun Q, Tamir I, et al. The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J. 2011;30:2388–2404. doi: 10.1038/emboj.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.von Freeden-Jeffry U, Vieira P, Lucian LA, McNeil T, Burdach SE, Murray R. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kovanen PE, Leonard WJ. Cytokines and immunodeficiency diseases: critical roles of the gamma(c)-dependent cytokines interleukins 2, 4, 7, 9, 15 and 21, and their signaling pathways. Immunol Rev. 2004;202:67–83. doi: 10.1111/j.01052896.2004.00203.x. [DOI] [PubMed] [Google Scholar]
- 74.Sudo T, Nishikawa S, Ohno N, Akiyama N, Tamakoshi M, Yoshida H. Expression and function of the interleukin 7 receptor in murine lymphocytes. Proc Natl Acad Sci USA. 1993;90:9125–9129. doi: 10.1073/pnas.90.19.9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kikuchi K, Kasai H, Watanabe A, Lai AY, Kondo M. IL-7 specifies B cell fate at the common lymphoid progenitor to pre-proB transition stage by maintaining early B cell factor expression. J Immunol. 2008;181:383–392. doi: 10.4049/jimmunol.181.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kikuchi K, Lai AY, Hsu CL, Kondo M. IL-7 receptor signaling is necessary for stage transition in adult B cell development through upregulation of EBF. J Exp Med. 2005;201:1197–1203. doi: 10.1084/jem.20050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miller JP, Izon D, DeMuth W, Gerstein R, Bhandoola A, Allman D. The earliest step in B lineage differentiation from common lymphoid progenitors is critically dependent upon interleukin 7. J Exp Med. 2002;196:705–711. doi: 10.1084/jem.20020784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hennighausen L, Robinson GW. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev. 2008;22:711–721. doi: 10.1101/gad.1643908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol. 2010;11:171–179. doi: 10.1038/ni.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Malin S, McManus S, Busslinger M. STAT5 in B cell development and leukemia. Curr Opin Immunol. 2010;22:168–176. doi: 10.1016/j.coi.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 81.Fleming HE, Paige CJ. Pre-B cell receptor signaling mediates selective response to IL-7 at the pro-B to pre-B cell transition via an ERK/MAP kinase-dependent pathway. Immunity. 2001;15:521–531. doi: 10.1016/S1074-7613(01)00216-3. [DOI] [PubMed] [Google Scholar]
- 82.Bergman Y, Cedar H. Epigenetic control of recombination in the immune system. Semin Immunol. 2010;22:323–329. doi: 10.1016/j.smim.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hewitt SL, Chaumeil J, Skok JA. Chromosome dynamics and the regulation of V(D)J recombination. Immunol Rev. 2010;237:43–54. doi: 10.1111/j.1600-065X.2010.00931.x. [DOI] [PubMed] [Google Scholar]
- 84.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11:251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 85.Vettermann C, Schlissel MS. Allelic exclusion of immunoglobulin genes: models and mechanisms. Immunol Rev. 2010;237:22–42. doi: 10.1111/j.1600-065X.2010.00935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oracki SA, Walker JA, Hibbs ML, Corcoran LM, Tarlinton DM. Plasma cell development and survival. Immunol Rev. 2010;237:140–159. doi: 10.1111/j.1600-065X.2010.00940.x. [DOI] [PubMed] [Google Scholar]
- 87.Hjalgrim LL, Rostgaard K, Schmiegelow K, Soderhall S, Kolmannskog S, Vettenranta K, et al. Age- and sex-specific incidence of childhood leukemia by immunophenotype in the Nordic countries. J Natl Cancer Inst. 2003;95:1539–1544. doi: 10.1093/jnci/djg064. [DOI] [PubMed] [Google Scholar]
- 88.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 89.LeBien TW. Fates of human B-cell precursors. Blood. 2000;96:9–23. [PubMed] [Google Scholar]
- 90.Sánchez I, San Miguel JF, Corral J, Martin C, Perez R, Gonzalez M, et al. Gene rearrangement in acute non-lymphoblastic leukaemia: correlation with morphological and immunophenotypic characteristics of blast cells. Br J Haematol. 1995;89:104–109. doi: 10.1111/j.1365-2141.1995.tb08903.x. [DOI] [PubMed] [Google Scholar]
- 91.Sánchez-García I, San Miguel JF, Gonzalez-Sarmiento R. Immunoglobulin lambda chain gene rearrangement in a case of acute nonlymphoblastic leukemia. Leukemia. 1999;13:485–487. doi: 10.1038/sj.leu.2401204. [DOI] [PubMed] [Google Scholar]
- 92.Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer. 2003;3:639–649. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- 93.Cobaleda C, Sanchez-Garcia I. B-cell acute lymphoblastic leukaemia: towards understanding its cellular origin. Bioessays. 2009;31:600–609. doi: 10.1002/bies.200800234. [DOI] [PubMed] [Google Scholar]
- 94.Sánchez-García I, Vicente-Duenas C, Cobaleda C. The theoretical basis of cancer-stem-cell-based therapeutics of cancer: can it be put into practice? Bioessays. 2007;29:1269–1280. doi: 10.1002/bies.20679. [DOI] [PubMed] [Google Scholar]
- 95.Vormoor HJ. Malignant stem cells in childhood acute lymphoblastic leukemia: the stem cell concept revisited. Cell Cycle. 2009;8:996–999. doi: 10.4161/cc.8.7.7984. [DOI] [PubMed] [Google Scholar]
- 96.Crooks GM, Weinberg K. The unpredictable stem cell. Nat Immunol. 2006;7:1129–1130. doi: 10.1038/ni1106-29. [DOI] [PubMed] [Google Scholar]
- 97.McKenzie JL, Gan OI, Doedens M, Wang JC, Dick JE. Individual stem cells with highly variable proliferation and self-renewal properties comprise the human hematopoietic stem cell compartment. Nat Immunol. 2006;7:1225–1233. doi: 10.1038/ni1393. [DOI] [PubMed] [Google Scholar]
- 98.Hope KJ, Jin L, Dick JE. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat Immunol. 2004;5:738–743. doi: 10.1038/ni1080. [DOI] [PubMed] [Google Scholar]
- 99.Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 100.Kuster L, Grausenburger R, Fuka G, Kaindl U, Krapf G, Inthal A, et al. ETV6/RUNX1-positive relapses evolve from an ancestral clone and frequently acquire deletions of genes implicated in glucocorticoid signaling. Blood. 2011;117:2658–2667. doi: 10.1182/blood-2010-03-275347. [DOI] [PubMed] [Google Scholar]
- 101.Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van Delft FW, Horsley S, Colman S, Anderson K, Bateman C, Kempski H, et al. Clonal origins of relapse in ETV6-RUNX1 acute lymphoblastic leukemia. Blood. 2011;117:6247–6254. doi: 10.1182/blood-2010-10-314674. [DOI] [PubMed] [Google Scholar]
- 103.Notta F, Mullighan CG, Wang JC, Poeppl A, Doulatov S, Phillips LA, et al. Evolution of human BCRABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–367. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 104.Greaves M. Cancer stem cells: back to Darwin? Semin Cancer Biol. 2010;20:65–70. doi: 10.1016/j.semcancer.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 105.Barabé F, Kennedy JA, Hope KJ, Dick JE. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007;316:600–604. doi: 10.1126/science.1139851. [DOI] [PubMed] [Google Scholar]
- 106.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/S0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 107.Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Feldman AL, Arber DA, Pittaluga S, Martinez A, Burke JS, Raffeld M, et al. Clonally related follicular lymphomas and histiocytic/dendritic cell sarcomas: evidence for transdifferentiation of the follicular lymphoma clone. Blood. 2008;111:5433–5439. doi: 10.1182/blood-2007-11-124792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shao H, Xi L, Raffeld M, Feldman AL, Ketterling RP, Knudson R, et al. Clonally related histiocytic/dendritic cell sarcoma and chronic lymphocytic leukemia/small lymphocytic lymphoma: a study of seven cases. Mod Pathol. 2011 doi: 10.1038/modpathol.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Janz M, Dorken B, Mathas S. Reprogramming of B lymphoid cells in human lymphoma pathogenesis. Cell Cycle. 2006;5:1057–1061. doi: 10.4161/cc.5.10.2737. [DOI] [PubMed] [Google Scholar]
- 111.Mathas S, Janz M, Hummel F, Hummel M, Wollert-Wulf B, Lusatis S, et al. Intrinsic inhibition of transcription factor E2A by HLH proteins ABF-1 and Id2 mediates reprogramming of neoplastic B cells in Hodgkin lymphoma. Nat Immunol. 2006;7:207–215. doi: 10.1038/ni1285. [DOI] [PubMed] [Google Scholar]
- 112.le Viseur C, Hotfilder M, Bomken S, Wilson K, Rottgers S, Schrauder A, et al. In childhood acute lymphoblastic leukemia, blasts at different stages of immunophenotypic maturation have stem cell properties. Cancer Cell. 2008;14:47–58. doi: 10.1016/j.ccr.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Deshpande AJ, Buske C. Lymphoid progenitors as candidate cancer stem cells in AML: new perspectives. Cell Cycle. 2007;6:543–545. doi: 10.4161/cc.6.5.3968. [DOI] [PubMed] [Google Scholar]
- 114.Menendez P, Catalina P, Rodriguez R, Melen GJ, Bueno C, Arriero M, et al. Bone marrow mesenchymal stem cells from infants with MLL-AF4+ acute leukemia harbor and express the MLL-AF4 fusion gene. J Exp Med. 2009;206:3131–3141. doi: 10.1084/jem.20091050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sutcliffe MJ, Shuster JJ, Sather HN, Camitta BM, Pullen J, Schultz KR, et al. High concordance from independent studies by the Children's Cancer Group (CCG) and Pediatric Oncology Group (POG) associating favorable prognosis with combined trisomies 4, 10 and 17 in children with NCI Standard-Risk B-precursor Acute Lymphoblastic Leukemia: a Children's Oncology Group (COG) initiative. Leukemia. 2005;19:734–740. doi: 10.1038/sj.leu.2403673. [DOI] [PubMed] [Google Scholar]
- 116.Harrison CJ, Moorman AV, Broadfield ZJ, Cheung KL, Harris RL, Reza Jalali G, et al. Three distinct subgroups of hypodiploidy in acute lymphoblastic leukaemia. Br J Haematol. 2004;125:552–559. doi: 10.1111/j.1365-2141.2004.04948.x. [DOI] [PubMed] [Google Scholar]
- 117.Hock H, Meade E, Medeiros S, Schindler JW, Valk PJ, Fujiwara Y, et al. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 2004;18:2336–2341. doi: 10.1101/gad.1239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cleary ML. A new angle on a pervasive oncogene. Nat Genet. 1999;23:134–135. doi: 10.1038/13761. [DOI] [PubMed] [Google Scholar]
- 119.Goyama S, Mulloy JC. Molecular pathogenesis of core binding factor leukemia: current knowledge and future prospects. Int J Hematol. 2011 doi: 10.1007/s12185-011-0858-z. [DOI] [PubMed] [Google Scholar]
- 120.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/S0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 121.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hiebert SW, Sun W, Davis JN, Golub T, Shurtleff S, Buijs A, et al. The t(12;21) translocation converts AML-1B from an activator to a repressor of transcription. Mol Cell Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 124.Horton SJ, Williams O. The mechanism of hematopoietic progenitor cell immortalization by MLLENL. Cell Cycle. 2006;5:360–362. doi: 10.4161/cc.5.4.2464. [DOI] [PubMed] [Google Scholar]
- 125.Meyer C, Schneider B, Jakob S, Strehl S, Attarbaschi A, Schnittger S, et al. The MLL recombinome of acute leukemias. Leukemia. 2006;20:777–784. doi: 10.1038/sj.leu.2404150. [DOI] [PubMed] [Google Scholar]
- 126.Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev Cell. 2004;6:437–443. doi: 10.1016/S1534-5807(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 127.Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr Biol. 2004;14:2063–2069. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 128.Slany RK. The molecular biology of mixed lineage leukemia. Haematologica. 2009;94:984–993. doi: 10.3324/haematol.2008.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu H, Takeda S, Cheng EH, Hsieh JJ. Biphasic MLL takes helm at cell cycle control: implications in human mixed lineage leukemia. Cell Cycle. 2008;7:428–435. doi: 10.4161/cc.7.4.5426. [DOI] [PubMed] [Google Scholar]
- 130.Aspland SE, Bendall HH, Murre C. The role of E2APBX1 in leukemogenesis. Oncogene. 2001;20:5708–5717. doi: 10.1038/sj.onc.1204592. [DOI] [PubMed] [Google Scholar]
- 131.Seidel MG, Look AT. E2A-HLF usurps control of evolutionarily conserved survival pathways. Oncogene. 2001;20:5718–5725. doi: 10.1038/sj.onc.1204591. [DOI] [PubMed] [Google Scholar]
- 132.Selleri L, Depew MJ, Jacobs Y, Chanda SK, Tsang KY, Cheah KS, et al. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development. 2001;128:3543–3557. doi: 10.1242/dev.128.18.3543. [DOI] [PubMed] [Google Scholar]
- 133.Kamps MP, Murre C, Sun XH, Baltimore D. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell. 1990;60:547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- 134.LeBrun DP, Cleary ML. Fusion with E2A alters the transcriptional properties of the homeodomain protein PBX1 in t(1;19) leukemias. Oncogene. 1994;9:1641–1647. [PubMed] [Google Scholar]
- 135.Lu Q, Wright DD, Kamps MP. Fusion with E2A converts the Pbx1 homeodomain protein into a constitutive transcriptional activator in human leukemias carrying the t(1;19) translocation. Mol Cell Biol. 1994;14:3938–3948. doi: 10.1128/mcb.14.6.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Van Dijk MA, Voorhoeve PM, Murre C. Pbx1 is converted into a transcriptional activator upon acquiring the N-terminal region of E2A in pre-B-cell acute lymphoblastoid leukemia. Proc Natl Acad Sci USA. 1993;90:6061–6065. doi: 10.1073/pnas.90.13.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hunger SP, Ohyashiki K, Toyama K, Cleary ML. Hlf, a novel hepatic bZIP protein, shows altered DNA-binding properties following fusion to E2A in t(17;19) acute lymphoblastic leukemia. Genes Dev. 1992;6:1608–1620. doi: 10.1101/gad.6.9.1608. [DOI] [PubMed] [Google Scholar]
- 138.Hitzler JK, Soares HD, Drolet DW, Inaba T, O'Connel S, Rosenfeld MG, et al. Expression patterns of the hepatic leukemia factor gene in the nervous system of developing and adult mice. Brain Res. 1999;820:1–11. doi: 10.1016/S0006-8993(98)00999-8. [DOI] [PubMed] [Google Scholar]
- 139.Cobaleda C, Perez-Caro M, Vicente-Duenas C, Sanchez-Garcia I. Function of the zinc-finger transcription factor SNAI2 in cancer and development. Annu Rev Genet. 2007;41:41–61. doi: 10.1146/annurev.genet.41.110306.130146. [DOI] [PubMed] [Google Scholar]
- 140.Inukai T, Inoue A, Kurosawa H, Goi K, Shinjyo T, Ozawa K, et al. SLUG, a ces-1-related zinc finger transcription factor gene with antiapoptotic activity, is a downstream target of the E2A-HLF oncoprotein. Mol Cell. 1999;4:343–352. doi: 10.1016/S1097-2765(00)80336-6. [DOI] [PubMed] [Google Scholar]
- 141.Mancini M, Scappaticci D, Cimino G, Nanni M, Derme V, Elia L, et al. A comprehensive genetic classification of adult acute lymphoblastic leukemia (ALL): analysis of the GIMEMA 0496 protocol. Blood. 2005;105:3434–3441. doi: 10.1182/blood-2004-07-2922. [DOI] [PubMed] [Google Scholar]
- 142.Clark SS, McLaughlin J, Crist WM, Champlin R, Witte ON. Unique forms of the abl tyrosine kinase distinguish Ph1-positive CML from Ph1-positive ALL. Science. 1987;235:85–88. doi: 10.1126/science.3541203. [DOI] [PubMed] [Google Scholar]
- 143.Schwartzberg PL, Stall AM, Hardin JD, Bowdish KS, Humaran T, Boast S, et al. Mice homozygous for the ablm1 mutation show poor viability and depletion of selected B and T cell populations. Cell. 1991;65:1165–1175. doi: 10.1016/0092-8674(91)90012-N. [DOI] [PubMed] [Google Scholar]
- 144.Tybulewicz VL, Crawford CE, Jackson PK, Bronson RT, Mulligan RC. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-M. [DOI] [PubMed] [Google Scholar]
- 145.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5:172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 146.Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 147.Martinelli G, Iacobucci I, Storlazzi CT, Vignetti M, Paoloni F, Cilloni D, et al. IKZF1 (Ikaros) deletions in BCR-ABL1-positive acute lymphoblastic leukemia are associated with short disease-free survival and high rate of cumulative incidence of relapse: a GIMEMA AL WP report. J Clin Oncol. 2009;27:5202–5207. doi: 10.1200/JCO.2008.21.6408. [DOI] [PubMed] [Google Scholar]
- 148.Virely C, Moulin S, Cobaleda C, Lasgi C, Alberdi A, Soulier J, et al. Haploinsufficiency of the IKZF1 (IKAROS) tumor suppressor gene cooperates with BCR-ABL in a transgenic model of acute lymphoblastic leukemia. Leukemia. 2010;24:1200–1204. doi: 10.1038/leu.2010.63. [DOI] [PubMed] [Google Scholar]
- 149.Trageser D, Iacobucci I, Nahar R, Duy C, von Levetzow G, Klemm L, et al. Pre-B cell receptor-mediated cell cycle arrest in Philadelphia chromosome-positive acute lymphoblastic leukemia requires IKAROS function. J Exp Med. 2009;206:1739–1753. doi: 10.1084/jem.20090004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Iacobucci I, Lonetti A, Paoloni F, Papayannidis C, Ferrari A, Storlazzi CT, et al. The PAX5 gene is frequently rearranged in BCR-ABL1-positive acute lymphoblastic leukemia but is not associated with outcome. A report on behalf of the GIMEMA Acute Leukemia Working Party. Haematologica. 2010;95:1683–1690. doi: 10.3324/haematol.2009.020792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 152.Bousquet M, Broccardo C, Quelen C, Meggetto F, Kuhlein E, Delsol G, et al. A novel PAX5-ELN fusion protein identified in B-cell acute lymphoblastic leukemia acts as a dominant negative on wild-type PAX5. Blood. 2007;109:3417–3423. doi: 10.1182/blood-2006-05-025221. [DOI] [PubMed] [Google Scholar]
- 153.Cazzaniga G, Daniotti M, Tosi S, Giudici G, Aloisi A, Pogliani E, et al. The paired box domain gene PAX5 is fused to ETV6/TEL in an acute lymphoblastic leukemia case. Cancer Res. 2001;61:4666–4670. [PubMed] [Google Scholar]
- 154.Nebral K, Denk D, Attarbaschi A, Konig M, Mann G, Haas OA, et al. Incidence and diversity of PAX5 fusion genes in childhood acute lymphoblastic leukemia. Leukemia. 2009;23:134–143. doi: 10.1038/leu.2008.306. [DOI] [PubMed] [Google Scholar]
- 155.Strehl S, Konig M, Dworzak MN, Kalwak K, Haas OA. PAX5/ETV6 fusion defines cytogenetic entity dic(9;12)(p13;p13) Leukemia. 2003;17:1121–1123. doi: 10.1038/sj.leu.2402923. [DOI] [PubMed] [Google Scholar]
- 156.Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 157.Hentges KE, Weiser KC, Schountz T, Woodward LS, Morse HC, Justice MJ. Evi3, a zinc-finger protein related to EBFAZ, regulates EBF activity in B-cell leukemia. Oncogene. 2005;24:1220–1230. doi: 10.1038/sj.onc.1208243. [DOI] [PubMed] [Google Scholar]
- 158.Mega T, Lupia M, Amodio N, Horton SJ, Mesuraca M, Pelaggi D, et al. Zinc finger protein 521 antagonizes early B-cell factor 1 and modulates the B-lymphoid differentiation of primary hematopoietic progenitors. Cell Cycle. 2011;10:2129–2139. doi: 10.4161/cc.10.13.16045. [DOI] [PubMed] [Google Scholar]
- 159.Warming S, Suzuki T, Yamaguchi TP, Jenkins NA, Copeland NG. Early B-cell factor-associated zinc-finger gene is a frequent target of retroviral integration in murine B-cell lymphomas. Oncogene. 2004;23:2727–2731. doi: 10.1038/sj.onc.1207452. [DOI] [PubMed] [Google Scholar]
- 160.Yamasaki N, Miyazaki K, Nagamachi A, Koller R, Oda H, Miyazaki M, et al. Identification of Zfp521/ZNF521 as a cooperative gene for E2A-HLF to develop acute B-lineage leukemia. Oncogene. 2010;29:1963–1975. doi: 10.1038/onc.2009.475. [DOI] [PubMed] [Google Scholar]
- 161.Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8:945–954. [PubMed] [Google Scholar]
- 162.Hoelbl A, Kovacic B, Kerenyi MA, Simma O, Warsch W, Cui Y, et al. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood. 2006;107:4898–4906. doi: 10.1182/blood-2005-09-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Heltemes-Harris LM, Willette MJ, Ramsey LB, Qiu YH, Neeley ES, Zhang N, et al. Ebf1 or Pax5 haploinsufficiency synergizes with STAT5 activation to initiate acute lymphoblastic leukemia. J Exp Med. 2011;208:1135–1149. doi: 10.1084/jem.20101947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hosking FJ, Papaemmanuil E, Sheridan E, Kinsey SE, Lightfoot T, Roman E, et al. Genome-wide homozygosity signatures and childhood acute lymphoblastic leukemia risk. Blood. 2010;115:4472–4477. doi: 10.1182/blood-2009-09-244483. [DOI] [PubMed] [Google Scholar]
- 165.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Prasad RB, Hosking FJ, Vijayakrishnan J, Papaemmanuil E, Koehler R, Greaves M, et al. Verification of the susceptibility loci on 7p12.2, 10q21.2 and 14q11.2 in precursor B-cell acute lymphoblastic leukemia of childhood. Blood. 2010;115:1765–1767. doi: 10.1182/blood-2009-09-241513. [DOI] [PubMed] [Google Scholar]
- 167.Sherborne AL, Hosking FJ, Prasad RB, Kumar R, Koehler R, Vijayakrishnan J, et al. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat Genet. 2010;42:492–494. doi: 10.1038/ng.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Treviño LR, Yang W, French D, Hunger SP, Carroll WL, Devidas M, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1001–1005. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Henry CJ, Marusyk A, DeGregory J. Aging-associated changes in hematopoiesis and leukemogenesis: What's the connection? Aging (Albany NY) 2011;3:643–656. doi: 10.18632/aging.100351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 171.Nakada D, Levi BP, Morrison SJ. Integrating physiological regulation with stem cell and tissue homeostasis. Neuron. 2011;70:703–718. doi: 10.1016/j.neuron.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Dorshkind K, Montecino-Rodriguez E, Signer RA. The ageing immune system: is it ever too old to become young again? Nat Rev Immunol. 2009;9:57–62. doi: 10.1038/nri2471. [DOI] [PubMed] [Google Scholar]
- 173.Linton PJ, Dorshkind K. Age-related changes in lymphocyte development and function. Nat Immunol. 2004;5:133–139. doi: 10.1038/ni1033. [DOI] [PubMed] [Google Scholar]
- 174.Mehr R, Melamed D. Reversing B cell aging. Aging (Albany NY) 2011;3:438–443. doi: 10.18632/aging.100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Miller JP, Allman D. The decline in B lymphopoiesis in aged mice reflects loss of very early B-lineage precursors. J Immunol. 2003;171:2326–2330. doi: 10.4049/jimmunol.171.5.2326. [DOI] [PubMed] [Google Scholar]
- 176.Mori H, Colman SM, Xiao Z, Ford AM, Healy LE, Donaldson C, et al. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc Natl Acad Sci USA. 2002;99:8242–8247. doi: 10.1073/pnas.112218799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zuna J, Madzo J, Krejci O, Zemanova Z, Kalinova M, Muzikova K, et al. ETV6/RUNX1 (TEL/AML1) is a frequent prenatal first hit in childhood leukemia. Blood. 2011;117:368–369. doi: 10.1182/blood-2010-09-309070. [DOI] [PubMed] [Google Scholar]
- 178.Lausten-Thomsen U, Madsen HO, Vestergaard TR, Hjalgrim H, Nersting J, Schmiegelow K. Prevalence of t(12;21)[ETV6-RUNX1]-positive cells in healthy neonates. Blood. 2011;117:186–189. doi: 10.1182/blood-2010-05-282764. [DOI] [PubMed] [Google Scholar]
- 179.Vicente-Dueñas C, Abollo-Jimenez F, Ruiz-Roca L, Alonso-Escudero E, Jimenez R, Cenador MB, et al. The age of the target cell affects B-cell leukaemia malignancy. Aging (Albany NY) 2010;2:908–913. doi: 10.18632/aging.100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Henry CJ, Marusyk A, Zaberezhnyy V, Adane B, DeGregori J. Declining lymphoid progenitor fitness promotes aging-associated leukemogenesis. Proc Natl Acad Sci USA. 2011;107:21713–21718. doi: 10.1073/pnas.1005486107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cobaleda C, Sanchez-Garcia I. Stem cell aging and cancer: Immortal but vulnerable. Cell Cycle. 2011;10 doi: 10.4161/cc.10.17.16748. In press. [DOI] [PubMed] [Google Scholar]
- 182.Greaves M. Leukemogenesis and ageing: 'fit for transformation'? Aging (Albany NY) 2011;3:79–80. doi: 10.18632/aging.100274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Steliarova-Foucher E, Stiller C, Kaatsch P, Berrino F, Coebergh JW, Lacour B, et al. Geographical patterns and time trends of cancer incidence and survival among children and adolescents in Europe since the 1970s (the ACCISproject): an epidemiological study. Lancet. 2004;364:2097–2105. doi: 10.1016/S0140-6736(04)17550-8. [DOI] [PubMed] [Google Scholar]
- 184.Schüz J, Ahlbom A. Exposure to electromagnetic fields and the risk of childhood leukaemia: a review. Radiat Prot Dosimetry. 2008;132:202–211. doi: 10.1093/rpd/ncn270. [DOI] [PubMed] [Google Scholar]
- 185.Henderson MJ, Choi S, Beesley AH, Sutton R, Venn NC, Marshall GM, et al. Mechanism of relapse in pediatric acute lymphoblastic leukemia. Cell Cycle. 2008;7:1315–1320. doi: 10.4161/cc.7.10.5885. [DOI] [PubMed] [Google Scholar]