Abstract
Background
Angiogenesis is regarded as a hallmark in cancer development, and anti-angiogenic treatment is presently used in non-small cell lung cancer (NSCLC) patients. MicroRNAs (miRs) are small non-coding, endogenous, single stranded RNAs that regulate gene expression. In this study we aimed to identify significantly altered miRs related to angiogenesis in NSCLC.
Methods
From a large cohort of 335 NSCLC patients, paraffin-embedded samples from 10 patients with a short disease specific survival (DSS), 10 with a long DSS and 10 normal controls were analyzed. The miRs were quantified by microarray hybridization and selected miRs were validated by real-time qPCR. The impacts of different pathways, including angiogenesis, were evaluated by Gene Set Enrichment Analysis (GSEA) derived from Protein ANalysis THrough Evolutionary Relationship (PANTHER). One of the most interesting candidate markers, miR-155, was validated by in situ hybridization (ISH) in the total cohort (n = 335) and correlation analyses with several well-known angiogenic markers were done.
Results
128 miRs were significantly up- or down-regulated; normal versus long DSS (n = 68) and/or normal versus short DSS (n = 63) and/or long versus short DSS (n = 37). The pathway analysis indicates angiogenesis-related miRs to be involved in NSCLC. There were strong significant correlations between the array hybridization and qPCR validation data. The significantly altered angiogenesis-related miRs of high interest were miR-21, miR-106a, miR-126, miR-155, miR-182, miR-210 and miR-424. miR-155 correlated significantly with fibroblast growth factor 2 (FGF2) in the total cohort (r = 0.17, P = 0.002), though most prominent in the subgroup with nodal metastasis (r = 0.34, P<0.001).
Conclusions
Several angiogenesis-related miRs are significantly altered in NSCLC. Further studies to understand their biological functions and explore their clinical relevance are warranted.
Introduction
Lung cancer is the number one cause of cancer related mortality in both men and women, and approximately 80% are non-small cell lung cancer (NSCLC) [1]. Despite emerging improvements in early diagnosis and treatment modalities, the overall 5-year survival is at meager 15%. TNM (Tumor, Nodes, Metastasis) stage has been regarded the most important prognostic variable as early stage disease is a prerequisite for complete surgical resection necessary for potential cure. Treatment responses and side effects from new treatment options have been closely related to different histological entities of NSCLC [2]–[4]. However, targeted therapies directed against specific cellular alterations require a precise sub-classification of NSCLC which is largely beyond today's staging and routine diagnostic histopathological techniques [5].
MicroRNAs (miRs) are small non-coding, endogenous, single stranded RNAs that regulate gene expression [6]–[9]. By regulating the gene expression at a posttranscriptional level, miRs have a large impact on a wide variety of pathways, including different pathways related to cancer development. Several studies have explored the deregulation of different miRs in NSCLC and their potential oncogenic and tumor suppressor functions as well as their prognostic impact [10]–[20].
Some miRs appear to be involved in angiogenesis [21]–[24]. Angiogenesis is a process of new blood vessel formation from pre-existing ones, and plays a key role in tumor development [25]. Angiogenic inhibitors are used in NSCLC treatment. The monoclonal antibody bevacizumab in combination with chemotherapy is FDA approved as a treatment option in metastatic non-squamous NSCLC patients, and many tyrosine kinase inhibitors targeting angiogenic pathways are promising [3], [26].
The first evidence showing miRs in the regulation of angiogenesis came from Dicer knockout mice. Dicer is a critical enzyme in miR synthesis. The Dicer-deficit mice died early during development due to the thinning of vascular walls and severe disorganization of the network of blood vessels [27]. Besides, another murine study showed that a subset of miRs altered during the angiogenic switch became oppositely regulated in response to anti-angiogenic treatment [23].
In a large unselected NSCLC cohort we have previously studied the prognostic impact of several key angiogenic growth factors and receptors as well as the prognostic role of miR-155 and miR-126 by in situ hybridization [28]–[38]. Herein, we evaluated the miR expression profiles in NSCLC by using tissue from selected patients from this cohort. We aimed to identify interesting angiogenesis-related miR candidates for further potential large-scale and in-depth studies.
Results
Patients
Demographic, clinical, and histopathological variables in the patient groups (long DSS, short DSS and controls) are shown in Table 1. The median DSS were 8.0 (range 6.5–9.9) months and 160.5 (range 60.5–184.3) months in the low and high DSS group, respectively. There were four adenocarcinomas and six squamous cell carcinomas in each group.
Table 1. Characteristics of 20 non-small cell lung cancer (NSCLC) patients and ten normal controls; NSCLC_01-10 with a short disease-specific survival (DSS), NSCLC_11-20 with a long DSS and 10 normal controls (Norm_01-10).
| Patient ID | Histology | DSS (months) | Status | Stage | T | N | M | Differentiation | Age | Sex |
| NSCLC_01 | AC | 6.5 | LC Dead | IIa | 2a | 1 | 0 | Moderate | 53 | M |
| NSCLC_02 | AC | 7.0 | LC Dead | IIa | 2a | 1 | 0 | Poor | 73 | M |
| NSCLC_03 | SCC | 7.3 | LC Dead | IIIa | 1b | 2 | 0 | Poor | 62 | M |
| NSCLC_04 | AC | 7.4 | LC Dead | IIa | 1b | 1 | 0 | Moderate | 74 | M |
| NSCLC_05 | AC | 7.6 | LC Dead | IIb | 3 | 0 | 0 | Poor | 54 | M |
| NSCLC_06 | SCC | 8.4 | LC Dead | IIb | 3 | 0 | 0 | Well | 53 | M |
| NSCLC_07 | SCC | 8.4 | LC Dead | IIb | 3 | 0 | 0 | Poor | 74 | M |
| NSCLC_08 | SCC | 9.3 | LC Dead | IIa | 2a | 1 | 0 | Poor | 74 | M |
| NSCLC_09 | SCC | 9.6 | LC Dead | IIb | 3 | 0 | 0 | Poor | 68 | F |
| NSCLC_10 | SCC | 9.9 | LC Dead | IIb | 3 | 0 | 0 | Poor | 70 | F |
| NSCLC_11 | SCC | 147.8 | Alive | IIa | 2a | 1 | 0 | Moderate | 53 | F |
| NSCLC_12 | AC | 74.1 | Alive | IIa | 2b | 0 | 0 | Moderate | 75 | M |
| NSCLC_13 | AC | 60.5 | Alive | Ia | 1b | 0 | 0 | Moderate | 75 | F |
| NSCLC_14 | SCC | 147.8 | Alive | IIa | 2b | 0 | 0 | Moderate | 60 | M |
| NSCLC_15 | SCC | 163.8 | Alive | IIIa | 3 | 1 | 0 | Poor | 68 | M |
| NSCLC_16 | AC | 170.5 | Alive | IIa | 2b | 0 | 0 | Moderate | 47 | M |
| NSCLC_17 | SCC | 158.7 | Non-LC Dead | Ib | 2a | 0 | 0 | Moderate | 69 | M |
| NSCLC_18 | AC | 177.4 | Alive | IIb | 3 | 0 | 0 | Moderate | 64 | M |
| NSCLC_19 | SCC | 184.3 | Non-LC Dead | Ia | 1b | 0 | 0 | Moderate | 61 | M |
| NSCLC_20 | SCC | 162.3 | Alive | Ia | 1b | 0 | 0 | Moderate | 53 | M |
| *NORM_01 | AC | 6.5 | LC Dead | IIa | 2a | 1 | 0 | Moderate | 53 | M |
| *NORM_02 | AC | 7.0 | LC Dead | IIa | 2a | 1 | 0 | Poor | 73 | M |
| *NORM_03 | SCC | 8.4 | LC Dead | IIb | 3 | 0 | 0 | Well | 53 | M |
| *NORM_04 | SCC | 147.8 | Alive | IIa | 2a | 1 | 0 | Moderate | 53 | F |
| *NORM_05 | AC | 74.1 | Alive | IIa | 2b | 0 | 0 | Moderate | 75 | M |
| *NORM_06 | AC | 60.5 | Alive | Ia | 1b | 0 | 0 | Moderate | 75 | F |
| *NORM_07 | SCC | 163.8 | Alive | IIIa | 3 | 1 | 0 | Poor | 68 | M |
| *NORM_08 | AC | 170.5 | Alive | IIa | 2b | 0 | 0 | Moderate | 47 | M |
| *NORM_09 | AC | 177.4 | Alive | IIb | 3 | 0 | 0 | Moderate | 64 | M |
| *NORM_10 | SCC | 184.3 | Non-LC Dead | Ia | 1b | 0 | 0 | Moderate | 61 | M |
Abbreviations: SCC, squamous cell carcinoma; AC, adenocarcinoma; DSS; LC, lung cancer related; T, tumor; N, nodal status; M, metastasis.
*Samples from normal lung tissue sites in ten NSCLC patients (NSCLC_01, NSCLC_02, NSCLC_06, NSCLC_11, NSCLC_12, NSCLC_13, NSCLC_15, NSCLC_16, NSCLC_18 and NSCLC_19) were included in the study (NORM_01 - NORM_10).
Micro RNA expression analysis
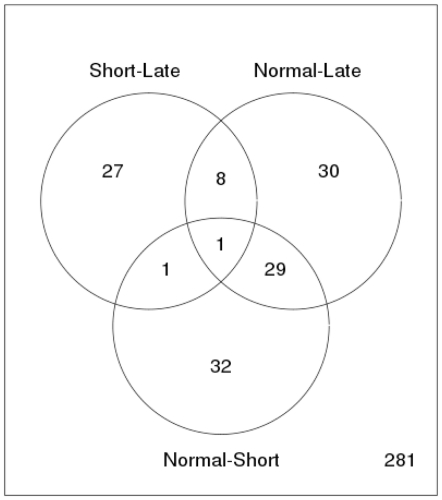
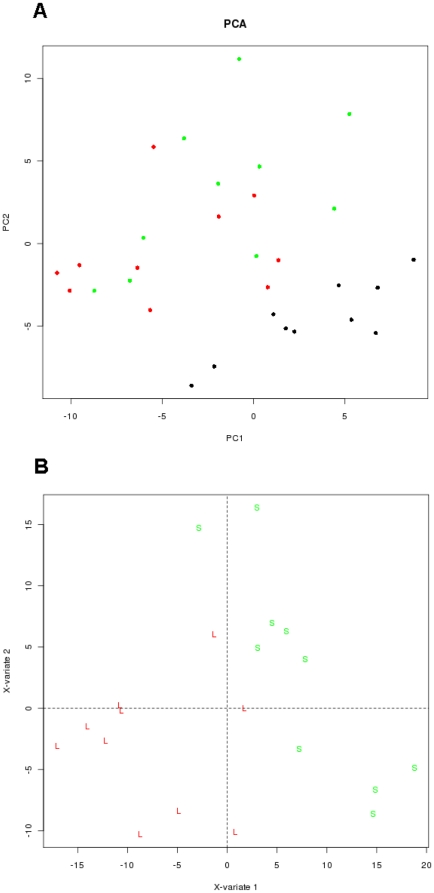
Out of 281 miRs, Figure 1 shows the number and distribution of 128 differentially expressed miRs significant across different comparisons in the study (P<0.1 adjusted for false discovery rate, FDR). The respective -up and down-regulated (−1,1) miRs are presented in Table S1. miR-182 was the only miR altered in all three combinations; normal versus short DSS, normal versus long DSS and short versus long DSS. A Principal component analysis (PCA) (Figure 2A) on the entire sample set of expression values revealed that the main principle component of variation separates normal samples from NSCLC samples. To highlight the difference between the groups of NSCLC tumor samples a bridge partial least squares (PLS) model was used to extract a subspace where the group differences are more obvious than in PCA (Figure 2B). Table 2 shows the ten miRs with highest fold change stratified into the groups long survival versus normal control and short survival versus normal control. Table 3 shows the miRs with highest fold change in short versus long survival groups.
Figure 1. Comparison of survival groups of NSCLC patients.
The Venn diagram shows the number of all differentially expressed miRNAs across different comparisons: Tissue from NSCLC patients with short survival versus tissue from NSCLC patients with long survival, tissue from normal lung versus tissue from NSCLC patients with long survival and tissue from normal lung versus tissue from NSCLC patients with short survival. Out of 281 miRs evaluated, the number of differential expressed miRs with FDR adjusted P<0.1 (total n = 128) of each comparison is indicated.
Figure 2. Principal component analysis (PCA) and partial least square analysis (PLS) on different NSCLC patient samples.
(A) Two-dimensional PCA of miRs, derived from 20 patients with NSCLC and 10 tissue samples from normal lung tissue, showing separation of the two sample groups. (B) The plot depicts components 1 and 2 of PLS model which used survival time as a scoring criteria. The analysis clearly separates the tissue sample groups for short and long survival NSCLC patients. All samples are colour coded according to group: Black: Normal patient samples; green: Samples from patients with short survival; red: Samples from patients with long survival.
Table 2. Ten each of the differentially expressed miRs that are up- or down-regulated the most ranged by fold change (long survival versus normal control and short survival versus normal control).
| Long survival versus normal | Short survival versus normal | |||||
| miR | Fold change | P-adjusted* | miR | Fold change | P-adjusted* | |
| Up- | miR-205 | 8.75 | 0.0273 | miR-1308 | 3.92 | 0.0117 |
| regulated | miR-21 | 3.68 | 0.0090 | miR-21 | 3.63 | 0.0094 |
| miR-1308 | 2.89 | 0.0532 | miR-182 | 3.32 | 0.0008 | |
| miR-93 | 2.11 | 0.0443 | miR-31 | 2.87 | 0.0362 | |
| miR-1274a | 1.99 | 0.0675 | miR-205 | 2.75 | 0.2917 | |
| miR-182 | 1.80 | 0.0939 | miR-193b | 2.31 | 0.0062 | |
| miR-708 | 1.75 | 0.0039 | miR-1259 | 2.28 | 0.0008 | |
| miR-210 | 1.74 | 0.0070 | miR-93 | 2.23 | 0.0337 | |
| miR-1259 | 1.64 | 0.0427 | miR-106a | 2.23 | 0.0933 | |
| miR-106b | 1.62 | 0.1644 | miR-183 | 2.20 | 0.0304 | |
| Down-regulated rrregulatedregulated | miR-451 | 5.74 | 0.0427 | miR-451 | 5.85 | 0.0397 |
| regulated | miR-126 | 4.53 | 0.0005 | miR-126 | 4.09 | 0.0008 |
| miR-30a | 3.14 | 0.00006 | miR-30a | 2.81 | 0.0003 | |
| miR-30b | 3.07 | 0.0124 | miR-140-3p | 2.68 | 0.0035 | |
| miR-30c | 2.45 | 0.0035 | miR-143 | 2.33 | 0.0787 | |
| miR-140-3p | 2.45 | 0.0070 | miR-126* | 2.28 | 0.0035 | |
| miR-126* | 2.41 | 0.0016 | miR-145 | 2.22 | 0.0070 | |
| miR-145 | 2.28 | 0.0061 | miR-30b | 2.07 | 0.0958 | |
| let-7a | 2.25 | 0.0480 | miR-29c | 2.01 | 0.0934 | |
| miR-125a-5p | 2.19 | 0.00001 | miR-30d | 1.89 | 0.0200 | |
*P-adjusted; corrected for false discovery rate (FDR).
Table 3. Ten each of the differentially expressed miRs that are up- or down-regulated the most ranged by fold change (short survival versus long survival).
| Short survival versus long survival | |||
| miR | Fold change | P-adjusted* | |
| Up- | miR-31 | 2.00 | 0.161 |
| regulated | miR-182 | 1.85 | 0.084 |
| miR-106a | 1.73 | 0.245 | |
| miR-183 | 1.72 | 0.094 | |
| let-7a | 1.68 | 0.194 | |
| miR-151-5p | 1.64 | 0.093 | |
| miR-138-1* | 1.61 | 0.073 | |
| miR-98 | 1.56 | 0.089 | |
| miR-424 | 1.53 | 0.044 | |
| miR-193b | 1.50 | 0.178 | |
| Down- | miR-205 | 3.16 | 0.232 |
| regulated | miR-142-3p | 1.96 | 0.084 |
| miR-557 | 1.65 | 0.084 | |
| miR-720 | 1.52 | 0.078 | |
| miR-378 | 1.44 | 0.070 | |
| miR-708 | 1.41 | 0.075 | |
| miR-29c | 1.39 | 0.076 | |
| miR-552 | 1.35 | 0.084 | |
| miR-27a* | 1.32 | 0.035 | |
| miR-155 | 1.31 | 0.095 | |
*P-adjusted; corrected for false discovery rate (FDR).
Pathway analysis
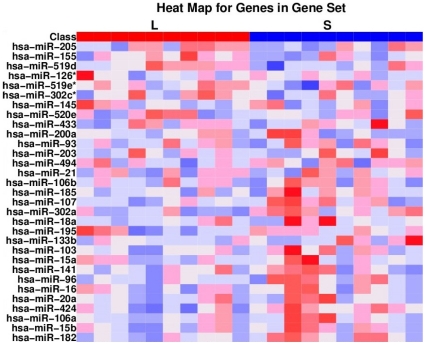
Table 4 shows results from the GSEA. The gene set connected to 31 angiogenesis-related miRs revealed the highest gene set nominal enrichment score (NES = −1.17, nominal p-value 0.28, FDR 0). Figure 3 shows the heat map representing 31miRs connected to the angiogenic gene set.
Table 4. Impacts of the different pathways evaluated by Gene Set Enrichment Analysis (GSEA) derived from Protein Analysis THrough Evolutionary Relationship (PANTHER).
| GS | Size | Pathway | NES | NOM p-value | FDR |
| P00005 | 31 | Angiogenesis | −1.17 | 0.28 | 0 |
| P00027 | 17 | Heterotrimeric G-protein signaling pathway-Gq alpha and Go alpha mediated pathway | −1.16 | 0.28 | 0 |
| P00004 | 42 | Alzheimer disease-presenilin pathway | −1.12 | 0.33 | 0.35 |
| P00047 | 37 | PDGF signaling pathway | −1.12 | 0.32 | 0.26 |
| P00029 | 37 | Huntington disease | −1.08 | 0.41 | 0.35 |
| P00059 | 21 | p53 pathway | −1.06 | 0.43 | 0.31 |
| P00006 | 28 | Apoptosis signaling pathway | −1.06 | 0.50 | 0.28 |
| P00048 | 18 | PI3 kinase pathway | −1.04 | 0.46 | 0.27 |
| P00031 | 45 | Inflammation mediated by chemokine and cytokine signaling pathway | −1.03 | 0.49 | 0.26 |
| P00019 | 19 | Endothelin signaling pathway | −1.00 | 0.50 | 0.33 |
| P00036 | 35 | Interleukin signaling pathway | −0.98 | 0.51 | 0.37 |
| P00057 | 62 | Wnt signaling pathway | −0.98 | 0.54 | 0.34 |
| P00052 | 44 | TGF-beta signaling pathway | −0.89 | 0.66 | 0.56 |
| P00034 | 43 | Integrin signalling pathway | −0.89 | 0.68 | 0.53 |
| P00012 | 40 | Cadherin signaling pathway | −0.87 | 0.66 | 0.55 |
| P00046 | 22 | Oxidative stress response | −0.86 | 0.67 | 0.55 |
| P00026 | 33 | Heterotrimeric G-protein signaling pathway-Gi alpha and Gs alpha mediated pathway | −0.85 | 0.68 | 0.57 |
| P04398 | 28 | p53 pathway feedback loops 2 | −0.83 | 0.74 | 0.58 |
| P00016 | 23 | Cytoskeletal regulation by Rho GTPase | −0.81 | 0.71 | 0.59 |
| P00060 | 44 | Ubiquitin proteasome pathway | −0.73 | 0.81 | 0.81 |
| P00007 | 16 | Axon guidance mediated by semaphorins | −0.67 | 0.87 | 0.90 |
GS, gene set; Size, numbers of miRs included; NES, nominal enrichment score; NOM p-value, nominal p-value; FDR, false discovery rate.
Figure 3. Heat map showing expression of 31 microRNAs (miRs) included in angiogenesis pathway gene set.
The difference in miR expression between tumor samples from NSCLC patients with long (L) and short (S) survival is shown. miR expression values are shown in a spectrum where down-regulated is blue and up-regulated is red.
PCR validation
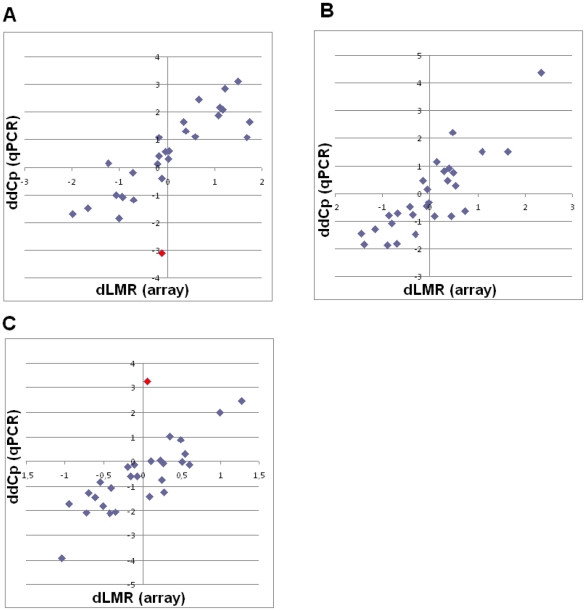
Scatter plots comparing microarray hybridization and qPCR data from 28 selected miRs are shown in Figure 4. The included miRs, and data from all three combinations (both hybridization and qPCR) are listed in Table S2. There were strong and significant correlation between the hybridization and qPCR data: Short versus normal r = 0.81, P<0.001; long versus normal r = 0.85, P<0.001; short versus long r = 0.85, P<0.001.
Figure 4. Scatter plot comparing microarray hybridization (all ten samples in each group) and qPCR data (five selected samples from each group: NSCLC_03, NSCLC_05, NSCLC_08, NSCLC_10, NSCLC_15, NSCLC_16, NSCLC_17, NSCLC_18, NSCLC_19, NSCLC_20, NORM_03, NORM_04, NORM_08, NORM_09 and NORM_10) and 28 miRs according to Table S2.
Comparison between microarray hybridization (dLMR, difference in average expression levels between sample groups, log2 scale) and qPCR (ddCP, difference in average expression levels between sample groups, log2 scale) data, correlation coefficient = r (Pearson): (A) low versus normal r = 0.81, P<0.001; (B) high versus normal r = 0.85, P<0.001; (C) high versus low r = 0.85, P<0.001. In red; miR-150.
Correlation analysis between angiogenic markers and miR-155
We have previously published on the prognostic impact of miR-155 in our large (335 patients) NSCLC population [37]. In this study we explored the correlation between miR-155 and several well-known angiogenic markers. The same cut-off values as previously published were used for FGF2 and miR-155 [33], [37]. Figure S1 shows in situ hybridization (ISH) analysis of NSCLC representing strong and weak intensities for tumor cell miR-155 expression as well as negative and positive controls. Table 5 shows the correlation between angiogenic markers [vascular endothelial growth factor - A (VEGF-A), platelet derived factor – B (PDGF-B), HIF-1α and fibroblast growth factor 2 (FGF2)] and miR-155. miR-155 correlated significantly with FGF2 in the total cohort (r = 0.17, P = 0.002), though most prominent in the subgroup with nodal metastasis (r = 0.34, P<0.001). The crosstabs in Table 6 and Table 7 show the correlation and distribution of high and low expression of FGF2 and miR-155.
Table 5. Correlation analyses between angiogenic markers and miR-155 expression in NSCLC patients.
| miR-155 | |||
| Total cohort (n = 335) | N0 (n = 232) | N+ (n = 103) | |
| VEGF-A | r = 0.09, P = 0.11 | r = 0.05, P = 0.50 | r = 0.18, P = 0.07 |
| PDGF-B | r = 0.04, P = 0.49 | r = 0.06, P = 0.37 | r = 0.01, P = 0.96 |
| HIF-1α | r = 0.02, P = 0.68 | r = 0.02, P = 0.75 | r = −0.1, P = 0.27 |
| FGF2 | r = 0.17, P = 0.002 | r = 0.06, P = 0.35 | r = 0.34, P<0.001 |
Abbreviations: VEGF-A, vascular endothelial growth factor-A; PDGF-B, platelet derived factor -B; HIF-1α, hypoxia inducible factor 1α; FGF2, fibroblast growth factor 2; N0, lymph node negative patients; N+, lymph node positive patients.
Table 6. Crosstab showing the correlation between miR-155 and FGF2 in the total cohort.
| miR-155 | ||||
| Low expression | High expression | Total | ||
| FGF2 | Low expression | 156 | 137 | 293 |
| High expression | 6 | 21 | 27 | |
| Total | 162 | 158 | 320 | |
Spearman correlation, r = 0.17, P = 0.002.
Table 7. Crosstab showing the correlation between miR-155 and FGF2 in patients with lymph node metastasis (N+).
| miR-155 | ||||
| Low expression | High expression | Total | ||
| FGF2 | Low expression | 48 | 37 | 85 |
| High expression | 1 | 13 | 14 | |
| Total | 49 | 50 | 99 | |
Spearman correlation, r = 0.34, P<0.001.
Discussion
To our knowledge, this is the first NSCLC miR expression profiling study showing a gene set connected to angiogenesis-related miRs with the highest impact in pathway analysis. Consistent with previously published studies we observe significant differences in miR profiles between normal and tumor tissue and between samples from patients with a poor versus a favorable DSS. Several of these significant altered miRs are associated with angiogenesis and are interesting candidate markers for further evaluation. By validating one of these markers, miR-155, in a cohort of 335 NSCLC patients, we find this miR for the first time to be significantly associated with the well-known angiogenic marker FGF2.
The major weaknesses of the study in the search for novel candidate markers are the relatively heterogeneous study population and the low number of patients included in the analysis. Many of the results presented herein are therefore only borderline significant (or some only showing a trend) and further validation in a larger NSCLC set, as done for miR-155, is a prerequisite for firmer conclusions. This is especially the case when comparing significant differences between tumor tissues (long versus short survival) as there, in general, are larger differences between tumor tissues and normal controls. The fold changes are in average higher and the p-values stronger in Table 2 (tumor tissue versus normal controls) compared with Table 3 (long versus short survival). From a clinical point of view, at least when it comes to miRs as potential prognostic or predictive markers, miRs with altered expression between short versus long survival group are of special interest. We have therefore explored the literature on four angiogenesis related miRs from Table 3 (miR-106a, miR-155, miR-182 and miR-424) and validated miR-155 in a large set of NSCLC patients. In addition we wanted to highlight the potential impact of miR-21, miR-126 and miR-210 from Table 2 as there are convincing reports indicating these miRs to have potential impact in angiogenesis. Besides, we have previously shown miR-126 to have prognostic impact in our NSCLC cohort [38].
Strengthening our results are strong and significant correlations between the microarray hybridization data and the same miRs validated by qPCR. Further, the altered miR expressions in tumor versus normal tissue are to a large degree consistent with previously published NSCLC miR studies [15]–[17], [20] and as discussed in the following there are constantly new literature added, supporting our candidate miRs to be involved in angiogenesis.
Few studies have explored the impact of pathways related to miR gene targets in NSCLC. The interpretation of such analyses is not straight-forward as the results are highly dependent on which target genes are linked to the miRs and which miRs are connected to the different pathways. In addition miRs are often multifunctional and the same miR may target diverse genes in different pathways, and a gene may be targeted by several miRs. Although not statistically significant, the GSEA ranged angiogenesis as the most important pathway in our NSCLC samples. There are several novel reports connecting specific miRs to angiogenesis supporting our findings [23], [24], [39]–[42]. Besides, Raponi et al. have shown that the angiogenesis-related fibroblast growth factor (FGF) pathway had a significant gene enrichment in squamous NSCLC [17].
In a comprehensive murine study, Olson et al. identified specific miR expression signatures associated with steps in tumorigenesis and different hallmarks of cancer, including angiogenesis [23]. Angiogenesis–inhibiting signature miRs were evaluated by qPCR technique and defined as those miRs significantly altered in normal versus sunitinib-treated primary tumors by >1.3 fold change [23]. Sunitinib is a tyrosine kinase inhibitor targeting vascular endothelial growth factor receptor (VEGFR), platelet-derived growth factor (PDGFR) and c-Kit. In accordance with our qPCR validation results, PCR data in general often show an increased fold change compared with hybridization data. Many of the angiogenesis–related miRs observed by Olson and coworkers are also significantly altered in our material [23]. Among the most interesting angiogenesis-related miRs significantly altered in both studies we find miR-126, miR-155, and miR-21 and miR-424.
miR-126 has recently been reviewed as an important player in angiogenesis [43] and Wang et al. showed in a murine model that it enhances the proangiogenic actions of VEGF and FGF by repressing the expression of Spred-1, an intracellular inhibitor of angiogenic signaling [44]. miR-126 has also been located within the epidermal growth factor-like domain 7 (EGFL7) gene. EGFL7 may have a major role in angiogenesis by promoting VEGF signaling and vascular integrity [45]. Moreover, we have previously reported that miR-126 expression is significantly associated with VEGF-A in NSCLC [38]. Further, we observed that the prognostic impact of miR-126 is related to histological subtypes and nodal status. In addition the expression and roles of miR-126 may be different in various malignancies [23].
miR-155 is one of the miRs most consistently involved in neoplastic disease [46], but to our knowledge, not been related to angiogenesis prior to the study by Olson and coworkers [23]. It has been reported to be up-regulated in several human NSCLC studies [15], [17], [20], and down-regulated in a murine model when using the angiogenic inhibitor sunitinib [23]. In our array data, miR-155 tended to be significantly altered between the short and long survival groups (Table 3) and was significantly over-expressed in tumor versus normal tissue in the qPCR validation set. We have reported the prognostic impact of miR-155 to be associated with histological subtypes and nodal status but did not link these results to angiogenic markers [37]. Based on these new data we now examined whether miR-155 correlated to well-known angiogenic/hypoxic parameters as VEGF-A, PDGF-B, HIF-1α and FGF2. Interestingly, we observed, for the first time, miR-155 to be significantly associated with FGF2 with the highest impact in the N+ NSCLC subgroup. Though FGF2 has several functions, it is an important player in angiogenesis. In fact, miR-155 also tended to correlate with VEGF-A in the N+ subgroup. miR-155 was one of the first miRs to be associated with NSCLC development and further studies will be important in order to explore its potential angiogenic function.
In this study, miR-21 is significantly up-regulated in tumor versus normal tissue (Table 2), supporting previous NSCLC reports [15], [17], [20] and the observed down-regulation after anti-angiogenic treatment [23]. miR-21 has also been found to be highly expressed in endothelial cells [24]. In a recent study by Liu et al., miR-21 was overexpressed by transfecting pre-miR-21 into human prostate cancer cells and tumor angiogenesis was assayed using chicken chorioallantoic membrane [47]. They found that overexpression of miR-21 in DU145 cells increased the expression of HIF-1α and VEGF, and induced tumor angiogenesis. They conclude that miR-21 induces tumor angiogenesis through targeting PTEN, leading to activation of the AKT and ERK1/2 signaling pathways, and thereby enhancing HIF-1α and VEGF expression. The same pathways are described as important in NSCLC development as well [48], and further studies to address this relationship with miR-21 in lung cancer would be of interest.
The Olson study indicates miR-424 to be involved in angiogenesis as this miR was down-regulated after sunitinib treatment [23]. In addition, Ghosh et al. have studied this miR and described that hypoxia-induced miR-424 expression in human endothelial cells regulates HIF-α isoforms and promotes angiogenesis [49]. They further suggest miR-424 to play an important physiological role in post-ischemic angiogenesis. In contrast, Nakshima et al. reported down-regulation of miR-424 to contribute to the abnormal angiogenesis via MEK1 and cyclin E1 in senile hemangioma, demonstrating the potential tissue and stage specific impact of the same miR [50]. In our study, miR-424 was not altered when comparing tumor with normal tissue, but was significantly up-regulated in tissue from patients with poor prognosis compared to tissue from patients with a favorable prognosis (Table 3). There is to our knowledge no published study which has explored the prognostic impact of miR-424 in cancer. However, as there is a significant miR-424 up-regulation in the poor prognostic group it would be of interest to further evaluate this marker in a large scale NSCLC study.
Another significant up-regulated marker in our material is miR-210 (Table 2), which previously has been related to angiogenesis [41], [51], [52]. miR-210 up-regulation is believed to be a crucial element of endothelial cell response to hypoxia and therefore potentially important in tumor angiogenesis. This miR has recently been reviewed by Devlin et al. [51], concluding that miR-210 is a robust target of hypoxia-inducible factor and that its overexpression has been detected in a variety of cardiovascular diseases and solid tumors. In NSCLC cell lines, miR-210 is reported to mediate mitochondrial alterations associated with modulation of HIF-1 activity [52]. However, much of the functional and prognostic impact of miR-210 in NSCLC still remains unresolved.
miR-182 is included in the angiogenesis pathway in the GSEA, partly because fibroblast growth factor receptor substrate 2 (FRS2) is one of its target genes. FRS2 is a major regulator of the fibroblast growth factor pathway that we and others have shown to be important in NSCLC progression, potentially by stimulating angiogenesis [33], [53]. Interestingly, miR-182 was the only miR significantly altered in all three combinations as shown in the Venn diagram (Figure 1). Its up-regulation in tumor (both poor and favorable prognostic group, Table 2) when compared to normal tissue, is consistent with the previous study by Raponi et al. [17]. Moreover, miR-182 is significantly up-regulated in short DSS versus long DSS patients (Table 3). However, Barchack et al. observed higher miR-182 expression in primary tumors than in the metastatic lung tumors, indicating that miR-182 expression may reach a peak in early stage NSCLC.
We observe the same trend for miR-106a, which has been shown to be up-regulated during hypoxia in breast and colon cancer cell lines [40]. As previously reported, miR-106a was up-regulated in NSCLC [16], [17], [20]. We also observe its expression to be significantly augmented in tumors versus normal controls (Table 2). As for miR-182, miR-106a was further increased in tissue from patients with a short survival rather than long survival (Table 3).
As expected, many miRs known to be associated with angiogenesis were not significantly altered in our study. Cluster miR-17-92 (miR-17-5p, miR-17-3p, miR-18a, miR-19a, miR-19b, miR-20a and miR92-1), let-7, let-7f, miR-27b, miR-15b, miR-16, miR-92a, miR-99a, miR-130a, miR-214, miR-221, miR-222 and miR-296 as well as some of the miRs presented in the heat map (Figure 3), were not significantly altered or they exhibited only minor fold changes [24], [39], [41], [42]. This lack of significant alterations may be due to false negative results as there are few patients in each group. Furthermore, as many miRs appear to be tissue and stage specific, some of these miRs may alternatively exert an impact limited to subgroups of resected NSCLC or may be important in other malignancies.
Angiogenic inhibitors in combination with chemotherapy are established as NSCLC treatment, but a further knowledge on biology, effectors mechanisms and prognostic and predictive markers are warranted. miRs are stable and potentially measurable in both tissue and serum. Several miRs are linked to angiogenesis and this study supports the assumption that a number of angiogenesis-related miRs are potentially important in NSCLC progression. We propose miR-21, miR-106a, miR-126, miR-155, miR-182, miR-210 and miR-424 to be among the most interesting candidates. Since miRs often are stage and tissue specific, large scale studies are warranted to further explore their prognostic and predictive impact. Besides the same miR often has multiple target genes and different functions. Therefore functional studies are highly needed to gain better insight into their biological functions.
Materials and Methods
Ethics statement
The study is approved by The National Data Inspection Board, The Regional Committee for Research Ethics and Biobank Board Collection of Tissue. Information and subsequent written consent from patients or next of kin was considered, but as this was a retrospective study with more than half of patients deceased, some more than ten years ago, The Regional Committee for Research Ethics specifically waived the need for consent.
Patients and tissue samples
From a previously well-described large NSCLC cohort [30], twenty formalin-fixed and paraffin-embedded (FFPE) tumor tissue samples were obtained from patients diagnosed with stage I-IIIA at the University Hospital of Northern Norway (UNN) and Nordland Central Hospital (NLCH). These were ten patients with a short disease-specific survival (DSS) (NSCLC_01 - NSCLC_10), and ten patients with long DSS (NSCLC_11 - NSCLC_20). In addition, samples from normal lung tissue sites in ten NSCLC patients (NSCLC_01, NSCLC_02, NSCLC_06, NSCLC_11, NSCLC_12, NSCLC_13, NSCLC_15, NSCLC_16, NSCLC_18 and NSCLC_19) were included in the study (NORM_01 - NORM_10). Four sample cores, each 0.6 mm in diameter, from viable tumor areas from each tumor and normal specimen were taken and pooled prior to RNA isolation in order to obtain a representative sample of each patient. Long and short DSS groups were stratified in order to get equal distribution of gender and histology. The tumor specimens were stored at the Departments of Pathology at UNN and NLCH, staged according to The International Union Against Cancer (UICC) TNM classification, and histologically subtyped according to the World Health Organization guidelines [54], [55]. All patient material was reviewed by two independent and trained pathologists (SAS and KAS). None of the patients received chemo- or radiotherapy prior to surgery. Complete patient data was obtained from medical records, including demographic, clinical, treatment and outcome data.
RNA isolation
RNA was isolated from the collected core samples by using the Recover All™ Total Nucleic Acid Isolation Kit for FFPE Tissues (Ambion, Austin, TX), according to the manufacturer's instructions, with some minor adjustments. Xylene treatment was increased from 3 min to 8 min, and protease treatment was increased from 3 hrs to approximately 20 hrs. RNA quality and quantity was assessed by using the NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE) and further verified by an Agilent 2100 Bioanalyzer profile.
Microarray procedures
Total RNA (700 ng) from sample and reference was labelled with Hy3™ and Hy5™ fluorescent label using the miRCURYTm LNA Array power labelling kit (Exiqon, Vedbaek, Denmark) following the procedure described by the manufacturer. The Hy3™-labeled samples and a Hy5™-labelled reference RNA sample (containing an equal aliquot of all RNA species included in this study) were mixed pair-wise and hybridized to the miCURYTm LNA Array version 5th generation (Exiqon, Denmark), which contains capture probes targeting all human miRNAs registered in the miRBASE version 14.0 at the Sanger Institute. The hybridization was performed according to the miRCURY™ LNA array manual using a Tecan HS4800 hybridization station (Tecan, Austria). After hybridization the microarray slides were scanned and stored in an ozone free environment (ozone level below 2.0 ppb) in order to prevent potential bleaching of the fluorescent dyes. The miCURY™ LNA Array microarray slides were scanned using the Agilent G2565BA Microarray Scanner System (Agilent technologies Inc., USA) and the image analysis was carried out using the ImaGene 8.0 software (BioDiscovery Inc., USA). The quantified signals were background corrected with offset value 10 and normalized using the global Lowess (Locally Weighted Scatterplot Smoothing) regression algorithm [56].
Database submission of microarray data
The microarray data were prepared according to minimum information about a microarray experiment (MIAME) recommendations [57] and deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/). The GEO accession number for the platform is GSE27705 and the study may be viewed at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE27705.
Quantification of mature miRNAs by real-time qPCR
One or several of the following criteria were used when selecting miRs for PCR validation; significant P-value, high fold change and/or angiogenesis-related miRs. Five samples from each group with sufficient tumor miR remaining after the array analysis were used in the PCR validation. Out of the 41 tested miRs, 28 miRs were detected in 7 or more samples and analysed. The stability of the miRNAs detected in all samples was tested by Exicon using the Normfinder software. miR-423-5p was one of the recommended reference miRNAs together with miR-150, miR-423-5p and miR-300. miR-300 was excluded due to very low levels detected. Further Normfinder found large variation in miR- 150, however miR-423-5p was found to be the most stable miRNA in the data set and consequently used as reference. All real-time qPCR experiments were performed by Exiqon, Vedbaek, Denmark. 10 ng RNA was reverse transcribed in 10 µl reactions using the miRCURY LNA™ Universal RT miR PCR, Polyadenylation and cDNA synthesis kit (Exiqon); each sample was processed in triplicates. cDNA was diluted 100× and 4 ul was used in 10 ul PCR reactions according to the protocol for miRCURY LNA™ Universal RT miR PCR; each miR was assayed once by qPCR in triplicate cDNA. The amplification was performed in a LightCycler® 480 Real-Time PCR System (Roche) in 384 well plates. LinRegPCR (version 11.5) software was used to determine the qPCR amplification efficiency. The average amplification efficiency was used to correct the Raw Cp values.
Pathway analysis
The impacts of the different pathways were evaluated by Gene Set Enrichment Analysis (GSEA) derived from Protein ANalysis THrough Evolutionary Relationship (PANTHER). The PANTHER classification system is a unique resource that classifies genes by their functions, using published scientific experimental evidence and evolutionary relationships to predict function even in the absence of direct experimental evidence. The gene set was taken from the MirDB (http://www.mirdb.org) (http://rnajournal.cshlp.org/content/14/6/1012.full) database who has compiled gene lists from pathways curated in the PANTHER database.
A full list is available at http://mirdb.org/cgi-bin/pathway.cgi. miRNA annotation is based on miRBase version 13 (http://www.mirbase.org/). GSEA analysis was carried out using the R statistical language (www.r-project.org) and the GSEA package for R available at http://www.broadinstitute.org/gsea/downloads.jsp.
Data analysis and statistical analysis
The data were analyzed using Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray experiments (LIMMA) [58], and significance was determined at the 0.1 level corrected for false discovery rate (FDR) using the method of Benjamini & Hochberg [59]. Principal component analysis (PCA) and Partial least squares (PLS) were carried out on the data in order to visualize the data structure and look for potential outlier samples [60].
The enrichment score (ES) reflects the degree to which a gene set is overrepresented at the top or bottom of a ranked list of genes. Normalized enrichment scores (NES) are used to compare analysis results across gene sets. GSEA accounts for differences in gene set size and in correlations between gene sets and the expression dataset [61]. The false discovery rate (FDR) is the estimated probability that a gene set with a given NES represents a false positive finding. The nominal p- value estimates the statistical significance of the enrichment score for a single gene set.
Pearson's correlation coefficient, r, was used to describe the association between array data and results from qPCR. Pearson Chi-Square and Fisher Exact Test were used to correlate angiogenic markers with miR-155.
Supporting Information
In situ hybridization (ISH) analysis of NSCLC representing strong and weak intensities for tumor cell miR-155 expression. Negative (scramble-miR) and positive (U6) controls from the same tissue area are shown. Strong miR-155 staining (A) with corresponding negative (C) and positive (E) controls to the left. Weak miR-155 staining (B) with corresponding negative (D) and positive (F) controls to the right. ISH positive signals (miR-155 and U6) stain blue, while nuclei stain red.
(TIF)
The 128 significant -up and down-regulated (−1, 1) miRs are presented with FDR adjusted p-values and respective fold changes (log2 scale).
(XLS)
Comparison between the array and the qPCR data in validation set. The differences in average expression levels between the sample groups in the array study are shown in columns 2–4 (dLMR values = log2 scale). The differences in average expression levels between the sample groups in the qPCR study are shown in columns 5–7 (ddCp values = log2 scale). Positive numbers >1 are shown in red ( = fold change >2), negative numbers <−1 ( = fold change >−2).
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The study was solely funded by the Northern Norway Regional Health Authority (Helse Nord RHF), which is responsible for the public hospitals in northern Norway. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Lebanony D, Benjamin H, Gilad S, Ezagouri M, Dov A, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009;27:2030–7. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 3.Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–50. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 4.Scagliotti G, Hanna N, Fossella F, Sugarman K, Blatter J, et al. The differential efficacy of pemetrexed according to NSCLC histology: a review of two Phase III studies. Oncologist. 2009;14:253–63. doi: 10.1634/theoncologist.2008-0232. [DOI] [PubMed] [Google Scholar]
- 5.Lin PY, Yu SL, Yang PC. MicroRNA in lung cancer. Br J Cancer. 2010;103:1144–8. doi: 10.1038/sj.bjc.6605901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 8.Wu X, Piper-Hunter MG, Crawford M, Nuovo GJ, Marsh CB, et al. MicroRNAs in the pathogenesis of Lung Cancer. J Thorac Oncol. 2009;4:1028–34. doi: 10.1097/JTO.0b013e3181a99c77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42:1273–81. doi: 10.1016/j.biocel.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 10.Jiang L, Huang Q, Zhang S, Zhang Q, Chang J, et al. Hsa-miR-125a-3p and hsa-miR-125a-5p are downregulated in non-small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells. BMC Cancer. 2010;10:318. doi: 10.1186/1471-2407-10-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Y, Bai Y, Zhang F, Wang Y, Guo Y, et al. miR-126 inhibits non-small cell lung cancer cells proliferation by targeting EGFL7. Biochem Biophys Res Commun. 2010;391:1483–9. doi: 10.1016/j.bbrc.2009.12.098. [DOI] [PubMed] [Google Scholar]
- 12.Cho WC. Role of miRNAs in lung cancer. Expert Rev Mol Diagn. 2009;9:773–6. doi: 10.1586/erm.09.57. [DOI] [PubMed] [Google Scholar]
- 13.Bishop JA, Benjamin H, Cholakh H, Chajut A, Clark DP, et al. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin Cancer Res. 2010;16:610–9. doi: 10.1158/1078-0432.CCR-09-2638. [DOI] [PubMed] [Google Scholar]
- 14.Hu Z, Chen X, Zhao Y, Tian T, Jin G, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–6. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 15.Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16:430–41. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patnaik SK, Kannisto E, Knudsen S, Yendamuri S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010;70:36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- 17.Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–83. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 18.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009;106:12085–90. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voortman J, Goto A, Mendiboure J, Sohn JJ, Schetter AJ, et al. MicroRNA expression and clinical outcomes in patients treated with adjuvant chemotherapy after complete resection of non-small cell lung carcinoma. Cancer Res. 2010;70:8288–98. doi: 10.1158/0008-5472.CAN-10-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 21.Du L, Pertsemlidis microRNAs and lung cancer: tumors and 22-mers. Cancer Metastasis Rev. 2010;29:109–22. doi: 10.1007/s10555-010-9204-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulshreshtha R, Davuluri RV, Calin GA, Ivan M. A microRNA component of the hypoxic response. Cell Death Differ. 2008;15:667–71. doi: 10.1038/sj.cdd.4402310. [DOI] [PubMed] [Google Scholar]
- 23.Olson P, Lu J, Zhang H, Shai A, Chun MG, et al. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–65. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suarez Y, Sessa WC. MicroRNAs as novel regulators of angiogenesis. Circ Res. 2009;104:442–54. doi: 10.1161/CIRCRESAHA.108.191270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 26.Nguewa PA, Calvo A, Pullamsetti SS, Banat GA, Grimminger F, et al. Tyrosine kinase inhibitors with antiangiogenic properties for the treatment of non-small cell lung cancer. Expert Opin Investig Drugs. 2011;20:61–74. doi: 10.1517/13543784.2011.541153. [DOI] [PubMed] [Google Scholar]
- 27.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 28.Andersen S, Donnem T, Al-Saad S, Al-Shibli K, Busund LT, et al. Angiogenic markers show high prognostic impact on survival in marginally operable non-small cell lung cancer patients treated with adjuvant radiotherapy. J Thorac Oncol. 2009;4:463–71. doi: 10.1097/JTO.0b013e3181991d18. [DOI] [PubMed] [Google Scholar]
- 29.Andersen S, Eilertsen M, Donnem T, Al-Saad S, Al-Shibli K, et al. Diverging prognostic impacts of hypoxic markers according to NSCLC histology. Lung Cancer. 2011;72:294–302. doi: 10.1016/j.lungcan.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Donnem T, Al-Saad S, Al-Shibli K, Delghandi MP, Persson M, et al. Inverse prognostic impact of angiogenic marker expression in tumor cells versus stromal cells in non small cell lung cancer. Clin Cancer Res. 2007;13:6649–57. doi: 10.1158/1078-0432.CCR-07-0414. [DOI] [PubMed] [Google Scholar]
- 31.Donnem T, Al-Saad S, Al-Shibli K, Andersen S, Busund LT, et al. Prognostic impact of platelet-derived growth factors in non-small cell lung cancer tumor and stromal cells. J Thorac Oncol. 2008;3:963–70. doi: 10.1097/JTO.0b013e3181834f52. [DOI] [PubMed] [Google Scholar]
- 32.Donnem T, Al-Shibli K, Al-Saad S, Delghandi MP, Busund LT, et al. VEGF-A and VEGFR-3 correlate with nodal status in operable non-small cell lung cancer: inverse correlation between expression in tumor and stromal cells. Lung Cancer. 2009;63:277–83. doi: 10.1016/j.lungcan.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 33.Donnem T, Al-Shibli K, Al-Saad S, Busund LT, Bremnes RM. Prognostic impact of fibroblast growth factor 2 in non-small cell lung cancer: coexpression with VEGFR-3 and PDGF-B predicts poor survival. J Thorac Oncol. 2009;4:578–85. doi: 10.1097/JTO.0b013e31819f2e38. [DOI] [PubMed] [Google Scholar]
- 34.Donnem T, Andersen S, Al-Shibli K, Al-Saad S, Busund LT, et al. Prognostic impact of Notch ligands and receptors in nonsmall cell lung cancer: Coexpression of Notch-1 and vascular endothelial growth factor-A predicts poor survival. Cancer. 2010;116:5676–85. doi: 10.1002/cncr.25551. [DOI] [PubMed] [Google Scholar]
- 35.Donnem T, Al-Shibli K, Andersen S, Al-Saad S, Busund LT, et al. Combination of low vascular endothelial growth factor A (VEGF-A)/VEGF receptor 2 expression and high lymphocyte infiltration is a strong and independent favorable prognostic factor in patients with nonsmall cell lung cancer. Cancer. 2010;116:4318–25. doi: 10.1002/cncr.25333. [DOI] [PubMed] [Google Scholar]
- 36.Donnem T, Al-Saad S, Al-Shibli K, Busund LT, Bremnes RM. Co-expression of PDGF-B and VEGFR-3 strongly correlates with lymph node metastasis and poor survival in non-small-cell lung cancer. Ann Oncol. 2010;21:223–31. doi: 10.1093/annonc/mdp296. [DOI] [PubMed] [Google Scholar]
- 37.Donnem T, Eklo K, Berg T, Sorbye SW, Lonvik K, et al. Prognostic Impact of MiR-155 in Non-Small Cell Lung Cancer Evaluated by in Situ Hybridization. J Transl Med. 2011;9:6. doi: 10.1186/1479-5876-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donnem T, Lonvik K, Eklo K, Berg T, Sorbye SW, et al. Independent and tissue-specific prognostic impact of miR-126 in nonsmall cell lung cancer: Coexpression with vascular endothelial growth factor-A predicts poor survival. Cancer. 2011;15:3193–200. doi: 10.1002/cncr.25907. [DOI] [PubMed] [Google Scholar]
- 39.Kuehbacher A, Urbich C, Dimmeler S. Targeting microRNA expression to regulate angiogenesis. Trends Pharmacol Sci. 2008;29:12–5. doi: 10.1016/j.tips.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 40.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–67. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu F, Yang Z, Li G. Role of specific microRNAs for endothelial function and angiogenesis. Biochem Biophys Res Commun. 2009;386:549–53. doi: 10.1016/j.bbrc.2009.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang C. MicroRNAs in vascular biology and vascular disease. J Cardiovasc Transl Res. 2010;3:235–40. doi: 10.1007/s12265-010-9164-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meister J, Schmidt MH. miR-126 and miR-126*: new players in cancer. ScientificWorldJournal. 2010;10:2090–100. doi: 10.1100/tsw.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S, Aurora AB, Johnson BA, Qi X, McAnally L, et al. The endothelial specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikolic I, Plate KH, Schmidt MH. EGFL7 meets miRNA-126: an agiogenesis alliance. J Angiogenesis Res. 2010;2:9. doi: 10.1186/2040-2384-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene a typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792:497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 47.Liu LZ, Li C, Chen Q, Jing Y, Carpenter R, et al. MiR-21 induced angiogenesis through AKT and ERK activation and HIF-1α expression. PLoS ONE. 2011;22:e19139. doi: 10.1371/journal.pone.0019139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Al-Saad S, Al-Shibli K, Donnem T, Al-Shibli K, Persson M, et al. Diverse prognostic roles of Akt isoforms, PTEN and PI3K in tumor epithelial cells and stromal compartment in non-small cell lung cancer. Anticancer Res. 2009;29:4175–83. [PubMed] [Google Scholar]
- 49.Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, et al. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-α isoforms and promotes angiogenesis. J Clin Invest. 2010;120:4141–54. doi: 10.1172/JCI42980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakashima T, Jinnin M, Etoh T, Fukushima S, Masuguchi S, et al. Down-regulation of mir-424 contributes to the abnormal angiogenesis via MEK1 and cyclin E1 in senile hemangioma: its implications to therapy. PLoS ONE. 2010;5:e14334. doi: 10.1371/journal.pone.0014334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Devlin C, Greco S, Martelli F, Ivan M. miR-210: More than a silent player in hypoxia. IUBMB Life. 2011;63:94–100. doi: 10.1002/iub.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puissêgur MP, Mazure NM, Bertero T. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18:465–78. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou W, Feng X, Wu Y, Benge J, Zhang Z, et al. FGF-receptor substrate 2 functions as a molecular sensor integrating external regulatory signals into the FGF pathway. Cell Res. 2009;19:1165–77. doi: 10.1038/cr.2009.95. [DOI] [PubMed] [Google Scholar]
- 54.World Health Organization. Histological Typing of Lung and Pleural Tumours. 3 edn. Geneva, Switzerland: Springer-Verlag; 1999. 3 [Google Scholar]
- 55.Goldstraw P. The 7th Edition of TNM in Lung Cancer: what now? J Thorac Oncol. 2009;4:671–3. doi: 10.1097/JTO.0b013e31819e7814. [DOI] [PubMed] [Google Scholar]
- 56.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–7. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 57.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 58.Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–6. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 59.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–84. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 60.Gidskehaug l, Stodkilde-Jorgensen H, Martens M, Martens H. Bridge-PLS regression:two-block bilinear regression without deflation. Journal of Chemometrics. 2004;18:208–15. [Google Scholar]
- 61.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In situ hybridization (ISH) analysis of NSCLC representing strong and weak intensities for tumor cell miR-155 expression. Negative (scramble-miR) and positive (U6) controls from the same tissue area are shown. Strong miR-155 staining (A) with corresponding negative (C) and positive (E) controls to the left. Weak miR-155 staining (B) with corresponding negative (D) and positive (F) controls to the right. ISH positive signals (miR-155 and U6) stain blue, while nuclei stain red.
(TIF)
The 128 significant -up and down-regulated (−1, 1) miRs are presented with FDR adjusted p-values and respective fold changes (log2 scale).
(XLS)
Comparison between the array and the qPCR data in validation set. The differences in average expression levels between the sample groups in the array study are shown in columns 2–4 (dLMR values = log2 scale). The differences in average expression levels between the sample groups in the qPCR study are shown in columns 5–7 (ddCp values = log2 scale). Positive numbers >1 are shown in red ( = fold change >2), negative numbers <−1 ( = fold change >−2).
(DOC)