Abstract
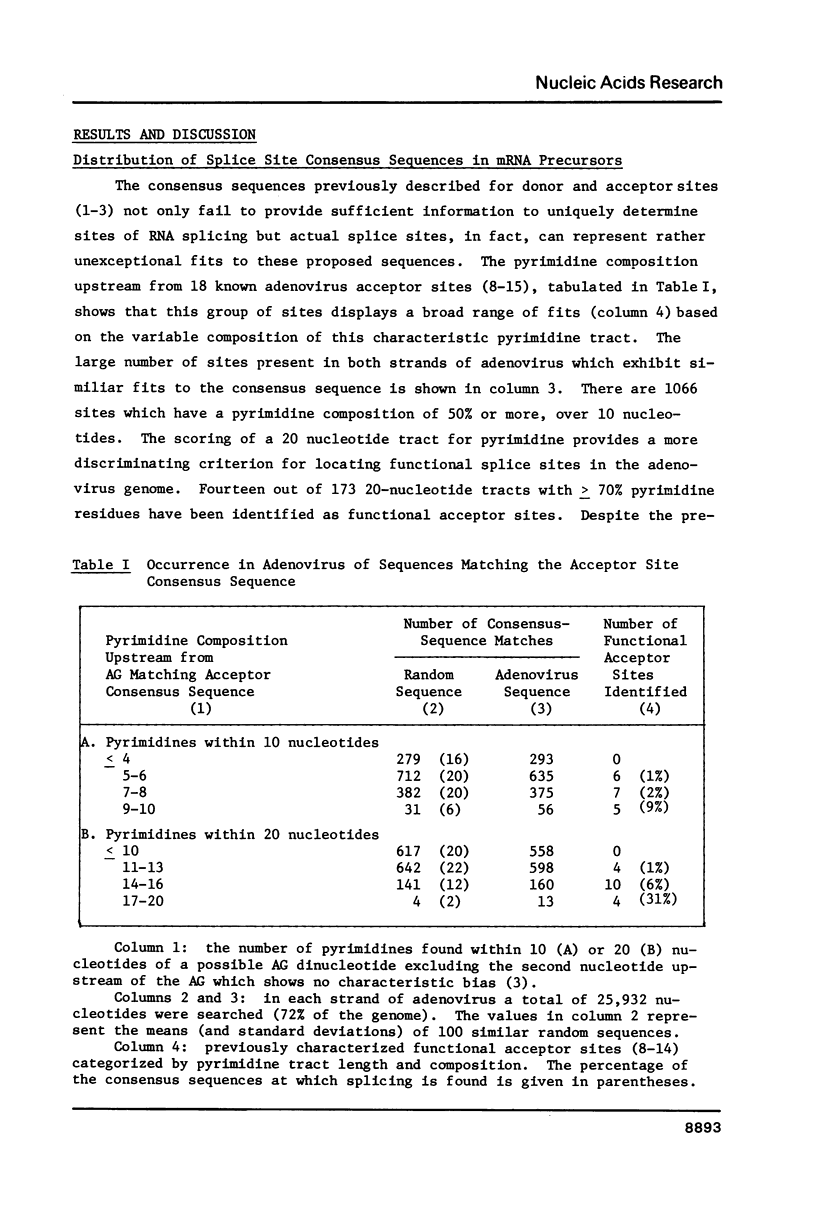
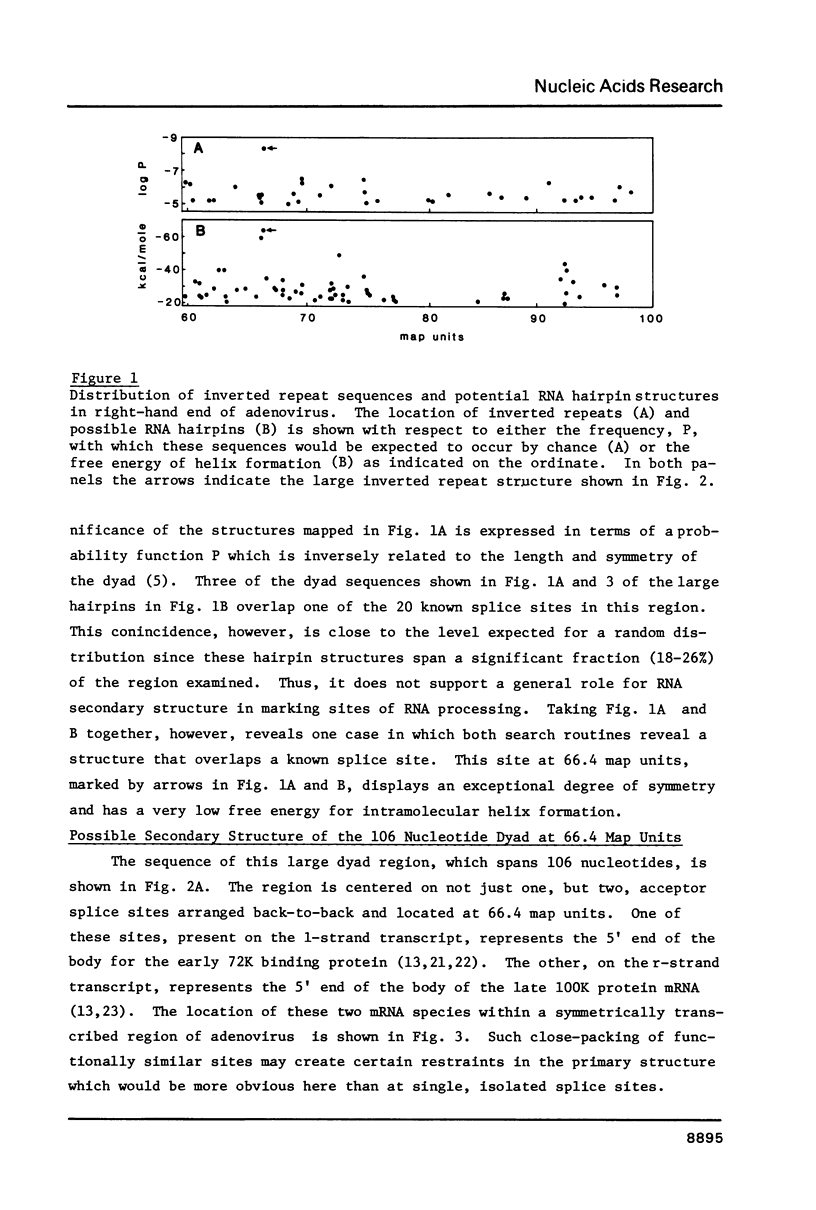
The distribution of nucleotide sequences resembling functional sites for mRNA splicing was examined by computer-directed searches in order to determine what factors may influence splice site selection in nuclear precursors. In particular, the distribution of large potentially stable hairpin structures or regions of extensive dyad symmetry was studied in adenovirus sequences. One region, spanning 106 nucleotides, was found at 66.4 map units, overlapping back-to-back acceptor sites for two mRNA molecules, those coding for the 100K protein and the 72K DNA binding protein, which are transcribed from opposite strands. This region displays exceptional dyad symmetry and is potentially capable of forming a single, highly stable hairpin when transcribed. It seems likely that the secondary structure as well as the primary structure of RNA plays a role in determining the correct splicing of these mRNA molecules.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abelson J. RNA processing and the intervening sequence problem. Annu Rev Biochem. 1979;48:1035–1069. doi: 10.1146/annurev.bi.48.070179.005131. [DOI] [PubMed] [Google Scholar]
- Akusjärvi G., Zabielski J., Perricaudet M., Pettersson U. The sequence of the 3' non-coding region of the hexon mRNA discloses a novel adenovirus gene. Nucleic Acids Res. 1981 Jan 10;9(1):1–17. doi: 10.1093/nar/9.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Structure of the adenovirus 2 early mRNAs. Cell. 1978 Jul;14(3):695–711. doi: 10.1016/0092-8674(78)90252-0. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Burke J. M., RajBhandary U. L. Intron within the large rRNA gene of N. crassa mitochondria: a long open reading frame and a consensus sequence possibly important in splicing. Cell. 1982 Dec;31(3 Pt 2):509–520. doi: 10.1016/0092-8674(82)90307-5. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg M. D., Kelly T. J. Eukaryotic DNA replication: viral and plasmid model systems. Annu Rev Biochem. 1982;51:901–934. doi: 10.1146/annurev.bi.51.070182.004345. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Broker T. R., Lewis J. B. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J Mol Biol. 1979 Oct 25;134(2):265–303. doi: 10.1016/0022-2836(79)90036-6. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Broker T. R. The spliced structures of adenovirus 2 fiber message and the other late mRNAs. Cell. 1978 Oct;15(2):497–510. doi: 10.1016/0092-8674(78)90019-3. [DOI] [PubMed] [Google Scholar]
- Davies R. W., Waring R. B., Ray J. A., Brown T. A., Scazzocchio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982 Dec 23;300(5894):719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- Fitch W. M. The large extent of putative secondary nucleic acid structure in random nucleotide sequences or amino acid derived messenger-RNA. J Mol Evol. 1974;3(4):279–291. doi: 10.1007/BF01796043. [DOI] [PubMed] [Google Scholar]
- Flint S. J. Expression of adenoviral genetic information in productively infected cells. Biochim Biophys Acta. 1982 Apr 29;651(2-3):175–208. doi: 10.1016/0304-419x(82)90011-7. [DOI] [PubMed] [Google Scholar]
- Galibert F., Hérissé J., Courtois G. Nucleotide sequence of the EcoRI-F fragment of adenovirus 2 genome. Gene. 1979 May;6(1):1–22. doi: 10.1016/0378-1119(79)90081-7. [DOI] [PubMed] [Google Scholar]
- Gingeras T. R., Sciaky D., Gelinas R. E., Bing-Dong J., Yen C. E., Kelly M. M., Bullock P. A., Parsons B. L., O'Neill K. E., Roberts R. J. Nucleotide sequences from the adenovirus-2 genome. J Biol Chem. 1982 Nov 25;257(22):13475–13491. [PubMed] [Google Scholar]
- Goad W. B., Kanehisa M. I. Pattern recognition in nucleic acid sequences. I. A general method for finding local homologies and symmetries. Nucleic Acids Res. 1982 Jan 11;10(1):247–263. doi: 10.1093/nar/10.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérissé J., Courtois G., Galibert F. Nucleotide sequence of the EcoRI D fragment of adenovirus 2 genome. Nucleic Acids Res. 1980 May 24;8(10):2173–2192. doi: 10.1093/nar/8.10.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérissé J., Galibert F. Nucleotide sequence of the EcoRI E fragment of adenovirus 2 genome. Nucleic Acids Res. 1981 Mar 11;9(5):1229–1240. doi: 10.1093/nar/9.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérissé J., Rigolet M., de Dinechin S. D., Galibert F. Nucleotide sequence of adenovirus 2 DNA fragment encoding for the carboxylic region of the fiber protein and the entire E4 region. Nucleic Acids Res. 1981 Aug 25;9(16):4023–4042. doi: 10.1093/nar/9.16.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. I., Goad W. B. Pattern recognition in nucleic acid sequences. II. An efficient method for finding locally stable secondary structures. Nucleic Acids Res. 1982 Jan 11;10(1):265–278. doi: 10.1093/nar/10.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Gruss P., Dhar R., Lai C. J. Processing and expression of early SV40 mRNA: a role for RNA conformation in splicing. Cell. 1979 Sep;18(1):85–92. doi: 10.1016/0092-8674(79)90356-8. [DOI] [PubMed] [Google Scholar]
- Kruijer W., van Schaik F. M., Speijer J. G., Sussenbach J. S. Structure and function of adenovirus DNA binding protein: comparison of the amino acid sequences of the Ad5 and Ad12 proteins derived from the nucleotide sequence of the corresponding genes. Virology. 1983 Jul 15;128(1):140–153. doi: 10.1016/0042-6822(83)90325-2. [DOI] [PubMed] [Google Scholar]
- Kruijer W., van Schaik F. M., Sussenbach J. S. Structure and organization of the gene coding for the DNA binding protein of adenovirus type 5. Nucleic Acids Res. 1981 Sep 25;9(18):4439–4457. doi: 10.1093/nar/9.18.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühne T., Wieringa B., Reiser J., Weissmann C. Evidence against a scanning model of RNA splicing. EMBO J. 1983;2(5):727–733. doi: 10.1002/j.1460-2075.1983.tb01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Lerner M. R., Boyle J. A., Mount S. M., Wolin S. L., Steitz J. A. Are snRNPs involved in splicing? Nature. 1980 Jan 10;283(5743):220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. D. Escherichia coli ribonuclease III cleavage sites. Cell. 1982 Oct;30(3):669–672. doi: 10.1016/0092-8674(82)90270-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Schmelzer C., Schmidt C., Schweyen R. J. Identification of splicing signals in introns of yeast mitochondrial split genes: mutational alterations in intron bI1 and secondary structures in related introns. Nucleic Acids Res. 1982 Nov 11;10(21):6797–6808. doi: 10.1093/nar/10.21.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A. Speculations on RNA splicing. Cell. 1981 Mar;23(3):643–646. doi: 10.1016/0092-8674(81)90425-6. [DOI] [PubMed] [Google Scholar]
- Stillman B. W., Lewis J. B., Chow L. T., Mathews M. B., Smart J. E. Identification of the gene and mRNA for the adenovirus terminal protein precursor. Cell. 1981 Feb;23(2):497–508. doi: 10.1016/0092-8674(81)90145-8. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Raziuddin, Sobota A., Boublik M., Szer W. A RNA helix-destabilizing protein is a major component of Artemia salina nuclear ribonucleoproteins. Proc Natl Acad Sci U S A. 1981 May;78(5):2888–2892. doi: 10.1073/pnas.78.5.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Wollenzien P. L., Cantor C. R., Grant D. M., Lambowitz A. M. RNA splicing in neurospora mitochondria: structure of the unspliced 35S precursor ribosomal RNA detected by psoralen cross-linking. Cell. 1983 Feb;32(2):397–407. doi: 10.1016/0092-8674(83)90459-2. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]


