Abstract
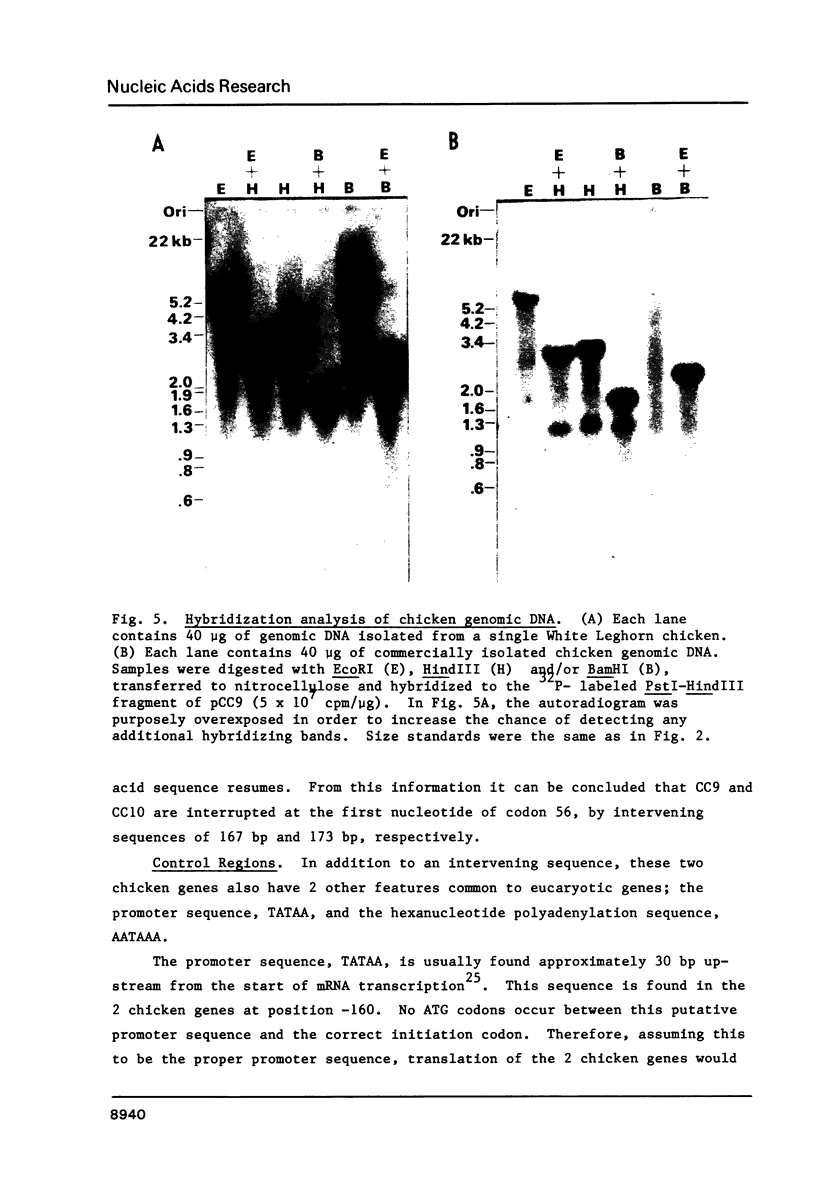
Analysis of total chicken DNA by genomic blot hybridization indicates that only one cytochrome c gene exists in the chicken genome. The two alleles of this single cytochrome c gene have been isolated from a Charon 4A-chicken genomic library. This isolation made use of the yeast CYC1 cytochrome c gene as a specific hybridization probe. The 2 chicken alleles, CC9 and CC10, have been sequenced. The amino acid sequence predicted by these 2 alleles is identical, and agrees with the published chicken cytochrome c protein sequence. The flanking regions of these 2 alleles exhibit approximately 1% divergence, indicating a very limited polymorphism. Comparative sequence analysis with the flanking regions of previously isolated cytochrome c genes (yeast and rat) indicate no significant regions of homology. The presence of only one cytochrome c-like sequence in the chicken genome is in striking contrast with mammalian genomes, which contain as many as 20-30 cytochrome c-like sequences.
Full text
PDF

















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F. R., Williams B. G., Blechl A. E., Denniston-Thompson K., Faber H. E., Furlong L., Grunwald D. J., Kiefer D. O., Moore D. D., Schumm J. W. Charon phages: safer derivatives of bacteriophage lambda for DNA cloning. Science. 1977 Apr 8;196(4286):161–169. doi: 10.1126/science.847462. [DOI] [PubMed] [Google Scholar]
- Boss J. M., Darrow M. D., Zitomer R. S. Characterization of yeast iso-1-cytochrome c mRNA. J Biol Chem. 1980 Sep 25;255(18):8623–8628. [PubMed] [Google Scholar]
- Chan S. K., Margoliash E. Amino acid sequence of chicken heart cytochrome c. J Biol Chem. 1966 Jan 25;241(2):507–515. [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. The structures of cytochrome c and the rates of molecular evolution. J Mol Evol. 1971;1(1):26–45. doi: 10.1007/BF01659392. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Fyrberg E. A., Bond B. J., Hershey N. D., Mixter K. S., Davidson N. The actin genes of Drosophila: protein coding regions are highly conserved but intron positions are not. Cell. 1981 Apr;24(1):107–116. doi: 10.1016/0092-8674(81)90506-7. [DOI] [PubMed] [Google Scholar]
- Gojobori T., Li W. H., Graur D. Patterns of nucleotide substitution in pseudogenes and functional genes. J Mol Evol. 1982;18(5):360–369. doi: 10.1007/BF01733904. [DOI] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Guo L. H., Wu R. New rapid methods for DNA sequencing based in exonuclease III digestion followed by repair synthesis. Nucleic Acids Res. 1982 Mar 25;10(6):2065–2084. doi: 10.1093/nar/10.6.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978 Dec;15(4):1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- Lewin B. Alternatives for splicing: recognizing the ends of introns. Cell. 1980 Nov;22(2 Pt 2):324–326. doi: 10.1016/0092-8674(80)90340-2. [DOI] [PubMed] [Google Scholar]
- Margoliash E., Schejter A. Cytochrome c. Adv Protein Chem. 1966;21:113–286. doi: 10.1016/s0065-3233(08)60128-x. [DOI] [PubMed] [Google Scholar]
- Matner R. R., Sherman F. Differential accumulation of two apo-iso-cytochromes c in processing mutants of yeast. J Biol Chem. 1982 Aug 25;257(16):9811–9821. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConaughy B. L., Laird C. D., McCarthy B. J. Nucleic acid reassociation in formamide. Biochemistry. 1969 Aug;8(8):3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- Montgomery D. L., Hall B. D., Gillam S., Smith M. Identification and isolation of the yeast cytochrome c gene. Cell. 1978 Jul;14(3):673–680. doi: 10.1016/0092-8674(78)90250-7. [DOI] [PubMed] [Google Scholar]
- Montgomery D. L., Leung D. W., Smith M., Shalit P., Faye G., Hall B. D. Isolation and sequence of the gene for iso-2-cytochrome c in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jan;77(1):541–545. doi: 10.1073/pnas.77.1.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perler F., Efstratiadis A., Lomedico P., Gilbert W., Kolodner R., Dodgson J. The evolution of genes: the chicken preproinsulin gene. Cell. 1980 Jun;20(2):555–566. doi: 10.1016/0092-8674(80)90641-8. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Russell P. R., Hall B. D. Structure of the Schizosaccharomyces pombe cytochrome c gene. Mol Cell Biol. 1982 Feb;2(2):106–116. doi: 10.1128/mcb.2.2.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R. C., Agne K. M., Wu R. Cytochrome c gene-related sequences in mammalian genomes. Proc Natl Acad Sci U S A. 1982 Feb;79(3):739–743. doi: 10.1073/pnas.79.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R. C., Agne K. M., Wu R. Isolation and structure of a rat cytochrome c gene. J Biol Chem. 1981 Jun 25;256(12):6480–6486. [PubMed] [Google Scholar]
- Scarpulla R. C., Wu R. Nonallelic members of the cytochrome c multigene family of the rat may arise through different messenger RNAs. Cell. 1983 Feb;32(2):473–482. doi: 10.1016/0092-8674(83)90467-1. [DOI] [PubMed] [Google Scholar]
- Smith M., Leung D. W., Gillam S., Astell C. R., Montgomery D. L., Hall B. D. Sequence of the gene for iso-1-cytochrome c in Saccharomyces cerevisiae. Cell. 1979 Apr;16(4):753–761. doi: 10.1016/0092-8674(79)90091-6. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Stiles J. I., Bahl C. P., Wu R. Specific binding of a synthetic oligodeoxyribonucleotide to yeast cytochrome c mRNA. Nature. 1977 Jan 6;265(5589):61–63. doi: 10.1038/265061a0. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]