Abstract
The clinicopathological heterogeneity of glioblastoma (GBM) and the various genetic and phenotypic subtypes in GBM stem cells (GSCs) are well described. However, the relationship between GSCs and the corresponding primary tumor from which they were isolated is poorly understood. We have established GSC-enriched neurosphere cultures from 15 newly diagnosed GBM specimens and examined the relationship between the histopathological and genomic features of GSC-derived orthotopic xenografts and those of the respective patient tumors. GSC-initiated xenografts recapitulate the distinctive cytological hallmarks and diverse histological variants associated with the corresponding patient GBM, including giant cell and gemistocytic GBM, and primitive neuroectodermal tumor (PNET)–like components. This indicates that GSCs generate tumors that preserve patient-specific disease phenotypes. The majority of GSC-derived intracerebral xenografts (11 of 15) demonstrated a highly invasive behavior crossing the midline, whereas the remainder formed discrete nodular and vascular masses. In some cases, GSC invasiveness correlated with preoperative MRI, but not with the status of PI3-kinase/Akt pathways or O6-methylguanine methyltransferase expression. Genome-wide screening by array comparative genomic hybridization and fluorescence in situ hybridization revealed that GSCs harbor unique genetic copy number aberrations. GSCs acquiring amplifications of the myc family genes represent only a minority of tumor cells within the original patient tumors. Thus, GSCs are a genetically distinct subpopulation of neoplastic cells within a GBM. These studies highlight the value of GSCs for preclinical modeling of clinically relevant, patient-specific GBM and, thus, pave the way for testing novel anti-GSC/GBM agents for personalized therapy.
Keywords: genomic profile, glioblastoma, glioblastoma stem cells, invasion, phenotype
Glioblastoma (GBM), World Health Organization (WHO) grade IV, is the most malignant and common form of primary brain tumor in adults.1 Despite increased understanding of the molecular alterations associated with disease pathogenesis and the use of current multimodal treatment, consisting of surgery, radiation, and temozolomide chemotherapy, prognosis for patients with GBM remains grim, with median overall survival of ∼15 months.2,3 Histopathologically, GBM has long been recognized as exhibiting striking heterogeneity between tumors and within a tumor, including diverse histological patterns and cytological features, such as giant cell GBM or PNET-like components.4 In line with this, recent studies using large-scale genomic analysis, such as The Cancer Genome Atlas (TCGA) project, have identified multiple subtypes of GBM and associated treatment prognoses.5,6 These findings suggest that a better understanding of the phenotype/genotype of each GBM will be necessary to design optimal therapies for individual patients.
A growing body of evidence suggests that many cancers are organized by a cellular hierarchy in which only a subpopulation of undifferentiated neoplastic cells drive tumor progression and give rise to proliferative and more differentiated cancer cells.7 GBM is one of a number of solid malignancies that contain such cells termed cancer stem cells or tumor-initiating cells. GBM stem cells (GSCs) are characterized by their ability to efficiently generate tumors upon transplantation into the brains of immunocompromised mice.8,9 In addition, GSCs possess stem cell-like properties, sharing with normal neural stem cells the characteristics of neurosphere formation in serum-free culture conditions, self-renewal, and differentiation to multiple neural cell lineages. GSCs also express genes associated with neural stem cells, such as nestin and CD133, although cells fulfilling GSC criteria also exist in a CD133-negative population.10–12 From a clinical point of view, the cancer stem cell hypothesis implies that long-term cancer remission or cure depends on elimination of the highly tumorigenic cancer stem cell subpopulation. In fact, GSCs have been reported to resist radiation and chemotherapy,13,14 lending support to the concept that residual GSCs that have survived therapy might be responsible for the nearly inevitable recurrence of GBM. Therefore, better characterization of GSCs, both biologically and molecularly, is crucial for the development of effective therapeutic strategies for GBM.
We previously reported that neurosphere cultures isolated from human GBM specimens were enriched for GSCs that could self-renew and very efficiently generate orthotopic tumors in immunodeficient mice.15 The GSCs generated highly invasive or more circumscribed tumors with hypervascularity and intratumoral bleeding.15 Multiple human GSCs have been established that give rise to invasive and localized orthotopic tumors,9,11,16,17 and the xenograft phenotypes sometimes correlate with CD133 expression in GSCs.11,18 Thus, GSCs can display varying degrees of invasiveness and can reproduce the important pathological features of GBM, which serum-cultured glioma cells or commonly used glioma cell lines do not.19,20 This ability of GSCs to recapitulate the pathological hallmarks of GBM provides a preclinical GBM model potentially representative of the disease. Nevertheless, it has not been previously determined whether human GSCs can reproduce the pathological characteristics exhibited by the particular tumor from which the GSCs were isolated. There have been sporadic reports illustrating instances of histological resemblance of CD133+ xenografts to the original patient tumor,8,16 but evidence for GSCs' representation of individual patient tumors has been insufficient.
In accordance with the heterogeneous nature of GBM as a disease entity, molecular genetic studies have increasingly uncovered different genotypes and gene expression profiles present in GSC populations from different patients.11,21 GSCs have been shown to maintain the genomic alterations seen in the primary tumor, whereas serum-cultured cells typically lose such genomic features over time.20,22 However, it is not known whether there are genomic aberrations that are specifically carried by the GSC subpopulations. Discovery of such GSC-specific genomic alterations will have a significant impact on the precise identification of possibly rare GSC populations within the tumor and exploitation of the related signaling pathways that might regulate important GSC properties and, thus, have therapeutic implications.
In this study, we have established a panel of human GSCs from 15 newly diagnosed GBMs. To clarify the extent of GSC representation to the corresponding primary GBM, we have conducted phenotypic and genomic characterization of the GSCs and GSC-generated xenografts and, when available, compared this data with that obtained from the original patient tumors.
Materials and Methods
Isolation and Culture of GSC
Resection specimens of newly diagnosed GBM were collected at Massachusetts General Hospital with approval of the Institutional Review Board. Methods for primary culture of GBM tissue were previously described.15 In brief, tissue was minced and trypsin-digested, and cells were grown in neurobasal medium (Invitrogen) supplemented with L-glutamine (3 mM; Mediatech), B27 supplement (Invitrogen), N2 supplement (Invitrogen), heparin (5 µg/mL; Sigma), EGF (20 ng/mL; R and D systems), and FGF2 (20 ng/mL; Peprotec) to establish neurosphere cultures enriched for GSCs. We succeeded in establishing neurosphere cultures that could be passaged at least 5 times from 16 of 30 specimens.
Immunocytochemistry
Staining of GSCs and serum-treated cells for differentiation markers were performed as described.15 The primary antibodies used were anti-nestin (Santa Cruz Biotechnology), anti-GFAP (Sigma), anti-MAP2 (Millipore, and anti-GalC (Chemicon).
In Vivo Tumorigenicity Assay
GSC-enriched neurospheres grown in vitro for 2–10 days (passage 1–2) since the initiation of culture were collected, and 20 000–50 000 cells were stereotactically implanted into the right striatum of the brains of 7–10-week-old female SCID mice as described.15 Mice were monitored for status twice per week and sacrificed when neurological deficits became significant. Brains were removed for pathological studies or dissected to excise intracerebral tumors to re-establish cultures, which were re-implanted to new mice within 7 days. All mouse procedures were approved by the Subcommittee on Research Animal Care at Massachusetts General Hospital.
Pathological Analysis
Hematoxylin and eosin staining and immunohistochemistry were performed on formalin-fixed paraffin-embedded sections as described.15 Primary antibodies used for immunohistochemistry were anti-human nestin (Santa Cruz Biotechnology), anti-GFAP (Sigma), anti-NeuN (Millipore), anti-olig2 (DF308), and MIB-1 (anti-Ki67, Dako). Except for GFAP, all sections were microwave-treated in 10mM sodium citrate buffer (pH = 6) for antigen retrieval. Sections were reviewed independently by 2 neuropathologists (A.O.S. and D.N.L.).
Immunoblots
Cell pellets were lysed in radioimmunoprecipitation buffer (Boston Bioproducts) with a cocktail of protease and phosphatase inhibitors (Roche); 12.5 μg of protein was separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes by electroblotting. After blocking with 5% nonfat dry milk in TBST (20 mM Tris [pH, 7.5], 150 mM NaCl, 0.1% Tween20), membranes were incubated at 4°C overnight with antibodies against PTEN, Akt, phosphorylated-Akt (Ser473), phosphorylated-Akt (Thr308; all Cell Signaling Technology), MGMT (Sigma), and actin (Sigma). After washing and incubation with horseradish peroxidase-conjugated secondary antibodies (Promega), blots were washed, and signals were visualized with an ECL kit (Amersham Bioscience).
In Vitro Cell Viability Assays
Dissociated GSCs were seeded to 96-well plates at 7000–8000 cells per well. Serially diluted temozolomide was added to wells, and cells were further cultured for 5 days. Viability of cells was measured by MTS cell viability assay (The CellTiter 96 AQueous One Solution Cell Proliferation Assay; Promega). Percentage of viability was calculated using cells without the drug as a control, and the EC50 values were determined.
Epigenetic Analysis
Genomic DNA was isolated from cultured GSCs (passages 3–10) using the Gentra Puregene Cell kit (Qiagen) according to the manufacturer's protocol. Promoter methylation analysis of MGMT (O6-methylguanine methyltransferase) was accomplished by bisulfite conversion of 500 ng of genomic DNA using the EpiTect bisulfite conversion kit (Qiagen). This was followed by methylation-specific PCR (MSP) of the converted DNA with methylated-and unmethylated-specific PCR using primers previously described and validated.23 Genomic DNA from the Jurkat cell line methylated excessively by CpG methyltransferase (New England Biolabs) and genomic DNA from normal male donor (Promega) were used as positive and negative controls, respectively. The PCR products were separated in 1.5% agarose gel and visualized under UV illumination.
Array Comparative Genomic Hybridization (aCGH)
Oligonucleotide aCGH was performed to determine DNA copy number changes in GSCs and xenograft tumors derived from the GSCs following a published protocol.24
Fluorescence In Situ Hybridization (FISH)
Genomic alterations identified by aCGH were validated by FISH both in GSCs and formalin-fixed paraffin-embedded (FFPE) sections from original patient tumors as described elsewhere.17,25 The following BAC clones were used as probes: CTD-2014F22 (NMYC), RP11-307A11 (2q35, control probe for NMYC), RP11-626H4 (PDGFRA), RP11-572O17 (4p16.3, control probe for PDGFRA), CTD-3056O22 (CMYC), RP11-301H15 (8p12, control probe for CMYC), RP11-611O2 (MDM2), and RP11-264F23 (12p13.32, control probe for MDM2). Gene-specific probes were labeled in Cy3-dCTP, and control probes were labeled in FITC-dUTP for all hybridizations. Gene-amplified cells were counted in at least 3 different high-power fields (>50 total cells per field), and the proportion of amplification-positive per total cells was calculated.
Statistics
Responses to temozomomide by MGMT methylated and unmethylated GSCs were compared using a 2-tailed Student's t test (unpaired). P values <.05 were considered to be statistically significant.
Results
GSC-Derived Xenografts Recapitulate Histological Hallmarks of Respective Patient GBM
In our previous report, a small set of primary neurosphere cultures enriched for GSCs generated intracerebral tumors after orthotopic implantation into SCID mice.15 Neurosphere culture enriched for cells possessing multilineage differentiation potential, as illustrated in Supplementary Fig. S1. These cells were typically tumorigenic in immune-deficient mice15 except for the culture isolated from a GBM specimen (MGG15) that was not able to generate intracerebral tumors after implantation of 5 × 105 cells into SCID mice (5 of 5 mice). Here, we sought to extend our previous work by asking whether GSC-derived xenografts recapitulate the histological features of the respective GBM tumors from which the GSCs were established.
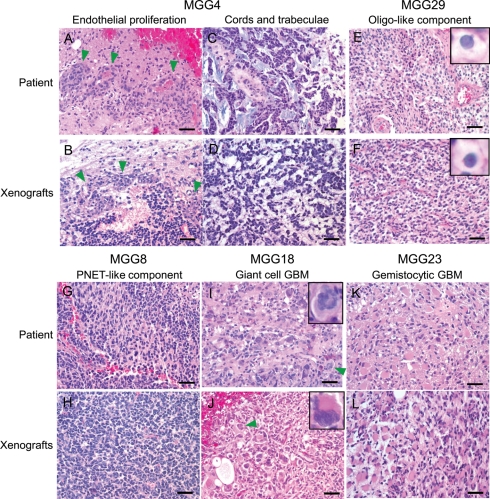
We retrieved FFPE blocks of the patient tumors that were used to generate GSCs and compared the histopathology of patient GBMs and GSC-derived orthotopic xenografts on hematoxylin and eosin–stained sections. Microvascular endothelial proliferation is a characteristic of GBM-associated angiogenesis and constitutes one of the important diagnostic criteria for GBM. This pathological feature seen in the MGG4 primary tumor was reproduced in its GSC-derived xenograft (Fig. 1A and B), which, of interest, is one of the most hypervascular and hemorrhagic xenografts in our GSC series. Neoplastic glioma cells within the MGG4 primary tumor were arranged in cords and trabeculae (Fig. 1C), a cellular architecture that was recapitulated in the MGG4 xenografts (Fig. 1D). Primary tumor MGG29 featured an oligodendroglial component characterized by cells with clear cytoplasm (perinuclear halo) and round nuclei (Fig. 1E), features that were also present in the GSC-derived xenografts (Fig. 1F). The MGG8 primary tumor contained foci that display PNET-like nodules characterized by densely cellular foci composed of large nuclei with fine chromatin and scant cytoplasm (Fig. 1G).26 The same histological feature was easily recognized in corresponding MGG8 xenografts (Fig. 1H).
Fig. 1.
GSC-derived xenografts recapitulate histopathological features of the original patient GBM. Top and third rows, primary tumors from patients; second and bottom rows, intracerebral xenografts derived from GSCs. (A and B) MGG4 showing endothelial proliferation (arrowheads). (C and D) Tumor cells arranged in cords and trabeculae in MGG4. (E and F) Oligodendroglial component observed in MGG29 was recapitulated in its respective xenografts. Insets, cells with perinuclear halo. (G and H) Many MGG8 tumor cells display undifferentiated cytological features like primitive neuroectodermal tumors (PNETs). (I and J) MGG18 tumor is characterized by marked cellular pleomorphism and giant, bizarre cells, features of giant cell GBM. Arrowheads and insets show multinucleated giant cells. (K and L) MGG23 contains tumor cells with abundant eosinophilic cytoplasm indicative of the gemistocytic phenotype. Bars, 50 µm.
MGG18 is a giant cell GBM currently categorized by the WHO as a distinct variant of GBM, which is characterized histologically by the presence of multinucleated giant cells.27 Consistent with its relative rarity, MGG18 was the only case diagnosed with this entity in our series of 15 cases of GBM (Fig. 1I). MGG18 xenografts demonstrated histological characteristics very similar to the primary tumor, with marked pleomorphism and the presence of bizarre-looking large cells, some of which were multinucleated (Fig. 1J). Of note, cellular heterogeneity, a mixture of neoplastic cells with a variety of sizes and morphology, was striking in the MGG18 xenografts (Fig. 1J). The primary MGG23 tumor was composed of malignant gemistocytic astrocytes, displaying abundant eosinophilic cytoplasm and eccentrically placed nuclei (Fig. 1K). The MGG23 xenografts similarly displayed the gemistocytic phenotype with strong expression of astrocyte marker GFAP (Figs 1L and 2B, arrows), thus presenting another example of the faithful recapitulation of histological features by GSC. We also observed histopathological similarity comparing conventional GBMs that lack unique characteristics and corresponding GSC xenografts (Supplementary material, Fig. S2). To objectively evaluate the histopathological resemblance between parental and the respective GSC xenograft tumors and identify which pathological characteristics are preserved in GSC tumors, label-masked 12 slides consisting of 6 primary GBM specimens and the corresponding GSC-derived tumors were subjected to an unbiased blinded review by neuropathologists (A.O.S. and D.N.L.), scoring for histological features associated with malignant gliomas (Table 1). Overall, pathological similarities between the patient-xenograft pairs were evident, especially distinct cytological features, including gemistocytic-like cells, oligo-like cells, and giant cells, which matched perfectly for each pair, and undifferentiated cells in 3 of 4 pairs. However, endothelial proliferation was only observed in 1 GSC xenograft (MGG4) (Fig. 1B) of 4 cases in which the primary GBM specimens displayed this feature. Necrosis was absent in the examined GSC xenografts and present in 4 of 6 patient specimens. We postulate that the relatively small size of GSC tumors in the mouse brain limits the development of necrosis. GSCs retain the capacity to generate xenografts displaying their unique histopathological features after serial transplanation in mice for at least 3 passages (Supplementary Fig. S3), indicating the phenotypic stability of GSC-derived xenografts.
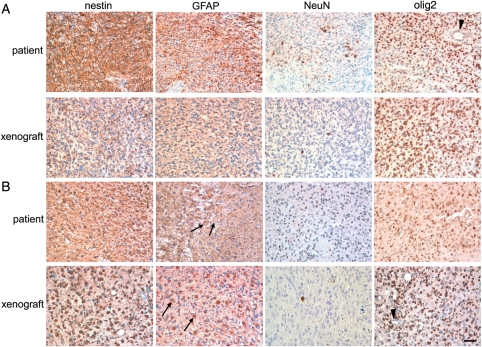
Fig. 2.
Immunohistochemical comparison between the original tumor and GSC-derived intracerebral xenografts. (A) MGG28 and (B) MGG23. Upper panels, primary patient tumor; Lower panels, intracerebral xenograft generated by GSCs. From left to right: staining for neural progenitor marker nestin, astrocytic marker GFAP, neuronal marker NeuN, glioma/oligodendroglial/stem cell marker olig2. Arrows show GFAP-positive gemistocytic cells present in both patient and GSC MGG23 tumors. Note negative staining of olig2 in blood vessels (arrowheads). Bar, 50 µm.
Table 1.
Unbiased evaluation and comparison of histopathological features in 6 pairs of primary GBM and GSC-derived intracerebral tumors
| Feature | MGG4 |
MGG7 |
MGG8 |
MGG18 |
MGG23 |
MGG29 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | X | P | X | P | X | P | X | P | X | P# | X | |
| Fibrillary processes | − | − | + | − | + | − | − | − | − | − | − | + |
| Gemistocytic-like cells | − | − | − | − | − | − | − | − | ++ | ++ | − | − |
| Oligo-like cells | − | − | − | − | − | − | − | − | − | − | ++ | + |
| Giant cells, multinucleation | − | − | − | − | − | − | + | ++ | − | − | − | − |
| Undifferentiated cells (e.g., PNET-like) | ++ | ++ | + | ++ | + | ++ | − | + | − | − | − | − |
| Epithelioid | − | − | − | − | − | − | + | − | − | − | − | − |
| Endothelial proliferation | + | + | + | − | + | − | + | − | − | − | − | − |
| Necrosis | + | − | − | − | + | − | + | − | + | − | − | − |
Abbreviations: P, Patient; X, Xenograft. Patient sections were from tissue used for clinical diagnosis, separate from that used for GSC isolation, except for MGG29 #, where a small piece of tumor from the sample used for GSC culture was sectioned. Official histopathological diagnosis of MGG29 on a larger separate tissue was GBM. −, Absent; +, Few or scarce; ++, Many or abundant.
We next examined the expression of proteins associated with different neural lineages using immunohistochemistry and compared the results between the primary tumor and corresponding xenografts (Fig. 2, Supplementary Fig. S4). In the MGG28 primary tumor, the neural progenitor marker nestin was diffusely immunopositive, suggesting the poorly differentiated nature of tumor cells (Fig. 2A, upper). Many tumor cells were also positive for GFAP, but the tumor staining was patchy. Transcription factor olig2, associated with gliomas, oligodendrocytes, and neural progenitor cells, was immunoreactive in the nuclei of most tumor cells, but not in the endothelial cells lining blood vessels (Fig. 2A, arrowhead). In contrast, NeuN, a marker for mature neurons, was expressed only in scattered individual cells. The immunostaining profiles were similar in the corresponding MGG28 GSC-generated xenografts (Fig. 2A, lower). Immunostaining results obtained with other pairs of primary GBMs (MGG23 and 18) and the respective GSC xenografts demonstrated resemblance of expression profiles between the patient tumors and the xenografts (Fig. 2B, Supplementary material, Fig. S4).
Collectively, these results demonstrate that GSCs are able to generate intracranial tumors that recapitulate the histological hallmarks, cytological characteristics, and cell lineage differentiation patterns of the original patient GBM from which the GSCs were derived. Also demonstrated is the ability of GSCs to form tumors that consist of a mixture of cytologically heterogenous cells, which is in clear contrast to the xenografts generated with commonly used glioma cell lines (e.g., U87MG), consisting of morphologically monotonous cells.
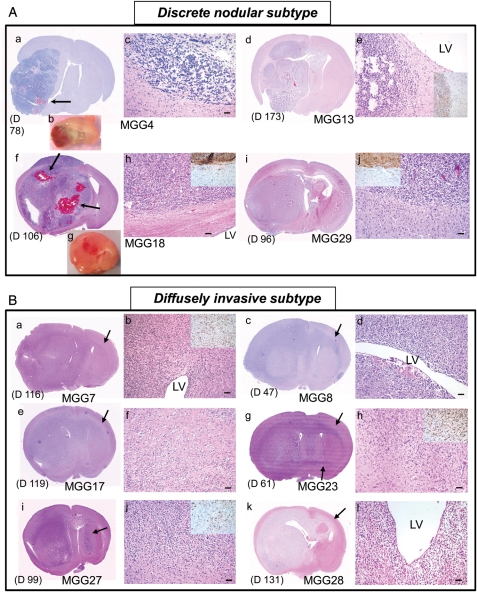
GSC-Derived Xenografts Display Highly Invasive or Discrete Nodular Phenotypes
Extensive invasiveness is a biological characteristic of GBM that renders this tumor extremely difficult to treat. We have previously shown that some GSC-derived xenografts exhibit extensive migratory and invasive behaviors in vivo.15 In the present study, histopathological analysis of the xenografts derived from our 15 GSC lines revealed that GSC-derived tumors can be categorized into 2 subtypes based on degree of invasiveness (Fig. 3, Table 2). First, the discrete nodular subtype involves 4 GSC lines forming a hemispheric tumor mass relatively well demarcated from the surrounding brain (Fig. 3A, Table 2). Although microinvasion a short distance away from the main mass was observed as illustrated by staining for human-specific nestin (insets in Fig. 3A), there was no evidence of extensive cellular migration. Accordingly, such tumors were nearly always confined to the ipsilateral hemisphere (i.e., cell implantation side) and only rarely crossed the midline at the terminal stage of disease. Of interest, these tumors were usually rich in vasculature and associated with frequent intratumoral hemorrhages (Fig. 3A). Except for one case (MGG29), there was no tendency for tumor cells to accumulate in the subventricular zones (Fig. 3A). Second, with the diffusely invasive subtype, 11 GSC lines were markedly invasive locally and diffusely infiltrative of the brain (Fig. 3B, Table 2), with indistinct brain-tumor borders that were validated by staining for human-specific nestin (insets in Fig. 3B). In contrast to the discrete nodular subtype, tumor cells belonging to this invasive subtype showed a consistent tendency to migrate along white matter tracts and spread in the subventricular zones (Fig. 3B). They always extended to the contralateral hemisphere through the corpus callosum or the anterior commissure, which is reminiscent of the characteristic growth pattern of advanced bilateral GBM, termed “butterfly GBM.” The vasculature was not prominent in these tumors, which lacked endothelial proliferation and intratumoral hemorrhage. Thus three-quarters of the GSC lines exhibited the capacity to extensively migrate and invade brain tissue, likely preserving molecular mechanisms underlying primary GBM-associated cellular migration/invasion. GSCs lacking such invasive capacity tended to promote tumor-associated angiogenesis.
Fig. 3.
Invasiveness subtyping of GSC-derived GBM xenografts. (A) Discrete nodular subtype. a-c, MGG4. d, e, MGG13. f-h, MGG18. i, j, MGG29. H & E staining of coronal brain sections, except b and g, which show coronally cut planes of freshly removed brains. These tumors are characterized by relatively delineated borders between tumor mass and brain parenchyma, which was confirmed by immunohistochemistry for human-specific nestin (insets in e, h, and j). This tumor subtype was associated with increased vascularity (b, g) and intratumoral bleeding (arrows in a and f). There is typically no tendency for cells to accumulate in the subventricular zones (adjacent LV) (e, h). (B) Diffusely invasive subtype. a, b, MGG7. c, d, MGG8. e, f, MGG17. g, h, MGG23. i, j, MGG27. k, l, MGG28. These tumors are diffuse and highly invasive, and always extend to the contralateral hemisphere through white matter tracts (shown by arrows). Indistinct tumor-brain interface (f, h, and j; insets in b, h, and j showing human-specific nestin-positive cells on infiltrative edge) and heavy neoplastic infiltration in the subventricular zone (b, d, and l) are other phenotypic characteristics of this subtype of tumor. The timing of animal sacrifice when tumors caused significant symptoms is shown in days. (D) For each panel. LV, lateral ventricle. Bars, 50 µm.
Table 2.
Summary of in vivo phenotype, and the status of PTEN, Akt and MGMT of GSCs
| MGG # | In vivo phenotype |
PTEN | p-Akt (Ser473) | p-Akt (Thr308) | MGMT |
||
|---|---|---|---|---|---|---|---|
| Invasiveness | Pathological feature | Methylation | Protein | ||||
| 4 | N | Endothel. prolif. | + | + | + | M | − |
| 6 | I | Conventional | + | + | + | M | − |
| 7 | I | Conventional | NT | NT | NT | M | NT |
| 8 | I | PNET-like component | + | − | + | M | − |
| 13 | N | Conventional | − | ++ | + | U | + |
| 17 | I | Conventional | NT | NT | NT | NT | NT |
| 18 | N | Giant cell | − | ++ | ++ | M | − |
| 22 | I | Conventional | + | ++ | ++ | U | + |
| 23 | I | Gemistocytic | − | ++ | + | U | + |
| 24 | I | Conventional | + | + | + | U/m | + |
| 27 | I | Conventional | + (weak) | ++ | ++ | U | + |
| 28 | I | Conventional | − | ++ | ++ | U | + |
| 29 | N | Oligo-like component | + | ++ | + | U | − |
| 32 | I | Conventional | NT | NT | NT | U | NT |
| 34 | I | Conventional | + | + | + | U | − |
Abbreviations: N, Nodular; I, Invasive; PNET, Primitive neuroectodermal tumor; +, positive; −, negative; U/m, a major unmethylated band with a weak methylated band; NT, not tested.
Patient MRI Correlates to GSC Phenotypes
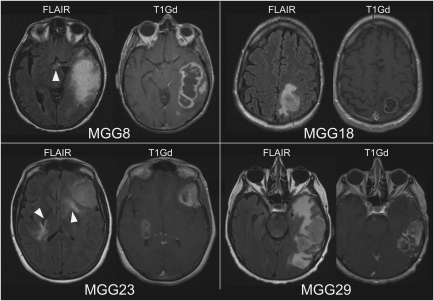
To assess the clinical significance of GSC xenograft phenotypes, we examined whether the preoperative patient MRI findings are predictive of the invasive or noninvasive phenotypes of the corresponding GSCs. Available fluid attenuation inversion recovery (FLAIR) images from the patients that gave rise to diffuse invasive GSCs (MGG8, 23) revealed abnormal increased signal intensities with indistinct borders present at the midline regions or bilateral cerebral hemispheres suggestive of striking diffuse infiltration (Fig. 4 left). In contrast, FLAIR MRI of the patients that produced discrete nodular GSCs showed abnormalities exclusively within the affected side of the hemisphere, with elevated signal intensity areas contiguous within white matter (Fig. 4 right). This indicates a correlation between patient MRI features and the respective GSCs phenotypes and suggests that GSC tumor invasiveness might be predictable by preoperative MRI.
Fig. 4.
Patient preoperative MRIs. MR images taken before surgeries showing MGG8 and MGG23 that produced GSCs of highly invasive phenotype, and MGG18 and MGG29, from which GSCs of discrete nodular phenotype were derived. Arrowheads show hyperintense abnormal lesions in FLAIR images at the midline or bilateral regions suggestive of tumor infiltration. T1Gd, T1 images after infusion of gadolinium contrast agent.
Status of PTEN/PI3kinase Pathways and MGMT in GSCs and Therapeutic Responses
Activation of PI3kinase/Akt signaling has been implicated in increased invasiveness of stem-like GBM cells.28 We examined the expression of PTEN and the activation status of its downstream signaling molecule Akt in 11 of our GSC lines (Table 2, Supplementary material, Fig. S5A). Expression levels of PTEN differed greatly between GSCs, with 4 GSC lines having undetectable protein, which generally correlated with phosphorylated Akt (Supplementary material, Fig. S5A), but GSC invasiveness did not correlate with either PTEN expression or Akt activation status (Table 2, Supplementary material, Fig. S5A).
Because epigenetic status of the gene encoding the DNA repair protein MGMT is associated with prognosis of patients with GBM,2,29 we assessed its status in 14 GSC cultures. Five GSC cultures were MGMT-methylated, 8 cultures were unmethylated, and 1 had a mixture of strong unmethylated band with a weak methylated band (Table 2). Methylation status of the promoter typically correlated with protein expression demonstrated by Western blot (Table 2, Supplementary material, Fig. S5B), but 2 unmethylated GSCs (MGG29 and 34) did not express MGMT protein. In vitro cytotoxicity assay revealed that GSCs with unmethylated MGMT were more resistant to temozolomide than were GSCs with methylated MGMT (P < .05) (Supplementary material, Fig. S5CD). We found no correlation between MGMT promoter methylation and invasiveness in our GSCs (Table 2).
Genomic profiling of GSCs and identification of GSC-associated genetic abnormalities in primary tumors
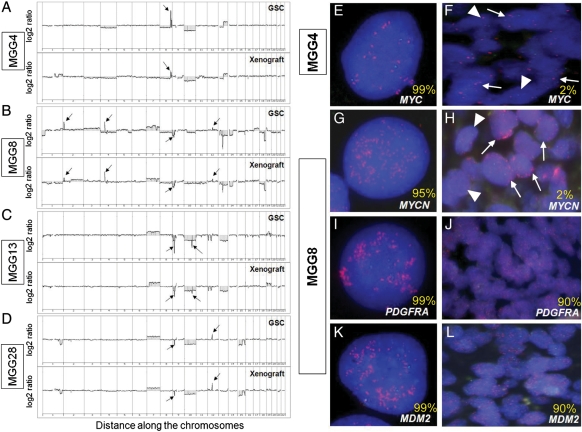
We next sought to identify genomic copy number alterations (CNAs) in GSCs and determine whether gene amplifications can be used to identify GSCs in patient tumor sections. We performed aCGH on 7 cultured GSC lines and the xenograft tumors generated from each line. Representative results for 4 GSCs are shown in Fig. 5A–D, with the CNAs listed in Tables 3 and S1. The genomic profiles varied across GSC lines, and each line harbored distinctive genomic profiles that typically involved focal genomic amplifications and deletions. Gain of chromosome 7 and loss of chromosome 10 and 13 were frequently seen (in 5, 5, and 4 lines, respectively) (Table 3). Homozygous deletion of CDKN2A/B genes on 9p21.3, the most common homozygous genetic loss in GBM and involved in its pathogenesis,5 was observed in 5 of the 7 GSCs tested. In MGG4, high-level amplification of the MYC gene locus on 8q24.2 was detected (Fig. 5A), whereas MGG8 had focal high-level amplification of MYCN (on 2p24.3), along with PDGFRA (on 4q12) and MDM2 (on 12q15) genes (Fig. 5B). Of interest, MYCN amplification has been associated with malignant gliomas with PNET-like components,26 which was the phenotype of MGG8 (Fig. 1H). Homozygous deletion of a locus on 10q23.31, downstream of the PTEN gene, was identified in MGG13 (Fig. 5C). MGG28 was found to carry an amplification of the CDK4 gene on 12q14.1 and homozygous deletion of CDKN2A/B genes (Fig. 5D), an infrequent combination in the commonly inactivated RB pathway in GBM.30 Genomic profiles of the GSC-derived xenograft tumors mirrored the profiles seen in the cultured GSCs (Fig. 5A–D), suggesting that the genetic alterations are necessary for maintenance and tumorigenicity of GSCs.
Fig. 5.
Genomic profiles of GSCs and identification of GSC-associated gene copy abnormalities in primary tumors. (A–D) aCGH analysis reveals gene copy number aberrations in cultured GSCs (upper panels) and xenografts derived from GSCs (lower panels). (A) MGG4, arrow shows amplification of CMYC on 8q24.2. (B) MGG8, arrows show amplifications of MYCN (2p24.3), PDGFRA (4q12), MDM2 (12q15), and homozygous deletion of CDKN2A/B (9p21.3) genes. (C) MGG13, arrows show homozygous deletion of CDKN2A/B genes (9p21.3) and 10q23.31 locus. (D) MGG28, arrows show homozygous deletion of CDKN2A/B (9p21.3) and amplification of CDK4 (12q14) genes. (E–L) FISH analysis of MYC in MGG4 GSCs. (E) and primary tumor (F), and MYCN (G), PDGFRA (I) and MDM2 (K) in MGG8 GSCs and primary tumors (H, J, L). Amplifications are shown by red spots in nuclei. Green spots indicate hybridization with a control probe. In F and H, arrows indicate cells with, and arrowheads without MYC/MYCN amplification. Numbers in yellow denote the percentage of gene-amplified cells in GSCs and primary tumors.
Table 3.
Summary of copy number alterations (CNAs) in GSCs
| GSCs | CNAs |
|---|---|
| MGG4 | −4, −8p, −8q11.21-q24.13, ++8q24.21 (MYC), −8q24.22-q24.23, ++8q24.24, −8q24.24, −10, −13q12.11-q21.31, +13q21.32-q33.4 |
| MGG6 | +1p36-p31.3, −1p31.1, +1p13.2-p12, +1q21.3-q25.3, −1q31.1-q31.3, ++1q32.1 (MDM4, PI3KC2B), +1q32.2-q44, −4q13.1-q13.2, +4q13.3, ++4q21.23-q22.1, +4q22.1-q23, −4q26.3, -6p25.3-p21.31, +6p21.2-p12.1, -6q, +7, ++7p11.2 (EGFR), −9p, −−9p22.3, −−9p21.3 (CDKN2A&B), +9q, +10p, −10q, −13, +20p |
| MGG8 | −1p36.33-p34.3, +1q42.12-q44, ++2p24.3 (MYCN), +2p24.2-p13.1, +2q, +3, −4, ++4q12 (PDGFRA), +5q11.2-q31.1, −5q31.2-q35.3, +7, −8, −9p24.1-p21.1, −−9p21.3 (CDKN2A&B), +10p14-p11.22, −10p11.21, −10q, ++12q15 (MDM2), +12q21.1-q24.22, −13q12.11-q12.12, −13q13.3-q34, −−13q21.1, −14q12-q23.1, −15, +17p11.2, +17q, −18p, 18q11.2-q21.32, −−18q21.33-q23, +19p13.3-p13.1, −20, −21 |
| MGG13 | −1p36.33-p36.22, −1p22.2, +7, −9p24.1-p13.2, −−9p21.3 (CDKN2A&B), −10, −−10q23.31, −11p15.5-p15.4, −11q12.3-q13.2, −12p13.2-p12.3, −12q12-q13.2, −13, +19p, +19q12-q13.31, −19q13.32-q13.33, +19q13.33-q13.43, −20q13.33 |
| MGG18 | +1p, +1q21.1-q32.2, −1q32.2-q42.3, −4p15.31-p13, −4q, −9p13.2-p23, −10, −11, −13, +14, +20, +21, −22 |
| MGG23 | +1p36.33-p31.2, −1p31.1-p22.1, +1p13.3-p12, +1q, −2q37.1, −4, −5p13.3-p13.2, −6, +7, +8, −9p24.3-p13.1, −−9p21.3 (CDKN2A&B), −10, +12, −13, −14q22.1-q24.2, −14q32.32-q32.33, +16p13.3-p12.3, −16p12.3-p12.1, +16p11.2, −16q, +17, −18p, +21 |
| MGG28 | −1q32.2-q42.2, +7, −−9p21.3 (CDKN2A&B), −10, ++12q14.1 (CDK4), −15q11.2-q22.3, −15q25.2-q26.3 |
Abbreviations: + gain, − loss, ++ amplification, −− homozygous deletion.
We then determined whether cells carrying the gene amplifications identified in vitro by aCGH in GSCs are present in the respective patient tumors. FISH analysis using specific probes targeting MYC in MGG4 GSCs, and MYCN, PDGFRA, and MDM2 genes in MGG8 GSCs identified high levels of gene amplification in 95%–99% of GSCs (Fig. 5E, G, I, and K), confirming the aCGH results. In situ analysis using FFPE primary tumor sections from MGG4 and MGG8 patient tumors revealed that amplification of MYC (in MGG4) and MYCN (in MGG8) was present, but only in scattered clusters of tumor cells (∼2%) within the tumor sections (Fig. 5F and H), with the majority of tumor cells not harboring these gene amplifications. However, FISH analysis of PDGFRA and MDM2 genes in the primary MGG8 tumor showed high-level amplifications in ∼90% of the tumor cells (Fig. 5J and L). Of note, the patient specimens were from different regions of the tumor than the regions used for GSCs isolation. These results indicate that (1) the gene copy number aberrations found in GSCs are the result of genetic abnormalities that have accumulated in the patient tumors, not an artifact produced by in vitro culture of cells, and (2) some of the genetic abnormalities in GSCs are limited to an infrequent population of tumor cells in the parent tumors.
Discussion
The cancer stem cell model posits a functional cellular hierarchy in cancer in which cancer stem cells are a subpopulation that gives rise to the bulk of tumor.7,31,32 In this model, the application of established principles from adult stem cells and developmental biology to cancer can provide an explanation for heterogeneous cell populations in GBM and for tumor relapses after apparently successful elimination of radiologically defined disease. During the past several years, knowledge about the molecular pathways regulating important functions of GSCs and the potential presence of molecularly distinct subtypes of GSCs has accumulated.11,21 One of the areas poorly explored thus far concerns the relationship between individual GSCs and the GBM tumor from which they were isolated: whether and to what extent GSCs embody a given GBM tumor. To address this, we performed both phenotypic and genomic characterization studies comparing GSCs and their orthotopic xenografts with their respective primary patient tumors.
In this study, we generated a panel of GSC lines that were isolated from newly diagnosed GBMs. Our phenotypic characterization of xenograft pathological features found that the majority (11 of 15) of GSCs display highly invasive behavior, whereas only a quarter displayed a nodular phenotype with minimal invasiveness. Similarly, Gunther et al. found that 5 of 7 GSCs generated tumors with a diffuse invasive phenotype, whereas the other 2 produced solid masses. They further described a positive correlation between CD133 expression and tumor invasiveness.18 Chen et al. reported that only GSCs that give rise to CD133-positive cells generate invasive intracerebral xenografts.11 We did not identify an association between levels of CD133 and invasiveness in our GSCs15 (data not shown). Nevertheless, these observations suggest that most GSCs preserve a highly migratory capacity, analogous to neural stem cells.33 This is in contrast to commonly used glioma cell lines cultured in serum that typically fail to display an invasive growth pattern in vivo.34 Examination of PTEN and the PI3K/Akt pathway status also did not reveal any correlation of this pathway to GSC invasiveness, but the role of this pathway in GSC invasion may differ between GSC lines. Among current imaging modalities, MRI offers a practical clinical tool to evaluate GBM invasiveness,35 and imaging features can be predictive for molecular subtypes of GBM.36 In this study, the preoperative MRIs of selected patients revealed features that correlated with the invasiveness of GSC-derived xenografts. This implicates a role for GSCs in shaping progression patterns or guiding invasion of patient GBMs, uncovering an important bench-to-bedside link in GSC biology, which warrants further investigation.
We showed that GSC-generated intracranial xenografts phenocopy histopathological characteristics displayed by the GBM from which the cells were isolated. Published work from our group and others suggest that GSCs are capable of recapitulating the cardinal pathological features of GBM, such as invasiveness and hypervascularity, and, thus, provide clinically relevant models of GBM. However, our findings uncovered an unknown aspect of GSC biology: GSC-derived tumors recapitulate the histopathology of individual GBMs, with resulting respective histological subtypes. We demonstrated this through a case-by-case examination of the histopathology of GSC-derived xenografts, compared with the original GBMs that were diagnosed as containing different histological components (e.g., oligodendroglial or PNET-like) or classified as GBM subtypes or variants because of specific cytological features (e.g., giant cell or gemistocytic). Typically, gemistocytic GBMs and GBMs with oligodendroglial features are highly invasive,4 like MGG23 and MGG7, whereas giant cell GBMs are, more often than other GBMs, well-circumscribed masses with a high frequency of PTEN deletion and rare CDKN2A deletion,4 as in MGG18. Furthermore, we demonstrated that MGG18 GSC xenografts exhibit extensive cellular heterogeneity comprising cells ranging from multinucleated large cells to much smaller glioma cells. This supports the view that GSCs are multipotent, with the ability to differentiate into multiple cell lineages or stages of cellular differentiation. Thus, GSCs are not only able to initiate tumors in the brains of mice, but likely play a major role in the propagation of human GBM, giving rise to cells with different morphology that ultimately define the pathological features and diagnosis of each GBM. In addition, our findings indicate that GSCs will be useful for creating models that are representative of the particular tumor from which the GSCs were isolated. Present work also revealed a limitation with regard to GSC recapitulation of human GBM pathology, because necrosis, a pathological feature important for the diagnosis of GBM, is typically lacking in GSC xenografts despite our previous report of a single example of GSC xenograft necrosis.15 We speculate that this is most likely a result of the size limitation of tumors in the mouse brains that hampers the development of a larger hypoxic region necessary for generating necrosis. Of importance, GSCs retain their specific histopathological characteristics after culturing in vitro and serial transplantation in vivo (Supplementary material, Fig. S3). Another GBM model, which retains invasiveness and amplification of EGFR and PDGFRA genes, was established by subcutaneous implantation of patient GBM tissue to nude mice and serial transplantation.37,38 The need to passage these xenograft lines as mouse flank tumors, and the heterogeneous nature of the material implanted may complicate its use as a preclinical model. Although the impact of prolonged in vitro culture of GSCs on the maintenance of GSC biology and genomic stability needs to be addressed,17 the GSC-based GBM models offer a superb opportunity to test novel therapeutic agents both in vitro and in vivo and to guide personalized treatment after surgical removal of tumor. Furthermore, we describe new models for less-frequent GBM variants, such as giant cell GBM, which will further our understanding of the biology of less characterized tumor types.
Genomic profiling of our GSCs identified selective gene CNAs that include those both unique to each GSC and widely recognized in GBM pathogenesis. Of note, we found high-level amplification of 2 genes belonging to the MYC transcription factor family, MYC and MYCN, in 2 of the 7 GSCs tested. The TCGA study and a recent report compiling published data on GBM genomics found high-level focal amplification of MYCN and MYC in only ∼2% of tumors.5,39 c-Myc has been shown to play a role in GSC maintenance40–42 and is one of the critical factors for reprogramming somatic cells to iPS cells.42 Therefore, it is conceivable that over-expression of c-Myc and N-myc proteins endows these GSCs with stem-like properties that promote GSC maintenance and/or survival. Our in situ analysis on primary GBM tumors demonstrated the presence of these gene amplifications in patient tumor cells, although only in a subset of tumor cells. This indicates that the amplifications are not an artificial product that arose in culture and suggests the preferential in vitro outgrowth of cells with amplification of MYC and MYCN in the neurosphere culture conditions.
We show that the CNAs highly enriched in GSCs can vary in frequency to those found in the patient GBM (Fig. 5E–L). There are a number of possibilities that may explain the observation that neurosphere-forming GSCs contain unique and infrequent CNAs, such as myc amplification (Supplementary material, Fig. S6). First, tumors including GBM may contain multiple genetically heterogeneous subsets of GSCs, and a particular subset(s) of GSC may be more efficient at proliferation in the culture conditions used. Supporting this, 2 types of GSCs with distinct genetic abnormalities and tumorigenic potential have been found in the same GBM tumor.43 Second, the myc amplification events may have occurred at a later stage of tumorigenesis and/or the bulk tumor cells were derived from an earlier and less malignant GSC with fewer genomic abnormalities. Cancer stem cells in leukemia and breast cancer are known to undergo clonal evolution through additional genetic mutation to more aggressive phenotypes.44,45 Recently, the presence of genetically and functionally diverse subclones of leukemia-initiating cells has been demonstrated in individual patients, and a complex evolutionary process seems to be operative in generating the diversity of cancer stem cell subclones.46,47 Third, intratumoral heterogeneity in distribution of cells with specific genotypes may have influenced the result because the tumor fragment used for GSC isolation is different from the pathology specimens used for FISH, and both are likely representing only a small portion of the whole tumor. Another possibility, although less likely, may be that as tumor progresses, certain genetic CNAs, such as myc, may be unstable and lost in many of the bulk tumor cells. Because cutting-edge research in the field suggests that the hierarchical model for cancer stem cells and the stochastic model of clonal evolution of cancer are not mutually exclusive, we speculate that our result reflects the possibility that the combination of clonal evolution and genetic heterogeneity of cancer stem cells is in effect in GBM (Supplementary Fig. S6). However, the limited number of cases tested and the genes identified (MYC and MYCN) prevent us from drawing definitive conclusions. It will be important to determine whether chromosomal instability is a feature of GSCs in situ and whether GSCs typically contain genetic alternations that are infrequent in the tumor or whether this feature is limited to gene amplifications of the myc family.
Our findings also indicate that unique genetic CNAs identifiable in GSCs can serve as genetic markers to identify putative GSCs within a particular tumor. Specific CNAs, focal amplification in particular, identified in GSCs from a given GBM will be useful to localize GSCs in situ on primary tumor sections, which will allow assessment of GSC frequency and their physical relation to other cells within a GSC niche microenvironment. Furthermore, activation or loss of the encoded proteins and downstream signaling pathways caused by these genetic alterations likely contributes to survival, proliferation, and maintenance of GSCs. Silencing or inhibition of the amplified genes and overexpression of the deleted genes in GSCs should elucidate their functional significance and identify patient-specific therapeutic targets. One caveat is the possibility that GSC populations within a tumor are genetically heterogeneous (as discussed in the previous paragraph),43 and our culturing method selectively expanded subpopulation(s) of GSCs that carry the genetic alterations we found in this study. However, there was no consistent genetic alteration across the majority of GSC lines, except for those frequently observed in GBM.
In conclusion, these studies highlight the value of using GSCs for establishing preclinical models of GBM that are representative of the disease and recapitulating the features of individual patient tumors. This paves the way for testing novel anti-GSC/GBM agents in the context of individual patients and, thus, may facilitate developing personalized therapy. Our work also identifies GSCs as a genetically distinct subpopulation of neoplastic cells in a given GBM tumor and suggests a need to validate the usefulness of such genetic alterations for diagnostic and therapeutic purposes.
Supplementary Material
Funding
This work was supported by the National Institute of Health (R01NS032677 to R.L.M., R21NS067541 to H.W., R01CA57683 to D.N.L., and P01NS024279 to A.O.S.) and the Department of Defense (W81XWH-07-1-0359 to S.D.R.).
Conflict of interest statement. None declared.
Supplementary Material
Acknowledgments
We thank James C. Kim for help with sample collection for this study.
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. doi:10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. doi:10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. doi:10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Miller CR, Perry A. Glioblastoma. Arch Pathol Lab Med. 2007;131:397–406. doi: 10.5858/2007-131-397-G. [DOI] [PubMed] [Google Scholar]
- 5.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. doi:10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. doi:10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 8.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. doi:10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 9.Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. doi:10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 10.Beier D, Hau P, Proescholdt M, et al. CD133(+) and CD133(-) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. doi:10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 11.Chen R, Nishimura MC, Bumbaca SM, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17:362–375. doi: 10.1016/j.ccr.2009.12.049. doi:10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 12.Joo KM, Kim SY, Jin X, et al. Clinical and biological implications of CD133-positive and CD133-negative cells in glioblastomas. Lab Invest. 2008;88:808–815. doi: 10.1038/labinvest.2008.57. doi:10.1038/labinvest.2008.57. [DOI] [PubMed] [Google Scholar]
- 13.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. doi:10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 14.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. doi:10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wakimoto H, Kesari S, Farrell CJ, et al. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res. 2009;69:3472–3481. doi: 10.1158/0008-5472.CAN-08-3886. doi:10.1158/0008-5472.CAN-08-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.deCarvalho AC, Nelson K, Lemke N, et al. Gliosarcoma stem cells undergo glial and mesenchymal differentiation in vivo. Stem Cells. 2010;28:181–190. doi: 10.1002/stem.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vik-Mo EO, Sandberg C, Olstorn H, et al. Brain tumor stem cells maintain overall phenotype and tumorigenicity after in vitro culturing in serum-free conditions. Neuro Oncol. 2010;12:1220–1230. doi: 10.1093/neuonc/noq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunther HS, Schmidt NO, Phillips HS, et al. Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene. 2008;27:2897–2909. doi: 10.1038/sj.onc.1210949. doi:10.1038/sj.onc.1210949. [DOI] [PubMed] [Google Scholar]
- 19.Li A, Walling J, Kotliarov Y, et al. Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol Cancer Res. 2008;6:21–30. doi: 10.1158/1541-7786.MCR-07-0280. doi:10.1158/1541-7786.MCR-07-0280. [DOI] [PubMed] [Google Scholar]
- 20.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. doi:10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 21.Lottaz C, Beier D, Meyer K, et al. Transcriptional profiles of CD133+ and CD133- glioblastoma-derived cancer stem cell lines suggest different cells of origin. Cancer Res. 2010;70:2030–2040. doi: 10.1158/0008-5472.CAN-09-1707. doi:10.1158/0008-5472.CAN-09-1707. [DOI] [PubMed] [Google Scholar]
- 22.Ernst A, Hofmann S, Ahmadi R, et al. Genomic and expression profiling of glioblastoma stem cell-like spheroid cultures identifies novel tumor-relevant genes associated with survival. Clin Cancer Res. 2009;15:6541–6550. doi: 10.1158/1078-0432.CCR-09-0695. doi:10.1158/1078-0432.CCR-09-0695. [DOI] [PubMed] [Google Scholar]
- 23.Esteller M, Hamilton SR, Burger PC, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 24.Gabeau-Lacet D, Engler D, Gupta S, et al. Genomic profiling of atypical meningiomas associates gain of 1q with poor clinical outcome. J Neuropathol Exp Neurol. 2009;68:1155–1165. doi: 10.1097/NEN.0b013e3181ba3952. doi:10.1097/NEN.0b013e3181ba3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohapatra G, Betensky RA, Miller ER, et al. Glioma test array for use with formalin-fixed, paraffin-embedded tissue: array comparative genomic hybridization correlates with loss of heterozygosity and fluorescence in situ hybridization. J Mol Diagn. 2006;8:268–276. doi: 10.2353/jmoldx.2006.050109. doi:10.2353/jmoldx.2006.050109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perry A, Miller CR, Gujrati M, et al. Malignant gliomas with primitive neuroectodermal tumor-like components: a clinicopathologic and genetic study of 53 cases. Brain Pathol. 2009;19:81–90. doi: 10.1111/j.1750-3639.2008.00167.x. doi:10.1111/j.1750-3639.2008.00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. World Health Organization Histological Classification of Tumours of the Central Nervous System. Lyon: International Agency for Research on Cancer; 2007. [Google Scholar]
- 28.Molina JR, Hayashi Y, Stephens C, et al. Invasive glioblastoma cells acquire stemness and increased Akt activation. Neoplasia. 2010;12:453–463. doi: 10.1593/neo.10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. doi:10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 30.Wiedemeyer WR, Dunn IF, Quayle SN, et al. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proc Natl Acad Sci USA. 2010;107:11501–11506. doi: 10.1073/pnas.1001613107. doi:10.1073/pnas.1001613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho RW, Clarke MF. Recent advances in cancer stem cells. Curr Opin Genet Dev. 2008;18:48–53. doi: 10.1016/j.gde.2008.01.017. doi:10.1016/j.gde.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 32.Shackleton M, Quintana E, Fearon ER, et al. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138:822–829. doi: 10.1016/j.cell.2009.08.017. doi:10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 33.Picard-Riera N, Nait-Oumesmar B, Baron-Van Evercooren A. Endogenous adult neural stem cells: limits and potential to repair the injured central nervous system. J Neurosci Res. 2004;76:223–231. doi: 10.1002/jnr.20040. doi:10.1002/jnr.20040. [DOI] [PubMed] [Google Scholar]
- 34.Candolfi M, Curtin JF, Nichols WS, et al. Intracranial glioblastoma models in preclinical neuro-oncology: neuropathological characterization and tumor progression. J Neurooncol. 2007;85:133–148. doi: 10.1007/s11060-007-9400-9. doi:10.1007/s11060-007-9400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramakrishna R, Barber J, Kennedy G, et al. Imaging features of invasion and preoperative and postoperative tumor burden in previously untreated glioblastoma: Correlation with survival. Surg Neurol Int. 2010;1:40. doi: 10.4103/2152-7806.68337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aghi M, Gaviani P, Henson JW, et al. Magnetic resonance imaging characteristics predict epidermal growth factor receptor amplification status in glioblastoma. Clin Cancer Res. 2005;11:8600–8605. doi: 10.1158/1078-0432.CCR-05-0713. doi:10.1158/1078-0432.CCR-05-0713. [DOI] [PubMed] [Google Scholar]
- 37.Pandita A, Aldape KD, Zadeh G, et al. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosomes Cancer. 2004;39:29–36. doi: 10.1002/gcc.10300. doi:10.1002/gcc.10300. [DOI] [PubMed] [Google Scholar]
- 38.Giannini C, Sarkaria JN, Saito A, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro Oncol. 2005;7:164–176. doi: 10.1215/S1152851704000821. doi:10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao SK, Edwards J, Joshi AD, et al. A survey of glioblastoma genomic amplifications and deletions. J Neurooncol. 2010;96:169–179. doi: 10.1007/s11060-009-9959-4. doi:10.1007/s11060-009-9959-4. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Wang H, Li Z, et al. c-Myc is required for maintenance of glioma cancer stem cells. PLoS One. 2008;3:e3769. doi: 10.1371/journal.pone.0003769. doi:10.1371/journal.pone.0003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suva ML, Riggi N, Janiszewska M, et al. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009;69:9211–9218. doi: 10.1158/0008-5472.CAN-09-1622. doi:10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- 42.Zheng H, Ying H, Yan H, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. doi:10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piccirillo SG, Combi R, Cajola L, et al. Distinct pools of cancer stem-like cells coexist within human glioblastomas and display different tumorigenicity and independent genomic evolution. Oncogene. 2009;28:1807–1811. doi: 10.1038/onc.2009.27. doi:10.1038/onc.2009.27. [DOI] [PubMed] [Google Scholar]
- 44.Barabe F, Kennedy JA, Hope KJ, et al. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007;316:600–604. doi: 10.1126/science.1139851. doi:10.1126/science.1139851. [DOI] [PubMed] [Google Scholar]
- 45.Park SY, Gonen M, Kim HJ, et al. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120:636–644. doi: 10.1172/JCI40724. doi:10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson K, Lutz C, van Delft FW, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. doi:10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 47.Notta F, Mullighan CG, Wang JC, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–367. doi: 10.1038/nature09733. doi:10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.