Abstract
While many cancers show a sex bias, the genetic basis and molecular mechanisms underlying sex bias are not always clear. Astrocytoma and glioblastoma show male predominance in humans. We have shown previously that glial tumors forming in the Nf1−/+; Trp53−/+cis (NPcis) mouse model also show a sex bias in some genetic contexts. Using cross-species comparisons we have identified candidate male-specific modifiers of astrocytoma/glioblastoma. Linkage analysis of B6X(B6X129)-NPcis mice identifies a modifier of astrocytoma resistance specific to males, named Arlm1, on distal mouse Chr 12. Arlm1 is syntenic to human Chr 7p15, 7p21, 7q36, and 14q32 regions that are altered in human glioblastoma. A subset of these genes shows male-specific correlations to glioblastoma patient survival time and represents strong candidates for the Arlm1 modifier gene. Identification of male-specific modifier genes will lead to a better understanding of the molecular basis of male predominance in astrocytoma and glioblastoma.
Keywords: astrocytoma, glioblastoma, modifier, sex differences
Astrocytic gliomas, including astrocytoma and glioblastoma multiforme (GBM), are the most common primary tumors of the CNS in adults and are currently incurable. Both astrocytoma and GBM show male predominance in the population, with a male to female ratio of 1.31:1 for astrocytoma and 1.26:1 for GBM.1 A better understanding of the susceptibility factors for astrocytoma/GBM and the differences between males and females may lead to better approaches for prevention and treatment. Genome-wide association studies (GWASs) are beginning to enlighten the genetic risk factors for astrocytoma and GBM2–4; however, the heterogeneity of the human population makes it difficult to find risk factors affecting subpopulations.
Mouse models of cancer offer a complementary approach to finding genetic susceptibility factors to human GWASs. Mice can be bred as homogeneous populations under controlled environmental conditions to identify cancer risk modifiers using smaller populations. These identified loci can be tested for their relevance to human disease. We have shown previously that tumorigenesis in the NPcis mouse model is dependent on genetic background and is additionally influenced by sex and parental inheritance of the NPcis mutation.5–9 We are using this model of both astrocytoma/GBM and malignant peripheral nerve sheath tumors to understand how genetic factors, epigenetic factors, and sex interact to determine an individual's risk for nervous system tumors. We can compare these factors with human data on GBM to identify promising candidates for further study.
The NPcis mouse model carries mutations in the Nf1 gene, encoding the RasGAP protein neurofibromin, and the Trp53 gene, encoding the tumor suppressor p53 (the TP53 gene in humans), together in cis on mouse Chr 11. Both NF1 and TP53 are implicated in sporadic human GBM,10,11 and this mouse model resembles human secondary GBM progressing from astrocytoma, as well as the mesenchymal subtype of GBM involving mutations in NF1.12,13 NPcis mice on the 129S4/SvJae (129) strain are resistant to astrocytoma and GBM compared with NPcis mice on the C57BL/6J (B6) strain.6,7 Although B6X129 F1 hybrids from reciprocal crosses are genetically identical, they show varying susceptibility depending on how the NPcis mutation and the strain are inherited from the parents.7 This suggests that susceptibility polymorphisms interact with the inheritance of the NPcis mutation on Chr 11 from the mother or the father or with additional parent-of-origin effects. These findings point to an interaction of genetic factors (strain background) with epigenetic factors (inheritance of genes from the mother or father) in determining the overall susceptibility to astrocytoma/GBM.
In addition to a genetic-by-epigenetic interaction, we have also found an interaction between the sex of the mouse and whether the mutant Chr 11 is inherited from the mother or the father (sex-by-epigenetic interaction). In inbred B6-NPcis mice where the NPcis mutation is inherited from the father (NPcispat), males and females have a similar incidence and grade spectrum of astrocytoma, with 52% of females and 49% of males developing astrocytoma/GBM.8,9 In B6-NPcis mice where the mutation is inherited from the mother (NPcismat), there is a gender bias in astrocytoma incidence and grade. Females develop 51% astrocytoma/GBM, with roughly a quarter of the tumors scored as GBM, whereas males develop more astrocytomas overall (71%) but no GBM.9 These data show that the increased incidence of astrocytoma in males is dependent on the inheritance of the parental NPcis allele. We therefore have examined the interaction between sex and genetic factors (sex-by-genetic interactions) in a backcross of B6 and 129 strains where the parental inheritance of the NPcis mutation is held constant.
Materials and Methods
Mouse Breeding
The C57BL/6J and 129S4/SvJae strains were used for breeding experiments. We described the B6-NPcispat and 129-NPcispat datasets previously.6–9 The B6-NPcispat cohort consists of 45 males and 47 females. The 129-NPcispat cohorts consist of 18 males and 16 females. We generated the F1(B6X129)-NPcispat cohort by crossing wild-type B6 females to 129-NPcis males and examined 25 males and 32 females. This cohort includes 23 F1 mice reported previously.7 We generated the B6X(B6X129)-NPcispat cohort by crossing wild-type B6 females to the B6X129-NPcispat F1 males. The B6X(B6X129)-NPcispat cohort consists of 28 males and 30 females. Mice were maintained at the National Cancer Institute–Frederick according to the guidelines and regulations of the Institutional Animal Care and Use Committee.
Phenotyping of Astrocytoma
We aged and euthanized mice according to predetermined criteria, as described previously,8 except that only visible masses, brain, and spinal cord were collected for histology. Sagittal sections of the brain and spinal cord, as well as cross sections of spinal cord at the cervical, thoracic, lumbar, and caudal levels, were scored for the presence of astrocytoma World Health Organization (WHO) grades II–IV by K.M.R. and independently scored by the Pathology/Histology Laboratory at Science Applications International Corporation.
Genome-wide SNP Genotyping
We prepared tail DNA using the Promega Wizard SV Genomic DNA Purification System. We concentrated the DNA by ethanol precipitation to a range of 5 ng/µL to 155 ng/µL, with 75 ng/µL to 150 ng/µL considered optimal. Thirty-five µL of DNA was plated according to Center for Inherited Disease Research (CIDR) standard procedures and sent to CIDR for genotyping using the Illumina 1440K single nucleotide polymorphism (SNP) panel containing 880 SNPs polymorphic for B6 and 129.
Statistical Analysis of Linkage
We performed binary trait interval mapping using the genetic mapping software R/qtl14 to identify the locations of quantitative trait loci (QTLs). Statistical significance was determined by permutation testing with 1,000 replicates. A significance threshold of 0.05 was used to declare statistical significance. We analyzed males and females separately.
Bioinformatic Analysis of Mouse and Human Data
We identified polymorphic loci between B6 and 129 in the Arlm1 region using the SNP Wizard of the Mouse Phenome Database (http://phenome.jax.org/SNP/), accessing data from mouse genome build 37.1/mm9, dbSNP128, and Ensembl 48. We considered loci that were polymorphic between B6 and either 129S1 or 129X1. We analyzed publicly available gene expression in 10- to 12-week-old male mouse brains (http://phenogen.ucdenver.edu/PhenoGen/web/datasets/geneData.jsp) using Excel. We calculated the difference in mean expression level between C57BL/6J and 129S1/SvImJ for each probe corresponding to a polymorphic gene in Arlm1. Probes that were up- or downregulated ≥1.5-fold were considered candidate hits. We identified human homologs of the polymorphic Arlm1 genes and chromosome cytobands using the National Center for Biotechnology Information (NCBI) Homology Maps Viewer (http://www.ncbi.nlm.nih.gov/projects/homology/maps/) and NCBI Unigene (http://www.ncbi.nlm.nih.gov/unigene), accessing mouse genome build 37.1 and human genome build 37.2. We assessed chromosome aberrations in human astrocytomas and GBM using NCBI Cancer Chromosomes Mittleman Data (http://www.ncbi.nlm.nih.gov/sites/entrez?db=cancerchromosomes), excluding cases of pilocytic astrocytoma. The effect of changes in gene expression levels on patient survival time or age of diagnosis was determined using Kaplan–Meier analysis in the Repository of Molecular Brain Neoplasia Data (REMBRANDT; https://caintegrator.nci.nih.gov/rembrandt/; Application Release 1.5.5, 7/27/10) and the Cancer Molecular Analysis (CMA) portal (https://cma.nci.nih.gov/cma-tcga/; Version 2.0, 2/23/09), subdividing patient data by sex and by whether the gene of interest was 2-fold over- or underexpressed. Of the 100 genes polymorphic between B6 and 129 in the Arlm1 locus, we identified 64 human homologs, of which 58 had probe sets available in REMBRANDT and 53 had probe sets available in The Cancer Genome Atlas (TCGA). We examined gene amplification or deletion using the Cancer Genome Workbench Heatmap Viewer (https://cgwb.nci.nih.gov/cgi-bin/heatmap; Update 9/2/10) to analyze the REMBRANDT AST project (18 female, 28 male, 10 sex unknown samples), the REMBRANDT GBM project (17 female, 59 male, 53 sex unknown samples), and the TCGA project (793 combined, unpaired samples), with an amplification threshold of 1. Copy number variation was also determined using Cancer Genome Workbench Heatmap Viewer.
Reverse Transcription qPCR for Crip2 and Cdca7l
RNA was isolated from the brains of age-matched wild-type B6 and 129 mice or from cultured tumor cell lines using TRIzol (Invitrogen Life Technologies) and treated with TURBO DNAfree (Ambion). Mouse tumor cell lines were described previously,15 and only NPcispat cell lines were used for analysis. RNA was reverse-transcribed using SuperScript II (Invitrogen Life Technologies). Quantitative (q)PCR was carried out on the Mx3000P qPCR machine (Agilent Technologies), using Brilliant SYBR Green master mix (Agilent Technologies): for Cdca7l (forward primer: 5′-GATAAGCCATGGTGTTTTGA-3′; reverse primer: 5′-CTACTGCTGGAAGCATGAAC-3′) and Crip2 (forward primer: 5′-GACTGGCACAAGTTCTGTCT-3′; reverse primer: 5′-CTGAGGCTTCTCGTAGATGT-3′). Whole brain experiments were normalized to Snrp70 levels (forward primer: 5′-CTCCTCCTCCAACAAGAGCAG-3′; reverse primer: 5′-CGATGAAGGCATAACCACG-3′), and cell line experiments were normalized to EIF4H levels (forward: 5′-GGCTAGTCAGAGACAAAGACACAG-3′; reverse: 5′-ATGTCCACACGAAGTGACCG-3′).
Results
Characterization of B6X(B6X129)-NPcis Progeny Compared with B6-NPcis, 129-NPcis, and B6X129-NPcis Progeny
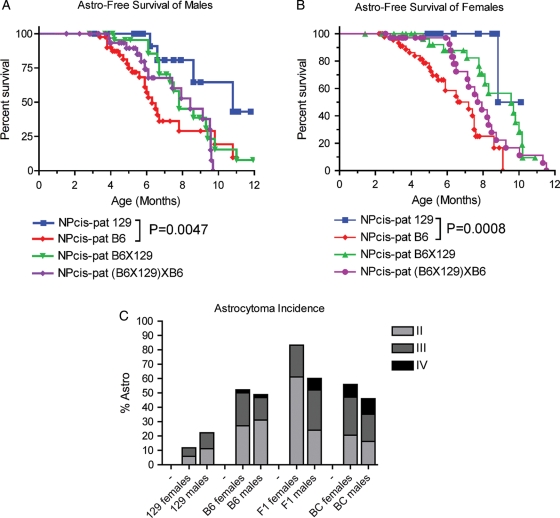
We compared the astrocytoma/GBM-free survival of the NPcispat F1(B6X129) hybrid (F1) and the B6X(B6X129) (BC) backgrounds with mice on the parental B6 and 129 strains to better understand the genetics of susceptibility. By comparing F1 animals with the parental lines, we can determine whether the B6 or 129 strain is dominant over the other strain. 129-NPcispat mice show a significant delay in astrocytoma latency compared with B6-NPcispat mice in both males (P = .0047, log-rank test) and females (P = .0008, log-rank test). When the B6 and 129 strains are intercrossed, both the F1 and BC backgrounds show intermediate survival in males (Fig. 1A) and females (Fig. 1B), suggesting co-dominance of the B6 and 129 strains in determining the timing of astrocytomas in the population. F1 mice show an increased incidence of astrocytoma compared with either parental strain (Fig. 1C). This suggests that the B6 strain also carries recessive resistance alleles for astrocytoma incidence, such that when only one B6 allele is present in the F1, susceptibility is increased.
Fig. 1.
Comparison of astrocytoma-free survival and incidence for male and female NPcis mice on the B6, 129, B6X129, and B6X(B6X129) strain backgrounds. (A) Astrocytoma-free survival for NPcis males. (B) Astrocytoma-free survival for females. (C) Astrocytoma incidence by grade for NPcis males and females.
In addition to the co-dominance of the B6 and 129 strains, we also observed sex-specific differences in the F1 mice. Whereas the NPcispat mice on the 129 or B6 backgrounds show similar incidence and grades of astrocytoma in males and females, the F1 mice show greater divergence between the sexes. F1 females develop more astrocytoma than the parental strains, whereas F1 males develop higher-grade tumors (Fig. 1C). BC mice show similar incidence of astrocytoma as the B6 parental strain, although the grades of astrocytoma may be somewhat higher in BC mice than in B6 mice. In summary, these data demonstrate the complex genetics underlying astrocytoma latency, incidence, and grade in the NPcis model on the 129 and B6 strains.
Mapping of Susceptibility to Astrocytoma in B6X(B6X129)-NPcispat Mice
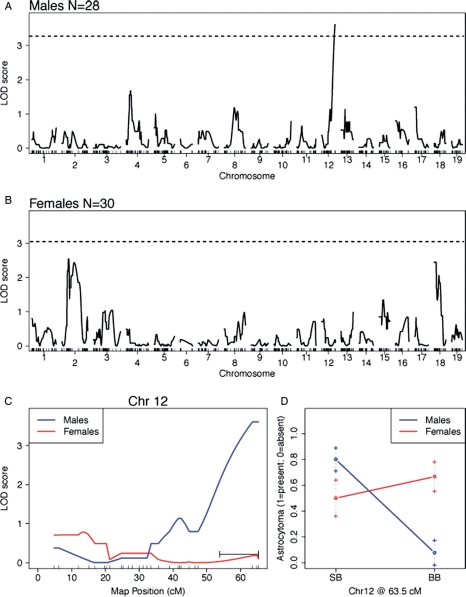
We mapped the presence or absence of astrocytoma (WHO grades II–IV, including GBM) as a binary trait in B6X(B6X129)-NPcispat mice. Males and females were considered separately and combined, but no increase in linkage was observed in the combined sexes (data not shown), suggesting that genetic susceptibility to astrocytoma is linked to different loci in males (Fig. 2A) and females (Fig. 2B). A significant linkage peak (LOD 3.68; P = .024 genome-wide) was found on distal Chr 12 in males (Fig. 2A and C). This locus has no effect on female susceptibility to astrocytoma (Fig. 2C and D), whereas males carrying the B6/B6 allele were resistant to astrocytoma (Fig. 2D) compared with males carrying the B6/129 allele. The peak is located at 63.5 cM on the far distal end of Chr 12. The 1.5-LOD support interval, which defines the region of interest, covers 53–63 cM on Chr 12. We have named this the astrocytoma resistance locus in males 1 (Arlm1).
Fig. 2.
Linkage analysis for astrocytoma resistance in B6X(B6X129)-NPcis males and females. A graph of the genome-wide linkage analysis LOD scores for males is shown in (A) and for females in (B). The dotted line indicates the 0.05 alpha thresholds for statistical significance using permutation testing. (C) Linkage analysis of Chr 12 for both males (blue) and females (red). (D) The effect size of Arlm1B6/129 (SB) and Arlm1B6/B6 (BB) at 63.5 cM for males and females.
Arlm1 Affects Astrocytoma Incidence, but not Latency, in Males
We analyzed the B6X(B6X129)-NPcispat progeny based on their genotype at Arlm1. Arlm1B6/B6 NPcispat males developed significantly fewer astrocytomas (8%; n = 13; P = .0002, Fisher's 2-tailed test) than Arlm1B6/129 NPcispat males (80%; n = 15) (Fig. 2D). Arlm1B6/B6 NPcispat females developed similar numbers of astrocytomas (65%; n = 17) as Arlm1B6/129 NPcispat females (54%; n = 13) (Fig. 2D). In contrast, the astrocytomas forming in Arlm1B6/B6 males arise at the same age as those in Arlm1B6/129 males, with no difference in astrocytoma-free survival in either males (Supplementary Fig. S1A) or females (Supplementary Fig. S1B). These data are consistent with Arlm1 being a recessive resistance locus in B6 acting on astrocytoma incidence in males.
Bioinformatic Analysis of Arlm1 in Mouse and Human to Prioritize Candidates
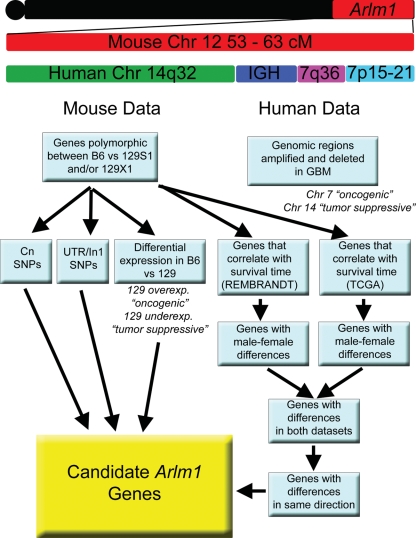
We are interested in comparing the Arlm1 locus identified in mouse with human risk factors for astocytoma and GBM. The region syntenic to Arlm1 has not yet been reported as a significant risk locus in GWASs of glioma (www.genome.gov/GWAStudies)16; however, this may be due to pooling of males and females in the analysis. We therefore took a broader approach to identify candidate Arlm1 genes likely to have a role in glioma based on a variety of data (Fig. 3). We used bioinformatic approaches combining mouse haplotype analysis, mouse strain–specific brain expression, human chromosomal synteny, and GBM patient data (summarized in Table 1 and described below) to prioritize Arlm1 candidate genes. For GBM patient data, we focused on genes within tumors that may alter the survival time of GBM patients as a surrogate for genes that affect GBM prior to tumor formation. These candidates can be tested directly in association studies for their role in glioma susceptibility in future studies.
Fig. 3.
Diagram of candidate gene analysis. The region of the Arlm1 locus was defined and compared to human syntenic genome regions. The genes in the region were examined in both mouse and human data. Cross-species comparisons were used to prioritize candidate Arlm1 genes.
Table 1.
Databases and samples numbers used to identify candidate Arlm1 genes
| Description | Database | # Samples | # Genes Examined | Results |
|---|---|---|---|---|
| Genes in Arlm1 locus | NCBI Map Viewer | NA | NA | 503 genes |
| Mouse genes polymorphic B6 vs 129 | Mouse Phenome Database | NA | 267 genes | 100 genes |
| Differential exp in male B6 vs 129 brains | PhenoGen | 4–6 per strain | 73 probe sets | 41 genes |
| Human amp/del of Chr 7 or 14 in astrocytoma | NCBI Cancer Chromosomes | 642 grade II-IV, NOS | NA | 39% Chr 7 |
| 15% Chr 14 | ||||
| Human amp/del of Chr 7 or 14 in astrocytoma | Cancer Genome Workbench (REMBRANDT) | 56 astrocytoma (28 males, 18 females, 10 NOS) | NA | 21% Chr 7 |
| 17% Chr 14 | ||||
| Human amp/del of Chr 7 or 14 in GBM | Cancer Genome Workbench (REMBRANDT) | 130 GBM (59 male, 17 female, 53 NOS) | NA | 49% Chr 7 |
| 22% Chr 14 | ||||
| Human amp/del of Chr 7 or 14 in GBM | Cancer Genome Workbench (TCGA) | 793 GBM | NA | 77% Chr 7 |
| 33% Chr 14 | ||||
| Correlation of expression with survival time | REMBRANDT | 248 glioma (153 males, 95 females) | 58 homologs | 30 genes |
| Correlation of expression with survival time | Cancer Molecular Analysis Portal | 210 GBM (131 males, 79 females) | 53 homologs | 11 genes |
Abbreviations: NOS, not otherwise specified.
Arlm1 Genes Are Differentially Expressed in B6 and 129 Male Mouse Brains
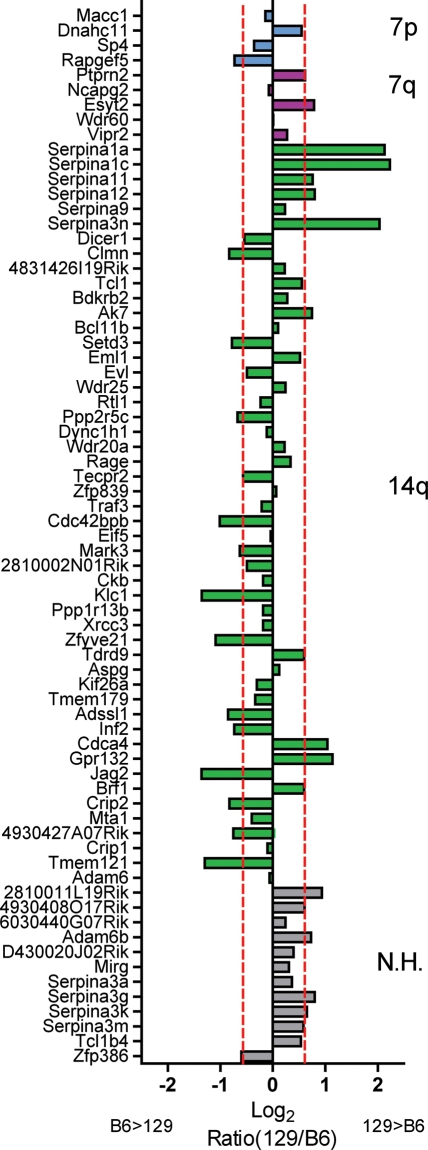
The Arlm1 locus covers the region of Chr 12 from 105 to 121 Mb and contains 100 genes that are polymorphic between B6 and 129S1/SvImJ and/or 129X1/SvJ (Mouse Phenome Database, http://phenome.jax.org/SNP/), including the Igh cluster. Of the polymorphic genes, 28 carry at least one nonsynonymous coding SNP predicted to change the sequence of the protein (Cn SNPs) (Table 2), and an additional 43 carry a SNP in the untranslated region or first intron sequence, which are most likely to play a role in transcription regulation or transcript stability. For 73 of the 100 polymorphic genes in the Arlm1 locus, expression data were available on 10- to 12-week-old male brains from the B6 and 129S1/SvImJ strains (http://phenogen.ucdenver.edu/PhenoGen/web/datasets/geneData.jsp). Forty-one genes showed a ≥1.5-fold difference in expression levels between B6 and 129S1/SvImJ (Fig. 4). In combining coding differences and expression level differences, at least 55 genes are likely to vary between the B6 and 129 strains in the Arlm1 locus.
Table 2.
Arlm1 genes with Cn SNPs
| Gene | Nonsynonymous coding SNPs |
|---|---|
| Adam6 | D298G; M415L; Y595F; T614A; K753R |
| Adam6b | E510D |
| Cdca7l | N113S |
| Crip2 | Q201P |
| Dicer1 | R1604G |
| Dnahc11 | D3491G; I2560V; P433S; E239G |
| Eml1 | Y206C |
| Esyt2 | N453S; A817V |
| Gm2698 | K10E |
| Gm5441 | Q66L; P43S |
| Igh locus | 83 Cn SNPs |
| Itgb8 | A651V |
| Macc1 | Q15R; H441P; S774F; T782M |
| Ncapg2 | E586D; V1057L; T1100A |
| Ptprn2 | T120A; A265S; I661V |
| Rapgef5 | A300V |
| Serpina11 | C218W; S141F |
| Serpina12 | Q182R; R109W; Q68R |
| Serpina1d | A262V; I52L; S7G |
| Serpina1e | A200V |
| Serpina3g | L384F |
| Serpina3i | L36V; R211C; Q242R; E244D; E245*; I313V; P338L; T353A; N381K; I386M |
| Serpina3j | I315V; S316F |
| Serpina3k | I14V; N319D; T348A; I385V; I397F |
| Serpina9 | N124K; A53S |
| Vipr2 | P396A |
| Wdr60 | Q731H |
| Zfp386 | N112D |
Fig. 4.
Graph of the expression level of Arlm1 candidate genes in B6 and 129 male mouse brains, taken from publicly available microarray data. Genes are organized along the Y-axis by their synteny with human Chr 7p (cyan), 7q (magenta), or 14q (green), corresponding to the cartoon in Fig. 3. Genes with no clear human homolog are shown in gray. The X-axis shows the ratio of 129/B6 on the log2 scale. Red dotted lines indicate the 1.5-fold up or down threshold used to prioritize candidates. The Igh locus showed a lot of variability between different probes in the dataset and was not included in the graph. Abbreviation: N.H., no homolog.
Arlm1 Is Syntenic with Human Chr 14q32, Chr 7q36, and Chr 7p15-21
We compared the Arlm1 locus on mouse Chr 12 with the human genome using the NCBI Homology Maps Viewer (http://www.ncbi.nlm.nih.gov/projects/homology/maps/). Arlm1 is syntenic with human Chr 14 98.8–106 Mb, corresponding to cytobands 14q32.1–0.33; Chr 7 20–22.4 Mb, corresponding to cytobands 7p21.2-7p15.3; and Chr 7 157.3–158.9 Mb, corresponding to cytoband 7q36.3. Importantly, it excludes the EGFR locus frequently amplified in human GBM at cytoband 7p12 (located on mouse Chr 11), suggesting that an alternate gene from Chr 7p is involved. It includes the human IGH cluster on Chr 14q32. We investigated gains and losses of these regions in human GBM using NCBI Cancer Chromosomes Mittelman Data (http://www.ncbi.nlm.nih.gov/sites/entrez?db=cancerchromosomes), and we used the Cancer Genome Workbench Heatmap Viewer (https://cgwb.nci.nih.gov/cgi-bin/heatmap) to examine the REMBRANDT glioma and TCGA GBM datasets. Details of the numbers of samples analyzed are given in Table 1. Data from NCBI Cancer Chromosomes and the Cancer Genome Workbench Heatmap show that, overall, human Chr 7 is commonly amplified in GBM, while Chr 14 is commonly deleted. In addition, at the cytoband level there was evidence for specific alterations in the NCBI Cancer Chromosomes data, with both amplification and deletion of 7p15, deletion of 7p21, and amplification of 14q32. The TCGA GBM dataset represents a homogeneous group of GBMs, in which both Chr 7p15-21 and 7q36 regions were amplified in 74% of samples and deleted in 3%. Chr 14q32 was deleted in 27% of samples and amplified in 6%. The REMBRANDT glioma dataset represents a more heterogeneous population but showed similar results. Chr 7p15-21 was amplified in 21% of astrocytoma and 49% of GBM. Chr 7q36 was amplified in 17% of astrocytoma and 34% of GBM, and deleted in 5% of astrocytoma. Chr 14q32 was deleted in 17% of astrocytoma and 22% of GBM. In summary, the Arlm1 locus overlaps regions of the human genome that are both amplified (Chr 7) and deleted (Chr 14) in GBM, and we hypothesize that modifier candidates syntenic with Chr 7 are most likely to act in an oncogenic manner, while candidates syntenic with Chr 14 are most likely to act in a tumor-suppressive manner. Because the 129 allele increases susceptibility to astrocytoma, we hypothesize that Arlm1 genes syntenic with human Chr 7 will have higher expression in 129 male brains and lower expression in B6 male brains, whereas genes syntenic with Chr 14 will have lower expression in 129 brains and higher expression in B6 brains. Figure 4 shows the ratio of 129 to B6 expression levels compared with whether genes are syntenic with Chr 7 or 14.
Arlm1 Genes Show Sex-Specific Effects on GBM Survival Time
Amplification or deletion of genomic regions in cancer can point to regions where there is selective pressure to maintain or lose genes that affect cell growth. However, this selective pressure can apply to broad regions of the genome such that neighboring genes can influence amplifications and deletions of any given gene in cancer, giving rise to a “bystander effect.” As an example, the high rate of amplification of human Chr 7 in GBM is likely driven by strong selective pressure to amplify the EGFR locus, although the chromosome carries many genes that may not all contribute to cancer. Gene expression analysis and comparisons of mouse and human syntenic regions where neighboring relationships are unlinked can help to dissect the varying roles of different loci. We took advantage of GBM gene expression data for males and females from REMBRANDT and TCGA to examine the effects of low and high expression of Arlm1 candidates on patient survival time. The REMBRANDT website (https://caintegrator.nci.nih.gov/rembrandt/menu.do) was used to analyze glioma survival data by gene expression, and the CMA portal (https://cma.nci.nih.gov/cma-tcga/) was used to analyze TCGA datasets. Details of the number of samples analyzed are given in Table 1. Thirty genes show significant differences in survival time based on different gene expression levels. Of these, 16 genes show different effects in males and females. While increases and decreases in expression level may be secondary to copy number changes, we found that not all linked genes have the same correlation to survival time. For example, NCAPG2 and VIPR2 are linked on 7q36.3, but overexpression of NCAPG2 correlates with reduced survival time in males, whereas VIPR2 overexpression correlates with increased survival time (Supplementary Fig. S2). This suggests that specific correlations between gene expression levels and patient survival time are more likely to reflect a causal relationship between a modifier candidate gene and tumorigenesis, compared with gene amplifications and deletions.
TCGA samples showed far less variation in expression levels of the different genes compared with REMBRANDT samples, likely due to the more homogeneous population being assessed. Because of this, there were very few samples in the overexpressed and underexpressed category for each gene, and the statistics were not as robust. Nonetheless, 11 genes showed a significant effect on survival times in males vs females when stratified by expression level. In comparing the 2 independent datasets, we find 22 genes in which male-female differences are detected in at least 1 human dataset, and 7 genes where male-female differences are significant in both datasets. Of these 7 genes, only 2—CRIP2 on Chr 14q32.3 (Fig. 5) and CDCA7L on Chr 7p15 (Fig. 6)—show similar effects in both datasets, correlating with shortened survival time when overexpressed, and show a significant effect in males, but not females. Because of the small variation in the TCGA dataset, we do not want to exclude promising candidates found in only the REMBRANDT data. These include NCAPG2 and WDR60 on Chr 7q36; and DICER1, WDR25, WDR20, XRCC3, TDRD9, and CDCA4 on Chr 14q32. Advanced GBMs, such as those in the TCGA dataset, may evolve to overcome the resistance mechanisms of modifier genes, thus making it more difficult to identify subtle effects. Finally, because of the complexity of the Igh/IGH cluster, we did not subject it to the same level of analysis; however, the association of the immune system with glioma risk is clear from previous studies,17–20 and IGH is also a strong candidate in the Arlm1 locus. Table 3 summarizes the evidence data from mouse and human for the highest priority candidates for a male-specific modifier at Arlm1. Individual Kaplan–Meier curves of top candidates are shown in Supplementary Figure S2.
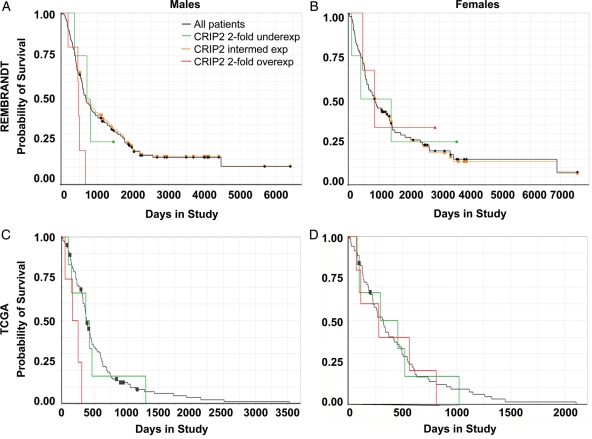
Fig. 5.
Males with 2-fold higher levels of CRIP2 in their tumors survive less time than males with intermediate or low levels of CRIP2. Graphs show survival curves for male GBM patients (A and C) and female GBM patients (B and D) stratified by expression level of CRIP2 for the REMBRANDT dataset (A and B) and the TCGA dataset (C and D). The survival curve for all pooled samples is shown in black. Survival curves for samples with 2-fold overexpression are shown in red and samples with 2-fold underexpression are shown in green. The gold line in the REMBRANDT data represents the survival of patients with intermediate levels of CRIP2 in their tumors. Data for which the survival curve for CRIP2-overexpressing GBMs is shifted to the right suggest an oncogenic effect of CRIP2 in human GBM.
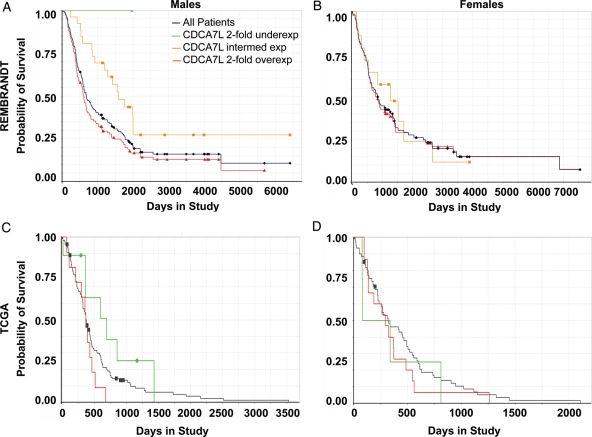
Fig. 6.
Males with 2-fold higher levels of CDCA7L in their tumors survive less time than males with intermediate or low levels of CDCA7L. Graphs show survival curves for male GBM patients (A and C) and female GBM patients (B and D) stratified by expression level of CDCA7L for the REMBRANDT dataset (A and B) and the TCGA dataset (C and D). The survival curve for all pooled samples is shown in black. Survival curves for samples with 2-fold overexpression are shown in red and samples with 2-fold underexpression are shown in green. The gold line in the REMBRANDT data represents the survival of patients with intermediate levels of CDCA7L in their tumors. Data for which the survival curve for CDCA7L-overexpressing GBMs is shifted to the right suggest an oncogenic effect of CDCA7L in human GBM.
Table 3.
Top candidates for the Arlm1 gene based on B6vs129 polymorphisms, differential expression in B6 and 129 male brains, hazard ratios for correlation to changes in GBM survival time in REMBRANDT (REMB) and TCGA datasets for males and females, and percentage of samples with copy number changes in human astrocytoma and GBM. Genes are listed in order of location in human genome
| Genes on Mouse Chr 12 105–121Mb polymorphic between B6 and 129 | Mouse B6 vs 129 Cn SNPs | Mouse B6 vs 129 UTR/Intron 1 SNPs | Mouse Brain Exp Fold Ratio 129S1/B6 | Human Homolog | Human Location | Hazard Ratio Males REMB n = 153 | Hazard Ratio Females REMB n = 95 | Hazard Ratio Males TCGA n = 131 | Hazard Ratio Females TCGA n = 79 | Percent Amp in REMB Astro | Percent Del in REMB Astro | Percent Amp in REMB GBM | Percent Del in REMB GBM | Percent Amp in TCGA GBM | Percent Del in TCGA GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Itgb8 | 1 | 7 | NP | ITGB8 | 7p21.1 | NS | 1.95c | 2.02a | NS | 3 | 0 | 15 | 0 | 70 | 3 |
| Dnahc11 | 4 | 10 | 1.5 | DNAH11 | 7p21 | 1.65b | 1.44e | NS | NS | 21 | 0 | 49 | 0 | 72 | 2 |
| Cdca7l | 1 | 48 | NP | CDCA7L | 7p15 | 1.76b | NS | 1.67f | NS | 19 | 0 | 45 | 0 | 72 | 2 |
| Ncapg2 | 3 | 21 | NC | NCAPG2 | 7q36.3 | 1.48b | NS | NS | 3.74a,g | 17 | 0 | 34 | 0 | 74 | 2 |
| Wdr60 | 1 | 12 | NC | WDR60 | 7q36.3 | 2.31d | NS | NS | NS | 17 | 0 | 23 | 0 | 73 | 3 |
| Vipr2 | 1 | 2 | NC | VIPR2 | 7q36.3 | 0.66a | 0.60b | 0.70e | NS | 16 | 5 | 23 | 0 | 74 | 2 |
| Serpina1a | 0 | 9 | 4.4 | SERPINA1 | 14q32.13 | 1.57c | 1.96b | NS | 2.19e | 0 | 5 | 0 | 0 | 4 | 27 |
| Serpina3n | 0 | 9 | 4.1 | SERPINA3 | 14q32.13 | 1.82c | 1.26e | NS | NS | 0 | 5 | 0 | 0 | 4 | 26 |
| Dicer1 | 1 | 2 | 0.7 | DICER1 | 14q32.13 | 0.47b | NS | NS | 0.46e | 0 | 7 | 0 | 9 | 3 | 27 |
| Tcl1 | 0 | 1 | 1.5 | TCL1A | 14q32.1 | NS | 0.65a | 3.91b,g | NS | 0 | 0 | 0 | 0 | 4 | 27 |
| Bcl11b | 0 | 1 | NC | BCL11b | 14q32.2 | 0.73a | 0.47b | 0.098d,g | NS | 0 | 0 | 0 | 3 | 4 | 27 |
| Evl | 0 | 2 | NC | EVL | 14q32 | 0.60b | 0.51c | 0.12c,g | NS | 0 | 5 | 0 | 3 | 4 | 26 |
| Wdr25 | 0 | 0 | NC | WDR25 | 14q32.2 | 0.39c | NS | 10.50b,g | NS | 7 | 5 | 0 | 8 | 4 | 26 |
| Ppp2r5c | 0 | 0 | 0.6 | PPP2R5c | 14q32.31 | 0.50c | 0.59b | NS | NS | 0 | 8 | 0 | 0 | 5 | 26 |
| Wdr20a | 0 | 3 | NC | WDR20 | 14q32.31 | 0.43a,g | NS | NS | NS | 0 | 8 | 0 | 0 | 5 | 26 |
| Cdc42bpb | 0 | 1 | 0.5 | CDC42BPB | 14q32.3 | 0.61a | 0.65a | NS | 0.43e,g | 0 | 14 | 0 | 21 | 6 | 26 |
| Xrcc3 | 0 | 3 | NC | XRCC3 | 14q32.3 | 1.66a | NS | NS | NS | 0 | 17 | 0 | 22 | 6 | 26 |
| Tdrd9 | 0 | 1 | 1.5 | TDRD9 | 14q32.33 | 0.76b | NS | NS | NS | 0 | 8 | 0 | 5 | 5 | 26 |
| Cdca4 | 0 | 0 | 2.1 | CDCA4 | 14q32.33 | 1.70c | NS | NS | NS | 0 | 7 | 5 | 3 | 5 | 25 |
| Jag2 | 0 | 0 | 0.4 | JAG2 | 14q32 | 4.97a | NS | 0.027d,g | NS | 0 | 7 | 5 | 3 | 5 | 26 |
| Crip2 | 1 | 0 | 0.6 | CRIP2 | 14q32.3 | 2.41a | NS | 4.01b,g | NS | 0 | 7 | 5 | 3 | 5 | 25 |
| Crip1 | 0 | 8 | NC | CRIP1 | 14q32.33 | 1.63b | NS | NS | NS | 0 | 7 | 5 | 3 | 5 | 25 |
Abbreviations: NP, no probe; NC, no change; NS, not significant.
aP < 0.05.
bP < 0.01.
cP < 0.001.
dP < 0.0001.
eP < 0.1.
fUnderexp vs Intermed (2-fold change) is not significant (P = .18); Overexp vs Underexp (4-fold change) is significant (HR = 2.66, P = .020).
gn < 5 for one group in comparison.
The genotype at the Arlm1 locus does not affect astrocytoma-free survival in the NPcis mice (Supplementary Fig. S1A and B); however, the top candidates we have chosen from the human datasets correlate with changes in the length of survival after diagnosis in patients. To more directly compare Arlm1 with the human gene candidates, we examined whether patients with different levels of CDCA7L or CRIP2 in their tumors were diagnosed at different average ages (more analogous to the identification of mouse tumors that are not treated). Using Kaplan–Meier time-to-event analysis, we found no difference in the age of diagnosis in either male or female patients with different levels of CDCA7L or CRIP2, consistent with the findings in mice (Supplementary Fig. S1C–F).
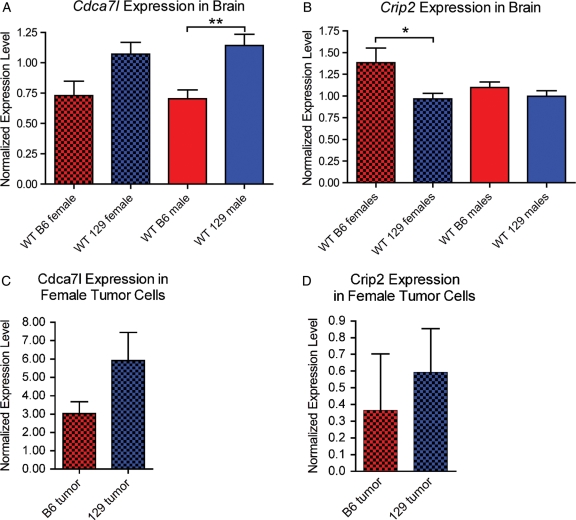
Cdca7l Is Differentially Expressed between B6 and 129 Brains in both Males and Females
To verify the strain differences in expression of Cdca7l and Crip2, we used reverse transcription qPCR to measure the levels of the 2 genes in male and female B6 and 129 brains. There were no probes for Cdca7l in the publicly available microarray dataset for the B6 and 129S1/SvImJ (http://phenogen.ucdenver.edu/PhenoGen/web/datasets/geneData.jsp) (Table 3). We found that the 129S4/SvJae (129) strain that we used for our mapping studies expressed more Cdca7l than the B6 strain (1.6-fold difference for males, t-test P = .0030; and 1.5-fold difference for females, t-test P = .056) (Fig. 7A). Crip2 expression in the publicly available microarray dataset was 1.8-fold higher in B6 than in 129S1/SvImJ males; however, we found no difference between B6 and 129 male brains. B6 females brains expressed 1.4-fold higher Crip2 than 129 female brains (t-test P = .049) (Fig. 7B). Taken together these data suggest that Cdca7l is a stronger candidate for the Arlm1 locus and that strain differences in Cdca7l between B6 and 129 may have different effects depending on male or female physiology.
Fig. 7.
Strain differences in expression of Cdca7l and Crip2 in mouse brains. Wt B6 female brains (n = 5) and wt B6 male brains (n = 7) express lower levels of Cdca7l than wt 129 female brains (n = 5) and wt 129 male brains (n = 8) (A). The difference between B6 and 129 females is suggestive (t-test P = .056) and the difference between B6 and 129 males is significant (t-test P = .0030). Wt B6 female brains express higher levels of Crip2 than wt 129 female brains (t-test P = .049), but males show no difference in Crip2 expression between B6 and 129 (B). Brackets indicate which data comparison is statistically significant (*P < .05; **P < .005). Female B6-NPcis astrocytoma lines (n = 2) express lower levels of Cdca7l than female 129-NPcis astrocytoma lines (n = 4) (C), consistent with results in normal brains (A); however, the number of available tumor lines is too low to draw statistically significant conclusions. The level of Crip2 in female B6-NPcis astrocytoma lines (n = 2) and 129-NPcis astrocytoma lines (n = 4) (D) is variable and does not reflect the expression differences seen in normal brain (B). Red bars indicate B6 samples and blue bars indicate 129 samples. Solid bars represent male samples and checked bars represent female samples.
We extended the Cdca7l and Crip2 expression studies to mouse and human tumor cell lines. Mouse astrocytoma cell lines have been isolated from male and female B6-NPcis mice and from female 129-NPcis mice.15 We have not yet been able to derive astrocytoma cell lines from male 129-NPcis mice for reasons that are unclear, so we were not able to directly compare male tumors on the B6 and 129 strain background. Expression of Cdca7l in astrocytoma cells from B6-NPcis females and 129-NPcis females supports our findings in whole brain, in that 129 tumor lines express higher levels of Cdca7l compared to B6 tumor line (Fig. 7C), although we noted wide variablility between lines from the same strain (Supplementary Fig. S3A). Crip2 expression was even more variable between lines (Supplementary Fig. S3B) and did not show the same trend we found in normal brains (Fig. 3D), again supporting Cdca7l as a stronger candidate for the Arlm1 locus than Crip2.
To examine whether there were male and female differences in Cdca7l in tumors, we compared B6-NPcis male and female tumor lines and human GBM male and female tumor lines. We found variable levels of Cdca7l expression between different tumor lines (Supplementary Fig. S3A and C), but for both mouse and human the highest expressing lines were male and the lowest expressing lines were female. Additional samples are needed to determine whether male astrocytomas/GBMs have a greater propensity for upregulating Cdca7l than female tumors, but this data presents the possibility that the strain-specific increase in Cdca7l in 129 mice interacts with male-specific increases in Cdca7l in tumors to affect tumor susceptibility.
Discussion
We show using an unbiased mouse screen that a polymorphic modifier locus on mouse Chr 12 affects resistance to astrocytoma in males. Although this modifier region includes 100 polymorphic candidate genes, it is difficult to assess all of these individually, and cross-species comparison was used to prioritize the candidates for further study. Two genes, Cdca7l and Crip2, are of particular interest due to their correlation to GBM patient survival time in 2 separate human datasets; however, an additional 14 genes (Itgb8, Dnah11, Ncapg2, SerpinA, Dicer1, Bcl11b, Evl, Ppp2r5c, Xrcc3, Tdrd9, Cdca4, Jag2, Crip1 and the Igh locus) are also of great interest due to a variety of bioinformatic data as well as what is already known about these genes in cancer and the nervous system. Furthermore, it is important to note that in addition to these genes, other polymorphisms in the DNA in the region of Arlm1, such as those affecting miRs or chromatin structure, could also affect astrocytoma resistance and were not specifically examined in this study. Nevertheless, prioritization of genes within the region allows a direct comparison with human GWASs and a more rapid test for the role of these genes in human astrocytoma/GBM.
CDCA7L encodes the transcription factor R1 (RAM2/JPO2) with links to both nervous system function and cancer. R1 was first identified as interacting with Myc21 and being involved in medulloblastoma malignant transformation. It was further shown to inhibit apoptosis downstream of Bcl2 and to enhance proliferation downstream of Myc and upstream of cyclinD1/E2F1.22 This oncogenic role for R1 is consistent with its being within the 7p15 amplicon and being correlated with poorer prognosis in males when overexpressed in GBM. We show that the B6 brains carrying the resistance allele of Arlm1 express lower levels of Cdca7l, again consistent with an oncogenic role for Cdca7l. R1 functions in the brain to repress transcription of monoamine oxidase A23 and B24 involved in neurotransmitter turnover. Although we did not see male vs female differences in Cdca7l levels in mouse brains, sex-specific effects on neurotransmitter pathways may explain why CDCA7L and Arlm1 show male-specific effects in humans and mice, respectively.
CRIP2 encodes a molecular adaptor protein in the LIM domain family of proteins that has recently been identified as a tumor suppressor25 in nasopharyngeal carcinoma. CRIP2 was originally identified as a binding partner of the protein tyrosine phosphatase PTP-BL, localized to the cell cortex with a potential role in regulating the actin cytoskeleton.26 It is highly expressed in brain and shows specific expression patterns in subsets of neurons27 and during neural crest patterning and migration.28 In nasopharyngeal carcinoma cells CRIP2 is found in the nucleus and inhibits NF-kB (nuclear factor kappa-light-chain-enhancer of activated B cells), resulting in the inhibition of angiogenesis and resulting tumor suppression.25 Although Crip2 is expressed at higher levels in the brains of B6 males compared with 129S1/SvImj males, we were not able to confirm this difference between B6 and 129S4/SvJae (129) males. Higher expression in the B6 strain carrying the Arlm1-resistant allele would be consistent with it acting in a tumor-suppressive way; however, overexpression of CRIP2 in human GBM correlates with shorter survival time, suggesting a pro-oncogenic effect. Further experiments are needed to examine these contradictions. The differences in expression of Crip2 in our data and the public dataset could be due to differences between the 129S4/SvJae and 129S1/SvImJ strains, or to temporal changes in expression levels as mice age. The conflicting data on Crip2 acting as a tumor suppressor or oncogene may be due to conflicting roles of CRIP2 in cell migration, through cytoskeletal dynamics, and in angiogenesis, through transcriptional repression that affects tumorigenesis.
Additional candidates in the Arlm1 region have roles in the processes identified as hallmarks of cancer,29 specifically cell cycle regulation, DNA damage repair, angiogenesis, and inflammation. Functions in cell proliferation and cell cycle checkpoints have been shown for CDCA4,30,31 JAG2,32–34 PPP2R5C,35–37 and BCL11B.38 Epidemiological studies support the role of DNA damage repair in glioma prevention,19 and several Arlm1 candidates have been implicated in chromatin dynamics and DNA repair. NCAPG239,40 and DNAHC1141 encode structural proteins important for chromosome condensation and segregation, and XRCC342–45 and EVL46,47 encode proteins involved in homologous recombination and genome stability. XRCC3 has been studied in human association studies for its potential role in susceptibility to glioma.42,44,45 Intriguingly, the rs3212092 SNP in XRCC3 affects males more strongly than females in one study of glioma risk.45 Progression of gliomas requires an angiogenic switch and high-grade glioblastomas are characterized by microvascular proliferation.1 The Arlm1 candidate ITGB8 regulates tumor angiogenesis in brain cancer 48 and is upregulated in GBM.49,50 The association of immune function and glioma risk is now well established,19 with an inverse association between glioma and allergies or asthma,51–53 associations of cytokine signaling and invasion pathways with glioma risk,54 and conflicting roles for infiltrating T-cells in glioma grade and prognosis.55–57 Four of the Arlm1 candidates are linked to immune system function: the IGH locus, SERPINA1,58,59 SERPINA3,60–62 and CRIP1.63–65 In addition to the genes directly involved in different hallmarks of cancer, genes involved in regulation of non–protein coding RNAs can have indirect effects on cancer. Non–protein coding RNAs are increasingly recognized as major tumor suppressors and oncogenes in cancer, including GBM.66 DICER167 and TDRD968 regulate microRNAs and Piwi-interacting RNAs, respectively, and could have wide-ranging, complex effects on cancer.
Through cross-species comparison, we identify Cdca7l as a high priority candidate as well as 15 additional strong candidates for a male-specific modifier of brain cancer. These candidates can be tested directly in human GWAS data for glioblastoma risk as they become available. Future studies of the mechanism of action of Arlm1 will give new insights into the male predominance of both astrocytoma and GBM, and may suggest new therapeutic or preventative approaches.
Supplementary Material
Funding
Intramural Research Program of the National Institutes of Health, National Cancer Institute [ZIA BC 010539 to K.M.R.]; federal funds from the National Cancer Institute to SAIC Frederick [contract N01-CO-12400]; federal contract from the National Institutes of Health to The Johns Hopkins University [contract HHSN268200782096C]; extramural funding from the National Institutes of Health [R01 GM074244 to K.W.B.].
Conflict of interest statement. None declared.
Supplementary Material
Acknowledgments
We thank M. Anvers, R. Tuskan, K. Smith, K. Fox, and E. Truffer for technical assistance and B. Mock, J. Simmons, N. Colburn, and K. Hunter for helpful discussions and comments on the manuscript. J.C.A.-V.S. is a predoctoral student in the Graduate Partnership Program of the N.I.H. and the Institute for Biomedical Sciences at George Washington University. This work is from a dissertation to be prepared in partial fulfillment of the requirements for the PhD degree. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsements by the US Government.
References
- 1.Kleihues P, Cavenee W. Pathology and Genetics of Tumours of the Nervous System. Lyon: International Agency for Research on Cancer; 2000. [Google Scholar]
- 2.Malmer B, Adatto P, Armstrong G, et al. GLIOGENE an International Consortium to Understand Familial Glioma. Cancer Epidemiol Biomarkers Prev. 2007;16(9):1730–1734. doi: 10.1158/1055-9965.EPI-07-0081. doi:10.1158/1055-9965.EPI-07-0081. [DOI] [PubMed] [Google Scholar]
- 3.Shete S, Hosking FJ, Robertson LB, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41(8):899–904. doi: 10.1038/ng.407. doi:10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wrensch M, Jenkins RB, Chang JS, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–908. doi: 10.1038/ng.408. doi:10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reilly KM, Broman KW, Bronson RT, et al. An imprinted locus epistatically influences Nstr1 and Nstr2 to control resistance to nerve sheath tumors in a neurofibromatosis type 1 mouse model. Cancer Res. 2006;66(1):62–68. doi: 10.1158/0008-5472.CAN-05-1480. doi:10.1158/0008-5472.CAN-05-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reilly KM, Loisel DA, Bronson RT, McLaughlin ME, Jacks T. Nf1;Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects. Nat Genet. 2000;26(1):109–113. doi: 10.1038/79075. doi:10.1038/79075. [DOI] [PubMed] [Google Scholar]
- 7.Reilly KM, Tuskan RG, Christy E, et al. Susceptibility to astrocytoma in mice mutant for Nf1 and Trp53 is linked to chromosome 11 and subject to epigenetic effects. Proc Natl Acad Sci USA. 2004;101(35):13008–13013. doi: 10.1073/pnas.0401236101. doi:10.1073/pnas.0401236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walrath JC, Fox K, Truffer E, Gregory Alvord W, Quinones OA, Reilly KM. Chr 19(A/J) modifies tumor resistance in a sex- and parent-of-origin-specific manner. Mamm Genome. 2009;20(4):214–223. doi: 10.1007/s00335-009-9179-4. doi:10.1007/s00335-009-9179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reilly KM. The Nf1−/+;Trp53−/+cis mouse model of anaplastic astrocytoma and secondary glioblastoma: Dissecting genetic susceptibility to brain cancer. In: Van Meir EG, editor. CNS cancer, Models, Prognostic Factors and Targets. New York: Humana Press; 2009. pp. 93–118. [Google Scholar]
- 10.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. doi:10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. doi:10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. doi:10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 14.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19(7):889–890. doi: 10.1093/bioinformatics/btg112. doi:10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 15.Gursel DB, Connell-Albert YS, Tuskan RG, et al. Control of proliferation in astrocytoma cells by the receptor tyrosine kinase/PI3K/AKT signaling axis and the use of PI-103 and TCN as potential anti-astrocytoma therapies. Neuro Oncol. 2011;13(6):610–621. doi: 10.1093/neuonc/nor035. doi:10.1093/neuonc/nor035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. doi:10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartzbaum J, Ahlbom A, Malmer B, et al. Polymorphisms associated with asthma are inversely related to glioblastoma multiforme. Cancer Res. 2005;65(14):6459–6465. doi: 10.1158/0008-5472.CAN-04-3728. doi:10.1158/0008-5472.CAN-04-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wigertz A, Lonn S, Schwartzbaum J, et al. Allergic conditions and brain tumor risk. Am J Epidemiol. 2007;166(8):941–950. doi: 10.1093/aje/kwm203. doi:10.1093/aje/kwm203. [DOI] [PubMed] [Google Scholar]
- 19.Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol. 2006;2(9):494–503. doi: 10.1038/ncpneuro0289. quiz 491 p following 516 doi:10.1038/ncpneuro0289. [DOI] [PubMed] [Google Scholar]
- 20.Zhou M, Wiemels JL, Bracci PM, et al. Circulating levels of the innate and humoral immune regulators CD14 and CD23 are associated with adult glioma. Cancer Res. 2010;70(19):7534–7542. doi: 10.1158/0008-5472.CAN-10-0815. doi:10.1158/0008-5472.CAN-10-0815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang A, Ho CS, Ponzielli R, et al. Identification of a novel c-Myc protein interactor, JPO2, with transforming activity in medulloblastoma cells. Cancer Res. 2005;65(13):5607–5619. doi: 10.1158/0008-5472.CAN-05-0500. doi:10.1158/0008-5472.CAN-05-0500. [DOI] [PubMed] [Google Scholar]
- 22.Ou XM, Chen K, Shih JC. Monoamine oxidase A and repressor R1 are involved in apoptotic signaling pathway. Proc Natl Acad Sci USA. 2006;103(29):10923–10928. doi: 10.1073/pnas.0601515103. doi:10.1073/pnas.0601515103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ou XM, Chen K, Shih JC. Glucocorticoid and androgen activation of monoamine oxidase A is regulated differently by R1 and Sp1. J Biol Chem. 2006;281(30):21512–21525. doi: 10.1074/jbc.M600250200. doi:10.1074/jbc.M600250200. [DOI] [PubMed] [Google Scholar]
- 24.Chen K, Ou XM, Wu JB, Shih JC. Transcription factor E2F-associated phosphoprotein (EAPP), RAM2/CDCA7L/JPO2 (R1), and simian virus 40 promoter factor 1 (Sp1) cooperatively regulate glucocorticoid activation of monoamine oxidase B. Mol Pharmacol. 2011;79(2):308–317. doi: 10.1124/mol.110.067439. doi:10.1124/mol.110.067439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung AK, Ko JM, Lung HL, et al. Cysteine-rich intestinal protein 2 (CRIP2) acts as a repressor of NF-{kappa}B-mediated proangiogenic cytokine transcription to suppress tumorigenesis and angiogenesis. Proc Natl Acad Sci USA. 2011;108(20):8390–8395. doi: 10.1073/pnas.1101747108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Ham M, Croes H, Schepens J, Fransen J, Wieringa B, Hendriks W. Cloning and characterization of mCRIP2, a mouse LIM-only protein that interacts with PDZ domain IV of PTP-BL. Genes Cells. 2003;8(7):631–644. doi: 10.1046/j.1365-2443.2003.00660.x. doi:10.1046/j.1365-2443.2003.00660.x. [DOI] [PubMed] [Google Scholar]
- 27.Bourane S, Mechaly I, Venteo S, et al. A SAGE-based screen for genes expressed in sub-populations of neurons in the mouse dorsal root ganglion. BMC Neurosci. 2007;8:97. doi: 10.1186/1471-2202-8-97. doi:10.1186/1471-2202-8-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun X, Zhang R, Lin X, Xu X. Wnt3a regulates the development of cardiac neural crest cells by modulating expression of cysteine-rich intestinal protein 2 in rhombomere 6. Circ Res. 2008;102(7):831–839. doi: 10.1161/CIRCRESAHA.107.166488. doi:10.1161/CIRCRESAHA.107.166488. [DOI] [PubMed] [Google Scholar]
- 29.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. doi:10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi R, Goto Y, Ikeda R, Yokoyama KK, Yoshida K. CDCA4 is an E2F transcription factor family-induced nuclear factor that regulates E2F-dependent transcriptional activation and cell proliferation. J Biol Chem. 2006;281(47):35633–35648. doi: 10.1074/jbc.M603800200. doi:10.1074/jbc.M603800200. [DOI] [PubMed] [Google Scholar]
- 31.Tategu M, Nakagawa H, Hayashi R, Yoshida K. Transcriptional co-factor CDCA4 participates in the regulation of JUN oncogene expression. Biochimie. 2008;90(10):1515–1522. doi: 10.1016/j.biochi.2008.05.014. doi:10.1016/j.biochi.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 32.Gwak JW, Kong HJ, Bae YK, et al. Proliferating neural progenitors in the developing CNS of zebrafish require Jagged2 and Jagged1b. Mol Cells. 2010;30(2):155–159. doi: 10.1007/s10059-010-0101-4. doi:10.1007/s10059-010-0101-4. [DOI] [PubMed] [Google Scholar]
- 33.Yeo SY, Chitnis AB. Jagged-mediated Notch signaling maintains proliferating neural progenitors and regulates cell diversity in the ventral spinal cord. Proc Natl Acad Sci USA. 2007;104(14):5913–5918. doi: 10.1073/pnas.0607062104. doi:10.1073/pnas.0607062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yustein JT, Liu YC, Gao P, et al. Induction of ectopic Myc target gene JAG2 augments hypoxic growth and tumorigenesis in a human B-cell model. Proc Natl Acad Sci USA. 2010;107(8):3534–3539. doi: 10.1073/pnas.0901230107. doi:10.1073/pnas.0901230107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen W, Possemato R, Campbell KT, Plattner CA, Pallas DC, Hahn WC. Identification of specific PP2A complexes involved in human cell transformation. Cancer Cell. 2004;5(2):127–136. doi: 10.1016/s1535-6108(04)00026-1. doi:10.1016/S1535-6108(04)00026-1. [DOI] [PubMed] [Google Scholar]
- 36.Li HH, Cai X, Shouse GP, Piluso LG, Liu X. A specific PP2A regulatory subunit, B56gamma, mediates DNA damage-induced dephosphorylation of p53 at Thr55. Embo J. 2007;26(2):402–411. doi: 10.1038/sj.emboj.7601519. doi:10.1038/sj.emboj.7601519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shouse GP, Nobumori Y, Panowicz MJ, Liu X. ATM-mediated phosphorylation activates the tumor-suppressive function of B56gamma-PP2A. Oncogene. 30(35):3755–3765. doi: 10.1038/onc.2011.95. Apr 4 2011. [DOI] [PubMed] [Google Scholar]
- 38.Kamimura K, Mishima Y, Obata M, Endo T, Aoyagi Y, Kominami R. Lack of Bcl11b tumor suppressor results in vulnerability to DNA replication stress and damages. Oncogene. 2007;26(40):5840–5850. doi: 10.1038/sj.onc.1210388. doi:10.1038/sj.onc.1210388. [DOI] [PubMed] [Google Scholar]
- 39.Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115(1):109–121. doi: 10.1016/s0092-8674(03)00724-4. doi:10.1016/S0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- 40.Wood JL, Liang Y, Li K, Chen J. Microcephalin/MCPH1 associates with the Condensin II complex to function in homologous recombination repair. J Biol Chem. 2008;283(43):29586–29592. doi: 10.1074/jbc.M804080200. doi:10.1074/jbc.M804080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Armakolas A, Klar AJ. Left-right dynein motor implicated in selective chromatid segregation in mouse cells. Science. 2007;315(5808):100–101. doi: 10.1126/science.1129429. doi:10.1126/science.1129429. [DOI] [PubMed] [Google Scholar]
- 42.Kiuru A, Lindholm C, Heinavaara S, et al. XRCC1 and XRCC3 variants and risk of glioma and meningioma. J Neurooncol. 2008;88(2):135–142. doi: 10.1007/s11060-008-9556-y. doi:10.1007/s11060-008-9556-y. [DOI] [PubMed] [Google Scholar]
- 43.Thacker J. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 2005;219(2):125–135. doi: 10.1016/j.canlet.2004.08.018. doi:10.1016/j.canlet.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 44.Wang LE, Bondy ML, Shen H, et al. Polymorphisms of DNA repair genes and risk of glioma. Cancer Res. 2004;64(16):5560–5563. doi: 10.1158/0008-5472.CAN-03-2181. doi:10.1158/0008-5472.CAN-03-2181. [DOI] [PubMed] [Google Scholar]
- 45.Zhou K, Liu Y, Zhang H, et al. XRCC3 haplotypes and risk of gliomas in a Chinese population: a hospital-based case-control study. Int J Cancer. 2009;124(12):2948–2953. doi: 10.1002/ijc.24307. doi:10.1002/ijc.24307. [DOI] [PubMed] [Google Scholar]
- 46.Hu LD, Zou HF, Zhan SX, Cao KM. EVL (Ena/VASP-like) expression is up-regulated in human breast cancer and its relative expression level is correlated with clinical stages. Oncol Rep. 2008;19(4):1015–1020. [PubMed] [Google Scholar]
- 47.Takaku M, Machida S, Hosoya N, et al. Recombination activator function of the novel RAD51- and RAD51B-binding protein, human EVL. J Biol Chem. 2009;284(21):14326–14336. doi: 10.1074/jbc.M807715200. doi:10.1074/jbc.M807715200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tchaicha JH, Mobley AK, Hossain MG, Aldape KD, McCarty JH. A mosaic mouse model of astrocytoma identifies alphavbeta8 integrin as a negative regulator of tumor angiogenesis. Oncogene. 2010;29(31):4460–4472. doi: 10.1038/onc.2010.199. doi:10.1038/onc.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riemenschneider MJ, Mueller W, Betensky RA, Mohapatra G, Louis DN. In situ analysis of integrin and growth factor receptor signaling pathways in human glioblastomas suggests overlapping relationships with focal adhesion kinase activation. Am J Pathol. 2005;167(5):1379–1387. doi: 10.1016/S0002-9440(10)61225-4. doi:10.1016/S0002-9440(10)61225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rossi MR, La Duca J, Matsui S, Nowak NJ, Hawthorn L, Cowell JK. Novel amplicons on the short arm of chromosome 7 identified using high resolution array CGH contain over expressed genes in addition to EGFR in glioblastoma multiforme. Genes Chromosomes Cancer. 2005;44(4):392–404. doi: 10.1002/gcc.20256. doi:10.1002/gcc.20256. [DOI] [PubMed] [Google Scholar]
- 51.Wiemels JL, Wiencke JK, Patoka J, et al. Reduced immunoglobulin E and allergy among adults with glioma compared with controls. Cancer Res. 2004;64(22):8468–8473. doi: 10.1158/0008-5472.CAN-04-1706. doi:10.1158/0008-5472.CAN-04-1706. [DOI] [PubMed] [Google Scholar]
- 52.Wiemels JL, Wilson D, Patil C, et al. IgE, allergy, and risk of glioma: update from the San Francisco Bay Area Adult Glioma Study in the temozolomide era. Int J Cancer. 2009;125(3):680–687. doi: 10.1002/ijc.24369. doi:10.1002/ijc.24369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wrensch M, Wiencke JK, Wiemels J, et al. Serum IgE, tumor epidermal growth factor receptor expression, and inherited polymorphisms associated with glioma survival. Cancer Res. 2006;66(8):4531–4541. doi: 10.1158/0008-5472.CAN-05-4032. doi:10.1158/0008-5472.CAN-05-4032. [DOI] [PubMed] [Google Scholar]
- 54.Schwartzbaum JA, Xiao Y, Liu Y, et al. Inherited variation in immune genes and pathways and glioblastoma risk. Carcinogenesis. 2010;31(10):1770–1777. doi: 10.1093/carcin/bgq152. doi:10.1093/carcin/bgq152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang I, Tihan T, Han SJ, et al. CD8+ T-cell infiltrate in newly diagnosed glioblastoma is associated with long-term survival. J Clin Neurosci. 2010;17(11):1381–1385. doi: 10.1016/j.jocn.2010.03.031. doi:10.1016/j.jocn.2010.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jacobs JF, Idema AJ, Bol KF, et al. Prognostic significance and mechanism of Treg infiltration in human brain tumors. J Neuroimmunol. 2010;225(1–2):195–199. doi: 10.1016/j.jneuroim.2010.05.020. doi:10.1016/j.jneuroim.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 57.Lohr J, Ratliff T, Huppertz A, et al. Effector T-cell infiltration positively impacts survival of glioblastoma patients and is impaired by tumor-derived transforming growth factor-betas. Clin Cancer Res. 17(15):4296–4308. doi: 10.1158/1078-0432.CCR-10-2557. Apr 8 2011. [DOI] [PubMed] [Google Scholar]
- 58.El-Akawi ZJ, Al-Hindawi FK, Bashir NA. Alpha-1 antitrypsin (alpha1-AT) plasma levels in lung, prostate and breast cancer patients. Neuro Endocrinol Lett. 2008;29(4):482–484. [PubMed] [Google Scholar]
- 59.Kohnlein T, Welte T. Alpha-1 antitrypsin deficiency: pathogenesis, clinical presentation, diagnosis, and treatment. Am J Med. 2008;121(1):3–9. doi: 10.1016/j.amjmed.2007.07.025. doi:10.1016/j.amjmed.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 60.Arion D, Unger T, Lewis DA, Levitt P, Mirnics K. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol Psychiatry. 2007;62(7):711–721. doi: 10.1016/j.biopsych.2006.12.021. doi:10.1016/j.biopsych.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boer K, Crino PB, Gorter JA, et al. Gene expression analysis of tuberous sclerosis complex cortical tubers reveals increased expression of adhesion and inflammatory factors. Brain Pathol. 2010;20(4):704–719. doi: 10.1111/j.1750-3639.2009.00341.x. doi:10.1111/j.1750-3639.2009.00341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saetre P, Emilsson L, Axelsson E, Kreuger J, Lindholm E, Jazin E. Inflammation-related genes up-regulated in schizophrenia brains. BMC Psychiatry. 2007;7:46. doi: 10.1186/1471-244X-7-46. doi:10.1186/1471-244X-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cousins RJ, Lanningham-Foster L. Regulation of cysteine-rich intestinal protein, a zinc finger protein, by mediators of the immune response. J Infect Dis. 2000;182(Suppl 1):S81–84. doi: 10.1086/315917. doi:10.1086/315917. [DOI] [PubMed] [Google Scholar]
- 64.Hao J, Serohijos AW, Newton G, et al. Identification and rational redesign of peptide ligands to CRIP1, a novel biomarker for cancers. PLoS Comput Biol. 2008;4(8):e1000138. doi: 10.1371/journal.pcbi.1000138. doi:10.1371/journal.pcbi.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lanningham-Foster L, Green CL, Langkamp-Henken B, et al. Overexpression of CRIP in transgenic mice alters cytokine patterns and the immune response. Am J Physiol Endocrinol Metab. 2002;282(6):E1197–1203. doi: 10.1152/ajpendo.00508.2001. [DOI] [PubMed] [Google Scholar]
- 66.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. doi:10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 67.Merritt WM, Bar-Eli M, Sood AK. The dicey role of Dicer: implications for RNAi therapy. Cancer Res. 2010;70(7):2571–2574. doi: 10.1158/0008-5472.CAN-09-2536. doi:10.1158/0008-5472.CAN-09-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shoji M, Tanaka T, Hosokawa M, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17(6):775–787. doi: 10.1016/j.devcel.2009.10.012. doi:10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.