Abstract
Somatic mutations in the catalytic domain of isocitrate dehydrogenase (IDH) 1/2 and accumulation of the oncometabolite 2-hydroxyglutarate (2-HG) appear to be among the earliest events in gliomagenesis and may contribute to malignant transformation. The lack of cell lines with endogenous mutations has been one of the major challenges in studying IDH1/2-mutant glioma and developing novel therapeutics for these tumors. Here, we describe the isolation of a glioma brain tumor stem cell line (BT142) with an endogenous R132H mutation in IDH1, aggressive tumor-initiating capacity, and 2-HG production. The neurosphere culture method was used to establish a brain tumor stem cell line from an IDH1-mutant anaplastic oligoastrocytoma sample, and an orthotopic xenograft system was developed to allow its rapid expansion. Production of 2-HG by glioma cells with endogenous IDH1 mutations was confirmed by mass spectrometry. BT142 retained an endogenous R132H IDH1 mutation in culture and possessed aggressive tumor-initiating capacity, allowing it to be readily propagated in orthotopic xenografts of nonobese diabetic/severe combined immune deficiency (NOD SCID) mice. Endogenous 2-HG production by BT142 was detectable in both cell culture medium and xenograft animal serum. BT142 is the first brain tumor cell line with an endogenous IDH1 mutation and detectable 2-HG production both in vitro and in vivo, which thus provides a unique model for studying the biology of IDH1-mutant glioma and in vivo validation of compounds targeting IDH1-mutant cells.
Keywords: glioma, isocitrate dehydrogenase, 2-hydroxyglutarate
The recent discovery of IDH1/2 point mutations has generated renewed interest in the abnormal metabolism of cancer described several decades ago by Otto Warburg. IDH1/2 mutations have been found in the majority of World Health Organization (WHO) grade II and III gliomas of oligodendroglial, astroglial, and mixed histopathologies and in most secondary glioblastomas (GBMs) and rare primary GBMs.1,2 IDH1 and IDH2 are highly homologous enzymes that catalyze the conversion of isocitrate to 2-ketoglutarate (2-KG). Mutations in codons 132/172 of IDH1/2 significantly decrease conversion of isocitrate to 2-KG2 and instead confer a neomorphic ability to convert 2-KG to 2-HG.3 Although accumulation of 2-HG in IDH1/2-mutant cells appears to alter their metabolic profile and this effect is partially reproduced by treatment of wild-type cells with 2-HG,4 the precise functional significance to tumor biology remains an item of debate.2,3,5–7 However, elevated 2-HG levels appear to be a consistent finding and can be readily detected within IDH1-mutant glioma tissue3 and IDH2-mutant acute myelogenous leukemia samples,8 leading to speculation that 2-HG is an oncometabolite. Detection of GBM and low-grade gliomas at the earliest stages of tumor development, when treatment may be of the most long-term benefit, remains challenging; as such, 2-HG could become a biomarker, serving diagnostic, prognostic, and therapeutic purposes in IDH1/2-mutant gliomas.9
One of the major challenges in studying the implications of the IDH1 mutations and increased 2-HG production has been the dearth of human patient-derived cell lines endogenously expressing this mutant enzyme. In fact, others have reported the inability to propagate patient-derived glioma cell lines containing this mutation,10 suggesting that current in vitro culture conditions are suboptimal for IDH1-mutant cells. Our group has previously reported the isolation of 2 BTSC lines, BT054 and BT088, cultured from patients with IDH1-mutant oligodendroglioma.11 BT088, which was derived from a recurrent previously treated oligodendroglioma, grew in vivo, but neither the neurospheres nor the xenografts retained the IDH1 mutation. The neurosphere culture of BT054 retained the IDH1 mutation, making it the first reported cell line to carry an endogenous R132H IDH1 mutation. However, BT054 was slow to expand in culture and lacked the ability to initiate tumors in NOD SCID mice,11 properties that we reasoned might be attributable to its derivation from a previously untreated oligodendroglioma.
We reasoned that the neurosphere system could be used to culture IDH1-mutant cells from more aggressive subtypes of glioma and that the NOD SCID brain might prove to be a more amenable environment for expansion of these cells. Furthermore, we hypothesized that 2-HG would be detectable in cell culture medium and serum of animals bearing intracranial xenografts of endogenous IDH1-mutant cells, thus demonstrating a means of conveniently monitoring mutant IDH1 enzymatic activity both in vitro and in vivo.
Materials and Methods
Brain Tumor Stem Cell Culture
Fresh tissue samples were obtained from a 38-year-old male patient during resection of an anaplastic oligoastrocytoma (oligoastrocytoma WHO grade III). Informed consent was obtained from the patient through the Brain Tumor and Tissue Bank at the University of Calgary. Cell culture of the fresh tissue sample was performed using the neurosphere assay, as described elsewhere.12 Neurospheres were evident approximately 2 weeks following plating and were grown for several weeks until they reached a size adequate for differentiation, serial passaging, and orthotopic xenografts. Neurospheres were cultured in serum-free culture medium (SFM) supplemented with EGF (20 ng/mL; Peprotech), FGF2 (20 ng/mL; R&D Systems), heparan sulfate (HS; 2 mg/mL; R&D Systems), and PDGF-AA (100 ng/mL; Peprotech), as described elsewhere.11,12 IDH1/2 wild-type GBM (BT012 and BT074) and IDH1 wild-type and mutant oligodendroglioma (BT088 and BT054) BTSCs were from previously established cultures.11,12
Differentiation Assay and Triple-Label Immunostaining
Tumor spheres were plated on poly-L-ornithine (Sigma-Aldrich)–coated glass coverslips and allowed to differentiate for 4 or 7 days in SFM only or in SFM supplemented with 1% fetal bovine serum (Invitrogen). Cells were fixed using 4% paraformaldehyde, and triple-label immunostaining was performed as previously described12 for glial fibrillary acidic protein (GFAP; rabbit anti-GFAP; 1:400; BTI), β-III-tubulin (1:1000; Sigma), and O4 (1:20; Chemicon).
DNA Sequencing
Genomic DNA was extracted from patient tumors, cell lines, and xenograft tumors using DNeasy (Qiagen) according to the manufacturer's instructions; 30 ng of DNA were used in 25 µL polymerase chain reaction (PCR) to amplify exon 4 of IDH1 and exon 2 of IDH2, as described elsewhere.1,2 RNA was also extracted from cell lines and xenograft tumors using the RNeasy kit (Qiagen) according to the manufacturer's instructions; 500 ng of RNA were reverse transcribed with the Superscript III First-Strand Synthesis System (Invitrogen) using oligo-dT primers (Invitrogen). Two microliters of cDNA were used in a 50 µL Platinum Hi-Fidelity PCR (Invitrogen) to amplify the IDH1 coding region using the primers 5′-GCAAGACTGGGAGGAACTGGGG-3′ and 5′GCCTTTATCCTTGAGTGTAACACAG-3′ for amplification and 5′-ACTCCTGATGAGAAGAGGGTTGAGG-3′ for sequencing. The EGFR open reading frame was amplified in 3 fragments (fragment 1: forward, 5′-GCCCCCTGACTCCGTCCAGT-3′; reverse, 5′-GTTGCTTGGTCCTGCCGCGT-3′; fragment 2: forward, 5′-CTCCACATCCTGCCGGTGGC-3′; reverse, 5′-GCACCAAGCGACGGTCCTCC-3′; fragment 3: forward, 5′-CTGGTGGTGGCCCTGGGGAT-3′; reverse, 5′-GCGGGCATGGCTGTTGGGAT -3′). Each fragment was then sequenced bidirectionally (fragment 1: forward strand, 5′-AGCTCTTCGGGGAGCAGCGA-3′; reverse strand, 5′-AGGCATGGAGGTCCGTCCTGT-3′; fragment 2: forward strand, 5′-GGGGTGACTCCTTCACACATACTCC-3′; reverse strand, 5′-TGCCCTTTGCGATCTGCACACA-3′; fragment 3: forward strand, 5′-CCAGCGTGGACAACCCCCAC-3′; reverse strand, 5′-TGCTGTGGCTTGGTCCTGGG-3′. PTEN and TP53 were amplified and sequenced as described elsewhere.13 PCR and reverse-transcription PCR products were purified by agarose gel electrophoresis and isolated with the QIAquick Gel Extraction Kit (Qiagen). Automated DNA sequencing was performed at the University of Calgary Core DNA Services facility.
1p/19q Codeletion Analyses
FISH to evaluate for 1p and 19q codeletion was performed on paraffin sections of tumor tissue as previously described.11 For SNP-based copy number analysis, 2 µg of cell line DNA were sent to Expression Analysis (Durham, NC) for Affymetrix SNP Array 6.0 analysis.
Orthotopic Xenografts in NOD SCID Immunocompromised Mice
Sphere cultures of BT142 were enzymatically dissociated (Accumax; Innovative Cell Technologies). Cells were washed in SFM, and viable cells were counted on a hemocytometer using Trypan blue exclusion. Ten thousand viable cells in 3 μL of SFM were stereotactically implanted into the right striatum of 6–8-week-old CB-17 NOD SCID mice (The Jackson Laboratory, Bar Harbor, ME) as previously described.12 Mice were sacrificed after visible symptoms of distress and weight loss, as defined by University of Calgary animal care guidelines. Brains were removed and observed for gross morphological evidence of tumor formation. Each brain was cut in the coronal plane to obtain 2 segments each containing part of the tumor and either processed for cryopreservation or formalin fixation.
Histology and Immunohistochemistry
For histological examination of patient tumor tissue and of brains from orthotopically xenografted SCID mice, hematoxylin and eosin (H&E) staining was performed according to standard protocols. For immunohistochemistry, heat-induced antigen retrieval with sodium citrate was performed on deparaffinized sections according to standard protocols. Mouse antihuman R132H IDH1 antibody14 (1:20; Dianova), mouse antihuman GFAP (1:400; R&D Systems), mouse antihuman myelin basic protein (MBP, 1:100; Abcam), mouse antihuman NeuN (1:100; Millipore), and rabbit antihuman Ki67 (1:250; Novocastra) were used for immunohistochemistry. For antihuman antinuclear antigen (hANA) staining, 10 μM cryo-sections were incubated with mouse anti-hANA antibody (1:10, Chemicon). Staining was visualized with Vectastain Elite mouse IgG or rabbit IgG ABC kits (Vector Laboratories) and DAB substrate (Sigma-Aldrich), followed by hematoxylin counterstaining.
Immunoblotting
Frozen patient tumor tissue, xenograft tumors, or normal NOD SCID mouse brain samples were lysed in RIPA buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% Na deoxycholate, and 1% NP40) and Complete Protease Inhibitor Cocktail Tablets (Roche); 20 μg of each protein lysate was separated by SDS-PAGE and transferred onto a nitrocellulose membrane according to standard protocols. Membranes were blocked in Tris-buffered saline with 5% nonfat dry milk and incubated overnight with mouse antihuman IDH1 R132H antibody (1:100; Dianova) at 4°C. Blots were stripped and stained with rabbit antihuman IDH1 antibody and goat antihuman Actin (1:500 and 1:2000; Santa Cruz Biotechnology), followed by donkey antimouse, goat antirabbit, or donkey antigoat horseradish peroxidase–conjugated secondary antibodies (1:6000; Millipore).
Metabolite Analyses
Conditioned media was collected from triplicate T25 flasks with 2 × 105 cells plated in 10 mL SFM with growth factors. Serum samples were obtained from mice xenografted with an IDH1 wild-type GBM cell line or BT142 after presentation of clinical symptoms of distress and/or weight loss. Mice were euthanized using CO2 inhalation, and blood was collected by cardiac puncture. Blood was clotted at room temperature and centrifuged to obtain serum. Serum or conditioned media samples were flash frozen in liquid nitrogen and stored at −80°C until processing for mass spectrometry. The presence of tumor and IDH1-mutant status were confirmed by histology and immunohistochemistry in brains of mice from which serum samples were collected.
GCMS Experimentation
Fifty-microliter aliquots of conditioned cell media or serum were extracted and modified as previously described.15 Samples were analyzed with a Waters GCT Premier mass spectrometer using parameters as follows: injector temperature of 275°C, He carrier gas flow rate of 1.2 mL/min, DB5-MS column (Agilent; 30 m × 0.25 mm × 0.25 μm), GC oven ramp (12°C/min) from 80°C to 320°C, followed by 8 min hold at 320°C. Ions between m/z 50–800 were collected and analyzed. The 2-HG to 2-KG ratio was determined using 2-HG ions (m/z 349, 247, 203) eluting at 11.909 min and 2-KG ions (m/z 304, 288, 198, 186, 170) eluting at 11.918 min. Metabolites were identified using the NIST library and the GOLM metabolites database.
Statistical Analyses
Two-tailed and 1-tailed Student's t tests were used for comparisons of median survival and mean 2-HG to 2-KG and mean 2-HG to total ions ratios, respectively.
Results and Discussion
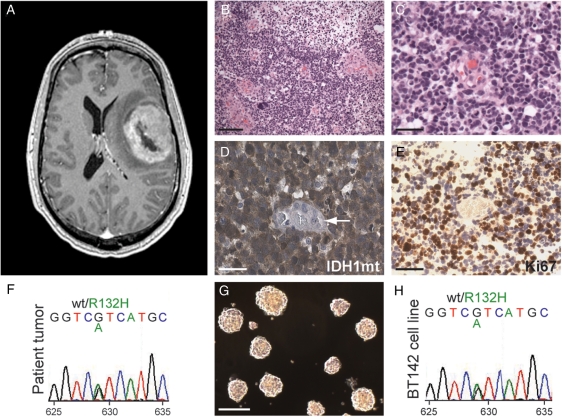
We obtained a tissue sample from a previously healthy 38-year-old man who presented to medical attention with a large ring-enhancing mass in the left frontal lobe (T1-weighted MR postgadolinium) (Fig. 1A). Microscopic examination showed a densely cellular tumor composed of tightly packed cells with rounded hyperchromatic nuclei and indistinct cytoplasm. In some areas of the tumor, increased nuclear pleomorphism and fibrillary cytoplasmic processes were noted. The resected tumor tissue also displayed high-grade features, including frequent mitoses, a high percentage of cells with Ki-67 labeling, microvascular proliferation, and extensive necrosis (Fig. 1B–D). Together, these histologic features led to a diagnosis of anaplastic oligoastrocytoma.
Fig. 1.
Culture of an endogenous IDH1-mutant BTSC line from an anaplastic oligoastrocytoma patient. (A) A large ring-enhancing lesion (T1-weighted MR postgadolinium) was present in the left frontal lobe of this patient. (B and C) H&E staining of resected tumor tissue showed dense cellularity, frequent mitotic figures, and microvascular proliferation leading to a diagnosis of anaplastic oligoastrocytoma (WHO grade III). Scale bar, 200 µm (B), 25 µm (C). (D and E) Strong expression of the R132H mutant IDH1 protein (D) and the proliferative marker Ki67 (E) was present by immunohistochemistry in tumor but not endothelial cells (arrow). Scale bars, 25 µm. (F) Sequencing of tumor DNA confirmed a heterozygous G to A transition at nucleotide 629 resulting in a substitution of histidine for arginine at codon 132 of IDH1. (G and H) A stable nonadherent, sphere-forming cell line (G) was established that retained the heterozygous R132H IDH1 mutation (H). Scale bar, 200 µm.
The relatively young age of presentation in this patient and oligoastrocytic morphology raised the possibility that this was an IDH1/2-mutant tumor. Immunohistochemistry with a mutant-specific antibody demonstrated strong expression of the R132H mutant IDH1 protein in tumor cells but not in nonneoplastic cells (Fig. 1E). Sequencing of tumor DNA confirmed the presence of a heterozygous G to A transition at nucleotide 629 resulting in a substitution of histidine for arginine at codon 132 of IDH1 (Fig. 1F). FISH analysis for codeletion of 1p/19q was inconclusive because of a high degree of chromosome 1 and 19 aneuploidy throughout the tumor.
A small portion of the resected tissue was placed into culture and, after 7–14 days, gave rise to nonadherent spheres (Fig. 1G) in multiple flasks that retained the heterozygous R132H IDH1 mutation observed in the parent tumor. The primary spheres were capable of self-renewal and gave rise to a stable cell line (BT142) that retained the IDH1 R132H mutation (Fig. 1H). Sequencing analysis showed no additional mutations in EGFR, PTEN, and TP53. SNP-based copy number analysis revealed small but detectable negative log-ratio values for multiple markers throughout 1p and 19q relative to markers on 1q and 19p that were consistent with relative whole arm losses of 1p and 19q in at least a subpopulation of BT142 cells (data not shown).
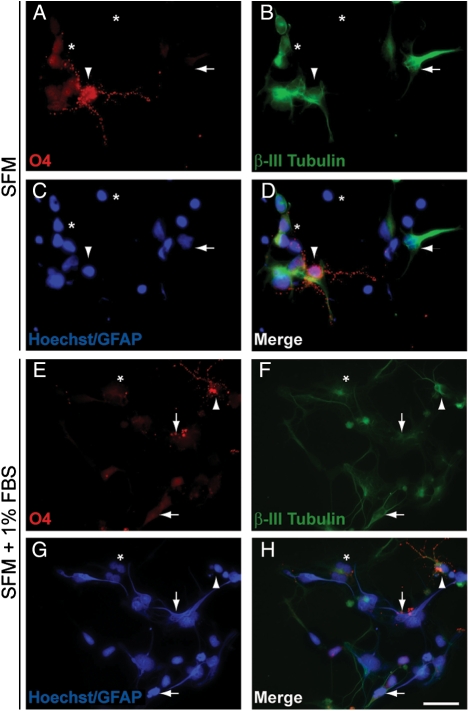
BT142 also demonstrated multilineage potential inherent to BTSCs. After 4 days of adherent culture in the absence of mitogens, a subpopulation of BT142 cells strongly expressed markers of oligodendroglial and neuronal lineages (Fig. 2A–D). Addition of 1% fetal bovine serum resulted in prominent expression of astrocytic lineage markers (Fig. 2E–H). Thus, we report the successful establishment of a stable line from a patient with IDH1-mutant anaplastic oligoastrocytoma, which retains the IDH1 mutation and displays the hallmark self-renewal and multipotency of BTSCs.
Fig. 2.
BT142 has multilineage potential. (A–D) After 4 days of adherent culture in the absence of mitogens, a subpopulation of BT142 cells expressed markers of oligodendroglial (arrowhead) and neuronal (horizontal arrow) lineages. Expression of cytoplasmic GFAP staining characteristic of astrocytes was weak and infrequent. Many cells (asterisks) also remained undifferentiated with minimal or no expression of any lineage markers. (E–H) With the addition of 1% FBS, a subpopulation of cells displayed cytoplasmic GFAP staining and astrocytic morphology (vertical arrow). Neuronal markers (horizontal arrow) were expressed in a subpopulation of cells and occasionally co-expressed with oligodendroglial markers (arrowhead). A subpopulation of cells (asterisk) also remained undifferentiated with no expression of any lineage markers. Nuclei were counterstained with Hoechst 33 258. Scale bar, 25 µm.
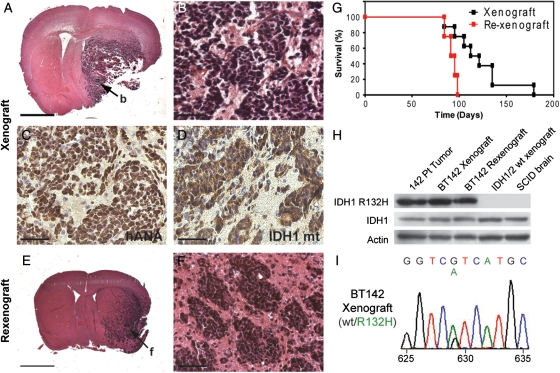
Although this cell line was stable, it remained consistently slow to expand after more than 6 months in culture. Because we and others10 have observed that existing in vitro culture conditions are suboptimal for IDH1/2-mutant cells, we reasoned that intracranial xenografts might provide a more amenable microenvironment for growth of these cells. A relatively small number of cells (1 × 104) were injected into the right striata of NOD SCID mice. After 12–14 weeks, mice began to lose weight and develop neurological symptoms. The tumors were uniformly lethal, and median survival in xenograft animals was 112 days after injection (Fig. 3G). At necropsy, the right hemisphere was grossly enlarged with abnormal soft grey tissue throughout the right striatum. On histology, large, densely cellular tumors expanded the right hemisphere and spread into the contralateral hemisphere (Fig. 3A and B). Morphologically, the tumor cells were poorly differentiated with enlarged hyperchromatic, irregularly round to angulated nuclei and scant cytoplasm. Immunohistochemistry confirmed that, within the tumor, the neoplastic cells, but not endothelial cells, were of human origin (Fig. 3C). The tumor cells also expressed the IDH1 R132H mutant protein (Fig. 3D). Further immunohistochemical studies revealed that xenograft tumors were intensively positive for Ki67 staining and were largely undifferentiated without evidence of expression of myelin basic protein, NeuN, or GFAP (data not shown). These results indicate that IDH1-mutant BT142 cells are tumor-initiating cells capable of aggressive growth in the brain microenvironment.
Fig. 3.
The NOD SCID brain provides an amenable environment that allows BT142 to rapidly expand while retaining its characteristic growth pattern and endogenous IDH1 mutation. (A and B) Orthotopic xenografts of BT142 cells resulted in large, densely cellular tumors with an angiocentric growth pattern. Scale bar, 2 mm (A), 50 µm (B). (C and D) Xenograft tumors expressed human nuclear antigen (C) and R132H mutant IDH1 protein confirming their origin from the implanted BT142 cells and retention of the IDH1 mutation. Scale bars, 50 µm. (E and F) Xenograft tumors were harvested, re-cultured as spheres, and re-xenografted to yield densely cellular tumors with prominent angiocentric and micronodular growth patterns. Scale bar, 2 mm (E), 50 µm (F). (G) BT142 xenograft tumors were uniformly lethal and re-xenograft tumors had a slightly shorter median survival (93 vs. 112 days) than the primary xenografts from which they were derived. (H) Both primary xenograft and re-xenograft tumors of BT142 retained strong expression of the IDH1 R132H mutant protein observed in the patient tumor. (I) Genomic sequencing of xenograft tumor DNA confirmed a heterozygous G to A transition in codon 132 of IDH1.
The stark contrast between the extremely slow in vitro growth and extremely aggressive in vivo growth of BT142 led us to ask whether serial passaging of BT142 through intracranial xenografts might be a more successful strategy for propagating this cell line. We found that tumors resulting from BT142 cells faithfully retained many of the phenoptypic and genomic characteristics of the parent tumor (Fig. 1B and C). Xenograft tumors were densely cellular with an extensive microvasculature and grew as large, aggressive, and relatively undifferentiated tumors with angiocentric growth (Fig. 3A and B). We were able to successfully reculture second-generation primary spheres out of xenografted tumors. Rexenografting 1 × 104 of these cells per mouse yielded tumors with dense cellularity and a prominent angiocentric growth pattern (Fig. 3E and F), thus phenocopying the histological appearance of the primary xenografts. Second-generation xenografts may have been modestly more aggressive than the primary xenograft tumors, resulting in a shorter median survival of 93 versus 112 days (Fig. 3G), but this was not statistically significant (P = .09). Strong expression of the R132H mutant IDH1 protein observed in the patient tumor was present in both primary and secondary xenografts (Fig. 3H) by Western blot analyses of tissue lysates. The heterozygous G to A transition in codon 132 was also confirmed by sequencing of primary and secondary xenografts (Fig. 3I). These results demonstrate that intracranial xenografts provide an effective means of propagating BT142 that also allows for faithful retention of its IDH1 mutation and characteristic growth pattern.
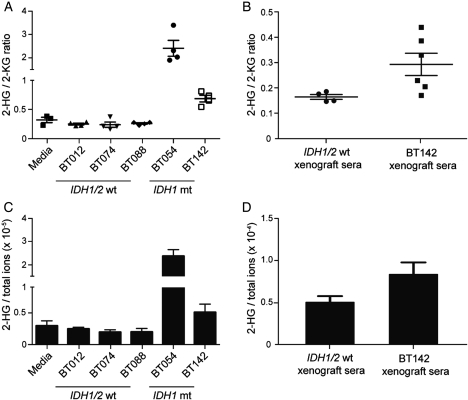
Ectopic expression studies have demonstrated that mutant IDH1/2 enzymes have a neomorphic ability to produce 2-HG3, and thus, 2-HG accumulation provides one of the few potential read-outs for mutant IDH1 enzymatic activity. Therefore, we investigated whether our endogenously IDH1-mutant BT142 cell line also produced excess 2-HG. We found that high levels of 2-HG could be detected in conditioned media from BT142 after 48 h with normalized 2-HG to 2-KG ratios significantly higher than those in conditioned medium from the IDH1/2 wild-type GBM BT074 (P = .0004, Fig. 4A). The 2-HG to 2-KG ratio was also significantly elevated in conditioned medium from our IDH1-mutant oligodendroglioma line BT054, compared with conditioned media from the IDH1/2 wild-type oligodendroglioma BT088 (P = .0004) (Fig. 4A). In contrast, 2-HG to 2-KG ratios in conditioned medium from 3 IDH1/2 wild-type lines were not different from those in fresh medium (BT012, P = .081; BT074, P = .141; BT088, P = .103). Furthermore, 2-HG accumulation could also be reliably detected in vivo with BT142 xenograft serum having significantly higher 2-HG to 2-KG ratios than those in serum from IDH1/2 wild-type xenograft mice (P = .025) (Fig. 4B). In further support of these findings, the ratio of 2-HG to the sum of all detected ions confirmed excess 2-HG in IDH1-mutant conditioned medium and xenograft serum (Fig. 4C and D). Thus, BT142 provides the first model system that recapitulates the genomic and metabolic alterations of IDH1/2-mutant glioma both in vitro and in vivo.
Fig. 4.
Endogenous IDH1–mutant BTSC lines produce 2-HG both in vitro and in vivo. (A) Elevated 2-HG to 2-KG ratios can be detected in conditioned medium from both BT054 and BT142 after 48 h when compared with conditioned medium from BT074 and BT088 (P = .0004). A similar accumulation of 2-HG was not detectable in IDH1/2 wild-type lines. (B) Serum samples from BT142 xenografted mice had a significantly higher mean ratio of 2-HG to 2-KG as compared to serum from IDH1/2 wild-type xenografted mice (P = .025). (C and D) 2-HG to total ions showed the same trend for IDH1-mutant lines in both conditioned media and xenograft serum.
The dramatic difference between the growth of BT142 in culture and in orthotopic xenografts suggests that the NOD SCID brain provides an amenable microenvironment for rapid expansion of these cells and retention of their IDH1 mutation. Further study of differences between BT142 in culture, compared with in the NOD SCID brain, may allow for more efficient propagation of IDH1-mutant cells in culture and improved modeling of IDH1-mutant glioma biology, leading to a better understanding of the mechanisms underlying IDH1-mediated gliomagenesis. Furthermore, BT142 fulfills a previously unmet role as the first in vivo model of endogenous IDH1-mutant glioma with enormous potential in drug development for IDH1-mutant tumors. Although BT142 may not supplant existing cell lines ectopically expressing the IDH1 mutation for initial high-throughput drug screens, it is an unrivaled, physiologically relevant model for critically important secondary in vitro and in vivo validation of any putative hits prior to phase I/II clinical trials.
Finally, we demonstrate here, for the first time to our knowledge, that elevated 2-HG production results from an endogenous IDH1 mutation in an in vivo model system. Although the functional consequences of 2-HG accumulation for tumor biology have yet to be fully elucidated,4 our finding that 2-HG permeates the systemic circulation validates 2-HG as a potential biomarker of glioma. A serum measurement of the 2-HG to 2-KG ratio or, possibly, the absolute 2-HG concentration may be a useful tool for noninvasive early diagnosis of IDH1/2-mutant gliomas below detection on MRI or monitoring treatment response, if the magnitude of the 2-HG to 2-KG ratio accurately reflects tumor extent. However, although a recent study has confirmed 2-HG accumulation in serum samples from patients with IDH1/2–mutant acute myeloid leukemia, it was unable to detect a similar correlation between tumor volume and absolute levels of 2-HG in serum samples from patients with IDH1/2-mutant gliomas.16 Of note, this study was performed on a relatively small cohort of WHO grade II and III gliomas. Analysis of a larger, more diverse set of IDH1/2-mutant gliomas and consideration of the effects of necrosis and radiotherapy-induced breakdown of the blood-brain barrier on systemic 2-HG permeation will likely be needed to fully evaluate serum 2-HG levels for use in diagnostic applications. Moreover, our study also used the ratio of 2-HG to 2-KG, which accentuates the imbalance between these 2 metabolites and, thus, may provide a more sensitive screening tool for IDH1/2-mutant gliomas than an increase in the absolute concentration of 2-HG. Our data confirm that 2-HG is present at high levels in conditioned medium from BT54 and BT142 cultures and can be consistently detected in serum from BT142 xenograft mice, thus providing an accessible means of validating drugs targeting endogenous IDH1-mutant enzymes. Therefore, BT142 provides an unparalleled model for investigation of IDH1-mutant tumor biology and development of novel therapeutics efficacious against this unique subgroup of glioma.
Conflict of interest statement. None declared.
Funding
This work was supported by an Alberta Cancer Foundation (ACF); grant to S.W. and J.G.C. O.S., J.K., J.C., and S.W. were recipients of Studentship, Fellowship, Clinical Investigator, and Scientist Awards, respectively, from the Alberta Heritage Foundation for Medical Research. The Metabolomics Research Centre is supported by the ACF, Alberta Innovates Health Solutions, and the University of Calgary.
Acknowledgments
We thank Rozina Hassam, Dorothea Livingstone, and Orsolya Cseh for technical help; Manpreet Kalkat and Jing Wen for help with mass spectrometry procedures; and Andrea Ramirez and Carmen Binding for help with mouse xenografts and monitoring. The patient samples were obtained through the Calgary Brain Tumour and Tissue Bank, generously supported by funds from the Clark H. Smith Family. H.A.L., O.D.S., J.G.C., and S.W. contributed equally to this work.
References
- 1.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. doi:10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. doi:10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. doi:10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reitman ZJ, Jin G, Karoly ED, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci USA. 2011;108:3270–3275. doi: 10.1073/pnas.1019393108. doi:10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green A, Beer P. Somatic mutations of IDH1 and IDH2 in the leukemic transformation of myeloproliferative neoplasms. N Engl J Med. 2010;362:369–370. doi: 10.1056/NEJMc0910063. doi:10.1056/NEJMc0910063. [DOI] [PubMed] [Google Scholar]
- 6.Zhao S, Lin Y, Xu W, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. doi:10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J Natl Cancer Inst. 2010;102:932–941. doi: 10.1093/jnci/djq187. doi:10.1093/jnci/djq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. doi:10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prensner JR, Chinnaiyan AM. Metabolism unhinged: IDH mutations in cancer. Nature Medicine. 2011;17:291–293. doi: 10.1038/nm0311-291. doi:10.1038/nm0311-291. [DOI] [PubMed] [Google Scholar]
- 10.Piaskowski S, Bienkowski M, Stoczynska-Fidelus E, et al. Glioma cells showing IDH1 mutation cannot be propagated in standard cell culture conditions. Br J Cancer. 2011;104:968–970. doi: 10.1038/bjc.2011.27. doi:10.1038/bjc.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly JJ, Blough MD, Stechishin OD, et al. Oligodendroglioma cell lines containing t(1;19)(q10;p10) Neuro Oncol. 2010;12:745–755. doi: 10.1093/neuonc/noq031. doi:10.1093/neuonc/noq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly JJ, Stechishin O, Chojnacki A, et al. Proliferation of human glioblastoma stem cells occurs independently of exogenous mitogens. Stem Cells. 2009;27:1722–1733. doi: 10.1002/stem.98. doi:10.1002/stem.98. [DOI] [PubMed] [Google Scholar]
- 13.Kato H, Kato S, Kumabe T, et al. Functional evaluation of p53 and PTEN gene mutations in gliomas. Clin Cancer Res. 2000;6:3937–3943. [PubMed] [Google Scholar]
- 14.Capper D, Reuss D, Schittenhelm J, et al. Mutation-specific IDH1 antibody differentiates oligodendrogliomas and oligoastrocytomas from other brain tumors with oligodendroglioma-like morphology. Acta Neuropathol. 2011;121:241–252. doi: 10.1007/s00401-010-0770-2. doi:10.1007/s00401-010-0770-2. [DOI] [PubMed] [Google Scholar]
- 15.Bligh E, Dyer W. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. doi:10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 16.Capper D, Simon M, Langhans CD, et al. 2-hydroxyglutarate concentration in serum from patients with gliomas does not correlate with IDH1/2 mutation status or tumor size. Int J Cancer. 2011 doi: 10.1002/ijc.26425. doi:10.1002/ijc.26425. [DOI] [PubMed] [Google Scholar]