Abstract
Metabolic memory (MM) is the phenomenon whereby diabetes complications persist and progress after glycemic recovery is achieved. Here, we present data showing that MM is heritable and that the transmission correlates with hyperglycemia-induced DNA hypomethylation and aberrant gene expression. Streptozocin was used to induce hyperglycemia in adult zebrafish, and then, following streptozocin withdrawal, a recovery phase was allowed to reestablish a euglycemic state. Blood glucose and serum insulin returned to physiological levels during the first 2 weeks of the recovery phase as a result of pancreatic β-cell regeneration. In contrast, caudal fin regeneration and skin wound healing remained impaired to the same extent as in diabetic fish, and this impairment was transmissible to daughter cell tissue. Daughter tissue that was never exposed to hyperglycemia, but was derived from tissue that was, did not accumulate AGEs or exhibit increased levels of oxidative stress. However, CpG island methylation and genome-wide microarray expression analyses revealed the persistence of hyperglycemia-induced global DNA hypomethylation that correlated with aberrant gene expression for a subset of loci in this daughter tissue. Collectively, the data presented here implicate the epigenetic mechanism of DNA methylation as a potential contributor to the MM phenomenon.
The results of several large-scale diabetes mellitus (DM) clinical trials suggest that once initiated, diabetes complications persist and continue to progress unimpeded even when glycemic control is achieved through pharmaceutical intervention (1–4). This persistence has been supported by multiple lines of experimental laboratory evidence (5–10) and collectively indicate that the initial hyperglycemic period results in permanent abnormalities in the target organs. This harmful phenomenon has been termed metabolic memory (MM) (11,12). The underlying molecular mechanisms of DM complications and MM may include the involvement of excess reactive oxygen species, the involvement of advanced glycation end products (AGEs), and/or alterations in tissue-wide gene expression patterns (13–16). However, the ability to sustain these complications in the absence of hyperglycemia invokes a role for the epigenome in perpetuating these complications through MM.
The study of heritable changes in gene expression that cannot be explained by changes in the DNA sequence is referred to as epigenetics. There are several epigenetic processes that allow for altered gene expression including histone modification, (de)methylation of cytosine residues within DNA, inclusion of histone variants in octomers, and control by noncoding RNAs (17–20). Altogether, these processes allow cells/organisms to quickly respond to changing environmental stimuli (21–23) and also confer the ability for the cell to “memorize” these encounters once the stimulus is removed (24,25). Recent in vitro investigations into the epigenome’s role in MM have documented specific hyperglycemia-induced histone modifications that persist in the posthyperglycemic (MM) state (26–32). Currently, these reports are somewhat limited to specific modifications or loci, but it is expected that a more global approach will reveal a wealth of information.
Many roles have been ascribed to the presence of 5-methyl cytosine residues in DNA: gene silencing, silencing of transposable elements, developmental regulation of transcription, cell-cycle control, differentiation, and, recently, the activation of genes (33–35). Not unexpectedly, because of the critical role in gene expression, aberrant DNA methylation has been associated with several human diseases, including the susceptibility to DM (36–39) or its complications (40,41). Furthermore, the induction of tissue-specific hypomethylation was reported in a type 1 DM rat model (42). Altered DNA methylation may become particularly important in the context of MM, as only DNA methylation is backed by strong mechanistic support for heritability of epigenetic changes (33,43). In this report, we present data documenting that hyperglycemia induces global demethylation and aberrant gene expression, which both persist following euglycemic restoration in a zebrafish model of type 1 DM.
RESEARCH DESIGN AND METHODS
Zebrafish husbandry, streptozocin injection, and fasting blood glucose determination.
The maintenance of zebrafish (Danio rerio) stocks, the induction of hyperglycemia, and blood glucose determinations were performed as previously described (44). All procedures were performed following the guidelines described in “Principles of Laboratory Animal Care” (National Institutes of Health publication no. 85-23, revised 1985) and the approved Institutional Animal Care and Use Committee animal protocol 08-19.
Serum insulin quantification.
Serum was collected from six fish and pooled in order to perform an insulin ELISA assay (Calbiotech, Spring Valley, CA) following the manufacturer’s protocol without exception. Triplicate samples were generated, and the mean and experimental errors were determined for the three samples.
Immunohistochemistry.
Pancreas and fin tissue sections were prepared and immunohistochemistry was performed as previously described (44). The primary antibody for AGEs and secondary antibodies were purchased from Abcam plc (Cambridge, MA). All images (×40) were collected using a Nikon Eclipse 80i microscope (Nikon, Melville, NY) equipped with a digital camera and NIS Elements software.
Fin regeneration and skin wound–healing assays.
The fin regeneration studies were performed as previously described (44). For skin wound healing, fish were anesthetized using 2-phenoxythethanol and a 1-mm-diameter circular wound was created using a skin biopsy punch (Xemax, Napa, CA) on the lateral posterior aspect of the body wall of the fish just proximal to the caudal fin. After wounding, the percentage of wound closure was determined by comparing the remaining open-wound area (24 h postwounding) with the original wound area. Fish were maintained in individual tanks for the duration of the observation period.
RNA extraction.
Triplicate samples from 15 caudal fins were collected (from acute DM, MM, and control [C] fish) and immediately placed into TRIzol (Life Technologies, Carlsbad, CA). The samples were homogenized with 20 strokes of both A and B pestles using a 2-mL Dounce homogenizer (Fisher Scientific, Pittsburgh, PA). The debris was pelleted by centrifugation in a microcentrifuge at maximum speed. The supernatant was collected, and the RNA was extracted via the Purelink RNA mini-kit (Life Technologies) without exception. The final RNA concentration was determined by an absorbance reading at 260 nm.
Quantitative real-time RT-PCR.
RNA samples were used to generate cDNA using the superscript III first-strand synthesis kit (Life Technologies). Gene expression was analyzed using the cDNA as a template for real-time RT-PCR analysis using the SYBR green system based on real-time detection of fluorescence accumulation under the manufacturer’s recommended cycle conditions (Life Technologies). The primer sequences for rela (p65) used were as follows: forward-GGAGAAGCGCAAGAGAACTGA and reverse-CGTAGGGAATGGCCGTCTTT. To control for variation in the amount of cDNA that was available for PCR in each sample, expression of the target sequence was normalized in relation to an endogenous control 18S rRNA (Quantum RNA Universal 18S; Life Technologies). Each experiment was performed on three different samples with three replicates of each. Results were expressed relative to RNA from control-injected zebrafish, which were assigned an arbitrary value of 1.
DNA extraction and methylation DNA immunoprecipitation sequencing.
Samples from 10 caudal fins were collected from C, day-0 DM, and day-60 MM zebrafish caudal fin tissue and immediately processed via the Purelink genomic DNA mini-kit (Life Technologies) without exception.
We used a methylated DNA (MeDIP) immunoprecipitation–based technique to profile the genome-wide methylation levels. For sequencing the enriched fragments on the Illumina Genome Analyzer IIx (Illumina, San Diego, CA), a modified workflow was used to prepare the Genome Analyzer compatible sequencing library. In brief, 1 μg sonicated DNA was end repaired, A tailed, and ligated to single-end adapters following the standard Illumina genomic DNA protocol. Immunoprecipitation of MeDIP was performed using a mouse monoclonal anti–5-methylcytosine antibody (Diagenode, Newark, NJ). A nonspecific mouse IgG immunoprecipitation was performed in parallel to MeDIP immunoprecipitation as a negative control. MeDIP and supernatant DNA were purified using Qiagen MinElute columns (Qiagen, Valencia, CA), and this was followed by an adapter-mediated PCR step to enrich the adapter-ligated DNA fragments. The libraries were quality controlled using an Agilent DNA Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). An aliquot of each library was diluted in elution buffer to 5 ng/μL, and 1 μL was used in real-time PCR reactions to confirm the enrichment for methylated regions. Flow cells were prepared with 8 pmol/L DNA according to the manufacturer’s recommended protocol and sequenced for 36 cycles on Genome Analyzer IIx.
Image analysis and base calling were performed using Solexa pipeline V1.8 (Illumina) with default settings provided by Illumina (Off-Line Basecaller Software v1.8) and short reads (a pass on the Solexa CHASTITY quality filter) were aligned to the zebrafish genome (Zv8 in the UCSC Genome Browser) using alignment software Bowtie (V0.12.7). Approximately 21 million total uniquely mapped reads, which represent >768 million bases of sequence, were obtained for the three samples (C, 7,892,236; DM, 6,595,012; and MM, 6,864,707). Each read was considered a 250–base pair block extended from the reads mapping position, which represents an entry for an MeDIP-enriched DNA fragment. Then, the absolute methylation level for each CpG island or for each promoter region of the RefSeq genes (UCSC Genome Browser, Zv8) was determined by the number of extended reads per kilobases (kb) (33). To further quantify the DNA methylation level of any specific region, we calculated the MeDIP score. The MeDIP score is defined as the number of extended reads per kb. To compare the DNA methylation profiles of multiple samples, the total number of uniquely mapped reads of each sample was normalized by the total number of reference reads (median total number of uniquely mapped reads of all samples), and the raw extended read counts were normalized accordingly. After normalization, the MeDIP score was calculated for a specific region according to the length of that region. The DNA methylation status of a specific region was defined as unmethylated if its MeDIP score was <15.6 reads per kb, as partially methylated if its MeDIP score was between 15.6 and 60.2 reads per kb, and as completely methylated if its MeDIP score was >60.2 reads per kb.
Microarray analysis.
Triplicate RNA samples (extraction procedure described above) were used to probe the previously established Affymetrix GeneChip Zebrafish Genome Array (Affymetrix, Santa Clara, CA), which contains 15,509 probe sets designed to interrogate expression of 14,900 D. rerio transcripts. Microarray analysis was conducted according to the manufacturer’s instructions for the Affymetrix 3′ IVT Express kit, and RNA quality was assessed by the Agilent Bioanalyzer. All control parameters were confirmed to be within normal ranges before normalization, and data reduction was initiated. Partek GS 6.5 software (Partek, St Louis, MO) was used for microarray data analysis. The raw data (.cel files) were processed with the GC Robust Multi-array Average (GCRMA) algorithm, and differentially expressed genes were identified using ANOVA. A gene was considered differentially expressed if two conditions were met: 1) expression fold change between two groups was ≥1.5 and 2) the false discovery rate (FDR) step-up corrected P value was ≤0.05.
RESULTS
Cessation of streptozocin administration allows for a return to euglycemia in the zebrafish.
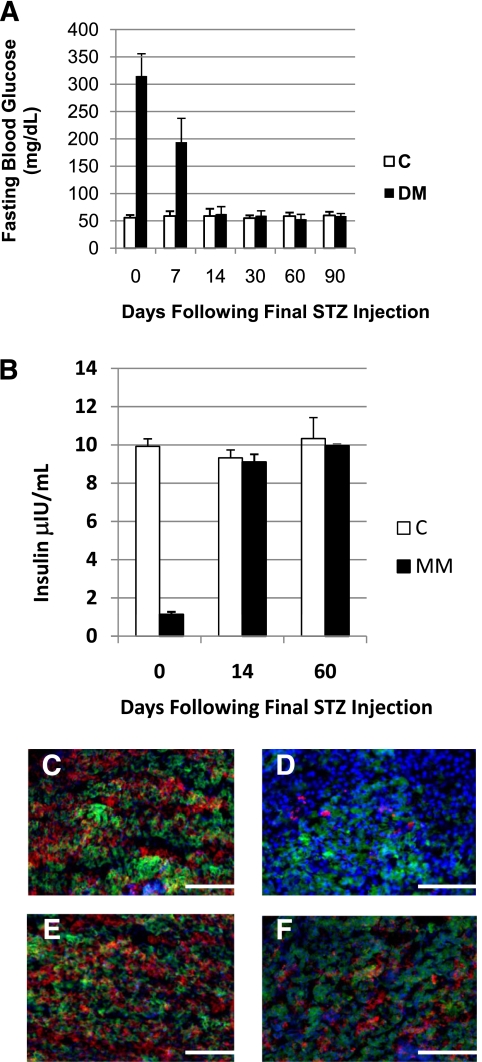
Our laboratory has previously reported that an adult zebrafish model of type 1 DM can be induced by administration of the diabetogenic drug streptozocin (44). Based on previous studies (45), we hypothesized that removal of streptozocin exposure would allow for the endogenous regeneration of pancreatic β-cells, resulting in a return to normal glycemia without the need for insulin injection or pharmaceutical intervention. In order to test this, hyperglycemia (∼315 mg/dL) was induced in a group of zebrafish by our standard 3-week induction protocol (the end of induction considered day 0) and fasting blood glucose levels (FBGLs) of these fish were assessed at various time points post–drug removal (Fig. 1A). Within the first 7 days of drug removal, the diabetic fish were still hyperglycemic but to a lesser extent (∼194 mg/dL); by 14 days, the fish had returned to a euglycemic state (∼60 mg/dL). Zebrafish that were previously hyperglycemic and recovered to normal FBGLs following streptozocin drug removal will be referred to as MM fish from here onward.
FIG. 1.
Fasting blood glucose normalizes and insulin production increases following cessation of streptozocin (STZ) administration. A: Graphic representation of fasting blood glucose concentration. Day 0: DM 315 ± 40.96 mg/dL, C 55.9 ± 4.91 mg/dL; P < 0.001. Day 7: DM 194.0 ± 43.7 mg/dL, C 58.89 ± 8.83 mg/dL; P < 0.05. Day 14: DM 62.5 ± 13.6 mg/dL, C 58.88 ± 13.36 mg/dL. Day 30: DM 59.5 ± 8.88 mg/dL, C 54.9 ± 5.19 mg/dL. Day 60: DM 53.1 ± 8.83 mg/dL, C 58.6 ± 6.70 mg/dL. Day 90: DM 58.66 ± 4.59 mg/dL, C 59.9 ± 6.65 mg/dL. n = 12 for all groups. B: Graphic comparison of insulin levels determined by enzyme-linked immunosorbent assay. Day 0: DM 1.13 ± 0.14 μIU, C 9.92 ± 0.40 μIU. Day 14: DM 9.10 ± 0.41 μIU, C 9.32 ± 0.42 μIU. Day 60: DM 9.51 ± 0.08 μIU, C 10.33 ± 1.1 μIU; n = 3 for all samples. Representative immunofluorescene microscopy images of pancreas tissue indicating presence of insulin (red), glucagon (green), and DAPI (blue). C: Control injected day 0. D: Streptozocin injected day 0. E: Control injected day 14. F: Streptozocin injected day 14. The scale bar indicates 50 μm. (All immunohistochemistry, n = 10.) (A high-quality digital representation of this figure is available in the online issue.)
The mechanism by which streptozocin induces diabetes is the selective ablation of pancreatic β-cells resulting in reduced serum insulin levels and, ultimately, poor glycemic control. As such, we determined whether the glycemic recovery observed in day-14 MM fish was accompanied by a repopulation of β-cells and increased serum insulin levels. Pooled samples of serum insulin (six fish per pool) and pancreas tissue were collected and analyzed from DM fish, day-14 MM zebrafish, and accompanying C fish. This analysis revealed that the serum insulin levels of DM fish were approximately 11% those of C fish and that by 14 days after drug removal, these levels had been restored to normal (Fig. 1B). Pancreas tissue was fixed, and serial 10-μm transverse cryosections were made. The sections were then processed using double indirect immunofluorescence to visualize insulin (red) and glucagon (green), and the slides were counterstained with DAPI to visualize nuclei. Epifluorescence microscopy indicated that the insulin signal was greatly reduced in DM islets when compared with C, as we have previously documented (Fig. 1B and C). Although full recovery was not seen (Fig. 1D and E) by day 14, there was a significant repopulation of insulin-producing β-cells in pancreas tissue taken from MM zebrafish versus zebrafish in the acute DM state (compare Fig. 1C and E). Nonetheless, serum insulin levels returned to normal within 14 days (Fig. 1B).
Caudal fin regeneration and skin wound healing are susceptible to MM.
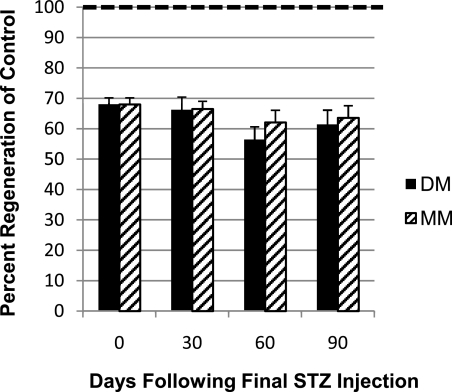
Type 1 DM zebrafish not only display the known secondary complications of retinopathy and nephropathy but also exhibit an additional complication: impaired caudal fin regeneration (44). Initially, we investigated whether this complication was susceptible to the MM phenomenon by observing the regenerative capacity of C, acute DM (fish that were maintained in the acute diabetic state via continued streptozocin injection beyond time 0), and MM fish. Regeneration in these groups was determined by measuring caudal fin outgrowth 48 h postamputation at 30, 60, and 90 days post–drug removal of the MM group (Fig. 2). Fin regeneration of DM fish was impaired compared with C fish, as previously described (44). Unexpectedly, the MM group’s ability to regenerate was reduced to levels nearly identical to those in the DM fish after their return to a euglycemic state.
FIG. 2.
Impaired fin regeneration persists after recovery from the diabetic state. Graphic representation of the relative fin regeneration (48 h postamputation) of DM and MM fish compared with C fish. Day 0: DM and MM 68.0 ± 2.18%. Day 30: DM 66.2 ± 4.24%, MM 66.5 ± 2.53%. Day 60: DM 56.4 ± 4.25%, MM 62.1 ± 3.95%. Day 90: DM 61.4 ± 4.71%, MM 63.6 ± 3.95%. The number of replicates is as follows. Day 0: C, n = 39; DM, n = 69. Day 30: C, n = 30; DM, n = 32; MM, n = 37. Day 60: C, n = 25; DM, n = 12; MM, n = 16. Day 90: C, n = 20; DM, n = 10; MM, n = 10. (In all cases, P < 0.001 compared with C.) The rate of C regeneration was set to be 100% and is indicated by the dotted line. STZ, streptozocin.
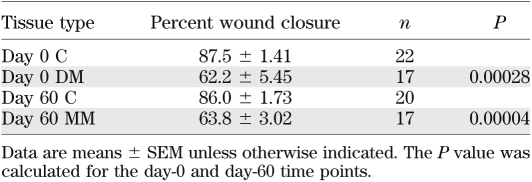
In order to ensure that this effect was not limited to caudal fins, we also investigated the rate of skin wound healing for the same three groups. Following wounding, the percentage of wound closure was determined by comparing the remaining open-wound area (24 h postwounding) with the original wound area (Table 1). These results indicated that wounds of DM fish heal slower than those of their C counterparts and that this reduction is maintained in 60-day MM fish. These data clearly demonstrate that fin regeneration and skin wound healing remain impaired after glycemic homeostasis is achieved and are susceptible to the MM phenomenon.
TABLE 1.
Wound healing is impaired in both the DM and MM states
MM is heritable to daughter cells in vivo.
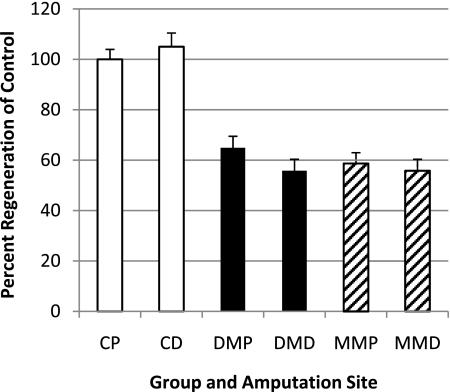
We took advantage of the zebrafish’s regenerative ability to examine the heritable nature of MM in vivo. These experiments required multiple rounds of caudal fin amputation, and the same three groups (C, DM, and MM) were used. At all time points presented below, a subset of fish was used for FBGL determination and yielded results as documented in Fig. 1 (data not shown). At day 0, all groups had their caudal fins amputated, their 48-h regenerative growth was documented, and they were allowed a 30-day period for regrowth. The caudal fins of these fish were then reamputated at 30 days, immediately proximal to the original transection line, and the fish were once again allowed a period of regenerative growth for an additional 30 days. We have termed the tissue grown from 30 to 60 days MM tissue (Supplementary Fig. 1, available in the Supplementary Data [hatched bars]). At 60 days, the caudal fins were once again amputated, but at this time the fish were split into two groups (for schematic, see Supplementary Fig. 1). For one-half, the caudal fin was cut immediately proximal to the original amputation site (Supplementary Fig. 1 [dotted line P, proximal cut]), and amputation for the other group was performed within the MM tissue (Supplementary Fig. 1 [dotted line D, distal cut, tissue never exposed to hyperglycemia]). The regeneration ability was determined at 48 h postamputation and was expressed as the percent regeneration of C (Fig. 3). First, these data document that there were not any differences detectable in the amount of regenerative growth, whether the amputation was performed proximally or distally. (Compare proximal versus distal for each group [Fig. 3].) Second, as expected the regenerative ability of DM fish was impaired (Fig. 3). Third, these experiments revealed that the MM tissue’s ability to regenerate was impaired to the same extent as both the tissue from which it was derived and DM tissue (Fig. 3) These results indicate that the effect of MM was transmitted to the daughter cells.
FIG. 3.
Daughter tissue inherits the regeneration deficit of the previous hyperglycemic state. Graphic representation of caudal fin regeneration 48 h postamputation. (C proximal [CP] was set as 100%.) C proximal: 100 ± 3.96%. C distal (CD): 105 ± 5.45%. DM proximal (DMP): 64.9 ± 4.58%. DM distal (DMD): 55.8 ± 4.54%. MM proximal (MMP): 58.7 ± 4.27%. MM distal (MMD): 55.8 ± 4.54%. In all cases, P < 0.01 compared with C. n = 12 for all groups except MM proximal and distal (n = 16).
MM tissue does not accumulate AGEs or express increased amounts of rela.
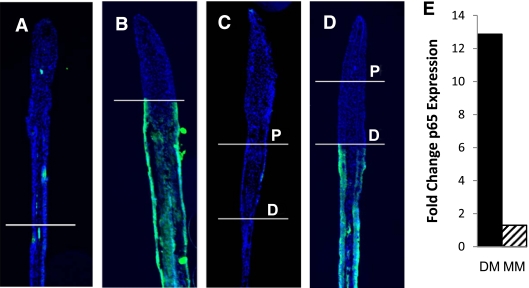
We then inspected MM tissue for the presence of AGEs and oxidative stress, as it has been reported that these may underlie diabetes complications and MM (13,15,16). Immunofluorescence microscopy was performed to detect the presence of AGEs present in C and DM tissue and 60-day MM regenerating caudal fins (48 h postamputation). Serial longitudinal 10-μm cryosections were processed by immunofluorescence to visualize AGEs (green) and were counterstained with DAPI to visualize nuclei (blue) (Fig. 4 A–D). Tissue in C caudal fins (Fig. 4A and C) and regenerating tissue of DM fins (tissue above top amputation line [Fig. 4B and D]) were not found to contain AGEs. In contrast, all of the tissue exposed to hyperglycemia (below the distal amputation line of DM and below the proximal line in MM fish) produced a significant AGE-specific signal (Fig. 4B and D). Importantly, the MM tissue (tissue between the two amputation lines) in the MM fins (Fig. 4D) did not produce any fluorescence and did not appear to accumulate AGEs.
FIG. 4.
MM tissue does not accumulate AGEs or markers of oxidative stress. Fluorescence microscopy images indicating the presence of AGEs (green) and DAPI (blue) in regenerating caudal fin tissue are presented as indicated. A: Day 0, control. B: Day 0, streptozocin. C: Day 60, C. D: Day 60, MM. The amputation sites are indicated on the images, and for the 60-day fish both the distal cut (D) and proximal cut (P) are specified. Scale bar represents 50 μm (n = 10). E: Graphic representation of quantitative PCR analysis of rela expression. DM: 12.87-fold ± 1.86-fold increase compared with C fish. MM: 1.3-fold ± 0.42-fold increase compared with C fish. (A high-quality digital representation of this figure is available in the online issue.)
The expression of nuclear factor-κB subunit p65 (rela) has been used as a marker of oxidative stress as well as stress signaling in DM and MM research (8,10,29). We analyzed the expression of this stress marker via RT–quantitative PCR with RNA that was extracted from caudal fin tissue of C, DM, and MM zebrafish. RNA from DM zebrafish was found to have a 12.8-fold increase in the expression of rela. However, RNA extracted from MM tissue of MM fish did not yield a significant increase in the amount of rela expressed over that of C fish (Fig. 4E). Hence, similar to the above AGE results, there does not appear to be increased reactive oxygen species–induced stress signaling in caudal fin tissue that has not been exposed to the hyperglycemic environment.
Hyperglycemia induces global DNA hypomethylation changes that are largely retained in MM tissue.
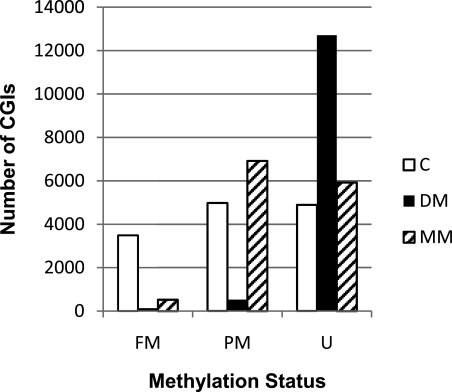
As MM is transmissible in fin tissue, and since we could not document the presence of AGEs or increased oxidative stress in metabolic memory tissue, we turned our focus to potential epigenetic changes. More specifically, we examined the presence of persistent DNA methylation changes induced by the hyperglycemic environment. Caudal fins were amputated and DNA was extracted from DM fish and their C fish. Sixty-day MM fish were then generated from this group, and DNA was extracted from the MM caudal fin tissue. The methylation status of 13,361 CpG islands within these DNAs was identified via the MeDIP-sequencing technique. This investigation revealed that hyperglycemia induced genome-wide demethylation, as the number of fully methylated CpG islands was reduced from 3,489 in C fish to 130 in the acute hyperglycemic state and the number of hypomethylated CpG islands concomitantly increased to 12,705 from 4,895 (Fig. 5). CpG island analysis of DNA from the MM fins revealed that a hypomethylated state persisted, as the number of fully methylated genes remained very low (C 3,489 vs. MM 526 [Fig. 5]), hypomethylated genes remained increased (C 4,895 vs. MM 5,924), and partially methylated genes increased (C 4,977 vs. MM 6,917) compared with C fish. These results were verified for a subset of loci via a MeDIP quantitative PCR approach (data not shown). Lastly, the induced demethylation appeared to be global, given that the results were similar irrespective of island location (intergenic, intragenic, and promoter located CpG islands [Supplementary Table 1]).
FIG. 5.
Hyperglycemia induces heritable persistent global DNA hypomethylation. Graphic representation of the methylation status of the 13,361 CpG islands (CGIs) examined. Fully methylated (FM): C, 3,489; DM, 130; MM, 520. Partially methylated (PM): C, 4,977; DM, 526; MM, 6,917. Unmethylated (U): C, 4,895; DM, 12,705; MM, 5,924.
Aberrant DNA methylation correlates with persistent gene expression changes.
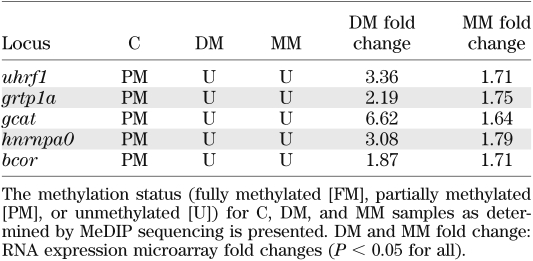
We used microarray analysis to examine gene expression changes that were induced by hyperglycemia and maintained in the MM state. We combined these data with the above-mentioned MeDIP-sequencing data and used Partek software to find genes whose changed expression correlated with induced promoter DNA hypomethylation. To do this, we used gene symbols to map genes to themselves in the two datasets. We then filtered the resulting datasets on three parameters: fold change (increased or decreased by 1.5-fold), FDR corrected P value (<0.05), and methylation status (unmethylated). From this analysis, of the approximately 4,207 genes altered in DM tissue, 60 (1.4%) were subjected to hyperglycemia-induced demethylation (Supplementary Table 2). When this was performed for MM tissue, 268 genes were identified as altered in their expression, of which 9 (3.3%) were demethylated in response to the previous hyperglycemic state (Supplementary Table 3). Most importantly, five of the nine loci (uhrf1, grtp1a, gcat, hnrnpa0, and bcor) identified in MM tissue were also identified in DM tissue, correlating a persistence of both hypomethylation and aberrant gene expression with this subset (Table 2).
TABLE 2.
MeDIP quantitative PCR and RNA expression analysis
DISCUSSION
Initial investigations into the pathogenesis of MM have revealed epigenetic changes, and more specifically, histone modifications that are maintained in the posthyperglycemic environment (26–32). Here, we provide evidence that the complications of reduced fin regeneration and impaired wound healing induced by hyperglycemia in a zebrafish model of DM are maintained through the MM phenomenon after a euglycemic state has been achieved. This analysis was possible because zebrafish are capable of regenerating insulin-producing pancreatic β-cells (45). Additionally, we have been able to separate a transmissible component of MM from potentially complicating factors of the previous hyperglycemic state such as AGE accumulation and oxidative stress in vivo. This has allowed us to examine epigenetic changes associated with aberrant DNA methylation and report for the first time that hyperglycemia induces genome-wide demethylation that is maintained in the MM state. In addition, these studies have revealed a correlation between induced DNA demethylation and induced gene expression changes for a subset of genes, including a member of the epigenetic code regulatory machinery complex.
To investigate the underlying mechanisms of MM in these fish, we developed a working hypothesis that the transmissible element in this process could be in the form of harmful products such as oxidative stress and AGEs or, alternatively, could be due to changes in epigenetic modifications. Our results ruled out oxidative stress and AGEs as contributing factors in MM of euglycemic zebrafish and led us to a detailed analysis of the most likely “transmissible” epigenetic process, DNA methylation.
Multiple epigenetic processes may mechanistically support persistent hyperglycemia-induced changes. Of these processes, DNA methylation has the strongest experimental support for heritability (33) and could contribute to the heritable transmission observed in impaired fin regeneration of MM tissue. CpG island methylation analysis via Me-DIP sequencing and Me-DIP quantitative PCR revealed a hyperglycemia-induced genome-wide demethylation of CpG islands relative to C fish. Moreover, the induced demethylation was transmitted to daughter cells, as hypomethylation was largely maintained in MM tissue. Our result is consistent with that of a previous study in which Williams et al. (42) performed a simple methylcytosine incorporation assay using a rat model of type 1 DM and concluded that hyperglycemia induced global hypomethylation in the acute state. They also indicated that CpG island methylation was unaffected, with data not shown. This is in contrast with our data that clearly revealed an equal distribution of demethylation at all genomic loci including promoter, intergenic, and intragenic sites. At this time, it is not clear what factor(s) initiates the demethylation process; however, we speculate that the increased oxidative stress present in the acute diabetic state could play a role, as correlations between oxidative stress and the induction of hypomethylation have been documented (46).
In an attempt to understand the consequences of hyperglycemia-induced demethylation, we examined global gene expression changes that correlate with DNA methylation alterations. Approximately 1.4% of genes with altered expression in the DM state and 3.3% of genes altered in the MM state correlated with DNA methylation differences. Most importantly, five of the nine (56%) genes identified in MM tissue (uhrf1, grtp1a, gcat, hnrnpa0, and bcor,) were also identified in DM tissue, indicating that the changes in DNA methylation correlate with changes in gene expression and that these changes persist in MM tissue. As these gene products are associated with a wide variety of pathways such as cell-cycle regulation, signaling, transcription regulation, RNA metabolism, ion transport, and amino acid metabolism, it is not clear at this time how these gene products are related to each other. Most interesting from an epigenetic perspective is the identification of the ubiquitin-like, containing PHD and ring finger (uhrf), domains 1 protein, Uhrf1. Uhrf1 has been documented to interact with the DNA methyltransferase Dnmt1 and also the histone deacetylase Hdac1 (47). This trio of proteins, along with others, is hypothesized to form a macromolecular protein complex called epigenetic code replication machinery (ECREM), which duplicates the epigenetic code during DNA replication (48). We have also observed increases in the expression of the dnmt1 and hdac1 genes in the diabetic state (data not shown) and speculate that alterations in the complex induced by hyperglycemia could provide a means to allow for epigenetic changes observed in DM and MM tissues. It is highly unlikely that epigenetic mechanisms are mutually exclusive; rather, they more likely work in concert to support the MM phenomena. The ECREM complex could provide a potential link for DNA methylation to the much larger body of evidence regarding gene expression control through histone modification alterations in MM.
When we consider our data in the broader context of disease, our results are not unlike those seen for a variety of cancers where genome-wide hypomethylation has been documented (49). In addition, increased uhrf1 expression and alterations of the ECREM complex function are also observed in several cancer tissues and are currently a focus of therapeutic drug discovery (47). The potential mechanistic commonality of what is seen in cancer and in the hyperglycemic state is unknown and warrants further investigation.
In summary, we have shown that the zebrafish is susceptible to MM and that its harmful effects can be inherited by daughter tissue in vivo. Additionally, we report for the first time an analysis of DNA methylation that revealed that hyperglycemia induced a genome-wide demethylation and aberrant gene expression that were inherited by daughter cells and may contribute to the phenomenon.
ACKNOWLEDGMENTS
This work was supported by a research grant from the Iococca Family Foundation, Rosalind Franklin University start-up funds, and National Institutes of Health Grant DK092721 (to R.V.I.).
No potential conflicts of interest relevant to this article were reported.
A.S.O. researched data, discussed data, and contributed to editing the manuscript. M.P.S. contributed to discussion of data and to writing and editing the manuscript. A.L. contributed to data analysis and to editing the manuscript. R.V.I. researched data, discussed data, contributed to writing and editing the manuscript, and is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank Gina Pisano (the Department of Basic Biomedical Sciences, Dr. William M. Scholl College of Podiatric Medicine) and Eric Vega (the Department of Cell Biology and Anatomy, Rosalind Franklin University of Medicine and Science) for technical support.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-0588/-/DC1.
REFERENCES
- 1.The Diabetes Control and Complications Trial Research Group The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N Engl J Med 1993;329:977–986 [DOI] [PubMed] [Google Scholar]
- 2.Turner RC, Cull CA, Frighi V, Holman RR; UK Prospective Diabetes Study (UKPDS) Group Glycemic control with diet, sulfonylurea, metformin, or insulin in patients with type 2 diabetes mellitus: progressive requirement for multiple therapies (UKPDS 49). JAMA 1999;281:2005–2012 [DOI] [PubMed] [Google Scholar]
- 3.Gaede PH, Jepsen PV, Larsen JN, Jensen GV, Parving HH, Pedersen OB. The Steno-2 study. Intensive multifactorial intervention reduces the occurrence of cardiovascular disease in patients with type 2 diabetes. Ugeskr Laeger 2003;165:2658–2661 [in Danish] [PubMed] [Google Scholar]
- 4.Holman RR, Paul SK, Bethel MA, Matthews DR, Neil HA. 10-year follow-up of intensive glucose control in type 2 diabetes. N Engl J Med 2008;359:1577–1589 [DOI] [PubMed] [Google Scholar]
- 5.Engerman RL, Kern TS. Progression of incipient diabetic retinopathy during good glycemic control. Diabetes 1987;36:808–812 [DOI] [PubMed] [Google Scholar]
- 6.Hammes HP, Klinzing I, Wiegand S, Bretzel RG, Cohen AM, Federlin K. Islet transplantation inhibits diabetic retinopathy in the sucrose-fed diabetic Cohen rat. Invest Ophthalmol Vis Sci 1993;34:2092–2096 [PubMed] [Google Scholar]
- 7.Kowluru RA. Effect of reinstitution of good glycemic control on retinal oxidative stress and nitrative stress in diabetic rats. Diabetes 2003;52:818–823 [DOI] [PubMed] [Google Scholar]
- 8.Kowluru RA, Chakrabarti S, Chen S. Re-institution of good metabolic control in diabetic rats and activation of caspase-3 and nuclear transcriptional factor (NF-kappaB) in the retina. Acta Diabetol 2004;41:194–199 [DOI] [PubMed] [Google Scholar]
- 9.Roy S, Sala R, Cagliero E, Lorenzi M. Overexpression of fibronectin induced by diabetes or high glucose: phenomenon with a memory. Proc Natl Acad Sci USA 1990;87:404–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li SL, Reddy MA, Cai Q, et al. Enhanced proatherogenic responses in macrophages and vascular smooth muscle cells derived from diabetic db/db mice. Diabetes 2006;55:2611–2619 [DOI] [PubMed] [Google Scholar]
- 11.Ihnat MA, Thorpe JE, Kamat CD, et al. Reactive oxygen species mediate a cellular ‘memory’ of high glucose stress signalling. Diabetologia 2007;50:1523–1531 [DOI] [PubMed] [Google Scholar]
- 12.Ceriello A, Ihnat MA, Thorpe JE. Clinical review 2: the “metabolic memory”: is more than just tight glucose control necessary to prevent diabetic complications? J Clin Endocrinol Metab 2009;94:410–415 [DOI] [PubMed] [Google Scholar]
- 13.Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes 2005;54:1615–1625 [DOI] [PubMed] [Google Scholar]
- 14.Giacco F, Brownlee M. Oxidative stress and diabetic complications. Circ Res 2010;107:1058–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pirola L, Balcerczyk A, Okabe J, El-Osta A. Epigenetic phenomena linked to diabetic complications. Nat Rev Endocrinol 2010;6:665–675 [DOI] [PubMed] [Google Scholar]
- 16.Reddy MA, Natarajan R. Epigenetic mechanisms in diabetic vascular complications. Cardiovasc Res 2011;90:421–429 [DOI] [PMC free article] [PubMed]
- 17.Blomen VA, Boonstra J. Stable transmission of reversible modifications: maintenance of epigenetic information through the cell cycle. Cell Mol Life Sci 2010;68:27–44 [DOI] [PMC free article] [PubMed]
- 18.Bogdanović O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma 2009;118:549–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem 2010;79:155–179 [DOI] [PubMed] [Google Scholar]
- 20.Kouzarides T. Chromatin modifications and their function. Cell 2007;128:693–705 [DOI] [PubMed] [Google Scholar]
- 21.Gluckman PD, Hanson MA, Beedle AS. Non-genomic transgenerational inheritance of disease risk. Bioessays 2007;29:145–154 [DOI] [PubMed] [Google Scholar]
- 22.Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet 2004;20:350–358 [DOI] [PubMed] [Google Scholar]
- 23.Whitelaw NC, Whitelaw E. Transgenerational epigenetic inheritance in health and disease. Curr Opin Genet Dev 2008;18:273–279 [DOI] [PubMed] [Google Scholar]
- 24.Morgan DK, Whitelaw E. The case for transgenerational epigenetic inheritance in humans. Mamm Genome 2008;19:394–397 [DOI] [PubMed] [Google Scholar]
- 25.Dolinoy DC, Jirtle RL. Environmental epigenomics in human health and disease. Environ Mol Mutagen 2008;49:4–8 [DOI] [PubMed] [Google Scholar]
- 26.Miao F, Gonzalo IG, Lanting L, Natarajan R. In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J Biol Chem 2004;279:18091–18097 [DOI] [PubMed] [Google Scholar]
- 27.Miao F, Wu X, Zhang L, Yuan YC, Riggs AD, Natarajan R. Genome-wide analysis of histone lysine methylation variations caused by diabetic conditions in human monocytes. J Biol Chem 2007;282:13854–13863 [DOI] [PubMed] [Google Scholar]
- 28.Miao F, Smith DD, Zhang L, Min A, Feng W, Natarajan R. Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation: an epigenetic study in diabetes. Diabetes 2008;57:3189–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Reddy MA, Miao F, et al. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J Biol Chem 2008;283:26771–26781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Osta A, Brasacchio D, Yao D, et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med 2008;205:2409–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brasacchio D, Okabe J, Tikellis C, et al. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes 2009;58:1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong Q, Kowluru RA. Role of histone acetylation in the development of diabetic retinopathy and the metabolic memory phenomenon. J Cell Biochem 2010;110:1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maunakea AK, Chepelev I, Zhao K, Bruneau B. Epigenome mapping in normal and disease States. Circ Res 2010;107:327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 2008;9:465–476 [DOI] [PubMed] [Google Scholar]
- 35.Illingworth RS, Bird AP. CpG islands—‘a rough guide’. FEBS Lett 2009;583:1713–1720 [DOI] [PubMed] [Google Scholar]
- 36.Laird PW. Cancer epigenetics. Hum Mol Genet 2005;14:R65–R76 [DOI] [PubMed] [Google Scholar]
- 37.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 2005;74:481–514 [DOI] [PubMed] [Google Scholar]
- 38.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415–428 [DOI] [PubMed] [Google Scholar]
- 39.Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet 2000;1:11–19 [DOI] [PubMed] [Google Scholar]
- 40.Sapienza C, Lee J, Powell J, et al. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics 2011;6:20–28 [DOI] [PubMed] [Google Scholar]
- 41.Bell CG, Teschendorff AE, Rakyan VK, Maxwell AP, Beck S, Savage DA. Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med Genomics 2010;3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams KT, Garrow TA, Schalinske KL. Type I diabetes leads to tissue-specific DNA hypomethylation in male rats. J Nutr 2008;138:2064–2069 [DOI] [PubMed] [Google Scholar]
- 43.Kaufman PD, Rando OJ. Chromatin as a potential carrier of heritable information. Curr Opin Cell Biol 2010;22:284–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olsen AS, Sarras MP, Jr, Intine RV. Limb regeneration is impaired in an adult zebrafish model of diabetes mellitus. Wound Repair Regen 2010;18:532–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moss JB, Koustubhan P, Greenman M, Parsons MJ, Walter I, Moss LG. Regeneration of the pancreas in adult zebrafish. Diabetes 2009;58:1844–1851 [DOI] [PMC free article] [PubMed]
- 46.Cerda S, Weitzman SA. Influence of oxygen radical injury on DNA methylation. Mutat Res 1997;386:141–152 [DOI] [PubMed] [Google Scholar]
- 47.Alhosin M, Sharif T, Mousli M, et al. Down-regulation of UHRF1, associated with re-expression of tumor suppressor genes, is a common feature of natural compounds exhibiting anti-cancer properties. J Exp Clin Cancer Res 2011;30:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bronner C. Control of DNMT1 abundance in epigenetic inheritance by acetylation, ubiquitylation, and the histone code. Sci Signal 2011;4:pe3. [DOI] [PubMed] [Google Scholar]
- 49.Alvarez H, Opalinska J, Zhou L, et al. Widespread hypomethylation occurs early and synergizes with gene amplification during esophageal carcinogenesis. PLoS Genet 2011;7:e1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]