Abstract
Knowledge regarding the genetic risk loci for gestational diabetes mellitus (GDM) is still limited. In this study, we performed a two-stage genome-wide association analysis in Korean women. In the stage 1 genome scan, 468 women with GDM and 1,242 nondiabetic control women were compared using 2.19 million genotyped or imputed markers. We selected 11 loci for further genotyping in stage 2 samples of 931 case and 783 control subjects. The joint effect of stage 1 plus stage 2 studies was analyzed by meta-analysis. We also investigated the effect of known type 2 diabetes variants in GDM. Two loci known to be associated with type 2 diabetes had a genome-wide significant association with GDM in the joint analysis. rs7754840, a variant in CDKAL1, had the strongest association with GDM (odds ratio 1.518; P = 6.65 × 10−16). A variant near MTNR1B, rs10830962, was also significantly associated with the risk of GDM (1.454; P = 2.49 × 10−13). We found that there is an excess of association between known type 2 diabetes variants and GDM above what is expected under the null hypothesis. In conclusion, we have confirmed that genetic variants in CDKAL1 and near MTNR1B are strongly associated with GDM in Korean women. There seems to be a shared genetic basis between GDM and type 2 diabetes.
There has been a marked increase in our understanding of the genetic predisposition to type 2 diabetes as a result of the technical advances in array-based genotyping and the knowledge derived from the Human Genome Project (1). Recent genome-wide association (GWA) studies and meta-analyses, including the Diabetes Genetics Replication and Meta-analysis+ (DIAGRAM+) Study (2), the Meta-analyses of Glucose and Insulin-Related Traits Consortium (MAGIC) Study (3), and the recent GWA studies of type 2 diabetes in Asians (4,5), enlisted up to 41 genetic risk loci for type 2 diabetes or glycemic traits (2,3,6–10). However, these loci explain only a limited part of the expected heritability of type 2 diabetes (11). A complementary approach to improve our insight into the genetics of diabetes might involve the identification of genetic risk loci in a different subtype of diabetes, such as gestational diabetes mellitus (GDM). This approach could enable us to compare the genetic risk factors between type 2 diabetes and GDM and relate the similarities and dissimilarities to the pathophysiology of the two closely related diseases.
GDM is defined as abnormal glucose tolerance first recognized during pregnancy (12). The estimated prevalence of GDM varies according to ethnicity (13), and in Korean women, 2–5 of 100 pregnancies are complicated by GDM (14). During pregnancy, women are faced with increased adiposity and increased insulin resistance. The insulin resistance that develops during pregnancy is explained in part by the increased production of human placental lactogen, estrogen, and prolactin (15–17). Those who have limited β-cell capacity for the compensation of insulin resistance are likely to develop GDM (18). Women with GDM are assumed to have decreased β-cell insulin secretory function similar to type 2 diabetes (18). After parturition, nearly one-half of these women progress to type 2 diabetes within 5 years (19–21). Therefore, GDM is often regarded as a herald of type 2 diabetes in later life.
Based on these findings, it can be hypothesized that GDM and type 2 diabetes share a common genetic background. We have previously reported that some of the genetic variants that are strongly associated with type 2 diabetes are similarly associated with GDM risk (22). However, genetic knowledge in the context of GDM is still limited, and systematic approaches such as GWA studies have not been applied to GDM. Thus, it is not known whether there are genetic risk loci specific to GDM. In this study, we performed a GWA study to investigate genetic risk factors for GDM in Korean women and we also compared the genetic basis of GDM and type 2 diabetes.
RESEARCH DESIGN AND METHODS
Stage 1 genome scan study subjects.
A total of 468 women with GDM and 1,242 nondiabetic control women were recruited for the stage 1 genome scan. The GDM group was selected from a hospital-based cohort enrolled between January 1996 and February 2003 from Cheil General Hospital. A 50-g 1-h glucose challenge test was performed during 24–28 weeks’ gestation in order to screen for GDM, and a glucose level of ≥7.2 mmol/L was considered positive and warranted a diagnostic 100-g oral glucose tolerance test. Glucose and insulin concentrations were measured at 0, 1, 2, and 3 h of the glucose challenge. GDM was diagnosed according to the criteria of the Third International Workshop Conference on GDM (12). The thresholds for the diagnosis of GDM were as follows: fasting ≥5.8 mmol/L, 1 h ≥10.6 mmol/L, 2 h ≥9.2 mmol/L, and 3 h ≥8.1 mmol/L. The clinical characteristics of stage 1 subjects are summarized in Table 1. The mean gestational age at GDM diagnosis was 27.9 ± 2.9 weeks.
TABLE 1.
Clinical characteristics of the study participants in the GDM GWA analysis
Nondiabetic control subjects were selected from two population-based cohort studies, the rural Ansung and the urban Ansan cohorts. The two cohorts comprised the Korean Genome Epidemiology Study and included 5,018 and 5,020 subjects, respectively (23). Only women were eligible for enrollment. From the Korean Genome Epidemiology Study subjects, we included 1,242 nondiabetic women according to the following criteria: age ≥50 years, no previous history of type 2 diabetes, no first-degree relatives with type 2 diabetes, fasting plasma glucose level <5.6 mmol/L, and HbA1c <6.0%. Information on parity and glucose tolerance status during pregnancy was not available for the control group.
Stage 2 follow-up study subjects.
A total of 931 women with GDM and 783 nondiabetic control women were included in the stage 2 follow-up study (Table 1). The GDM group consisted of two subgroups: 1) 426 women with GDM recruited from the Cheil General Hospital between January 1996 and February 2003 and 2) 505 women with GDM recruited as an independent cohort from the same hospital between March 2003 and February 2008. The diagnostic criteria of GDM in stage 2 were the same as those in stage 1. As control subjects, we enrolled nondiabetic women from four different institutes: 1) Seoul National University Hospital (n = 162), 2) Seoul National University Bundang Hospital (n = 96), 3) Kyungpook National University Hospital (n = 201), and 4) Korea National Institute of Health (from the Health T2D Study) (n = 324). The criteria for selecting control subjects were as follows: age ≥60 years, no previous history of type 2 diabetes, no first-degree relatives with type 2 diabetes, fasting plasma glucose level <5.6 mmol/L, and HbA1c <6.0%.
The institutional review board of the Clinical Research Institute at Seoul National University Hospital approved the study protocol (institutional review board no. H-0412-138-017), and written informed consent was obtained from each subject. All clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki.
Biochemical measurements.
The anthropometric and metabolic phenotypes of the women with GDM were measured at the time of the 100-g oral glucose tolerance test. Plasma glucose concentration was determined by YSI 2300 STAT (YSI, Yellow Springs, OH) using the glucose oxidase method. The insulin concentration was measured using a human-specific radioimmunoassay kit (Linco Research, St. Charles, MO). The homeostasis model assessment (HOMA) was used to estimate insulin resistance (HOMA-IR) and β-cell function (HOMA-B) with paired fasting glucose and insulin concentrations (24).
Genotyping.
We extracted genomic DNA from peripheral leukocytes. Genotyping for the stage 1 genome scan was performed using the Affymetrix Genome-Wide Human Single Nucleotide Polymorphism (SNP) Array 5.0, and genotypes were called using the Birdseed version 2.0 algorithm. Extensive quality-control protocols were applied. Markers with significant deviation from Hardy-Weinberg equilibrium (P < 1.0 × 10−6), genotype call rate < 97%, and minor allele frequency < 0.01 were excluded. The genotyping for the case and control subjects was conducted in the same laboratory but at a different time. To exclude the potential batch effect, we also excluded markers with significant differences (P < 1.0 × 10−3) in missingness between case and control subjects. Finally, 321,654 markers remained for analysis.
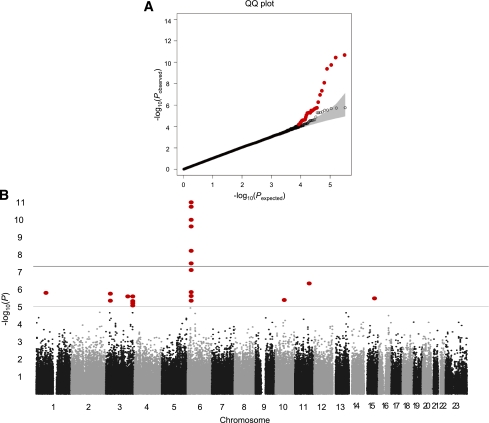
The genotype concordance rate in 10 replicated samples was >99.7% in the stage 1 control group. We excluded one sample with sex inconsistency and five samples with cryptic first-degree relatedness assessed by identity-by-descent analysis, resulting in 468 case and 1,242 control subjects. The overall genomic inflation factor (λ) of the stage 1 genome scan was 1.042, which suggests a low level of population stratification. A quantile-quantile (QQ) plot indicated that the observed distribution of the P values of the logistic regression analysis were similar to that expected under the null hypothesis, except for the deviation in the extremely low P values (Fig. 1A). These findings indicate that our study has good-quality data and that statistically significant signals are enriched.
FIG. 1.
Stage 1 genome scan results. A: QQ plot showing the distribution of the observed P values from the logistic regression analysis for the stage 1 genome scan against the expected distribution under the null hypothesis. The gray zone indicates the 95% CI. Colored circle, distribution of excess association signals driven by the known type 2 diabetes variants (listed in Table 4) and markers in LD (r2 > 0.8) with them. ○, distribution after excluding the known type 2 diabetes variants. B: Manhattan plot depicting the significance of all the association results of the stage 1 genome scan. SNP locations are plotted on the x-axis according to their chromosomal position. The negative log10 of P values derived from the logistic regression analysis under the additive model are plotted on the y-axis. (A high-quality color representation of this figure is available in the online issue.)
Genotype cluster plots were manually inspected for suggestive loci in the stage 1 genome scan. For stage 2 follow-up genotyping, the TaqMan assay (Applied Biosystems, Carlsbad, CA) was used, as previously described (25), except for the 324 subjects from the Health T2D Study, where genotyping was done using the Affymetrix Genome-Wide Human SNP Array 6.0 (26). Overall, the TaqMan genotyping success rate was 99.3%, and the concordance rate based on 15 blind duplicate comparisons was 100% in the stage 2 study. For the SNP genotyping in Health T2D Study, standard quality-control measures were applied and imputation was conducted using IMPUTE version 1.0 (http://mathgen.stats.ox.ac.uk/impute/impute.html) (27).
Statistical analyses.
Most of the association testing was performed using PLINK version 1.07 (http://pngu.mgh.harvard.edu/purcell/plink/) (28), and genotype imputation was conducted using MACH software (http://www.sph.umich.edu/csg/abecasis/mach/) (29). The genotype data that passed our quality-control criteria and had minor allele frequency >5% were used as input data for genotype imputation. The HapMap-phased genotype information of Japanese in Tokyo, Japan, and Han Chinese in Beijing, China (build 36 release 21) was used as a reference. After postimputation quality control using r2 ≥ 0.3 (squared correlation between imputed and true genotypes), a total of 2,188,613 SNPs were available for analysis. The association between genotypes and GDM risk was assessed by logistic regression analysis in an additive model. For imputed markers, an association test was performed with dosage data using mach2dat software (http://www.sph.umich.edu/csg/abecasis/mach/) (29).
Markers with suggestive genome-wide significance (P < 2.0 × 10−5) or markers near known type 2 diabetes risk loci with moderate significance (P < 1.0 × 10−3) in the stage 1 genome scan were further genotyped in stage 2 subjects. The significance threshold for the stage 2 follow-up study was P < 0.05. A joint analysis of stage 1 plus stage 2 results was performed with METAL (http://www.sph.umich.edu/csg/abecasis/metal/), using an inverse-variance method assuming fixed effects (30). An additional association analysis was performed using the EMMAX (http://genetics.cs.ucla.edu/emmax/), a variance component association method, to account for possible hidden relatedness and population stratification in the stage 1 genome scan (31). The association between genetic variants and fasting glucose, fasting insulin concentration, HOMA-IR, and HOMA-B were analyzed using linear regression in case and control subjects separately, assuming an additive genetic model. None of the women with GDM or the control subjects was using antidiabetes medications, including insulin, at the time of glucose and insulin measurement. The fasting insulin concentration, HOMA-IR, and HOMA-B was log10 transformed before analysis. The linear regression analysis was controlled for age in women with GDM and for age and BMI in control women.
The statistical power of the joint stage 1 plus stage 2 analysis was calculated using the CaTS power calculator (http://www.sph.umich.edu/csg/abecasis/cats/) (32). The QQ plot, which shows the distribution of the observed P values of the logistic regression analysis against the expected distribution under the null hypothesis, was generated using the R statistical package (http://www.r-project.org). The Manhattan plot, which depicts the negative log10 of P values derived from the logistic regression analysis, was plotted against the chromosomal position using the R statistical package. The dense regional association results of stage 1, stage 2, and the joint analysis of stage 1 plus stage 2 were plotted using LocusZoom software (http://csg.sph.umich.edu/locuszoom/) (33). To compare the effect size of known type 2 diabetes variants in GDM and type 2 diabetes, the β-coefficient of the logistic regression analysis derived from our stage 1 genome scan and from the stage 1 meta-GWA analysis of the Asian Genetic Epidemiology Network (AGEN) type 2 diabetes report (26) was plotted.
RESULTS
Stage 1 genome scan.
In the stage 1 genome scan, logistic regression analysis using an additive genetic model was used to test for the association between the genotypes and GDM. The negative log10 of the P values from the association test were plotted against their genomic position in Fig. 1B. The association test results for SNPs with P < 0.0001 in the stage 1 genome scan are listed in Supplementary Table 1. A total of nine independent (pairwise linkage disequilibrium [LD] r2 < 0.5) SNPs were suggestive of an association according to our predefined arbitrary threshold of P < 2.0 × 10−5. Among these, variants in CDKAL1 (rs7754840) and near MTNR1B (rs10830962) showed the strongest association with GDM risk. We added two additional variants (rs10757261 near CDKN2A/2B and rs10882066 in IDE) from the type 2 diabetes risk loci, which had suggestive P values in our stage 1 results. Among the 11 SNPs, 4 were substituted to imputed SNPs (rs6499500, rs12715106, rs9395950, and rs187230), which had more significant P values and had strong LD (r2 > 0.8) with the original SNPs. To eliminate hidden population stratification and cryptic relatedness, the EMMAX, a variance component approach accounting for hidden sample structure, was used to test the association between genetic variants and GDM. The P values of the EMMAX are listed in Supplementary Table 1. The results of the EMMAX were similar to those of the original logistic regression analysis. The Pearson correlation coefficient for the log10 of the P values of both analyses was 0.921 (P < 1.51 × 10−288). This implies that population stratification was minimal in our study samples.
Stage 2 follow-up and joint analysis.
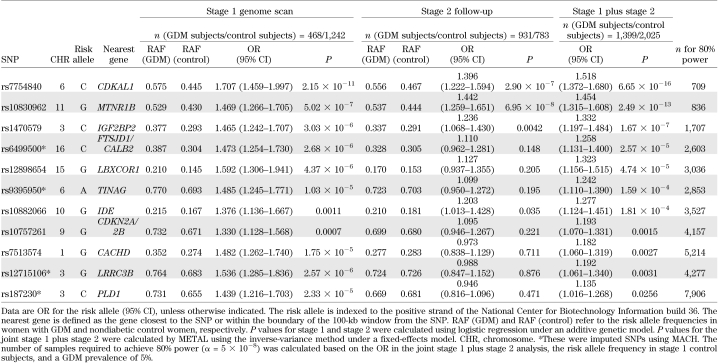
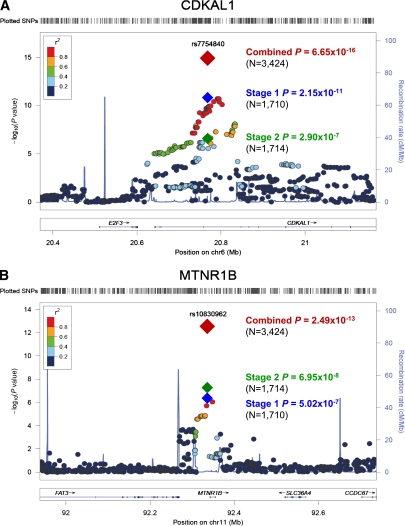
In stage 2 follow-up, we genotyped 11 SNPs in 931 case and 783 control subjects. The results of the stage 2 association test are shown in Table 2. Among these, rs7754840 in CDKAL1 (odds ratio [OR] 1.396 [95% CI 1.222–1.594]; P = 2.90 × 10−7), rs10830962 near MTNR1B (1.442 [1.259–1.651]; P = 6.95 × 10−8), rs1470579 in IGF2BP2 (1.236 [1.068–1.430]; P = 0.0042), and rs10882066 in IDE (1.203 [1.013–1.428]; P = 0.035) were significantly associated with GDM in the stage 2 samples. None of the other SNPs showed evidence of an association. In the joint analysis of stage 1 plus stage 2 results (Table 2), rs7754840 in CDKAL1 (1.518 [1.372–1.680]; P = 6.65 × 10−16) and rs10830962 near MTNR1B (1.454 [1.315–1.608]; P = 2.49 × 10−13) reached genome-wide significance for an association with GDM. One additional variant, rs1470579 in IGF2BP2 (1.332 [1.197–1.484]; P = 1.67 × 10−7), showed a near genome-wide significant association with GDM. The regional association plot of SNPs near CDKAL1 and MTNR1B, including those that have been imputed, are depicted in Fig. 2.
TABLE 2.
Association between SNPs and GDM in stage 1, stage 2, and joint stage 1 plus stage 2 analysis
FIG. 2.
Dense regional association plot near CDKAL1 (A) and MTNR1B (B). The hash marks above the panel represent the position of each SNP that was genotyped or imputed. The negative log10 of P values from logistic regression are shown in the panel. The blue diamond indicates the SNP with the most significant association in the stage 1 genome scan. The green and red diamonds represent the results of the SNP in stage 2 and joint stage 1 plus stage 2 analysis, respectively. Their corresponding P values are indicated on the right. Estimated recombination rates are plotted to reflect recombination hot spots. The SNPs in LD with the most significant SNP are color coded to represent their strength of LD. The locations of genes, exons and introns are shown in the lower panel (taken from the Human Genome hg18 build). (A high-quality color representation of this figure is available in the online issue.)
Association with glucose and insulin-related traits.
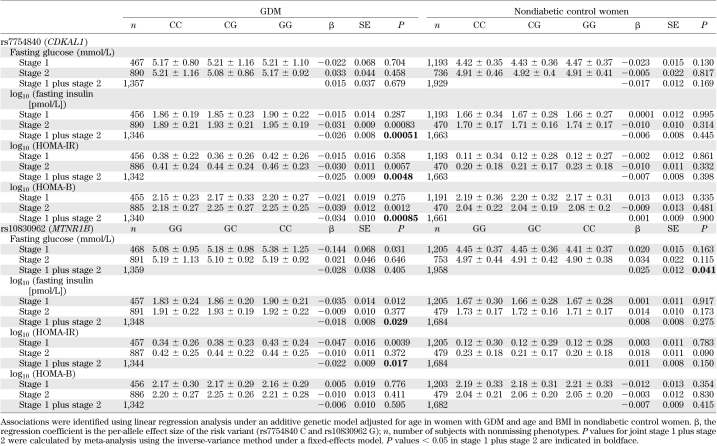
To obtain additional insight into the role of these two variants, we performed an association analysis between these variants and quantitative traits of fasting glucose, fasting insulin, HOMA-IR, and HOMA-B in women with GDM and control subjects, separately (Table 3). The rs7754840 C allele of CDKAL1, which was the risk variant of GDM, was significantly associated with decreased fasting insulin concentration (β = −0.026; P = 0.00051) and decreased HOMA-B (β = −0.034; P = 0.00085) in women with GDM. The rs10830962 G allele near MTNR1B was nominally associated with decreased fasting insulin concentrations (β = −0.018; P = 0.029) in women with GDM. This variant was also marginally associated with increased fasting glucose concentrations (β = 0.025; P = 0.041) in control subjects.
TABLE 3.
Association of rs7754840 and rs10830962 with fasting glucose, fasting insulin, HOMA-IR, and HOMA-B in women with GDM and control subjects
Comparison of genetic risk loci between GDM and type 2 diabetes.
Among 41 known type 2 diabetes loci, we were able to examine the association signals for 34 loci that were directly genotyped or imputed (Table 4). There was an excess of small P values compared with the expected distribution under the null hypothesis (Fig. 3). When these known type 2 diabetes risk variants and markers in LD (r2 > 0.8) were excluded, the QQ plot of our stage 1 genome scan was similar to that expected under the null hypothesis (Fig. 1A).
TABLE 4.
Association of confirmed type 2 diabetes loci with GDM risk in stage 1 genome scan results
FIG. 3.
QQ plot of the association between known type 2 diabetes variants and GDM. Comparison of the effect size of known type 2 diabetes variants in GDM and type 2 diabetes. The gray zone indicates the 95% CI. The known type 2 diabetes variants tested for association are listed in Table 4.
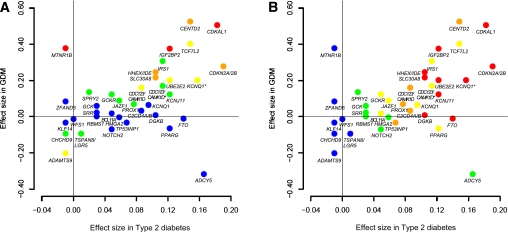
We compared the β-coefficient in the logistic regression analysis of known type 2 diabetes variants in GDM (our stage 1 genome scan) and type 2 diabetes (AGEN type 2 diabetes stage 1 meta-GWA results [26]) (Fig. 4 and Supplementary Table 2). There was a significant positive correlation between the β-coefficients of GDM and type 2 diabetes (Pearson correlation coefficient 0.442; P = 0.0062). However, some variants, such as rs10830963 of MTNR1B and rs11708067 of ADCY5, showed considerable differences in effect size between GDM and type 2 diabetes.
FIG. 4.
Comparison of the effect size of known type 2 diabetes variants in GDM and type 2 diabetes. Effect size (β-coefficient from logistic regression analysis) of the known type 2 diabetes variants in GDM (y-axis) and type 2 diabetes (x-axis) are plotted with their corresponding P values (A: P values in GDM; B: P values in type 2 diabetes): red, P < 0.0001; orange, 0.0001 ≤ P < 0.01; yellow, 0.01 ≤ P < 0.10; green, 0.10 ≤ P < 0.50; blue, 0.50 ≤ P. The β-coefficient for GDM was derived from our stage 1 genome scan and that for type 2 diabetes was derived from the AGEN type 2 diabetes study (26). The two CDC123/CAMK1D variants are distinguished by CDC123/CAMK1D for rs10906115 and CDC123/CAMK1D* for rs12779790. The two KCNQ1 variants are distinguished by KCNQ1 for rs231362 and KCNQ* for rs2237892. (A high-quality color representation of this figure is available in the online issue.)
Finally, we examined the association results for rs7754840 of CDKAL1 and rs10830962 of MTNR1B in the stage 1 meta-GWA study of the AGEN type 2 diabetes report (26). The C allele of rs7754840 was significantly associated with an increased risk of type 2 diabetes (n = 18,732; OR 1.20 [95% CI 1.14–1.25]; P = 9.63 × 10−15). However, the G allele of rs10830962 was not associated with type 2 diabetes risk (n = 10,754; 1.040 [0.98–1.11]; P = 0.209).
DISCUSSION
The main finding of this study is that variants in CDKAL1 and near MTNR1B are associated with GDM at a genome-wide significance level (P < 5.0 × 10−8). Our findings are confirmatory of previous candidate gene studies of GDM (22,34,35). Previously, we selected 18 SNPs, known to be associated with the risk of type 2 diabetes, for association testing in GDM (22). Among them, two SNPs (rs7756992 and rs7754840) in CDKAL1 were associated with GDM risk at a genome-wide significance level, and one SNP (rs10811661) near CDKN2A/2B also was strongly associated with GDM. Another study in Europeans tested 11 known SNPs of type 2 diabetes and found a modest association between GDM and TCF7L2, CDKAL1, and TCF2 variants (34). Recently, Kim et al. (35) reported that two SNPs in MTNR1B (rs1387153 and rs10830963) were strongly associated with GDM in Korean women. The study by Kim et al. was led by one of our investigators but was performed independently of this study, and 505 subjects with GDM in that study overlapped with our stage 2 follow-up subjects. However, only a limited number of genetic variants known to be associated with type 2 diabetes have been tested thus far, and a systematic approach such as GWA analysis has not been applied to GDM until now. To the best of our knowledge, this is the first study to investigate the genetic association of GDM using dense SNP markers across the whole genome.
Variants in CDKAL1 are known to be strongly associated with type 2 diabetes (8–10,36,37). In our previous studies, the variant rs7754840 in CDKAL1 was significantly associated with GDM risk in 1,501 Koreans (22) as well as type 2 diabetes in 6,719 Asians including Koreans (38). The precise mechanism by which this variant confers type 2 diabetes and GDM risk is not yet clearly understood. CDKAL1 (cyclin-dependent kinase 5 [CDK5] regulatory subunit-associated protein 1-like 1) has been suggested to interact with CDK5 because it has homology with CDK5RAP1, a known inhibitor of CDK5 (39). CDK5 is expressed in pancreatic β-cells, and it has recently been reported to promote β-cell survival (40). Pregnancy is a state in which increased insulin resistance results in stress that adversely affects β-cell survival (41). Therefore, it may be postulated that variants in CDKAL1 might alter the function of CDK5 in β-cell compensation during pregnancy. The rs7754840 C variant of CDKAL1 was significantly associated with decreased insulin concentration and HOMA-B in our subjects with GDM, which implies compromised β-cell compensation. It also should be noted that variants in CDKAL1 are associated with decreased birth weight, which could be explained by reduced fetal insulin secretion (42).
Variants in MTNR1B were first identified as genetic risk factors strongly associated with elevated fasting glucose (43–45). Although it also was associated with type 2 diabetes risk in Europeans, the strength of association was rather weak compared with its association with fasting glucose (44,45). It is interesting that the rs10830962 variant in MTNR1B was the second most strongly associated loci in GDM but was not associated with type 2 diabetes risk in a large-scale meta-GWA study of East Asians (AGEN type 2 diabetes meta-GWA study [26]). The effect size of the MTNR1B rs10830962 variant was considerably different in our GDM GWA study (β = 0.374) compared with the AGEN type 2 diabetes meta-GWA study (β = 0.040). The risk allele of rs10830962 G was enriched in women with GDM (risk allele frequency 0.529 and 0.537 in stage 1 and stage 2 subjects, respectively) compared with AGEN type 2 diabetic subjects (risk allele frequency range for participating studies 0.40–0.46). It is not clear why this variant is more strongly associated with the risk of GDM compared with type 2 diabetes. However, it could be cautiously speculated that there might be differences in genetic susceptibility between type 2 diabetes and GDM conferred by the variants of MTNR1B. Additional investigations are required to confirm these findings.
It was recently revealed that MTNR1B is expressed in pancreatic β-cells (44). Melatonin is thought to inhibit insulin secretion from β-cells through the activation of MTNR1B, which is a G-protein–coupled receptor (46). The rs10830962 variant is in strong LD (r2 = 0.98, in our study subjects) with rs10830963, which originally was reported to be associated with fasting glucose concentrations, insulin concentrations, and type 2 diabetes. It is interesting that the rs1083092 variant is located 4.36 kb upstream of MTNR1B and in the vicinity of the region where monomethylation of lysine 4 of histone 3 (H3K4Me1), known as an enhancer-associated histone mark, was enriched in the Encyclopedia of DNA Elements database (http://genome.ucsc.edu/encode/) (47). It would be of worth to study the functional role of this variant as well as to search for causative variants near this locus, especially in the promoter and/or enhancer region.
Because we performed a GWA analysis using 2.19 million genotyped or imputed markers, we were able to test the hypothesis that GDM and type 2 diabetes might share a similar genetic background. Among 11 variants listed in Table 2, five SNPs were located near the known type 2 diabetes loci. Three variants that showed an association at or near genome-wide significance, rs7754840 in CDKAL1, rs10830962 near MTNR1B, and rs1470579 in IGF2BP2, were identical or in strong LD with known type 2 diabetes variants. Two other variants, rs10757261 in CDKN2A/2B and rs10882066 in IDE, were not in LD with known type 2 diabetes variants (r2 = 0.013 between rs10757261 and rs10811661 and r2 = 0.002 between rs10882066 and rs5015480). Given that many of the type 2 diabetes risk loci were associated with GDM, unbiased GWA studies of GDM showed genome-wide significant associations confined to known type 2 diabetes loci, and the effect size of the type 2 diabetes risk variants in GDM and type 2 diabetes were mostly comparable, it would be acceptable to state that GDM and type 2 diabetes share a similar genetic background.
Among the known type 2 diabetes–related genes, those that are thought to modulate pancreatic β-cell function were preferentially associated with GDM (CDKAL1, MTNR1B, IGF2BP2, CDKN2A/2B, SLC30A8, IDE, KCNQ1, and CENTD2) (Table 4). In contrast, loci relevant to insulin resistance, such as FTO, PPARG, IRS1, KLF14, and GCKR were not significantly associated with GDM (Table 4). These findings support the previous notion that defective pancreatic β-cell compensation to overcome increased insulin resistance during pregnancy might be the core pathophysiology of GDM.
There are certain limitations in our study. First, the sample size was relatively modest for a GWA, and we had limited power in detecting possible variants with small effect size. Assuming a 2.2% prevalence of GDM (48), a disease allele frequency of 0.3, and a genotype relative risk of 1.5 in a multiplicative model, our study had 93% power. However, when genotype relative risk was assumed to be 1.4, the power dropped to 68%. It is possible that variants with a small effect would not have been detected in our study. Therefore, we might have missed genetic variants that have a small effect but are specific to GDM. Regarding the study design, the control group was not fully evaluated for their glucose tolerance status during pregnancy and parity. It is possible that women with GDM might have been included in the control group. However, because the prevalence of GDM was estimated to be 2.2% in Korean women (48), and approximately one-half of these women progress to type 2 diabetes within 10 years (22), only ~≤1% of women with GDM would have been included in the control group. Therefore, we speculate that the ascertainment bias would have been minimal. In studying genetic variants of GDM, the ideal control group would be pregnant women with normal glucose tolerance. However, we were only able to use nondiabetic control women as control subjects. This could be one of the reasons that variants in CDKAL1 and MTNR1B were most significantly associated with GDM in our study. In this regard, the results of our study should be interpreted cautiously.
In conclusion, we have performed a GWA study in the context of GDM for the first time and confirmed that variants in CDKAL1 and MTNR1B confer the risk of GDM with genome-wide significance. By comparing genetic variants in GDM and type 2 diabetes, we provide evidence that they share a similar genetic background. We hope that even larger GWA studies of GDM and meta-GWA studies could further reveal the genetic risk loci for GDM.
ACKNOWLEDGMENTS
This work was supported by the Korea Health 21 R&D Project, Korean Ministry of Health and Welfare (grant no. 00-PJ3-PG6-GN07-001); the Korea Healthcare Technology R&D Project, Korean Ministry of Health and Welfare (grant no. A102041); and the World Class University Project of the Ministry of Education, Science and Technology and the Korea Science and Engineering Foundation (grant no. R31-2008-000-10103-0). This work also was supported by grants from the Korea Center for Disease Control and Prevention (grant nos. 4845-301, 4851-302, and 4851-307) and an intramural grant from the Korea National Institute of Health (grant no. 2010-N73002-00), Republic of Korea.
No potential conflicts of interest relevant to this article were reported.
S.H.K. researched data, contributed to the discussion, and drafted the manuscript. S.-H.K., N.H.C., I.K.L., and B.-G.H. provided study samples. Y.M.C., S.H.C., M.K.M., H.S.J., H.D.S., and S.Y.K. contributed to the discussion. M.J.G. researched data. Y.S.C. provided study samples and researched data. H.M.K. provided statistical advice, contributed to the discussion, and edited the manuscript. H.C.J. and K.S.P. provided study samples, researched data, contributed to the discussion, and reviewed the manuscript. H.C.J. and K.S.P. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-1034/-/DC1.
REFERENCES
- 1.Lander ES, Linton LM, Birren B, et al. ; International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature 2001;409:860–921 [DOI] [PubMed] [Google Scholar]
- 2.Voight BF, Scott LJ, Steinthorsdottir V, et al. ; MAGIC Investigators; GIANT Consortium Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 2010;42:579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dupuis J, Langenberg C, Prokopenko I, et al. ; DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium; Anders Hamsten on behalf of Procardis Consortium; MAGIC Investigators New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010;42:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamauchi T, Hara K, Maeda S, et al. A genome-wide association study in the Japanese population identifies susceptibility loci for type 2 diabetes at UBE2E2 and C2CD4A-C2CD4B. Nat Genet 2010;42:864–868 [DOI] [PubMed] [Google Scholar]
- 5.Tsai FJ, Yang CF, Chen CC, et al. A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Genet 2010;6:e1000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeggini E, Scott LJ, Saxena R, et al. ; Wellcome Trust Case Control Consortium Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008;40:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007;445:881–885 [DOI] [PubMed] [Google Scholar]
- 8.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saxena R, Voight BF, Lyssenko V, et al. ; Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007;316:1331–1336 [DOI] [PubMed] [Google Scholar]
- 10.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007;316:1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature 2009;461:747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metzger BE. Summary and recommendations of the Third International Workshop-Conference on Gestational Diabetes Mellitus. Diabetes 1991;40(Suppl. 2):197–201 [DOI] [PubMed] [Google Scholar]
- 13.Jovanovic L, Pettitt DJ. Gestational diabetes mellitus. JAMA 2001;286:2516–2518 [DOI] [PubMed] [Google Scholar]
- 14.Jang HC. Gestational diabetes in Korea: incidence and risk factors of diabetes in women with previous gestational diabetes. Diabetes Metab J 2011;35:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H, Toyofuku Y, Lynn FC, et al. Serotonin regulates pancreatic beta cell mass during pregnancy. Nat Med 2010;16:804–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Cianni G, Miccoli R, Volpe L, Lencioni C, Del Prato S. Intermediate metabolism in normal pregnancy and in gestational diabetes. Diabetes Metab Res Rev 2003;19:259–270 [DOI] [PubMed] [Google Scholar]
- 17.Karnik SK, Chen H, McLean GW, et al. Menin controls growth of pancreatic beta-cells in pregnant mice and promotes gestational diabetes mellitus. Science 2007;318:806–809 [DOI] [PubMed] [Google Scholar]
- 18.Buchanan TA, Xiang AH. Gestational diabetes mellitus. J Clin Invest 2005;115:485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim C, Newton KM, Knopp RH. Gestational diabetes and the incidence of type 2 diabetes: a systematic review. Diabetes Care 2002;25:1862–1868 [DOI] [PubMed] [Google Scholar]
- 20.Lee H, Jang HC, Park HK, Metzger BE, Cho NH. Prevalence of type 2 diabetes among women with a previous history of gestational diabetes mellitus. Diabetes Res Clin Pract 2008;81:124–129 [DOI] [PubMed] [Google Scholar]
- 21.Metzger BE, Cho NH, Roston SM, Radvany R. Prepregnancy weight and antepartum insulin secretion predict glucose tolerance five years after gestational diabetes mellitus. Diabetes Care 1993;16:1598–1605 [DOI] [PubMed] [Google Scholar]
- 22.Cho YM, Kim TH, Lim S, et al. Type 2 diabetes-associated genetic variants discovered in the recent genome-wide association studies are related to gestational diabetes mellitus in the Korean population. Diabetologia 2009;52:253–261 [DOI] [PubMed] [Google Scholar]
- 23.Cho YS, Go MJ, Kim YJ, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet 2009;41:527–534 [DOI] [PubMed] [Google Scholar]
- 24.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985;28:412–419 [DOI] [PubMed] [Google Scholar]
- 25.Kwak SH, Kim TH, Cho YM, Choi SH, Jang HC, Park KS. Polymorphisms in KCNQ1 are associated with gestational diabetes in a Korean population. Horm Res Paediatr 2010;74:333–338 [DOI] [PubMed] [Google Scholar]
- 26.Cho YS, Chen CH, Hu C, et al. Meta-analysis of genome-wide association studies identifies 8 new loci for type 2 diabetes in East Asians. Nat Genet 2011;44:67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007;39:906–913 [DOI] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet 2009;10:387–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang HM, Sul JH, Service SK, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet 2010;42:348–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 2006;38:209–213 [DOI] [PubMed] [Google Scholar]
- 33.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010;26:2336–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauenborg J, Grarup N, Damm P, et al. Common type 2 diabetes risk gene variants associate with gestational diabetes. J Clin Endocrinol Metab 2009;94:145–150 [DOI] [PubMed] [Google Scholar]
- 35.Kim JY, Cheong HS, Park BL, et al. Melatonin receptor 1 B polymorphisms associated with the risk of gestational diabetes mellitus. BMC Med Genet 2011;12:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeggini E, Weedon MN, Lindgren CM, et al. ; Wellcome Trust Case Control Consortium (WTCCC) Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007;316:1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet 2007;39:770–775 [DOI] [PubMed] [Google Scholar]
- 38.Ng MC, Park KS, Oh B, et al. Implication of genetic variants near TCF7L2, SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, and FTO in type 2 diabetes and obesity in 6,719 Asians. Diabetes 2008;57:2226–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ching YP, Pang AS, Lam WH, Qi RZ, Wang JH. Identification of a neuronal Cdk5 activator-binding protein as Cdk5 inhibitor. J Biol Chem 2002;277:15237–15240 [DOI] [PubMed] [Google Scholar]
- 40.Daval M, Gurlo T, Costes S, Huang CJ, Butler PC. Cyclin-dependent kinase 5 promotes pancreatic β-cell survival via Fak-Akt signaling pathways. Diabetes 2011;60:1186–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buchanan TA. Pancreatic B-cell defects in gestational diabetes: implications for the pathogenesis and prevention of type 2 diabetes. J Clin Endocrinol Metab 2001;86:989–993 [DOI] [PubMed] [Google Scholar]
- 42.Freathy RM, Bennett AJ, Ring SM, et al. Type 2 diabetes risk alleles are associated with reduced size at birth. Diabetes 2009;58:1428–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prokopenko I, Langenberg C, Florez JC, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet 2009;41:77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lyssenko V, Nagorny CL, Erdos MR, et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet 2009;41:82–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouatia-Naji N, Bonnefond A, Cavalcanti-Proença C, et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet 2009;41:89–94 [DOI] [PubMed] [Google Scholar]
- 46.Peschke E, Mühlbauer E. New evidence for a role of melatonin in glucose regulation. Best Pract Res Clin Endocrinol Metab 2010;24:829–841 [DOI] [PubMed] [Google Scholar]
- 47.Raney BJ, Cline MS, Rosenbloom KR, et al. ENCODE whole-genome data in the UCSC genome browser (2011 update). Nucleic Acids Res 2011;39(Database issue):D871–D875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jang HC, Cho NH, Jung KB, Oh KS, Dooley SL, Metzger BE. Screening for gestational diabetes mellitus in Korea. Int J Gynaecol Obstet 1995;51:115–122 [DOI] [PubMed] [Google Scholar]