Abstract
In bacteria, the protein FtsZ is the principal component of a ring that constricts the cell at division. Though all mitochondria probably arose through a single, ancient bacterial endosymbiosis, the mitochondria of only certain protists appear to have retained FtsZ, and the protein is absent from the mitochondria of fungi, animals, and higher plants. We have investigated the role that FtsZ plays in mitochondrial division in the genetically tractable protist Dictyostelium discoideum, which has two nuclearly encoded FtsZs, FszA and FszB, that are targeted to the inside of mitochondria. In most wild-type amoebae, the mitochondria are spherical or rod-shaped, but in fsz-null mutants they become elongated into tubules, indicating that a decrease in mitochondrial division has occurred. In support of this role in organelle division, antibodies to FszA and FszA-green fluorescent protein (GFP) show belts and puncta at multiple places along the mitochondria, which may define future or recent sites of division. FszB-GFP, in contrast, locates to an electron-dense, submitochondrial body usually located at one end of the organelle, but how it functions during division is unclear. This is the first demonstration of two differentially localized FtsZs within the one organelle, and it points to a divergence in the roles of these two proteins.
Mitochondria and chloroplasts divide by fission, like their bacterial ancestors. In 1995, Osteryoung and Vierling (51) discovered a chloroplast-targeted version of the bacterial cell division protein FtsZ (AtFtsZ1-1) in Arabidopsis thaliana that was most similar to the FtsZs of cyanobacteria, the ancestors of chloroplasts. The implication of this finding was that all chloroplasts, and perhaps even mitochondria, might still use FtsZ to divide. FtsZ is the most widespread and important of a dozen or so bacterial division proteins (1, 13, 15, 36, 54). FtsZ is a GTPase that bears little amino acid sequence identity but a striking structural similarity to tubulins, a family of eukaryotic cytoskeletal proteins (30, 47). Like tubulin, FtsZ monomers can self-associate and have been observed to form microtubule-like filaments in vitro (14). FtsZ has, thus, been proposed to be the prokaryotic ancestor of tubulin (12, 38, 47).
FtsZ is associated with the invaginating inner margin of the bacterial cell membrane (7), and studies of FtsZ-green fluorescent protein (GFP) fusions showed the protein forms a ring at the division site (31). It is not known if the FtsZ cytokinetic ring, the Z-ring, generates the contractile force necessary to pull in the cell edges, or if it simply provides an assembly site for other constricting proteins.
FtsZs play a critical role in the division of chloroplasts. If the expression of either of two versions of FtsZ in Arabidopsis (AtFtsZ1-1 and AtFtsZ2-1) is inhibited by antisense RNA, chloroplasts fail to divide properly, if at all (52). Gene disruption of chloroplast FtsZ in the moss Physcomitrella patens produces a similar result (59). Immunofluorescence microscopy showed that AtFtsZ1-1 and AtFtsZ2-1 form coaligned rings at the chloroplast midpoint (65), and both are on the stromal side of the chloroplast inner membrane (39). A third Arabidopsis FtsZ, called AtFtsZ2-2, is closely related to AtFtsZ2-1 (52) and is also targeted to the chloroplast stroma (20, 39). Immunoelectron microscopy studies of plastids of the plant Lilium longifolium (45) and the red alga Cyanidioschyzon merolae (44) have shown that FtsZ localizes to the inner margin of the invaginating region of dividing chloroplasts.
With the discovery of FtsZ in chloroplasts, it was proposed that there might also be a specific class of FtsZs that divided mitochondria (51), since mitochondria too are believed to be derived from a bacterial endosymbiont. However, the complete genomes of yeast (Saccharomyces cerevisiae), animals (Caenorhabditis elegans, Drosophila melanogaster, and Homo sapiens), and A. thaliana contain no candidate FtsZ sequences, and it has become clear that members of the dynamin family of GTPases (63) act to divide the outer mitochondrial membrane of yeast, animals, and plants (4, 8, 27, 55-57). Since yeast, animals, and higher plants represent only a few highly derived branches of the tree of life, it was suspected there may be FtsZ relics from the mitochondrion's ancestor among the diverse and often more ancient eukaryotes within the protists. Any mitochondrial FtsZs would likely be most closely related to FtsZs of the α-proteobacteria—the probable ancestors of mitochondria (3, 28). This prediction proved correct when we identified MsFtsZ-mt, a gene from the chromophytic (stramenopile) alga Mallomonas splendens (6). MsFtsZ-mt is nuclearly encoded, it bears an N-terminal mitochondrial targeting peptide, and it is most closely related to FtsZs of the α-proteobacteria. An MsFtsZ-mt-GFP fusion was imported into yeast mitochondria, and immunolocalizations in M. splendens showed that MsFtsZ-mt was concentrated in regions of mitochondria that were presumptive or recent division sites. It seems, then, that the mitochondria of M. splendens, and probably many other eukaryotes, still use an FtsZ-based system of mitochondrial division inherited from their α-proteobacterial ancestors (6).
Despite initial expectations that the mitochondrial division systems used by eukaryotes might be mutually exclusive, i.e., a primitive one based on FtsZ in protists may have been replaced by another based on dynamin in higher eukaryotes, it has been recently discovered that both proteins apparently work together in some organisms. C. merolae not only produces FtsZs that are targeted to the interior of its mitochondrion (46, 62) and plastid (41, 43, 44) but also contains two dynamins. The first, CmDnm1, is targeted to the outside of the mitochondrion (46), and the second CmDnm2 (42) localizes to the outside of the plastid. During division, these dynamins overlay FtsZ rings formed on the inside of the organelles but probably act a little later than FtsZs (42, 46). The use of dynamins for dividing mitochondria from the outside is therefore probably quite ancient and may well be used by all eukaryotes not for only mitochondrial division but also for the division of plastids, where present (21). Though the observation that FtsZ forms rings of decreasing diameter during mitochondrial division constitutes important evidence that that protein is required for division (62), genetic data in support of this function are lacking. We therefore turned our attention to the genetically tractable, social amoeba Dictyostelium discoideum. Here we report the discovery of two mitochondrially targeted FtsZs, and through the use of gene knockouts, we study their possible roles in mitochondrial division.
MATERIALS AND METHODS
Strains and culture conditions.
D. discoideum strain AX3 (29) was cultured axenically in liquid HL5 medium (11) at 22°C in tissue culture plates or in conical flasks on an orbital shaker.
Isolation of fszA and fszB.
Degenerate PCR primers FtsZ1R (GGNGGNAAYGCNGTNAAYAAYATGAT) and FtsZ4L (ACRTCNGCRAARTCNARRTTDATNARNCC) were used to amplify a central fragment of fszA from 100 ng of Dictyostelium genomic DNA with the following PCR conditions: 35 cycles of 94°C for 30 s, 45°C for 30 s, and 72°C for 50 s. Internal gene-specific primers FszAR2 (TGCTCCTGCTCCTAATCC) and FszA2F (ATCAGCAGCAAAAGCCAAAGG) were used to amplify the 5′ and 3′ ends of the FszA gene, respectively, using random amplification of cDNA ends (RACE) (Gibco-BRL). PCR products were cloned into pGemT-easy (Promega) for sequencing. To isolate fszB, we used PCR primers DctZ2I (GGGGATCCATGACAATTTTAAATAGATTTTGTAGG) and DctZ2stop (CCGTCGACTTAATGTACAAATAAACAAG), which amplified the entire sequence of fszB from Dictyostelium genomic DNA or cDNA. PCR products were cloned between the BamHI and SalI sites of pBC (NdeI), a pBC (SK+) (Stratagene) derivative containing an NdeI site upstream of the SK polylinker. Genomic and cDNA clones were confirmed by sequencing.
Generation of fszA-null cells.
To construct a fszA knockout strain, a 1.1-kb internal portion of fszA genomic DNA was first amplified from Dictyostelium genomic DNA with primers DctZ1F (AAGGATCCAAGGTTTTCAATTCACCA) and DctZ1G (TTGTCGACTTGTGGTTCAATTGGTTT). The PCR product was cloned into BamHI-SalI-cleaved pBC (NdeI). The insert was then released from this plasmid by cleaving with XbaI and Asp718, and the fragment was cloned into pBAD18 (23) to create pWM1310. Into the PvuII site of the fszA gene within pWM1310, a uracil biosynthetic marker was inserted to create pWM1311. The uracil marker was a blunt-ended, 2.7-kb ClaI fragment of the pyr5-6 gene fragment isolated from pDH29.
The pDH29 plasmid had been made by amplifying the Pyr5-6 gene by PCR from pDU3B1 (9) using a vector primer upstream of the promoter and an antisense primer complementary to the Pyr5-6 sequence CCAGTCGAATGACTTTCAGAC, which lies ∼120 bp downstream from the stop codon. After cleavage of the PCR product with ClaI at an internal site 1 kb upstream of the start codon and one appended to the antisense primer's 5′ end, the 2.7-kb fragment was ligated into the ClaI site of pBluescript (Stratagene) to yield pDH29. This Pyr5-6 gene fragment corresponds precisely to the deletion in the DH1 strain (10) that is responsible for uracil auxotrophy.
The fszA knockout construct (i.e., uracil marker plus flanking fszA regions) was excised from pWM1311 by digestion with EcoRI and SalI and transformed into DH1 by electroporation (25). The transformants were selected for uracil prototrophy with FM medium (19). The resulting fszA-null cell line was designated the ΔfszA cell line.
Generation of fszB-null cells and fszA fszB double mutants.
To construct insertional knockouts of fszB, it was necessary to clone the gene as two halves to introduce an artificial XbaI site in the middle, because a suitable site for insertion of a Bsr gene cassette (which confers resistance to blasticidin S) was lacking. Primers dctZ2start (GGGAGCTCAAAAAATGACAATTTTAAATAG) and dctZ2knockA (CCTCTAGACCAGTACCACCACCC) were used to amplify the 5′ portion of fszB from genomic DNA of Dictyostelium. The PCR product was cloned as a SacI-XbaI fragment into pZG (31), replacing the Escherichia coli ftsZ gene. This plasmid was designated pDA1. Using primers DctZ2knockB (GGTCTAGATTATAGTTGGTGTTGTAAC) and DctZ2stop (CCGTCGACTTAATGTACAAATAAACAAG), the 3′ portion of fszB was amplified from Dictyostelium genomic DNA and cloned as an XbaI-SalI fragment into pDA1. The resulting plasmid was then cleaved with XbaI, and the Bsr cassette was isolated as an XbaI fragment from pBsR519 (53) and cloned into this site to make pWM1325. For introduction of the insertional knockout into Dictyostelium, a BsaBI-SalI fragment containing the knockout plus flanking regions was purified from pWM1325 and transformed into Dictyostelium strain AX3 as described above for the ΔfszA knockout. Transformants were selected with blasticidin S (5 μg/ml). In the same manner, this fszB knockout construct was introduced into the ΔfszA cell line described above to generate a double-knockout ΔfszA ΔfszB strain. All single- and double-knockout strains were verified by PCR and Southern blotting.
Microscopy of Dictyostelium mitochondria.
Wild-type and Δfsz vegetative amoebae were grown as log-phase cultures in six-well plates on sterile coverslips. The mitochondria were labeled by adding MitoTracker Red (CMX-Ros; Molecular Probes) to a final concentration of 100 nM to the HL5 growth medium. Unbound MitoTracker was removed by washing the cells in fresh HL5 medium. Prior to fixation, the coverslips were washed twice in phosphate buffer (12 mM Na2HPO4, 12 mM NaH2PO4 [pH 6.5]) and the cells were simultaneously flattened and fixed by placing the coverslips cell side down onto a layer of 1% agarose in phosphate buffer containing 3.5% paraformaldehyde for 30 min, and this was followed by permeabilization in 100% methanol prechilled to −20°C. The coverslips were then rehydrated in standard phosphate-buffered saline (PBS) and mounted for microscopy. All cells were observed on an Olympus Provis microscope fitted with a 100× objective lens. Digital images were captured with a cooled charge-coupled-device camera (Diagnostic Instruments).
Immunofluorescence.
Polyclonal rabbit antibodies were raised to a synthetic FszA peptide (PKPIIPGIFVEQELL) corresponding to amino acids P408 through to L422 of FszA, which was conjugated to diphtheria toxoid (Mimotopes). The specificity of the FszA antibodies was improved by ammonium sulfate precipitation followed by quenching against paraformaldehyde-fixed ΔfszA Dictyostelium cells. For immunofluorescence, the Dictyostelium cells were grown as described above, except 1 to 2 h prior to fixation the cells were settled onto 0.1% polyethylenimine (Sigma)-coated coverslips to help them more firmly attach. After being blocked in BB (1% bovine serum albumin [type V; Sigma], 1% cold-water fish-skin gelatin [Sigma], 0.05% Tween 20 in PBS), the cells were incubated with anti-FszA diluted to 1/10 in blocking buffer with 0.5% acetone powders prepared from the ΔfszA strain (24), 2% whole goat serum (Pierce), and 0.5% Tween 20 for 1 h. After incubation with the primary antibody, cells were washed three times in PBS with 0.05% Tween 20. Primary antibodies were detected with goat anti-rabbit 488- and 568-nm Alexa Fluor dye conjugates (Molecular Probes) diluted 1/2,000 in blocking buffer with 2% whole goat serum.
Immunoelectron microscopy.
Cells were prepared for immunoelectron microscopy by one of two methods. (i) Vegetative amoebae of the uracil auxotrophic strain DH1 were high-pressure frozen and freeze substituted. Cells were grown to early logarithmic phase in liquid culture (HL5 medium), centrifuged at 1,000 × g for 2 min, and transferred to 200-nm-deep, brass planchettes for immediate freezing in a Leica high-pressure freezing machine. Freeze substitution (in a Leica AFS machine) was in anhydrous acetone containing 0.1% uranyl acetate at −90°C for 76 h. Samples were then warmed to −45°C, washed in three changes of anhydrous acetone before stepwise infiltration with Lowicryl HM20 resin over 3 days, and then polymerized under UV light at −45°C. (ii) AX3 cells transformed with FszB-GFP (see below) were fixed in 2.5% glutaraldehyde and dehydrated in ethanol before being embedded in LR Gold resin.
For immunolabeling, ultrathin sections were cut and collected on Formvar-coated slot grids. Grids were blocked in BB overnight at 4°C and labeled with preadsorbed anti-FszA (described above) diluted 1/10 in BB and 0.5% acetone powders or labeled with affinity-purified rabbit anti-GFP diluted 1/100 in 2.5% normal goat serum in BB (goatBB) for 1 h at 22°C. Following incubation with the primary antibody, grids were washed four times for 10 min in PBS with 0.05% Tween 20, blocked for 10 min in goatBB, and incubated with goat anti-rabbit 15-nm-diameter gold particles (British BioCell International) diluted 1:100 in goatBB for 1 h at 22°C. Grids were stained with aqueous uranyl acetate (20 min) and lead citrate (5 min) before being viewed on a Philips CM12 transmission electron microscope. Control experiments, in which the primary antibody was omitted, did not specifically label any cellular structures (data not shown).
GFP-tagged Fsz constructs.
To construct the fszB-GFP fusion construct, primers dctz2start (GGGAGCTCAAAAAATGACAATTTTAAATAG) and dctz2nostop (CCCTCGAGATGTACAAATAAACAAG) were used to amplify the entire gene from Dictyostelium cDNA. This PCR product was cloned as a SacI-XhoI fragment into pWM694, a derivative of pZG containing a unique XhoI site just upstream of the GFP gene, to create the fszB-GFP fusion construct designated pSNC1. pSNC1 was cut with SacI and PstI, and the resulting fragment was cloned between the SacI and XhoI sites of pDXA3c (34) and designated pWM1324. To ensure that the proper upstream expression signals for fszB were present, a small sequence upstream of SacI was deleted by cutting pWM1324 with HindIII and Asp718 and filling in the overhangs with Klenow DNA polymerase. The resulting plasmid was designated pFszB-GFP and was used to transform AX3 Dictyostelium cells by electroporation (25). The transformants were selected with G418 (20 μg/ml; Gibco-BRL). Due to its being encoded on a multicopy plasmid, FszB-GFP was expressed at high levels in most cells. A similar strategy was used to clone FszA-GFP into the same plasmid, but the expression levels were so high that relatively little protein reached the mitochondria and most aggregated in the cytoplasm (data not shown). FszA-GFP was therefore expressed at much lower levels from a chromosomally integrating vector. ClaI restriction enzyme sites were introduced 5′ to the start codon and stop codons of a cDNA version of fszA by PCR with the primers FszACla1R (TATTTAATCGATAAATGAGTCAGTTCATG) and FszACla2L (TAATTTTTAATCGATATTTGAACCAGGAGT). The cDNA PCR product was cloned into pGemT-easy (Promega) before being ligated into the Dictyostelium GFP vector pA15GFP (16) to produce pFszA-GFP. This plasmid was transformed into amoebae as described above.
RESULTS
D. discoideum encodes two FtsZ genes.
To determine if Dictyostelium encoded FtsZs of mitochondrial origin, a PCR-based approach was used to amplify FtsZ gene sequences from amoebae. The protein sequence of MsFtsZ-mt from Mallomonas (6) was aligned with that of bacterial FtsZs, and regions that were identical between MsFtsZ-mt and the FtsZs of α-proteobacteria, but not other bacteria, were identified. The degenerate PCR primers FtsZ1R and FtsZ4L, each specific for the conserved regions G(G/C)NAVNNMI and LINLDFADV, respectively, amplified a product of 572 bp from Dictyostelium genomic DNA that was cloned and sequenced. BlastP searches of GenBank confirmed that the PCR product was an FtsZ, since it encoded amino acid sequence motifs typical of the FtsZ family. The 5′ and 3′ ends of the FtsZ gene, called fszA, were obtained by RACE. Northern analyses with a probe derived from fszA barely detected a transcript, although a cDNA was easily amplified by reverse transcriptase PCR (RT-PCR) (data not shown). PCR primers FszACla1R and FszACla2L, specific for either end of the coding region, amplified a coding sequence of 1,574 bp from cDNA and 1,655 bp from genomic DNA. Comparison of the two sequences indicated that the fszA gene contained an intron of 81 bp near the 5′ end of the gene.
While cloning fszA, we became aware of FtsZ gene data from the Dictyostelium Genome Sequencing Project (http://dicty.sdsc.edu). tBlastn searches of this database with fszA identified two sets of sequences. The first set corresponded to fszA, and the other set of sequences defined a second homolog that we called fszB. The fszB gene appeared to be complete in the database, and a full-length gene sequence for fszB was cloned by PCR amplification from Dictyostelium genomic DNA and cDNA. Comparison of the sequences derived from these clones indicated that fszB contained introns of 57 bp near the 5′ end and 108 bp in the middle of the gene.
FszA and FszB are 517 and 366 amino acids in length, respectively, and are 25.7% identical and 36.1% similar to each other. The N-terminal regions of both Dictyostelium FtsZs are positively charged, amphipathic helices predicted to comprise mitochondrial targeting presequences by the program MitoProt (http://www.mips.biochem.mpg.de/cgi-bin/proj/medgen/mitofilter). The presence of introns and poly(A) tails and the absence of fszA and fszB in the completely sequenced Dictyostelium mitochondrial genome (49) indicate that the genes are nuclearly encoded and that their proteins are likely to be imported into mitochondria from the cytoplasm. We predict that the sizes of FszA and FszB without their mitochondrial targeting sequences are about 53 and 35 kDa, respectively.
The bacterial ancestor of mitochondria is thought to be an α-proteobacterium (28), and phylogenetic analyses of mitochondrial FtsZ from M. splendens (6) and C. merolae (5, 62) indicated that the genes for these proteins were of α-proteobacterial origin. Similar analyses of the FszA and FszB proteins show that they also group with mitochondrial and α-proteobacterial FtsZs but bootstrap support for this relationship is, however, quite weak (data not shown).
Mitochondrial morphology in Dictyostelium.
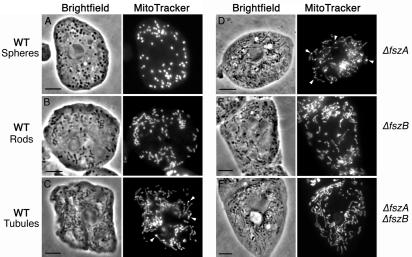
To determine if the fsz genes were necessary for mitochondrial division, we wanted to disrupt the coding sequences of the genes by insertional mutagenesis. Before we examined the effects of these mutations, it was first necessary to characterize the morphology of the organelles in wild-type cells of the AX3 strain. The mitochondria of Dictyostelium, when visualized by phase-contrast bright-field microscopy, appeared as small dark bodies in the cytoplasm of the amoebae (Fig. 1). Since mitochondria located towards the edges of the cells were not always visible under phase-contrast microscopy, the mitochondria were labeled with the mitochondrion-specific dye MitoTracker Red and were chemically fixed prior to microscopy (Fig. 1).
FIG. 1.
Mitochondrial morphology in wild-type Dictyostelium vegetative amoebae and the Δfsz mutants. The mitochondria are dark cytoplasmic organelles in bright-field (phase-contrast) images and bright fluorescent organelles when stained with MitoTracker Red. Arrowheads show examples of intense MitoTracker Red-staining regions within the mitochondria (SMBs). See the text for descriptions and Table 1 for frequencies of cells with mitochondrial spheres, rods, or tubules. Scale bar = 5 μm.
In our cultures, the width of mitochondria ranged between 0.5 and 1.0 μm but their lengths varied greatly, from 0.5 to >10 μm. We observed three classes or forms of mitochondrial shape: spheres (Fig. 1A), rod-like mitochondria with lengths two to four times their widths (Fig. 1B), and tubules with lengths many times greater than their widths (Fig. 1C). We note that, unlike the mitochondria of budding yeast (48), for example, the Dictyostelium mitochondrial tubules were never seen to be branched. In some cells, all three classes of mitochondria were present, suggesting that the different forms may represent a morphological continuum; however, usually only one type was prevalent in any individual cell. When classified by their dominant form of mitochondria, 66% of wild-type cells contained spheres, 24% of wild-type cells contained rods, and 10% of wild-type cells contained tubules (Table 1). At present, we do not know what determines this variation in length, but it may well be dependent on the frequency of mitochondrial division with respect to organelle growth and elongation—with numerous fission events creating spheres and progressively fewer fissions producing rods and then tubules. It is possible, too, that this length variation is random or else is linked to cell age. In order to reduce any potential effects of development and senescence, the cells used for our experiments were maintained as log-phase, vegetative cultures.
TABLE 1.
Mitochondrial morphology in wild-type (AX3) and fsz-null mutantsa
| Strain | No. of cells scored | Spheres (%) | Rods (%) | Tubules (%) |
|---|---|---|---|---|
| WT (AX3) | 569 | 66 | 24 | 10 |
| ΔfszA | 451 | 2 | 16 | 82 |
| ΔfszB | 505 | 4 | 25 | 71 |
| ΔfszA ΔfszB | 523 | 5 | 7 | 88 |
| ΔfszA fszA-GFP | 273 | 0 | 26 | 74 |
| ΔfszB fszB-GFP | 403 | 14 | 46 | 40 |
In each strain, the percentages of cells with predominantly spherical, rod-like (length one to four times the width), and tubular (length more than four times the width) mitochondria are given. The mitochondrial morphology of DH1, the background strain in which the ΔfszA and ΔfszA ΔfszB mutants were constructed, was similar to that of AX3.
To explore any effect that position in the cell cycle might have on mitochondrial morphology, we sought to obtain synchronized cultures. Preliminary observations suggested that all mitochondrial types were present in cells at different time points after the start of cell cycle synchronization (data not shown). However, the procedures for synchronizing cell division in Dictyostelium (32, 33) are not efficient, and in our hands, not all cells could be synchronized and synchronization lasted only one or two division cycles. At present then, we cannot rule out the possibility that the cell cycle and mitochondrial morphology are linked. In other cells that harbor many mitochondria or a mitochondrial network, no specific change in mitochondrial morphology linked to the cell cycle has been demonstrated though there are examples of physiological and developmental factors that influence morphology (22).
Knockouts of fszA and/or fszB promote tubule formation.
Since perturbation of chloroplast FtsZ expression in plants severely inhibits the division of chloroplasts (52, 59), it was anticipated that disruptions of either fszA or fszB in Dictyostelium would inhibit mitochondrial division and result in fewer, elongated mitochondria. In the AX3 wild-type strain, the coding sequence of fszB was interrupted with a blasticidin resistance gene cassette to create a null-allele strain called the ΔfszB strain. To produce a double-fsz gene knockout strain, the coding sequence of fszA was interrupted with a uracil biosynthetic gene in the uracil auxotrophic strain, DH1, to create the ΔfszA strain. The fszB gene of ΔfszA cells was then interrupted with a blasticidin resistance cassette to create the double-fsz knockout ΔfszA ΔfszB strain. The mutants were screened for fsz gene disruptions by Southern analyses and PCR (data not shown). To check that the fsz genes could not produce proteins, RT-PCR was performed with fsz gene-specific primers bordering the insertional cassette. No cDNA was amplified from the mutants with their respective fsz primers, indicating that no full-length fsz mRNA was being produced (data not shown). Finally, Western blots with anti-FszA antibodies raised to the synthetic FszA peptide PKPIIPGIFVEQELL detected a protein sized at 59 kDa in wild-type cells but not in ΔfszA and ΔfszA ΔfszB cells (data not shown).
The 59-kDa protein was slightly larger than the 53 kDa predicted for FszA. We believe the detected protein is FszA, because in cells expressing another FszA, tagged with GFP (27 kDa), an additional band of 86 kDa was present (data not shown). It is not clear why FszA and FszA-GFP electrophorese at weights higher than their predicted molecular weights, but this phenomenon was also noted for an FtsZ from Sinorhizobium meliloti (37).
A comparison of chemically fixed wild-type and Δfsz cells in at least two independent experiments revealed that the mutants always contained mostly tubular mitochondria and relatively few mitochondrial spheres or rods. The proportions of cells containing different mitochondrial types in one sampling period are shown in Table 1. Eighty-two percent of ΔfszA cells had tubular mitochondria, 16% had rod-shaped mitochondria, and only 2% had spheres (Fig. 1D; Table 1). Disruption of the fszB gene also increased the proportion of cells with tubular mitochondria. Seventy-one percent of ΔfszB cells contained tubular organelles, 25% contained rods, and 4% contained spheres (Fig. 1E; Table 1). Disruption of the fsz genes thus appears to have inhibited mitochondrial division—favoring the formation of longer, tubular organelles rather than the shorter spheres and rods that are presumably the products of more-frequent division events.
To test the possibility that the two Fsz proteins were to some degree functionally redundant, we created a double-fsz mutant. Eighty-eight percent of the ΔfszA ΔfszB cells contained tubules, and 7 and 5% contained rods and spheres, respectively (Fig. 1F; Table 1). Thus, there appears to be little or no additive effect on mitochondrial morphology by knocking out both fsz genes in comparison to the single-gene mutants.
The morphology of mitochondria was also examined in DH1, the strain in which the ΔfszA and the ΔfszA ΔfszB mutations were generated. Most of the DH1 cells, like the wild-type AX3 strain, contained spherical organelles, with fewer having rods and tubules. This further indicates that the increase in mitochondrial length in the Δfsz mutants is specifically caused by the loss of the fsz gene products.
We predicted that if mitochondrial fission was less frequent in the Δfsz mutants, they should contain fewer mitochondria per cell than the wild-type strain. Counts of the numbers of mitochondria per cell confirm this: wild-type cells each contained an average of 50 mitochondria, whereas the Δfsz cells had only about two-thirds this number. Apart from changes in mitochondrial length and numbers of organelles per cell in the fsz-null mutants, there appeared to be no other obvious changes in the cell or organelle morphology, such as any change in the total mass of the mitochondria per cell or in the fine structure of the inner membrane cristae (data not shown).
Loss of FszB causes growth defects.
To determine if the net increase in mitochondrial lengths in the Δfsz mutants could affect other aspects of cellular biology, such as cell growth, Dictyostelium amoebae were grown on bacterial lawns (Micrococcus luteus) and their growth rates were measured. As the amoebae consume bacteria and replicate, they produce plaques which increase in size as the colonies of amoebae grow. Comparisons of the growth rates of these Dictyostelium colonies revealed that the growth of the ΔfszA mutant was similar to that of the wild-type strain (data not shown); however, ΔfszB and ΔfszA ΔfszB colonies grew at about one-third this rate. Given that the ΔfszA mutant grows normally on bacteria, it appears that the loss of FszB in the double mutants is the cause of the slow-growth phenotype. To establish why the absence of FszB would produce this phenotype, the mitochondria of the mutants were examined. With light microscopy, it was difficult to differentiate mitochondria from phagocytosed bacteria, but electron microscopy of thin sections through the bacterially fed ΔfszB cells revealed no discernible changes in mitochondria compared to those of wild-type cells (data not shown).
To further explore the slow-growth phenotype, the amoebae were grown axenically in liquid media. When grown on a surface, such as the bottom of a tissue culture flask that was not being shaken, the ΔfszB and the double Δfsz mutants appeared to grow more slowly. To accurately measure this growth rate, the cultures were grown as a suspension and samples were removed periodically for cell counting. The cell doubling times for the ΔfszA mutant were about the same as those for the AX3 and DH1 parental strains (12 to 14 h). However, the doubling time of the FszB-lacking strains was much greater, and very often, shifting the ΔfszB and ΔfszA ΔfszB cells from surface to suspension growth resulted in the cultures ceasing to grow.
FszA forms belts and puncta within mitochondria.
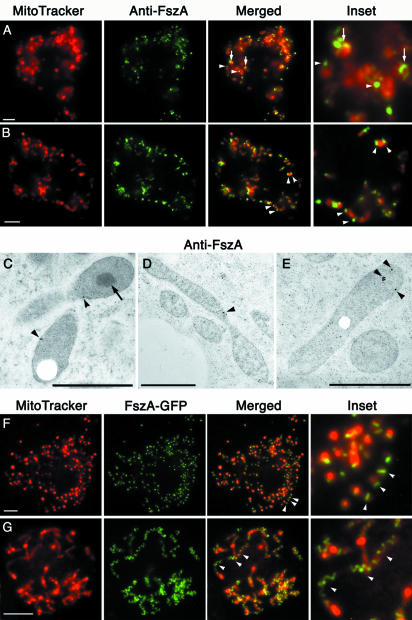
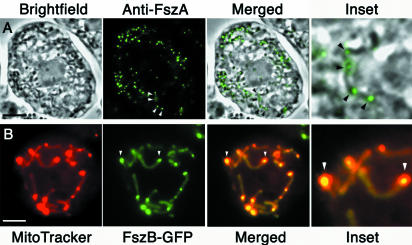
The localization of anti-FszA antibodies in wild-type, MitoTracker-labeled amoebae was consistent with FszA having role in mitochondrial division (Fig. 2A and B). FszA specifically labeled mitochondria, where it concentrated as punctate and belt-like foci presumably containing the polymerized protein (Fig. 2A and B). In some spherical mitochondria, a single FszA “belt” spanning the organelle was observed (Fig. 2A). These belts may be partially formed or labeled Z rings similar to those present in the mitochondria of C. merolae (46), in the chloroplasts of higher plants and algae (26, 39, 42, 44, 45, 65), and in bacteria (31, 60). In rod-shaped mitochondria, two or more FszA foci were observed; these foci varied from being punctate to belt-like and were often located at the ends of the organelle (Fig. 2B). Since FtsZ foci have been shown to persist after division in the daughter mitochondria of C. merolae (46), the FszA foci that we observed on the ends of Dictyostelium mitochondria may mark recently divided organelles. In addition to the punctate and belt-like foci, mitochondria usually displayed a low level of evenly distributed FszA labeling (Fig. 2A and B).
FIG. 2.
FszA formed belts and puncta within the mitochondria of Dictyostelium. (A and B) Immunofluorescence of Dictyostelium amoebae labeled with anti-FszA (green) and MitoTracker Red (red). (A) FszA formed belt-like (arrows) or punctate (arrowheads) structures in spherical mitochondria. (B) In rod-shaped mitochondria, one or two FszA foci were detected (arrowheads), often located near the ends of the organelles. Scale bar = 5 μm. (C to E) Immunoelectron microscopy with anti-FszA indicated that FszA (arrowheads) was located within the mitochondrion but did not concentrate within the electron-dense SMB (arrow) (C). Scale bars = 1 μm. (F and G) Expression of FszA-GFP in Dictyostelium. (F) At low expression levels, FszA-GFP was targeted to the mitochondria where it mostly formed a single belt in each spherical organelle (arrowhead). (G) At high expression levels, FszA-GFP formed extended helices (arrowheads) in long, tubular mitochondria. Scale bar = 5 μm.
In an attempt to obtain greater resolution of the localization of FszA, we performed immunoelectron microscopy on high-pressure-frozen, freeze-substituted Dictyostelium amoebae, and the patterns we observed concurred with the data from our immunofluorescence experiments. Although the levels of labeling were low, FszA predominately localized to narrowed regions within the mitochondria that might be presumptive regions of division (Fig. 2C and D) and to the ends of mitochondria (Fig. 2E). Analysis of serial sections will be required to determine if the narrowed regions of mitochondria are genuine sites of division and if FszA always localizes to them.
Unfortunately, in our preparations, we were unable to resolve the inner and outer mitochondrial membranes, and consequently the mitochondrial cristae were not resolved (Fig. 2C to E). We cannot confirm, then, if FszA is located in the mitochondrial matrix or the intermembrane space, though the matrix seems more likely given that this is where FtsZ is found in the mitochondrion of C. merolae algae (46, 61). Both FszA and FszB are unlikely to be membrane-spanning proteins, since they, like all other FtsZs, show no predicted transmembrane domains.
In Dictyostelium mitochondria, a submitochondrial body (SMB) of unknown function that, when examined by electron microscopy, appears as an electron-dense mass has been reported (64). We observed these bodies in our preparations (Fig. 2C), and unlike with FszB-GFP localization (see below), no anti-FszA gold particles ever localized to them.
Overexpression of FszA-GFP causes mitochondrial tubule formation.
In order to visualize FszA in vivo in Dictyostelium, we generated cells that expressed FszA-GFP. We immediately noticed dramatic differences in expression levels of the fusion protein. Since the FszA-GFP fusion was expressed from a chromosomally integrated construct, we expected some variation between transformants due to different copy numbers of the integrated pFszA-GFP construct; however, there were various levels of expression—even within single cloned lines (data not shown). To study the effects that the range of expression levels of FszA-GFP might have had on mitochondrial morphology, the cells were fixed for microscopic observation. In cells expressing lower levels of the FszA-GFP fusion protein (as judged by the intensity of GFP fluorescence), the protein formed belts and puncta similar to those observed by immunofluorescence (Fig. 2F). In cells expressing the highest levels of protein, bright helices were formed (Fig. 2G) and tubular mitochondria predominated, suggesting that the FszA-GFP helices inhibited mitochondrial fission. Intermediate levels of fusion protein expression corresponded to those of tubular mitochondria, with many FszA-GFP puncta being positioned at evenly spaced intervals along their lengths (data not shown). Similar inhibitory effects on division have been noted previously for bacteria and chloroplasts overexpressing FtsZ-GFP fusion proteins (31, 39).
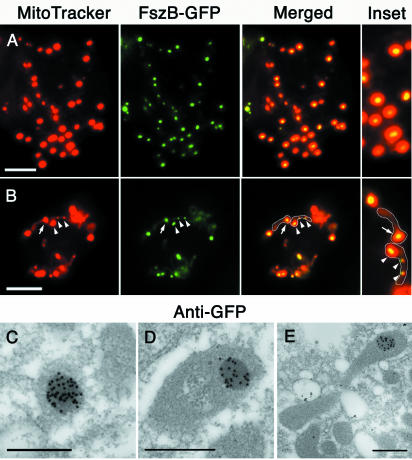
FszB-GFP localizes to the SMB.
Following several unsuccessful attempts to generate antibodies to FszB, we tagged FszB with GFP for expression and visualization in Dictyostelium amoebae. FszB-GFP was observed as a single body within mitochondrial spheres (Fig. 3A) and mostly as a single body located near one end of rod-shaped or tubular mitochondria (Fig. 3B). In mitochondrial rods and tubules, the FszB-GFP foci colocalized with regions of the mitochondrion that were brightly stained with MitoTracker. Usually only one of these bright regions was visible in mitochondria, but occasionally more than one was observed (Fig. 3B). These bright regions are likely to be identical to the electron-dense (Fig. 2C) SMBs of Dictyostelium that characteristically accumulate MitoTracker and contain the protein TorA (64).
FIG. 3.
FszB-GFP localizes to an SMB in Dictyostelium amoebae. (A and B) FszB-GFP accumulated within mitochondria to regions called SMBs that stained brightly with MitoTracker Red. SMBs are present in spherical (A) and tubular (B) organelles. (B) Mitochondria have been outlined. Usually, a single SMB was detected near the end of tubular mitochondria (arrow), but occasionally one or two additional SMBs are also visible within an organelle (arrowheads). (A and B) Scale bars = 5 μm. (C to E) Immunoelectron microscopy with anti-GFP gold demonstrates that FszB-GFP almost exclusively localized to the electron-dense SMB and in mitochondrial tubules this structure was often near one end (E). Scale bar = 0.5 μm.
In cells expressing FszB-GFP, the MitoTracker foci were brighter than those of both wild-type and ΔfszA cells; in addition, in contrast to wild-type cells, FszB-GFP cells contained detectable SMBs in most of their mitochondria (compare Fig. 3B to 1C and D). This finding may be due to the fusion protein being more highly expressed than endogenous FszB (see below). Consistent with this, in cells in which fszB was knocked out, the number and size of detectable MitoTracker foci were greatly reduced (Fig. 1E and F). Interestingly, when TorA was overexpressed as a TorA-GFP fusion protein, the size of the MitoTracker-staining SMBs also increased (64). We are confident that the FszB and not the GFP moiety is responsible for targeting to the SMB, because when GFP alone is targeted to the mitochondrion, the organelle is evenly labeled and there is no aggregation to the SMB (data not shown).
It was proposed that the mitochondrial regions that accumulate MitoTracker and TorA correspond to the electron-dense SMBs visualized by electron microscopy (64). To test this and also the ultrastructural localization of FszB-GFP, we prepared FszB-GFP cells for immunoelectron microscopy and labeled them with anti-GFP. Gold particles almost exclusively labeled the SMBs, demonstrating that FszB-GFP, and therefore MitoTracker and TorA-GFP, concentrate within these regions (Fig. 3C to E). The electron-dense nature of SMBs suggests that the concentration of proteins within this body is higher than in the surrounding matrix. Since MitoTracker Red cross-links to proteins, this may be why it accumulates within the SMBs.
The propensity of FtsZ proteins to mislocalize and form aberrant structures when expressed at high levels (e.g., FszA-GFP and chloroplast FtsZs) (65) prompted us to consider if this was happening for FszB-GFP. In the few cells expressing levels of FszB-GFP that were barely detectable (possibly at levels similar to the endogenous protein), FszB-GFP still appeared to form a single focus within mitochondria (data not shown). Furthermore, the fact that SMBs are present in wild-type cells (64) suggests that these structures are normal and contain FszB.
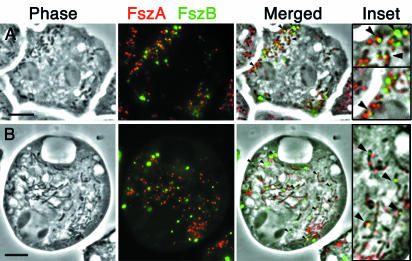
FszA partially colocalizes with FszB-GFP.
To determine if FszA and FszB formed any colocalizing structures, the location of FszA was resolved by immunofluorescence analysis in cells expressing FszB-GFP (Fig. 4). In spherical mitochondria, FszA localization was adjacent to or overlapped with an FszB punctum (Fig. 4A). In tubular mitochondria, where multiple FszA foci were present, one of the FszA foci was usually adjacent to, or overlapped with, the single FszB-GFP punctum (Fig. 4B). Considering our immunoelectron microscopic data, which showed FszB localizing to the SMBs and FszA-GFP not doing so, it appears that, although FszA and FszB can form adjacent structures, they do not coalign, in contrast to the rings composed of the two chloroplast FtsZs in Arabidopsis (39).
FIG. 4.
Double labeling of FszA and FszB in Dictyostelium cells. FszA (red) has been detected by immunofluorescence in cells transformed with FszB-GFP (green). (A) In more than half the spherical and rod-shaped mitochondria, one FszB punctum paired with or partially colocalized with an FszA punctum (arrowheads). (B) Tubular mitochondria usually contained a single FszB punctum and two or more FszA foci dispersed along the length of the organelle. FszA and FszB usually formed adjacent or overlapping puncta near one end of the mitochondrion (arrowheads). Scale bars = 5 μ m.
Complementation of the Δfsz mutants with the corresponding fsz strain.
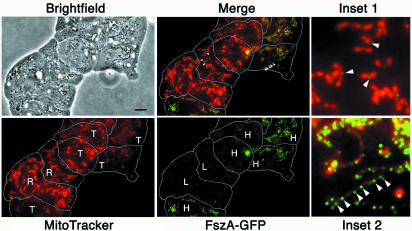
If knockouts of fszA and fszB reduce mitochondrial fission and promote tubule formation, then complementation with the wild-type versions of these genes should promote fission and increase the frequency of spheres and rods. Because overexpression of other FtsZs has been found to interfere with organelle division (65), we decided to complement the Δfsz strains with GFP-tagged versions of Fsz proteins so that the amount of the protein in individual cells could be easily observed by microscopy (Fig. 5).
FIG. 5.
At low expression levels, FszA-GFP partially complemented the loss of FszA in ΔfszA cells. A phase-contrast, bright-field image of several closely packed and flattened ΔfszA amoebae transformed with FszA-GFP is shown, and the outlines of the cells have been traced in white. Colabeling with MitoTracker Red showed that individual amoebae contained rod (R)- or tubule (T)- shaped mitochondria. The level of FszA-GFP expression, ascertained by intensity of GFP fluorescence, was high (H) or low (L). Provided FszA-GFP was expressed at low levels in ΔfszA cells, the fusion protein reduced the percentage of cells with tubular mitochondria by increasing the proportion of rods (Merge; Inset 1). FszA-GFP localized to the midpoints of mitochondria that may have been undergoing fission (Inset 1, arrowheads). When overexpressed, FszA-GFP (Inset 2, arrowheads) appeared to be unable to reduce mitochondrial tubule formation. Scale bar = 5 μm.
Expression of FszA-GFP only moderately reduced the number of ΔfszA cells with tubular mitochondria (from 82 to 74%) (Table 1). If, however, the levels of GFP fluorescence in individual cells were considered, the effect was more striking. Among cells expressing the lowest levels of FszA-GFP (roughly the lowest 20th percentile), the levels of those having predominately tubular mitochondria were reduced to 60%. In cells expressing the highest levels (upper 20th percentile), over 90% had tubular mitochondria. An example of this effect can be seen in Fig. 5, where cells expressing the lower levels of the fusion protein (inset 1) have rod-shaped mitochondria and cells with the higher levels have tubular mitochondria (inset 2). We conclude that FszA-GFP can partially complement ΔfszA cells provided the fusion protein is expressed at low levels.
Complementation of the ΔfszB strain with FszB-GFP reduced the percentage of tubules from 71 to 40% and increased the percentage of rods correspondingly (Table 1). Because FszB-GFP was expressed from a plasmid, rather than being chromosomally integrated, as FszA-GFP was, its expression was generally higher and less varied than that of FszA-GFP. Therefore no attempt was made to correlate the expression levels of FszB-GFP with mitochondrial morphology.
Though there were reductions in the ratios of tubular mitochondria in the complementation experiments, this was matched mostly by an increase in the number of rods and not spheres. It therefore seems that the Fsz-GFP fusion proteins may have a reduced ability to function normally and fully complement the losses of the native Fszs. Though the GFP-tagged Fsz proteins are still capable of polymerizing into structures within the mitochondria, the bulky GFP tags may have inhibited their ability to change conformation upon hydrolysis of GTP or to interact with other division proteins. Consistent with this was the observation that, in ΔfszB cells complemented with FszB-GFP, normal growth rates were not restored on bacterial lawns. The ability of FszB-GFP to allow ΔfszB cells to grow in suspension has not been tested.
Independence of FszA and FszB localization.
To establish whether FszB influences the distribution of FszA, immunolocalization with anti-FszA was performed on ΔfszB cells. In the absence of FszB, FszA continued to form punctate and/or belt-like foci within mitochondria (Fig. 6A). The number and spacing of these puncta in rod- and tubule-shaped ΔfszB mitochondria also appeared to be unchanged. The appearance of multiple, evenly spaced FszA foci in each elongated, tubular mitochondrion is reminiscent of those in bacterial non-ftsZ cell division mutants which contain numerous, equally spaced Z rings within their elongated cells, e.g., ftsW mutants (40).
FIG. 6.
Localizations of FszA and FszB appeared to be independent of one another. (A) In phase-contrast images of ΔfszB cells, mitochondrial tubules appear as dark, elongated shapes. Immunofluorescence of these cells with anti-FszA revealed that FszA still localized to the mitochondria and continued to form multiple foci in the tubular organelles (arrowheads). (B) In ΔfszA cells, FszB-GFP continued to concentrate in the submitochondrial structure, the SMB (arrowheads). Scale bar = 5 μm.
Any dependence of FszB localization on FszA was examined by introducing FszB-GFP into ΔfszA cells (Fig. 6B). In these cells, MitoTracker still accumulated as brightly fluorescent SMBs within the mitochondria and FszB-GFP continued to localize to these bodies but probably less efficiently than in wild-type cells (compare Fig. 3B and 6B). Despite the loss of FszA, most mitochondria continued to produce one FszB-GFP-labeled SMB per mitochondrion, although, as with wild-type cells, there was occasionally more than one SMB per organelle (Fig. 6B). We conclude that FszA and FszB do not depend on one another for normal localization.
DISCUSSION
We have identified the genes for two FtsZ proteins in amoebae of Dictyostelium. FtsZs are essential in bacterial cell division (35) and in the division of chloroplasts (50), and we propose that the Dictyostelium proteins FszA and FszB act in the division of mitochondria. FszA and FszB are likely to be derivatives of an FtsZ that was acquired from the ancestral mitochondrion, an endosymbiotic α-proteobacterium. It appears that FszA and FszB are most closely related to FtsZs of the α-proteobacteria, that they are targeted to mitochondria (with at least FszA displaying a localization pattern consistent with a direct role in mitochondrial division), and that disruptions of fszA and fszB result in elongated mitochondria, an effect that is likely to be due to a decreased ability of mutant cells to divide their mitochondria.
The morphological change from mitochondrial spheres and rods into tubules in the fsz knockout mutants of Dictyostelium resemble the changes induced in plant mitochondria when fission is blocked by disruption of the dynamin-like protein ADL2b (4). One difference, however, is that branched tubules are formed in fission-deficient plant mitochondria, and branches were never observed in the Dictyostelium wild-type or fsz knockout mutants. Branching of mitochondrial tubules is probably the result of mitochondrial fusion events that occur between the tips and the sides of organelles (56). Strains from animals and fungi with the genes for mitochondrial fission proteins knocked out and in which normal organelle morphology is maintained by balanced fusion and fission events produce networks of highly interconnected tubules (8, 55, 58). The fact that such networks were never observed in Dictyostelium fsz-null cells (neither were branches, even in wild-type cells) indicates that Dictyostelium may perform little, if any, mitochondrial fusion. We note that none of the fusion proteins identified in yeast or animals have obvious homologues in the Dictyostelium genome.
The localization patterns of FszA imply a direct role in mitochondrial division.
The belts and puncta of FszA and FszA-GFP observed in Dictyostelium mitochondria resemble the MtFtsZ (mitochondrial FtsZ) structures in Mallomonas (6). In both these organisms, FtsZ localizes to the ends of mitochondria that may have recently divided and to regions in the middle of mitochondria that might be future sites of division. In tubular Dictyostelium mitochondria, there can be several such sites evenly spaced along the length of the mitochondrion.
FtsZ characteristically forms rings in bacteria and chloroplasts, but we could observe no FszA rings in Dictyostelium mitochondria. FtsZ rings are almost certainly formed, though, since they have been observed within the mitochondria of C. merolae (46). It is possible that the FszA belts observed in Dictyostelium are partially formed rings (or even unresolved, complete rings) and that the FszA puncta are nucleation points from which FszA-GFP rings grow. Support for the latter idea comes from the fact that, in E. coli, GFP-tagged FtsZ rings have been observed to initiate as dots that grow into partial rings and then complete rings (2, 60).
FszB appears to be indirectly involved in mitochondrial division.
If FszB is restricted to SMBs, as our FszB-GFP fusion experiments indicate, it is not obvious how FszB could directly participate in organelle division. Since bacterial FtsZs characteristically form polymers (14) and cells that lack FszB produce smaller and fewer SMBs per mitochondrion, it may be that FszB produces a scaffold for SMB formation. How the loss of FszB and its subsequent effect upon SMBs go on to affect mitochondrial morphology, lower the rate of growth on bacteria, and cause surface-growth dependence is not known. Mutations of the other known SMB-resident protein, TorA, also result in slow growth on bacteria, and this has been shown to be due to a defect in chemotaxis (64). torA− cells appear to be able to detect external chemical gradients but cannot properly coordinate the extension of their pseudopods. Slow growth of torA− colonies on bacterial lawns is due to the reduced capacity of feeding amoebae to migrate into the surrounding lawn and expand the edges of the colony (64). Another curious similarity between ΔfszB and torA− mutants is that when the latter are complemented with a highly expressed TorA-GFP fusion protein, they become surface-growth dependent, like ΔfszB.
The loss of SMB proteins also produces altered mitochondrial morphologies, since the organelles of torA− mutants seem to be larger and rounder than those of wild-type cells (64). Though this phenotype is not exactly the same as for ΔfszB cells, it does indicate that Dictyostelium requires FszB and TorA to maintain normal mitochondrial morphology. Should ΔfszB, like torA−, cells be similarly chemotactically deficient, it would not appear to be mitochondrial size per se that affects chemotaxis since the colony growth rates of ΔfszA cells, which have elongated, tubular mitochondria, are normal.
Although it remains to be established if chemotaxis is responsible for the slow growth of ΔfszB cells on bacteria, other causes should be also considered. The slow growth of amoebae in axenic culture suggests that the impairment of some mitochondrion-specific functions, such as energy generation, may be responsible. Opposing this though is our observation that the ability of the mitochondria to take up MitoTracker Red, which depends upon the organelle's transmembrane potential, appears unaffected in ΔfszB cells. The arrest of growth when the cells are grown in suspension suggests that some apoptotic response may be triggered, conceivably by mechanical damage to the mitochondria and the leaking of their contents caused by the agitation of the cells. This damage would depend upon the loss of FszB structurally weakening the mitochondria.
Although the conserved N-terminal halves of FszA and FszB contain the residues characteristic of FtsZs that are involved in GTP binding and hydrolysis (50), their C-terminal halves are very different from one another. Like most other FtsZs, FszA contains a predicted C-terminal helix (data not shown) that is believed to interact with membrane-anchored proteins involved in division (50). However, the C-terminal half of FszB is largely truncated. These structural differences may account for the distinctly different distributions of the two proteins within mitochondria: FszA appears to form rings or puncta that are likely to be just inside the inner membrane, whereas FszB accumulates in SMBs that are deep within the mitochondrion and are seemingly devoid of membrane.
All eukaryotes probably use dynamin-like proteins for mitochondrial fission.
If FszA and FszB are involved in mitochondrial fission, why do a small proportion of ΔfszA and ΔfszB cells still contain rods and spheres, a characteristic of wild-type cells? One possibility is that other mitochondrial division proteins, such as dynamin-like proteins which divide the mitochondrial outer membrane (see below), may retain enough fission activity to prevent mitochondria from becoming tubular, even in the absence of inner membrane division. Extreme elongation of mitochondrial tubules may also be limited by organelle breakage that may arise from the shear forces generated by rapid cytoplasmic streaming. This finding is perhaps why most Δfsz cells do not contain a small number of extremely long tubules but instead have numerous tubules that are between 4 and 10 μm long.
The discovery that the dynamin-like protein CmDnm1 likely plays a role in dividing the mitochondrion of C. merolae from the outside indicates that dynamins probably have this function not only in higher eukaryotes but also in protists that contain mitochondrial FtsZ (46). The dynamin-like protein Dnm1p in S. cerevisiae and its orthologues, DRP1 in C. elegans and mammals, and ADL2b in A. thaliana are key proteins in mitochondrial outer membrane fission (4, 8, 27, 57, 58). Dnm1p appears to form patches, possibly rings, on the outside of a mitochondrion at sites of division. In C. merolae, CmDnm1 forms 10 to 20 patches around the mitochondrion's site of division but only in the later stages of division do these patches coalesce to form a ring around the constricted organelle (46). Since CmDnm1 remains until the organelle's membranes have finally severed, its main function may be to perform this final scission step (46).
Dictyostelium encodes two dynamin-like proteins, DymA (66) and DymB [H. Nöthe and D. J. Manstein, Abstr. Annu. Meet. Am. Soc. Cell Biol., Mol. Biol. Cell 10(Suppl.):314a, 1999], of which DymA is the most similar to Dnm1p and CmDnm1. The inhibition of mitochondrial fission in dymA− mutants appears to be more severe than that in the fsz mutants because the mitochondria of the former are not only tubular but are also clustered into interconnected sets of tubules (66). Rather than DymA having a direct role in mitochondrial division though, the alterations in mitochondrial morphology in dymA− cells was explained to be most likely due to other defects in membrane trafficking (66). However, in light of the discovery of CmDnm1's function, a reexamination of DymA's role in the division of Dictyostelium mitochondria is warranted.
Another protein known to influence mitochondrial morphology in Dictyostelium is CluA, the functional homologue of Clu1p in yeast (17). Disruptions of cluA caused the mitochondria of Dictyostelium to form clusters (17), and electron microscopy of these cells showed that the mitochondria were interconnected—apparently blocked in the final stages of mitochondrial division (18). Clearly, further studies are required to reveal how the potential mitochondrial division proteins FszA, FszB, DymA, and CluA act and perhaps interact with each other and as-yet-unidentified proteins in the maintenance of normal mitochondrial morphology in Dictyostelium.
Acknowledgments
We thank the contributors to the Dictyostelium Genome Sequencing Project: the Baylor Sequencing Center, Houston, Tex.; the Institute of Biochemistry, Cologne, Germany; the Institute of Molecular Biotechnology, Jena, Germany; and the Dictyostelium Sequencing Group at the Sanger Institute, Hixton, United Kingdom. We also thank Kimalee Weatherill and Mousumi Goswami for technical assistance.
P.R.G. gratefully acknowledges the award of an Australian Research Council Postdoctoral Fellowship. This work was supported by funds from the Australian Research Council to P.L.B.
REFERENCES
- 1.Addinall, S., and B. Holland. 2002. The tubulin ancester, FtsZ, draughtsman, designer and driving force for bacterial cytokinesis. J. Mol. Biol. 318:219-236. [DOI] [PubMed] [Google Scholar]
- 2.Addinall, S., and J. Lutkenhaus. 1996. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol. Microbiol. 22:231-237. [DOI] [PubMed] [Google Scholar]
- 3.Andersson, S. G. E., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Pontén, U. C. M. Alsmark, R. M. Podowski, A. K. Näslund, A.-S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-143. [DOI] [PubMed] [Google Scholar]
- 4.Arimura, S. S., and N. Tsutsumi. 2002. A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc. Natl. Acad. Sci. USA 99:5727-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beech, P. L., and P. R. Gilson. 2000. FtsZ and organelle division in protists. Protist 151:11-16. [DOI] [PubMed] [Google Scholar]
- 6.Beech, P. L., T. Nheu, T. Schultz, S. Herbert, T. Lithgow, P. R. Gilson, and G. I. McFadden. 2000. Mitochondrial FtsZ in a chromophyte alga. Science 287:1276-1279. [DOI] [PubMed] [Google Scholar]
- 7.Bi, E., and J. Lutkenhaus. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161-164. [DOI] [PubMed] [Google Scholar]
- 8.Bleazard, W., J. M. McCaffery, E. J. King, S. Bale, A. Mozdy, Q. Tieu, J. Nunnari, and J. M. Shaw. 1999. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1:298-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boy-Marcotte, E., F. Vilaine, J. Camonis, and M. Jacquet. 1984. A DNA sequence from Dictyostelium discoideum complements ura3 and ura5 mutations of Saccharomyces cerevisiae. Mol. Gen. Genet. 193:406-413. [DOI] [PubMed] [Google Scholar]
- 10.Caterina, M., J. Milne, and P. Devreotes. 1994. Mutation of the third intracellular loop of the cAMP receptor, cAR1, of Dictyostelium yields mutants impaired in multiple signaling pathways. J. Biol. Chem. 269:1523-1532. [PubMed] [Google Scholar]
- 11.Cocucci, S. M., and M. Sussman. 1970. RNA in cytoplasmic and nuclear fractions of cellular slime mold amebas. J. Cell Biol. 45:399-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erickson, H. 2000. Dynamin and FtsZ: missing links in mitochondrial and bacterial division. J. Cell Biol. 148:1103-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erickson, H. P. 1997. FtsZ, a tubulin homologue in prokaryote cell division. Trends Cell Biol. 7:362-367. [DOI] [PubMed] [Google Scholar]
- 14.Erickson, H. P., D. W. Taylor, K. A. Taylor, and D. Bramhill. 1996. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl. Acad. Sci. USA 93:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Errington, J., R. Daniel, and D. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fey, P., K. Compton, and E. Cox. 1995. Green fluorescent protein production in the cellular slime molds Polysphondylium pallidum and Dictyostelium. Gene 165:127-130. [DOI] [PubMed] [Google Scholar]
- 17.Fields, S., M. Conrad, and M. Clarke. 1998. The S. cerevisiae CLU1 and D. discoideum cluA genes are functional homologues that influence mitochondrial morphology and distribution. J. Cell Sci. 111:1717-1727. [DOI] [PubMed] [Google Scholar]
- 18.Fields, S. D., Q. Arana, J. Heuser, and M. Clarke. 2002. Mitochondrial membrane dynamics are altered in cluA− mutants of Dictyostelium. J. Musc. Res. Cell Motil. 23:829-838. [DOI] [PubMed] [Google Scholar]
- 19.Franke, J., and R. Kessin. 1977. A defined minimal medium for axenic strains of Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 74:2157-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujiwara, M., and S. Yoshida. 2001. Chloroplast targeting of chloroplast division FtsZ2 proteins in Arabidopsis. Biochem. Biophys. Res. Commun. 287:462-467. [DOI] [PubMed] [Google Scholar]
- 21.Gao, H., D. Kadirjan-Kalbach, J. Froehlich, and K. Osteryoung. 2003. ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. USA 100:4328-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griparic, L., and A. van der Bliek. 2003. The many shapes of mitochondrial membranes. Traffic 2:235-244. [DOI] [PubMed] [Google Scholar]
- 23.Guzman, L., D. Belin, M. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Knecht, D., and K. Pang. 1995. Electroporation of Dictyostelium discoideum. Methods Mol. Biol. 47:321-330. [DOI] [PubMed] [Google Scholar]
- 26.Kuroiwa, H., T. Mori, M. Takahara, S. Miyagishima, and T. Kuroiwa. 2002. Chloroplast division machinery as revealed by immunofluorescence and electron microscopy. Planta 215:185-190. [DOI] [PubMed] [Google Scholar]
- 27.Labrousse, A. M., M. D. Zappaterra, D. A. Rube, and A. M. van der Bliek. 1999. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell 4:815-826. [DOI] [PubMed] [Google Scholar]
- 28.Lang, B. F., M. W. Gray, and G. Burger. 1999. Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 33:351-397. [DOI] [PubMed] [Google Scholar]
- 29.Loomis, W. F. 1971. Sensitivity of Dictyostelium discoideum to nucleic acid analogues. Exp. Cell Res. 64:484-486. [DOI] [PubMed] [Google Scholar]
- 30.Löwe, J., and L. A. Amos. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203-206. [DOI] [PubMed] [Google Scholar]
- 31.Ma, X., D. Ehrhardt, and W. Margolin. 1996. Colocalization of cell division proteins in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. USA 93:12998-13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacWilliams, H., P. Gaudet, H. Deichsel, C. Bonfils, and A. Tsang. 2000. Biphasic expression of rnrB in Dictyostelium discoideum suggests a direct relationship between cell cycle control and cell differentiation. Differentiation 67:12-24. [DOI] [PubMed] [Google Scholar]
- 33.Maeda, Y. 1986. A new method for inducing synchronous growth of Dictyostelium discoideum cells using temperature shifts. J. Gen. Microbiol. 132:1189-1196. [Google Scholar]
- 34.Manstein, D., H. Schuster, P. Morandini, and D. Hunt. 1995. Cloning vectors for the production of proteins in Dictyostelium discoideum. Gene 162:129-134. [DOI] [PubMed] [Google Scholar]
- 35.Margolin, W. 2001. Spatial regulation of cytokinesis in bacteria. Curr. Opin. Microbiol. 4:647-652. [DOI] [PubMed] [Google Scholar]
- 36.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 37.Margolin, W., J. Corbo, and S. Long. 1991. Cloning and characterization of a Rhizobium meliloti homolog of the Escherichia coli cell division gene ftsZ. J. Bacteriol. 173:5822-5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Margolin, W., R. Wang, and M. Kumar. 1996. Isolation of an ftsZ homolog from the archaebacterium Halobacterium salinarium: implications for the evolution of FtsZ and tubulin. J. Bacteriol. 178:1320-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McAndrew, R., J. Froehlich, S. Vitha, K. Stokes, and K. Osteryoung. 2001. Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol. 127:1656-1666. [PMC free article] [PubMed] [Google Scholar]
- 40.Mercer, K., and D. Weiss. 2002. The Escherichia coli cell division protein FtsW is required to recruit its congate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184:904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyagishima, S., H. Kuroiwa, and T. Kuroiwa. 2001. The timing and manner of disassembly of the apparatuses for chloroplast and mitochondrial division in the red alga Cyanidioschyzon merolae. Planta 212:517-528. [DOI] [PubMed] [Google Scholar]
- 42.Miyagishima, S., K. Nishida, T. Mori, M. Matsuzaki, T. Higashiyama, H. Kuriowa, and T. Kuriowa. 2003. A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell 15:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyagishima, S., M. Takahara, and T. Kuroiwa. 2001. Novel filaments 5 nm in diameter constitute the cytosolic ring of the plastid division apparatus. Plant Cell 13:707-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyagishima, S., M. Takahara, T. Mori, H. Kuroiwa, T. Higashiyama, and T. Kuroiwa. 2001. Plastid division is driven by a complex mechanism that involves differential transition of the bacterial and eukaryotic division rings. Plant Cell 13:2257-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mori, T., H. Kuroiwa, M. Takahara, S. Miyagishima, and T. Kuroiwa. 2001. Visualization of an FtsZ ring in chloroplasts of Lilium longifolium leaves. Plant Cell Physiol. 42:555-559. [DOI] [PubMed] [Google Scholar]
- 46.Nishida, K., M. Takahara, S. Miyagishima, H. Kuroiwa, M. Matsuzaki, and T. Kuriowa. 2003. Dynamic recruitment of dynamin for final mitochondrial severence in a primitive red alga. Proc. Natl. Acad. Sci. USA 100:2146-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nogales, E., K. Dowling, L. Amos, and J. Löwe. 1998. Tubulin and FtsZ form a distinct family of GTPases. Nat. Struct. Biol. 5:451-458. [DOI] [PubMed] [Google Scholar]
- 48.Nunnari, J., W. Marshall, A. Straight, A. Murray, J. Sedat, and P. Walter. 1997. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell 8:1233-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogawa, S., R. Yoshino, K. Angata, M. Iwamoto, M. Pi, K. Kuroe, K. Matsuo, T. Morio, H. Urushihara, K. Yanagisawa, and Y. Tanaka. 2000. The mitochondrial DNA of Dictyostelium discoideum: complete sequence, gene content and genome organization. Mol. Gen. Genet. 263:514-519. [DOI] [PubMed] [Google Scholar]
- 50.Osteryoung, K., and R. McAndrew. 2001. The plastid division machine. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52:315-333. [DOI] [PubMed] [Google Scholar]
- 51.Osteryoung, K., and E. Vierling. 1995. Conserved cell and organelle division. Nature 376:473-474. [DOI] [PubMed] [Google Scholar]
- 52.Osteryoung, K. W., K. D. Stokes, S. M. Rutherford, A. L. Percival, and W. Y. Lee. 1998. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10:1991-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puta, F., and C. Zeng. 1998. Blasticidin resistance cassette in symmetrical polylinkers for insertional inactivation of genes in Dictyostelium. Folia Biol. (Prague) 44:185-188. [PubMed] [Google Scholar]
- 54.Rothfield, L., S. Justice, and J. García-Lara. 1999. Bacterial cell division. Annu. Rev. Genet. 33:423-448. [DOI] [PubMed] [Google Scholar]
- 55.Sesaki, H., and R. E. Jensen. 1999. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147:699-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaw, J., and J. Nunnari. 2002. Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 12:178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smirnova, E., L. Griparic, D. Shurland, and A. van der Bliek. 2001. Dynamin-like protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12:2245-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smirnova, E., D. L. Shurland, S. N. Ryazantsev, and A. M. van der Bliek. 1998. A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 143:351-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strepp, R., S. Scholz, S. Kruse, V. Speth, and R. Reski. 1998. Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc. Natl. Acad. Sci. USA 95:4368-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun, Q., and W. Margolin. 1998. FtsZ dynamics during the division cycle of live Escherichia coli cells. J. Bacteriol. 180:2050-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takahara, M., H. Kuriowa, S. Miyagishima, T. Mori, and T. Kuriowa. 2001. Localization of the mitochondrial FtsZ protein in a dividing mitochondrion. Cytologia 66:421-425. [Google Scholar]
- 62.Takahara, M., H. Takahashi, S. Matsunaga, S. Miyagishima, H. Takano, A. Sakai, S. Kawano, and T. Kuroiwa. 2000. A putative mitochondrial ftsZ gene is encoded in the unicellular primitive red alga Cyanidioschyzon merolae. Mol. Gen. Genet. 264:452-460. [DOI] [PubMed] [Google Scholar]
- 63.van der Bliek, A. 1999. Functional diversity in the dynamin family. Trends Cell Biol. 9:96-102. [DOI] [PubMed] [Google Scholar]
- 64.van Es, S., D. Wessels, D. Soll, J. Borleis, and P. Devreotes. 2001. Tortoise, a novel mitochondrial protein, is required for directional responses of Dictyostelium in chemotactic gradients. J. Cell Biol. 152:621-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vitha, S., R. McAndrew, and K. Osteryoung. 2001. FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 153:111-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wienke, D. C., M. L. Knetsch, E. M. Neuhaus, M. C. Reedy, and D. J. Manstein. 1999. Disruption of a dynamin homologue affects endocytosis, organelle morphology, and cytokinesis in Dictyostelium discoideum. Mol. Biol. Cell 10:225-243. [DOI] [PMC free article] [PubMed] [Google Scholar]