Abstract
Rapid postnatal growth is associated with increased risk of childhood adiposity. The aim of this study was to establish whether this pathway is mediated by altered DNA methylation and gene expression. Two distinct cohorts, one preterm (n = 121) and one term born (n = 6,990), were studied. Exploratory analyses were performed using microarrays to identify differentially expressed genes in whole blood from children defined as “slow” (n = 10) compared with “rapid” (n = 10) postnatal (term to 12 weeks corrected age) growers. Methylation within the identified TACSTD2 gene was measured in both cohorts, and rs61779296 genotype was determined by Pyrosequencing or imputation and analyzed in relation to body composition at 9–15 years of age. In cohort 1, TACSTD2 expression was inversely correlated with methylation (P = 0.016), and both measures were associated with fat mass (expression, P = 0.049; methylation, P = 0.037). Although associated with gene expression (cohort 1, P = 0.008) and methylation (cohort 1, P = 2.98 × 10−11; cohort 2, P = 3.43 × 10−15), rs61779296 was not associated with postnatal growth or fat mass in either cohort following multiple regression analysis. Hence, the lack of association between fat mass and a methylation proxy SNP suggests that reverse causation or confounding may explain the initial association between fat mass and gene regulation. Noncausal methylation patterns may still be useful predictors of later adiposity.
Both nutritional status and growth in early postnatal life have been shown to affect neurodevelopment and metabolic health into adulthood. Periods of rapid (or catch-up) growth in infancy are associated with adverse metabolic consequences in later life and are thought to lead to a subsequent increase in the risk of obesity and cardiovascular disease and the development of insulin resistance (1–3). Faster postnatal weight gain has been associated with subsequent adiposity at age 10 years in a prospective study of children mainly born at term (4,5) and a separate study of obese persons aged 5–22 years (6).
In the context of the preterm infant, rapid postnatal growth may be beneficial for neurodevelopment (7). A study of early postnatal and late infancy growth in a cohort of preterm infants showed that early weight gain from birth to 3 months, and to a lesser extent from 3 months to 1 year, was associated with a higher percentage of body fat, a greater abdominal pattern of fat distribution, and higher BMI SD scores at 19 years of age (8). Early growth and postnatal diet were also shown to have an effect on insulin sensitivity (9), indices of cardiovascular function (10), and other metabolic outcomes (11) in a preterm cohort aged 13–16 years. Evidence therefore shows that early growth in preterm children is associated with long-term influences on body composition.
The mechanisms underlying the association between early growth and later body composition are not yet clear. One potential mechanism explaining how early life exposures are memorized and persist throughout the life course is epigenetic modifications, whereby mitotically heritable changes in gene expression and chromatin organization occur independently of changes to the DNA sequence itself. The most widely examined epigenetic modification is DNA methylation where cytosine bases, primarily located next to guanine residues, are covalently bound at the carbon 5 position to a methyl group. A role for epigenetic processes in the programming of body composition has been proposed (12–14) and is supported by recent evidence from studies of DNA methylation in infants in relation to body composition in childhood. Two recent studies have reported associations between methylation in DNA extracted from cord blood (15) and umbilical cord tissue (16) and prepubertal adiposity in children.
The aim of the current study was to investigate whether growth during early postnatal life is associated with variability in gene expression and whether such variation is associated with later body composition in childhood and mediated by epigenetic mechanisms. Furthermore, in recognition of the strong potential in epigenetic epidemiological studies for both confounding (where a factor not on the causal pathway is associated with both exposure and outcome) and reverse causation (where phenotype impacts on the epigenetic marks being measured), additional strategies to strengthen causal inference were required. Hence, a “genetical epigenomics” approach (17), whereby a single nucleotide polymorphism (SNP) correlating highly with local DNA methylation levels and which cannot be subject to confounding and reverse causation was used as a proxy to interrogate causal relationships. Such approaches offer insight into causality, allowing more robust conclusions to be drawn about the mediating role of epigenetic mechanisms that are postulated to link postnatal growth with childhood adiposity.
RESEARCH DESIGN AND METHODS
A time line depicting the age from which measurements and samples were used in both cohorts is shown in Supplementary Fig. 1, available in the Supplementary Data. The analysis workflow for both cohorts and the number of samples used at each stage are shown in Supplementary Fig. 2A and B.
Cohort 1 (Newcastle Preterm Birth Growth Study).
Healthy preterm infants were recruited from the Special Care Baby Unit, Royal Victoria Infirmary (Newcastle upon Tyne, U.K.) between September 1993 and November 1999 (18,19). Anthropometric, biochemical, and nutrient markers were taken at birth, at intervals up to 18 months corrected age, and at ∼11 years of age. Whole blood (EDTA-Vacutainer and PAXgene Blood RNA tube; BD, Franklin Lakes, NJ) and saliva (Oragene·DNA; DNA Genotek, Kanata, Canada) samples were collected at age ∼11 years for DNA and RNA analysis. QIAamp DNA Blood Midi kits (Qiagen, West Sussex, U.K.) and Oragene DNA Purifier kits (DNA Genotek) were used for DNA extraction from blood and saliva samples, respectively. Total RNA was extracted using PAXgene Blood RNA System kits (Qiagen). Ethics approval for this study was obtained from the Joint Ethics Committee of Newcastle and North Tyneside.
Cohort 2 (Avon Longitudinal Study of Parents and Children).
Pregnant women from the Avon area in the southwest of England whose expected dates of delivery were between April 1991 and December 1992 were recruited. The Avon Longitudinal Study of Parents and Children (ALSPAC) is a prospective study, and the extensive data collected during pregnancy and throughout childhood have previously been described in detail (http://www.alspac.bristol.ac.uk) (20). Anthropometric data taken at ∼9 and ∼15 years of age along with DNA extracted from cord blood buffy coats and whole blood samples collected at ∼7 years of age (21) were provided for use in this study. All DNA samples were prepared using standard phenol-chloroform extraction methods. Ethics approval was obtained from the ALSPAC Law and Ethics Committee and from local research ethics committees.
Definition of postnatal growth and body composition.
Postnatal growth was defined as the difference in weight standardized score (SDS) between term and 12 weeks gestationally corrected age (i.e., 12 weeks from the expected term date) (range 10–16) in cohort 1 or between birth and 8 weeks of age (range 5–12) in cohort 2. These differing time points could not be avoided, as they were reliant on available data within these distinct cohorts. Weight SDS, adjusted for sex and age, was calculated using the British 1990 growth reference standards (22). Growth rate was then expressed and analyzed as both a continuous and dichotomized variable, in which children were split into slow and rapid growers based on the median weight change SDS across each study subgroup.
Body composition in childhood (cohort 1, ∼11 years of age; cohort 2, ∼9 and ∼15 years of age) was assessed by height (Harpenden stadiometer; Holtain, Crosswell, U.K.), weight (Tanita body fat analyzer; Tanita), BMI (kilograms per meters squared), fat mass, lean mass, and total mass (whole-body dual-energy X-ray absorptiometry) (cohort 1: software version 11, GE Lunar iDXA machine, GE Healthcare UK & Ireland; cohort 2: Prodigy scanner, Lunar Radiation, Madison, WI), using standard protocols (18–20). Data were expressed in absolute terms and as SDS (22).
Gene expression array analysis.
For cohort 1, RNA samples, extracted from whole blood from 20 individuals and selected on the basis of their rapid or slow growth and availability of RNA, were sent to ServiceXS (Leiden, the Netherlands) for gene expression analysis. Individuals used for expression analysis were matched for sex, and all were of white European ethnicity. Globin reduction was performed using a GeneChip Globin-Reduction kit (PreAnalytiX/Affymetrix). Human NuGO-Hs1a520180 GeneChip CEL files were normalized in BioConductor using the GCRMA package. Log-fold change was calculated using the RankProd package (percentage of false positive predictions [pfp] < 0.05 with 100 permutations of the class labels) to identify genes with differential expression. Annotations were attached to probe sets from the nugohs1a520180.db library (http://www.bioconductor.org/). Raw and normalized data from the experiment was deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with accession no. GSE22013. Real-time RT-PCR was performed with SuperScript III First Strand Synthesis System (Invitrogen) for validation of microarray data on the candidate gene selected for follow-up (primers in Table 1).
TABLE 1.
List of primers
In silico analysis of gene expression data.
After identification of differentially expressed loci, Gene2Promoter (Genomatix, Munich, Germany) was used to identify potential promoter regions of target genes. Promoter modules and CpG islands were identified using ModelInspector (version 5.4; Genomatix) and CpG Island Explorer, version 2.0 (23), respectively.
Methylation analysis by gene-specific Pyrosequencing.
For both cohorts, methylation of selected cytosine residues was quantified using Pyrosequencing. For cohort 1, all individuals with adequate blood-extracted DNA samples and postnatal growth data (n = 94) were included in the analysis and, where available, methylation was also quantified in DNA extracted from saliva samples (n = 68) for comparison. For cohort 2, DNA samples derived from cord blood (n = 148) and age 7 years whole blood (n = 161) with postnatal growth data were analyzed. Bisulphite conversion of 1 and 2 μg DNA was carried out using the EZ DNA Methylation kit (Zymo Research, Irvine, CA) (modification to CT conversion reagent–DNA mix incubation: 20 cycles of 95°C for 30 s; 50°C for 15 min) and the EZ DNA Methylation-Gold kit (Zymo Research) (following the manufacturer’s instructions), respectively. To determine the percentage of methylation at individual CpG sites, quantitative bisulphite Pyrosequencing (Qiagen) with Pyro Q-CpG software (version 1.0.6.) was used (primers in Table 1). Pyrosequencing PCR cycling conditions were as follows: denaturation at 95°C for 15 min and then fifty cycles of 95°C for 15 s, 50°C for 30 s, and 72°C for 15 s and extension at 72°C for 5 min followed by 4°C hold.
Quality-control assessment of methylation analysis.
Calibration curves were constructed to assess whether there was any PCR amplification bias between unmethylated and methylated sequences of TACSTD2. PCR reactions were run in duplicate. Pyrosequencing success rates in cohort 1 were 97 and 94% for DNA that was extracted from blood and saliva, respectively and, in cohort 2, 94% for DNA extracted from cord blood and 97% from year 7 blood.
Genotype analysis
Cohort 1.
SNPs located within the promoter region of the target gene were analyzed by Pyrosequencing (Qiagen). All available DNA samples were used (n = 122). Pyrosequencing PCR reactions (primers in Table 1) on genomic DNA, cycling conditions, and sequencing were performed as described above with automated genotype calling carried out using PyroMark MD (Qiagen). Genotype success rate was 98%.
Cohort 2.
Extensive genome-wide SNP data across large subgroups of the ALSPAC cohort have previously been generated and were available for analysis (http://www.alspac.bristol.ac.uk). Genotypes were imputed across ∼10,000 individuals using Illumina HumanHap550 and 1000 Genomes deep sequence data as reference sets. Imputations were carried out in Markov Chain Haplotyping software (version 1.0.16; MACH) (http://www.sph.umich.edu/csg/abecasis/MACH/). SNPs with imputation quality scores/r2 values <0.8 were rejected from further analysis. Of the 10,000 individuals included in the imputation analysis, 6,990 had data relating to postnatal growth and, hence, were included in subsequent analyses performed within this study.
Statistical analysis.
The two cohorts were analyzed using the same protocol. (See Supplementary Fig. 3 for an overview of relationships interrogated.) Associations between postnatal growth and TACSTD2 expression/methylation were assessed using nonparametric Mann-Whitney U and Spearman rank correlation tests, given the nonnormality of both exposure and outcome variables. Relationships between TACSTD2 expression/methylation and measures of body composition were initially explored using Spearman rank correlation followed by linear regression analysis to identify potential confounders. (Sex, age, height, and pubertal status at follow-up were considered.) For regression analyses, log transformations of the outcome measures were used where necessary. Associations between TACSTD2 genotypes and postnatal growth and between genotypes and expression/methylation were assessed by χ2 and Kruskal-Wallis tests, respectively. Relationships between genotypes and measures of body composition were also explored using Kruskal-Wallis tests followed by linear regression analysis as described above. Possible differences in baseline characteristics across study subgroups were tested by univariate analyses as appropriate. All statistical analyses were performed using STATA, version 10 (StataCorp, College Station, TX).
RESULTS
Cohort characteristics.
Two different cohorts were available for analysis: one of preterm children with follow-up data at a mean age of 11 years (cohort 1, n = 121) and the other composed of largely term-born children with follow-up data at mean ages of 9 and 15 years (cohort 2, n = 6,990). Gene expression and methylation analyses were performed across smaller subgroups in which adequate RNA and DNA samples were available. These subgroups were representative of the larger cohorts (Supplementary Table 1). Baseline characteristics are presented in Supplementary Table 1. Expected differences were observed in gestational age and birth weight between the two cohorts. Specifically, in cohort 1 the average gestation was 31 weeks and mean ± SD birth weight was 1.4 ± 0.3 kg compared with mean gestation 40 weeks and mean birth weight 3.5 ± 0.6 in cohort 2. The median change in weight SDS between term and ∼12 weeks corrected age in cohort 1 was 0.09 (range −1.10 to 2.66). In cohort 2, the median change in weight SDS between birth and ∼8 weeks of age was 0.06 (−3.64 to 8.25). These two postnatal growth distributions were comparable (Mann-Whitney U: z = 1.00, P = 0.316).
Postnatal growth was associated with differential gene expression in infants born preterm.
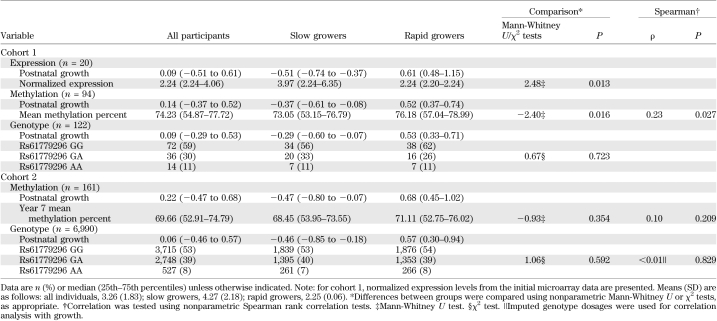
Genome-wide expression analysis of RNA samples from 10 slow and 10 rapid postnatal growers (cohort 1–expression) revealed 989 loci that were differentially expressed (data available in the Gene Expression Omnibus [http://www.ncbi.nlm.nih.gov/geo/], accession no. GSE22013). The 50 most differentially expressed loci (with a log-fold increase >1.8) were investigated using in silico approaches to define putative epigenetically regulated genes. TACSTD2 was identified as the top hit, being the most differentially expressed locus (with a 3.22 log-fold increase in slow compared with rapid growers) containing a CpG island within its promoter region. Four other loci showed greater differences in expression levels but did not contain a CpG island: DEFA4 and APOBEC3B, for which slow growers had higher expression levels compared with rapid growers, and HLADRB4 and IFI27, for which rapid growers had higher expression levels compared with slower growers. Table 2 presents associations between postnatal growth and TACSTD2 normalized expression levels. Validation of TACSTD2 expression measurement was successfully performed using RT-PCR (Spearman rank correlation between TACSTD2 array data and RT-PCR data: ρ = 0.64, P = 0.003).
TABLE 2.
Association analysis between postnatal growth and TACSTD2 genetic markers
TACSTD2 gene expression was associated with 11-year fat mass in a preterm cohort.
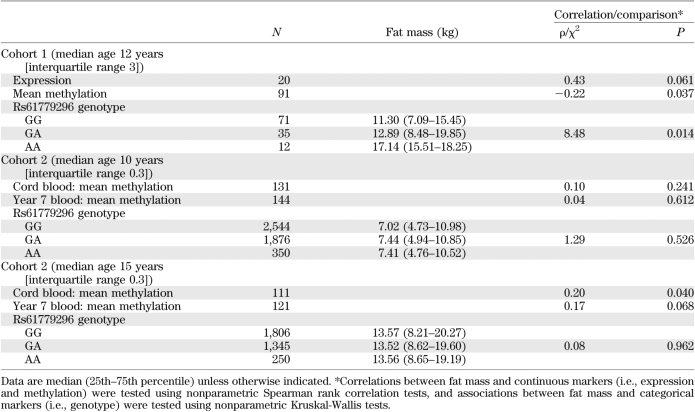
Childhood body composition measures across cohort 1 are available in Supplementary Table 2. There was weak evidence for a positive correlation between normalized TACSTD2 expression levels and fat mass (Spearman ρ = 0.44, P = 0.061) but not for other indices of body composition (Supplementary Table 2).
TACSTD2 promoter methylation patterns across childhood.
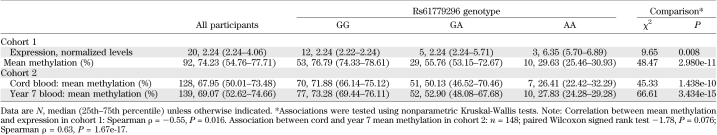
Whole blood samples, taken at ∼11 years of age, were available for DNA methylation analysis from 94 children born preterm (cohort 1–methylation). In addition, 148 cord blood and 161 whole blood samples taken at 7 years of age were available from cohort 2 (cohort 2–methylation) for validation analyses. Figure 1 illustrates the topography of the TACSTD2 gene region. Strong correlations in percentage methylation were demonstrated across the seven CpG sites investigated in three sources of DNA (average Spearman ρ: cohort 1–methylation 0.95, P = 1.05e-39; cohort 2–cord blood 0.97, P = 4.17e-71; cohort 2–age 7 blood 0.95, P = 3.24e-61) (Supplementary Table 3). Hence, all subsequent analyses were performed using the mean methylation across all seven CpG sites. Mean methylation levels derived from blood samples in cohort 1 were higher than but correlated with those estimated using DNA from available saliva samples (n = 68, Spearman ρ = 0.71, P = 1.12e-11) (Supplementary Table 4). In cohort 2, there was a strong positive correlation between mean methylation levels in cord blood and those in whole blood at age 7 years (n = 148, Spearman ρ = 0.63, P = 1.67e-17). Overall, methylation levels at 7 years of age were slightly higher (∼1.3%) than those measured in cord blood from the same children (Wilcoxon signed rank test for mean methylation −1.78, P = 0.076) (Supplementary Table 5).
FIG. 1.
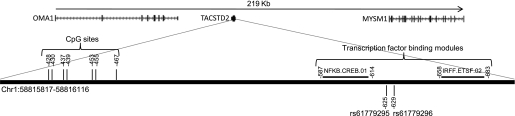
Topography of the TACSTD2 gene. DNA methylation analysis of TACSTD2 focused on seven CpG sites within the promoter region of the gene, located on chromosome (Chr)1. Positions are annotated with respect to the start codon. Two SNPs were identified at 162 and 158 base pairs from the closest CpG site analyzed (rs61779295 and rs61779296). Information was derived from UCSC Genome Browser on Human Mar. 2006 (NCBI36/hg18) Assembly.
TABLE 3.
Association analysis between childhood fat mass and TACSTD2 genetic markers
TABLE 4.
Multiple linear regression models for childhood fat mass (ln) and TACSTD2 genetic markers
TABLE 5.
Association analysis between TACSTD2 genetic markers
TACSTD2 promoter methylation was associated with postnatal growth and 11-year fat mass in a preterm cohort.
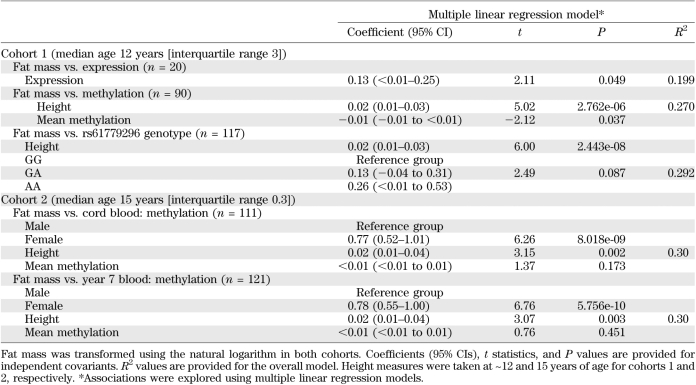
A negative correlation was demonstrated between mean methylation and TACSTD2 expression levels in cohort 1 (n = 19, Spearman ρ = −0.55, P = 0.016). In concordance with the expression data, methylation levels were associated with postnatal growth, with lower levels observed in the slow growers compared with rapid growers (Table 2). In addition, fat mass was negatively correlated with mean TACSTD2 methylation (Table 3). Multiple linear regression analyses were used to identify potential confounders. For this analysis, fat mass was log transformed. Height was identified as an independent predictor of log(fat mass). Upon adjustment, TACSTD2 methylation remained associated with fat mass (Table 4).
Associations between TACSTD2 promoter methylation, postnatal growth, and childhood fat mass were not replicated in a larger term cohort.
Childhood body composition measures across cohort 2 are presented in Supplementary Table 6. TACSTD2 methylation at 7 years of age was not associated with postnatal growth in this largely term-born cohort (Table 2). Fat mass at 15 years of age was positively correlated with DNA methylation in cord blood (Table 3). A similar pattern was also demonstrated between fat mass at 15 years and methylation measured at 7 years (Table 3). Associations were not evident between methylation in either DNA sources or fat mass at the age of 9 years (Table 3). The positive correlations between methylation and fat mass at 15 years of age observed in this term cohort were inconsistent with those observed in the preterm cohort at age 11 years. Furthermore, multiple regression analyses indicated that these initial associations were not independent of confounding factors, namely, height and sex (Table 4).
TACSTD2 promoter SNPs were associated with gene regulation.
Two SNPs (rs61779295 and rs61779296), situated 158 and 162 base pairs upstream of the first CpG site, respectively, were selected for analysis (Fig. 1). In cohort 1, these SNPs were genotyped directly in 122 children (cohort 1-genotype). Neither SNP had been genotyped previously within cohort 2. Hence, imputations using existing Illumina HumanHap550 quad chip and 1000 Genomes data were performed. The resulting quality scores and r2 values for both SNP genotypes were extremely robust at 0.994 and 0.998, respectively, enabling the imputed data to be used for subsequent investigations (cohort 2–genotype). Both SNPs conformed to Hardy-Weinberg equilibrium (P > 0.05) and were found to be in perfect linkage disequilibrium in both cohorts (D′ = 1, r2 = 1). Hence, only rs61779296 was used for subsequent analyses. Robust evidence of association between genotype and TACSTD2 gene expression and promoter methylation was demonstrated in both cohorts (Table 5).
TACSTD2 genotype was not associated with postnatal growth or fat mass in either cohort.
No evidence of association between postnatal growth and rs61779296 was observed in cohort 1–genotype or cohort 2–genotype (Table 2). In concordance with expression and methylation data, fat mass demonstrated evidence of association with genotype in cohort 1 (Table 3). However, after multiple regression analysis, rs61779296 was not predictive of log(fat mass) after adjustment for height (Table 4). In addition, there were no associations between this genetic marker and fat mass at either 9 or 15 years of age in cohort 2 (Table 3).
DISCUSSION
The current study investigated the molecular mechanisms underpinning previously reported associations linking rapid postnatal growth with increased adiposity in childhood (4–6,8). Gene expression analysis highlighted a novel, putative epigenetically regulated locus, TACSTD2, which demonstrated increased expression in preterm children who grew slowly compared with those who grew rapidly postnatally. Higher TACSTD2 expression levels were also associated with decreased promoter methylation at this locus and increased fat mass at age ∼11 years. These observations were not consistent with the expected pattern of rapid postnatal growth leading to increased adiposity in later life, although this inverse association could be a feature specific to preterm infants who have been shown to have higher resting energy expenditure and thus be protected from obesity (24).
The possible explanations for the observed, potentially causal, associations were probed in more detail. Regression analysis indicated that height explained much of the variation in fat mass and may have acted as a confounding variable in the association between methylation and fat mass, although this association did not disappear completely after adjustment. Height and sex were also shown to be confounding variables in the association between methylation at 7 years and fat mass at 15 years of age observed in the replication, albeit term, cohort. In addition, the initial univariate analyses were suggestive of a positive relationship between methylation and fat mass in cohort 2—the opposite of the inverse correlation observed in cohort 1. Although the unexpected trend in cohort 1 might be related to distinct growth trajectories of preterm infants, such inconsistencies in observed effects could also be evidence of spurious associations. Furthermore, given that the samples used in the expression analyses were also included in the methylation analysis, a false-positive effect would have been carried forward to the subsequent, albeit larger, methylation analysis. The potential for reverse causality, in which differences in childhood fat mass are responsible for changes in methylation, cannot be excluded from the findings observed in cohort 1, given that TACSTD2 methylation and childhood fat mass were measured at the same time point. Hence, the possibility remains that this finding could be spurious.
The availability of both cord and age 7 blood samples in cohort 2, along with longitudinal body composition data, enabled further exploration of the causal pathways among postnatal growth, DNA methylation, and later adiposity. Comparisons between DNA methylation in cord blood and later life may indicate whether observed differences in methylation detected in childhood result from poor postnatal growth (or other environmental factors) or preexist in the infant. However, no robust associations were observed between postnatal growth and methylation at birth or at age 7 years in this term cohort. Furthermore, analysis of TACSTD2 methylation in paired cord blood and whole blood DNA taken 7 years later showed little variation (1.3%). If growth were driving methylation changes during postnatal life, one might anticipate the change over this time period to be considerably greater.
Interest in the interplay between genetic variation and DNA methylation patterns has gained considerable momentum (25–27). A large majority of currently identified methylation-influencing SNPs occur in cis regions, usually within 5 kb of the methylation site (25,28), a feature that permits their potential application as a proxy for methylation measures. Such potential proxies within the TACSTD2 promoter region were identified. Rs61779296 correlated strongly with methylation levels in the promoter region and was therefore used as a surrogate to explore the relationship between methylation at this locus and childhood fat mass in the larger sample set. This approach, based on the principles of the now widely used Mendelian randomization method (29,30) and those similarly applied in the analysis of gene expression data (31,32), has been termed “genetical epigenomics” (17) and helps overcome the problems of reverse causation and confounding commonly encountered in (molecular) epidemiological studies.
Multiple regression analysis of rs61779296 with postnatal growth and measures of fat mass demonstrated no evidence of association in either instance, despite this SNP showing clear association with both DNA methylation and expression levels. An instrumental variable analysis using the rs61779296 SNP data was undertaken to further probe the relationship between methylation and fat mass demonstrated in cohort 1 (33). Although this analysis was underpowered, preventing robust conclusions, it suggested that confounding may be responsible for the observed association. We conclude, therefore, that the scenario depicted in Fig. 2 is the most likely; i.e., the observed associations between postnatal growth, TACSTD2 methylation and expression, and fat mass are explained largely by confounding. Much of the confounding could be explained by sex or height, although some residual confounding is likely. This correlation between fat mass and altered DNA methylation accords with other recent studies linking BMI with methylation status in adults (34,35), where the direction of causality and potential confounders remain undefined.
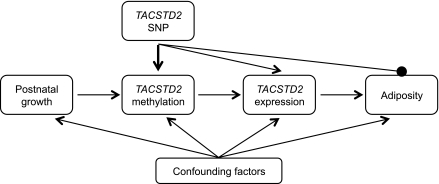
FIG. 2.
Defining the causal pathway linking postnatal growth with fat mass. The schematic depicts the postulated relationship among postnatal growth, TACSTD2 methylation, TACSTD2 gene expression, and fat mass. The figure provides a framework with which to infer the direction of causality between exposure (postnatal growth), outcome (childhood adiposity), and intermediate phenotypes (DNA methylation and gene expression). A SNP in the gene of interest can be used to infer causality. A robust correlation was observed between rs61779196 and DNA methylation (bold arrow). This permitted the use of the SNP as a surrogate for methylation levels. Genotype is not influenced by confounding factors and cannot be influenced by reverse causation (where the outcome, in this instance adiposity, influences methylation). Lack of association of the TACSTD2 SNP rs61779296 with fat mass provides evidence that DNA methylation is unlikely to have a causal influence on adiposity. Confounding is the most likely explanation for the observed association between fat mass and DNA methylation (and expression) in the TACSTD2 gene.
Despite our conclusion that TACSTD2 methylation is unlikely to provide a causal link between rapid postnatal growth and altered adiposity, this locus is clearly different in a subset of children with increased fat mass and may have particular relevance in a preterm setting. The TACSTD2 gene encodes a cell-surface glycoprotein that is implicated in cell proliferation (36). Increased expression of TACSTD2 correlates with poor prognosis in several cancers including those of the oral cavity (37), prostate (38), and ovary (39) and liver metastases of colorectal cancer (40). Our observed association of increased expression with higher fat mass, while novel, is consistent with such adipocyte hyperplasia and hypertrophy, mechanisms that have also been implicated in differential fat accumulation in adults (41).
The study findings focused on a single locus—an approach that has inherent limitations. Epigenetic control of development is likely to occur at a network/pathway level, so the analysis of an isolated gene is unlikely to give a full picture (42). However, focusing on one locus enabled a thorough assessment of the potential causal pathway. Specifically, this study combined analyses of the exposure of postnatal growth, methylation, expression, and genotype of TACSTD2 and body composition from the same individuals, providing a high level of insight into the molecular pathways linking postnatal growth to later adiposity. The same approach could be applied to numerous other loci, and it is our intention to follow up in the same way as for other differentially expressed genes identified from the exploratory analyses. Although RNA and DNA were extracted from peripheral blood and not adipose tissue, the gene expression profiles, in terms of pathways perturbed, have been shown to have considerable overlap (43). Replication in DNA and RNA extracted from adipose tissue would clearly be of interest; however, genotype will remain unchanged and the lack of association between genotype and fat mass would persist. A major strength of the current study is the use of a genetic proxy to probe causal relationships, and this genetical epigenomic approach may be valuable in future studies of this type.
The discovery cohort analyzed consisted of individuals born preterm, whereas the replication cohort was largely composed of term-born individuals. It is recognized that growth trajectories in these two groups differ markedly and that interpretation of conflated results could be misleading. Slight differences in the definition of postnatal growth between the cohorts could not be avoided, as these were reliant on available data points (cohort 1, term to term plus 12 weeks, and cohort 2, birth to 8 weeks). However, the distributions of postnatal growth did not differ statistically between the two cohorts and are therefore unlikely to have influenced the reported associations or conclusions drawn regarding methylation and fat mass.
In summary, postnatal growth and childhood fat mass were associated with altered TACSTD2 gene expression and promoter methylation. However, the lack of association between fat mass and a methylation proxy SNP in this gene suggests that confounding might explain the observed associations. However, the identification of a correlation between DNA methylation and childhood fat mass may have important predictive utility regardless of causality. The use of genetic proxies in epigenetic studies is advocated to strengthen causal inference.
ACKNOWLEDGMENTS
The study was supported by the Biotechnology and Biological Sciences Research Council (BB/F007981/1). Support for the follow-up studies was provided by the Special Trustees of Newcastle Hospitals. C.P. receives salary support from the National Institute for Health Research. V.T. was supported by Fonds québécois de la recherche sur la nature et les technologies and the Institute of Nutraceuticals and Functional Foods at Laval University (Québec, Canada) during her visiting tenure in the Relton laboratory, when she contributed to this work. The U.K. Medical Research Council (grant 74882), the Wellcome Trust (grant 076467), and the University of Bristol provide core support for ALSPAC. Support for the follow-up studies was also provided by Novo Nordisk. The Newcastle Preterm Birth Growth Study (NPBGS) was originally funded by Nutricia Advanced Medical Nutrition. No other potential conflicts of interest relevant to this study were reported.
A.G. undertook all laboratory analyses, assisted C.P. in the analysis and interpretation of data, and drafted the manuscript. C.P. analyzed and interpreted data and drafted the manuscript. D.C.S. provided bioinformatic support and contributed to drafting the manuscript. G.F. and D.M.E. imputed genotype information in the ALSPAC cohort and analyzed those data and contributed to drafting the manuscript. S.M.R. provided relevant data and biological samples from the ALSPAC study (cohort 2), and contributed to drafting the manuscript. V.T. identified the TACSTD2 SNPs, designed the assays for their analyses, and contributed to drafting the manuscript. M.S.P. provided statistical support to N.D.E. in their follow-up of the NPBGS (cohort 1). N.D.E. undertook the follow-up of the NPBGS (cohort 1), contributed to the study design and oversight of the experimental work, and contributed to drafting the manuscript. G.D.S. provided relevant data and biological samples from the ALSPAC study (cohort 2) and contributed to drafting the manuscript. J.C.M. contributed to the study design and oversight of the experimental work and contributed to drafting the manuscript. C.L.R. drafted the manuscript, conceptualized and designed the study, assisted C.P. with the analysis and interpretation of data, and is the guarantor for the contents of the article.
Parts of this study were presented in abstract form at the Developmental Origins of Health and Disease 7th World Congress, Portland, Oregon, 18–21 September 2011.
The authors thank all of the study members for their previous and continued involvement in the Newcastle Preterm Birth Growth Studies. They are extremely grateful to all of the families who took part in the ALSPAC study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. Dr. Srinivasa Korada, of Royal Victoria Infirmary, Newcastle upon Tyne, is acknowledged for recruitment and clinics of the Newcastle Preterm Birth Growth Study follow-up. Profs. Andy Ness and Jon Tobias, both of University of Bristol, are acknowledged for their contribution to the measurement of body composition in the ALSPAC cohort, and Dr. Kate Northstone of University of Bristol is acknowledged for her invaluable assistance in providing data from this resource. John Kemp of University of Bristol is acknowledged for help with the ALSPAC genotype imputation. Dr. Tom Palmer of University of Bristol is acknowledged for his guidance on performing and interpreting instrumental variable analyses. Prof. Heather Cordell of Newcastle University is acknowledged for her helpful contribution during the early study design phase. The authors also thank Cathy Elks of the Medical Research Council Epidemiology Unit Cambridge for her helpful discussions on deriving standardized scores of postnatal growth.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-1039/-/DC1.
The funders had no role in study design, data collection, analysis, the decision to publish, or the preparation of the manuscript.
REFERENCES
- 1.Hales CN, Ozanne SE. The dangerous road of catch-up growth. J Physiol 2003;547:5–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanigan J, Singhal A. Early nutrition and long-term health: a practical approach. Proc Nutr Soc 2009;68:422–429 [DOI] [PubMed] [Google Scholar]
- 3.Ong KK. Catch-up growth in small for gestational age babies: good or bad? Curr Opin Endocrinol Diabetes Obes 2007;14:30–34 [DOI] [PubMed] [Google Scholar]
- 4.Ong KK, Emmett P, Northstone K, et al. Infancy weight gain predicts childhood body fat and age at menarche in girls. J Clin Endocrinol Metab 2009;94:1527–1532 [DOI] [PubMed] [Google Scholar]
- 5.Ong KK, Loos RJ. Rapid infancy weight gain and subsequent obesity: systematic reviews and hopeful suggestions. Acta Paediatr 2006;95:904–908 [DOI] [PubMed] [Google Scholar]
- 6.Wells JC, Haroun D, Levene D, Darch T, Williams JE, Fewtrell MS. Prenatal and postnatal programming of body composition in obese children and adolescents: evidence from anthropometry, DXA and the 4-component model. Int J Obes (Lond) 2011;35:534–540 [DOI] [PubMed] [Google Scholar]
- 7.Wiedmeier JE, Joss-Moore LA, Lane RH, Neu J. Early postnatal nutrition and programming of the preterm neonate. Nutr Rev 2011;69:76–82 [DOI] [PubMed] [Google Scholar]
- 8.Euser AM, Finken MJ, Keijzer-Veen MG, Hille ET, Wit JM, Dekker FW; Dutch POPS-19 Collaborative Study Group Associations between prenatal and infancy weight gain and BMI, fat mass, and fat distribution in young adulthood: a prospective cohort study in males and females born very preterm. Am J Clin Nutr 2005;81:480–487 [DOI] [PubMed] [Google Scholar]
- 9.Singhal A, Fewtrell M, Cole TJ, Lucas A. Low nutrient intake and early growth for later insulin resistance in adolescents born preterm. Lancet 2003;361:1089–1097 [DOI] [PubMed] [Google Scholar]
- 10.Singhal A, Cole TJ, Fewtrell M, Deanfield J, Lucas A. Is slower early growth beneficial for long-term cardiovascular health? Circulation 2004;109:1108–1113 [DOI] [PubMed] [Google Scholar]
- 11.Singhal A, Farooqi IS, O’Rahilly S, Cole TJ, Fewtrell M, Lucas A. Early nutrition and leptin concentrations in later life. Am J Clin Nutr 2002;75:993–999 [DOI] [PubMed] [Google Scholar]
- 12.Simmons R. Epigenetics and maternal nutrition: nature v. nurture. Proc Nutr Soc 2011;70:73–81 [DOI] [PubMed] [Google Scholar]
- 13.Mathers JC, McKay JA. Epigenetics - potential contribution to fetal programming. Adv Exp Med Biol 2009;646:119–123 [DOI] [PubMed] [Google Scholar]
- 14.Mathers JC. Nutrition, epigenomics and the development of obesity: how the genome learns from experience. In Obesity Prevention: the Role of Brain and Society on Individual Behavior. 1st ed Dube LB, Dagher A, Drewnowski A, Lebel A, James J, Yada PRY, Eds. Amsterdam, the Netherlands, Elsevier/Academic Press; 2010, p. 191–199 [Google Scholar]
- 15.Relton CL, Groom A, Elliott HR, et al. Epigenetic epidemiology: evidence for the role of epigenetic variation in complex disease. Ann Nutr Metab 2009;55:190 [Google Scholar]
- 16.Godfrey KM, Sheppard A, Gluckman PD, et al. Epigenetic gene promoter methylation at birth is associated with child’s later adiposity. Diabetes 2011;60:1528–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Relton CL, Davey Smith G. Epigenetic epidemiology of common complex disease: prospects for prediction, prevention, and treatment. PLoS Med 2010;7:e1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooke RJ, Griffin IJ, McCormick K, et al. Feeding preterm infants after hospital discharge: effect of dietary manipulation on nutrient intake and growth. Pediatr Res 1998;43:355–360 [DOI] [PubMed] [Google Scholar]
- 19.Embleton ND, Cooke RJ. Protein requirements in preterm infants: effect of different levels of protein intake on growth and body composition. Pediatr Res 2005;58:855–860 [DOI] [PubMed] [Google Scholar]
- 20.Golding J, Pembrey M, Jones R; ALSPAC Study Team ALSPAC—the Avon Longitudinal Study of Parents and Children. I. Study methodology. Paediatr Perinat Epidemiol 2001;15:74–87 [DOI] [PubMed] [Google Scholar]
- 21.Jones RW, Ring S, Tyfield L, et al. ; ALSPAC Study Team A new human genetic resource: a DNA bank established as part of the Avon longitudinal study of pregnancy and childhood (ALSPAC). Eur J Hum Genet 2000;8:653–660 [DOI] [PubMed] [Google Scholar]
- 22.Cole TJ, Freeman JV, Preece MA. British 1990 growth reference centiles for weight, height, body mass index and head circumference fitted by maximum penalized likelihood. Stat Med 1998;17:407–429 [PubMed] [Google Scholar]
- 23.Wang Y, Leung FC. An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics 2004;20:1170–1177 [DOI] [PubMed] [Google Scholar]
- 24.Sipola-Leppänen M, Hovi P, Andersson S, et al. Resting energy expenditure in young adults born preterm—the Helsinki study of very low birth weight adults. PLoS ONE 2011;6:e17700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell JT, Pai AA, Pickrell JK, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol 2011;12:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hellman A, Chess A. Extensive sequence-influenced DNA methylation polymorphism in the human genome. Epigenetics Chromatin 2010;3:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shoemaker R, Deng J, Wang W, Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res 2010;20:883–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang D, Cheng L, Badner JA, et al. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet 2010;86:411–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davey Smith G. Use of genetic markers and gene-diet interactions for interrogating population-level causal influences of diet on health. Genes Nutr 2011;6:27–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davey Smith G, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 2003;32:1–22 [DOI] [PubMed] [Google Scholar]
- 31.Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet 2001;17:388–391 [DOI] [PubMed] [Google Scholar]
- 32.Schadt EE, Lamb J, Yang X, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet 2005;37:710–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawlor DA, Harbord RM, Tybjaerg-Hansen A, et al. Using genetic loci to understand the relationship between adiposity and psychological distress: a Mendelian Randomization study in the Copenhagen General Population Study of 53,221 adults. J Intern Med 2011;269:525–537 [DOI] [PubMed] [Google Scholar]
- 34.Feinberg AP, Irizarry RA, Fradin D, et al. Personalized epigenomic signatures that are stable over time and covary with body mass index. Sci Transl Med 2010;2:49ra67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Movérare-Skrtic S, Mellström D, Vandenput L, Ehrich M, Ohlsson C. Peripheral blood leukocyte distribution and body mass index are associated with the methylation pattern of the androgen receptor promoter. Endocrine 2009;35:204–210 [DOI] [PubMed] [Google Scholar]
- 36.Cubas R, Zhang S, Li M, Chen C, Yao Q. Trop2 expression contributes to tumor pathogenesis by activating the ERK MAPK pathway. Mol Cancer 2010;9:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fong D, Spizzo G, Gostner JM, et al. TROP2: a novel prognostic marker in squamous cell carcinoma of the oral cavity. Mod Pathol 2008;21:186–191 [DOI] [PubMed] [Google Scholar]
- 38.Ibragimova I, Ibáñez de Cáceres I, Hoffman AM, et al. Global reactivation of epigenetically silenced genes in prostate cancer. Cancer Prev Res (Phila) 2010;3:1084–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bignotti E, Todeschini P, Calza S, et al. Trop-2 overexpression as an independent marker for poor overall survival in ovarian carcinoma patients. Eur J Cancer 2010;46:944–953 [DOI] [PubMed] [Google Scholar]
- 40.Fang YJ, Lu ZH, Wang GQ, et al. Elevated expressions of MMP7, TROP2, and survivin are associated with survival, disease recurrence, and liver metastasis of colon cancer. Int J Colorectal Dis 2009;24:875–884 [DOI] [PubMed] [Google Scholar]
- 41.Tchoukalova YD, Votruba SB, Tchkonia T, Giorgadze N, Kirkland JL, Jensen MD. Regional differences in cellular mechanisms of adipose tissue gain with overfeeding. Proc Natl Acad Sci USA 2010;107:18226–18231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jablonka E, Raz G. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q Rev Biol 2009;84:131–176 [DOI] [PubMed] [Google Scholar]
- 43.Pietiläinen KH, Naukkarinen J, Rissanen A, et al. Global transcript profiles of fat in monozygotic twins discordant for BMI: pathways behind acquired obesity. PLoS Med 2008;5:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]