Abstract
Hypertension is an important determinant of cardiovascular morbidity and mortality and has a substantial heritability, which is likely of polygenic origin. The aim of this study was to assess to what extent multiple common genetic variants contribute to blood pressure regulation in both adults and children, and to assess overlap in variants between different age groups, using genome wide profiling. SNP sets were defined based on a meta-analysis of genome-wide association studies on systolic (SBP) and diastolic blood pressure (DBP) performed by the Cohort for Heart and Aging Research in Genome Epidemiology (CHARGE, n=29,136), using different P-value thresholds for selecting single nucleotide polymorphisms (SNPs). Subsequently, genetic risk scores for SBP and DBP were calculated in an independent adult population (n=2,072) and a child population (n=1,034). The explained variance of the genetic risk scores was evaluated using linear regression models, including sex, age and body mass index. Genetic risk scores, including also many non-genome-wide significant SNPs explained more of the variance than scores based only on very significant SNPs in adults and children. Genetic risk scores significantly explained up to 1.2% (P=9.6*10−8) of the variance in adult SBP and 0.8% (P=0.004) in children. For DBP, the variance explained was similar in adults and children (1.7% (P=8.9*10−10) and 1.4% (P=3.3*10−5) respectively). These findings suggest the presence of many genetic loci with small effects on blood pressure regulation both in adults and children, indicating also a (partly) common polygenic regulation of blood pressure throughout different periods of life.
Keywords: genome-wide association, genome-wide profiling, genetic risk scores, blood pressure, hypertension
INTRODUCTION
Elevated blood pressure is an important risk factor for stroke and ischemic heart disease, and is estimated to contribute to half of the global risk for cardiovascular disease 1–2. Anti-hypertensive treatment has been effective approach in reducing risks of cardiovascular events in people with hypertension 3.
The heritability of blood pressure levels is estimated to be 30–60% 4–5. Several genes with large effects have been identified in familial forms of hypertension, including salt-sensitivity genes 6. However, these explain a relatively small proportion of hypertension in the general population. Several common genetic variants associated with blood pressure have been identified through genome-wide association studies in adult populations, only explaining ~1% of the variance of blood pressure 7–8.
Despite the large size of the consortia used for gene discovery, many common variants with small effects on blood pressure remain unidentified. While their individual associations do not reach genome-wide significance, in combination these variants may nevertheless explain a substantial proportion of blood pressure. The extent to which unidentified common variants explain the missing heritability in blood pressure and other polygenic traits is an open and important question.
Recently, it has been shown that the presence of multiple variants affecting polygenic traits can be demonstrated by constructing genome wide prediction models (genetic risk scores) of common variants 9–10. In a polygenic disease model, the more markers are used in the model, the better the disease is predicted. Such a model also implies that everybody in the population carries a substantial number of risk variants with small effects on the disease, but patients carry more of these variants than non-diseased people. This has been demonstrated in a recent study of schizophrenia, showing that one can predict disease using both genome-wide significant and non-significant SNPs, covering a large part of the genome 9.
This approach can also be used to evaluate the evidence of overlap in genes affecting a continuous outcome as blood pressure, in different age groups. In metabolic diseases, such as diabetes and blood pressure, there is increasing interest in the role of genetic determinants of blood pressure and other risk factors of cardiovascular disease in order to improve prevention of chronic disease and identify targets for therapeutic interventions.
One may argue that many genes regulating blood pressure maintenance are similar across age groups. We used genome wide profiling to evaluate to what extent multiple common genetic variants influence blood pressure in adults and secondly and to test whether there is overlap in genes contributing to blood pressure levels in children and adults, which might indicate whether there is a common polygenic regulation of blood pressure throughout different periods of life.
METHODS
Genome-wide profiling
In genome-wide profiling, for a certain trait, genetic risk scores are constructed using data from a “discovery sample”. Sets of common variants to calculate genetic risk scores consist of all SNPs achieving a certain significance threshold (Pdiscovery threshold) in the discovery sample. In an independent “target sample”, a subject’s genetic risk score is computed across all SNPs with P-value lower than the Pdiscovery threshold. The subject’s genotype (coded 0/1/2) is multiplied by the regression coefficient for that SNP as estimated in the discovery sample, and divided by the total number of SNPs in that set. This risk score is calculated for all subjects in the independent target sample. Subsequently, an unbiased estimate of the variance explained by the genetic risk score is obtained by evaluating the increase in explained variance of the trait when adding the genetic risk score to a baseline model explaining that trait. The method has previously been described in detail 9.
In our study, we used systolic and diastolic blood pressure and hypertension as traits of interest. The discovery sample was the Cohort for Heart and Aging Research in Genome Epidemiology (CHARGE) consortium, with a total sample size of 29,136 participants 7. We used two Dutch target samples; one adult sample (Rotterdam Study III) and one child sample (Generation R Study).
In the discovery sample, within each cohort of CHARGE, regression models were fitted for systolic blood pressure (SBP) and diastolic blood pressure (DBP) separately, and allele dosage, using an additive genetic model. The models were adjusted for sex, age, age squared and body mass index (BMI). Subsequently, the within-study associations were combined by prospective meta-analysis. The methods have been described in detail previously 7.
Next, SNPs were selected using the results from the meta-analyses of GWAS on SBP in the discovery sample, on the basis of their nominal P-value (Pdiscovery) for association with SBP, using different Pdiscovery thresholds, ranging from 1.0×10−7 to 1.0. Subsequently, these sets of SNPs, with different Pdiscovery thresholds, were used to calculate the genetic scores in the target samples. The same was done using the results from the meta-analysis of GWAS on DBP, creating separate genetic risk scores for DBP.
For each individual in the two independent target samples, the genetic risk scores were calculated by multiplying the number of risk alleles per SNP (0, 1 or 2) with the effect size of that SNP in the discovery meta-analysis (weighted approach), summed over all SNPs in the set and divided by the number of SNPs in the considered set. The calculations of individual scores for each set of SNPs were performed using the PLINK (v1.07) software, specifically by “profile scoring” option.
Subsequently, linear regression models were used to test the association between the individual genetic risk scores and SBP and DBP in the target samples. For subjects using anti-hypertensive medication, we added 10 mmHg to the observed SBP values and 5 mmHg to the observed DBP values. Similarly to the discovery analysis the models were adjusted for sex, age BMI. For analyzing the explained variance for adult hypertension (SBP>=140 mmHg, DBP >=90 mmHg or use of antihypertensive medication) and childhood hypertension (SBP or DBP > p95 for age, gender and height 11), we used logistic regression. We performed sensitivity analyses in the adults excluding subjects meeting the criteria for hypertension in the Rotterdam Study III. We have repeated the main analyses after removing SNPs in high linkage disequilibrium, using the LD-pruning option in the Plink software12. We pruned the data considering a window of 200 SNPs, removing one of a pair of SNPs if the LD is greater than 0.25, and shifting the window 5 SNPs forward to repeat the procedure.
For both the Rotterdam Study and the Generation R study, regression analyses were performed in SPSS 17.0 for Windows (SPSS Inc., Chicago IL, USA). The difference of the explained variance in the null (without genetic risk score) and alternative model (including a genetic risk score) was considered as the variance explained by the genetic score. A genetic risk score with a P-value <0.05 in the model was considered as significantly associated with the trait. We also assessed the difference between two subsequent models both including a genetic risk score using the Akaike Information Criterion (AIC) 13.
In the online supplement, we describe the discovery and target samples, their respective data collection procedures and quality control. The approaches for sensitivity analyses using unrelated traits as outcome and analysis using randomly selected SNP sets or a SNP set based on a biological pathway to calculate a genetic risk score are also presented in the online supplement (please see http://hyper.ahajournals.org).
RESULTS
Demographic data of the CHARGE consortium have been published previously 7. Table 1 shows the baseline characteristics for the Rotterdam Study III (RS-III) and the Generation R Study. The median age in RS-III was 56.0 years (95% range: 47.7 – 62.3), in the Generation R Study the median age was 6.0 years (95% range: 5.8 – 6.7).
Table 1.
Subject characteristics of the Generation R Study and the Rotterdam Study III
| Characteristics | Rotterdam Study III (n = 2,078) | Generation R Study (n = 1,034) |
|---|---|---|
| Age (yrs) | 56.0 (47.7 – 62.3) | 6.0 (5.8 – 6.7) |
| Female (%) | 56.1 | 52.2 |
| BMI (weight(kg)/length(cm)2) | 26.9 (21.6 – 36.7) | 15.9 (13.9 – 18.4) |
| Mean systolic blood pressure (mmHg) | 132.7 (19.2) | 102.5 (8.1) |
| Mean diastolic blood pressure – (mmHg) | 82.6 (11.1) | 60.5 (7.2) |
| Subjects with hypertension (%)* | 47.3 | |
| Subjects using anti-hypertensive medication (%) | 20.8 | |
| Children referred to nephrology for hypertension (%)† | 0.5 | |
| Prevalent Type 2 Diabetes (%) | 7.3 | |
| Serum total cholesterol (mmol/l) | 5.6 (1.1) | |
| HDL cholesterol (mmol/l) | 1.5 (0.5) |
Values are means (sd) or medians (95%-range)
Adults: Defined as SBP>140mmHg, DBP>90 mmHg or use of anti-hypertensive medication
Children: SBP or DBP >95th percentile for gender, age and height 12
Children were referred to pediatric nephrology department when the lowest blood pressure measurement was higher than the P99 for their gender, length and age 12.
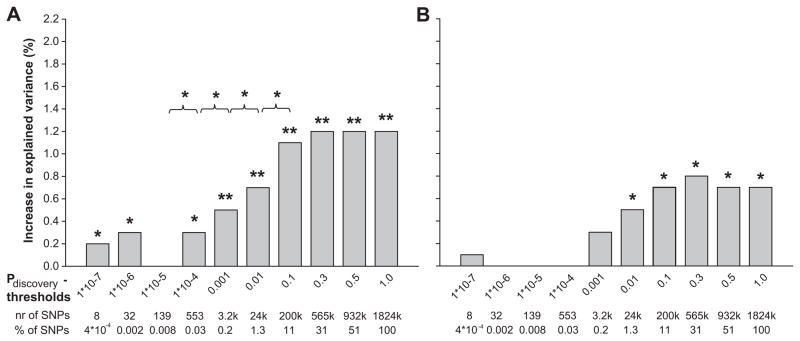
Figures 1a and 1b show the increase in explained variance of SBP by the genetic risk scores for a range of different Pdiscovery thresholds, in the adult and child target samples, respectively. When considering risk scores based on sets of SNPs with low P-values in the discovery sample (up to Pdiscovery <1.0×10−4), the risk scores significantly explained up to 0.3% of variance in SBP in adults (P=0.01). These scores were non-significant in children, increasing the explained variance by 0.1% (P=0.305). When adding more SNPs using greater Pdiscovery thresholds, the variance explained by the genetic risk scores increased, in both adults and children. Adding genetic risk scores based on SNPs with a Pdiscovery of <0.1, increased the explained variance of the model significantly with 1.2% in adults (P=9.6*10−8). In 6 year old children, the risk score based on SNPs with Pdiscovery of <0.3 significantly increased the explained variance of the model by 0.8% (P=0.004). The increase in explained variance in children was less pronounced compared to adults, but showed a similar pattern as the adult population.
Figure 1. Increase in explained variance in systolic blood pressure by genetic risk scores.
Bars represent the increase in explained variance (%) of systolic blood pressure, when adding the genetic risk scores for different p-value thresholds, to the base line model for systolic blood pressure including BMI, sex and age as covariates. The baseline model explained 12.8% and 5.2 % of the variance in systolic blood pressure for adults and children respectively. We evaluated the difference in explained variance between two subsequent models by calculating the Akaike Information Criterion (AIC) of each model. The difference in AIC follows a χ2 distribution with one degree of freedom, from which the P-value was derived.
* p < 0.05
** p < 0.001
1a. Rotterdam Study III
1b. Generation R Study
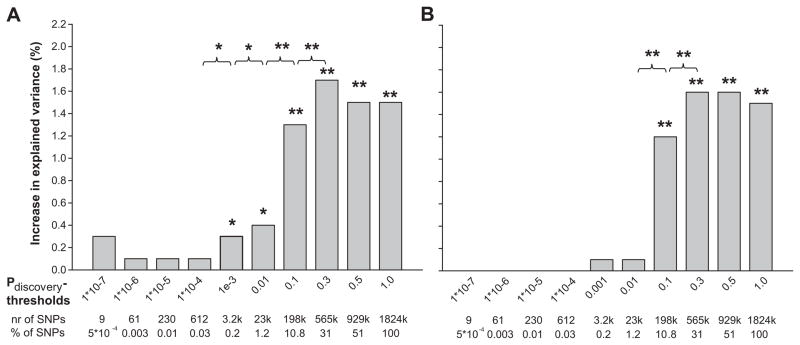
Figures 2a and 2b show the proportion of explained variance of DBP for the various genetic risk scores. The difference between adults and children considering genetic risk scores based on sets of SNPs with a low P-value in the discovery sample (up to Pdiscovery <1.0×10−4), was more marked. The SNP sets explained up to 0.3% (P = 0.006) of the variance of DBP in the adult population but no variance in DBP was explained in children (P = 0.544). The genetic risk score based on SNPs with a Pdiscovery <0.3 increased the explained variance significantly by 1.7% (P=8.9*10−10) when the score was added to the baseline model. Again, in children, a similar pattern was observed. The same genetic risk score (Pdiscovery threshold <0.3) significantly increased the explained variance of DBP by 1.4% (P=5.2*10−5), almost a similar increase as in adults.
Figure 2. Increase in explained variance in diastolic blood pressure by genetic risk scores.
Bars represent the increase in explained variance (%) of diastolic blood pressure, when adding the genetic risk scores for different p-value thresholds, to the base line model for diastolic blood pressure including BMI, sex and age as covariates. The baseline model explained 8.4% and 1.4 % of the variance in diastolic blood pressure for adults and children respectively. We evaluated the difference in explained variance between two subsequent models by calculating the Akaike Information Criterion (AIC) of each model. The difference in AIC follows a χ2 distribution with one degree of freedom, from which the P-value was derived.
* p < 0.05
** p < 0.001
2a. Rotterdam Study III
2b. Generation R Study
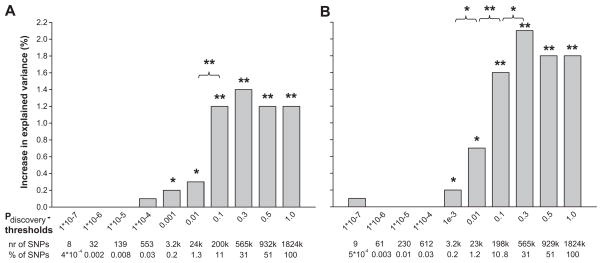
Genetic risk scores for systolic and diastolic blood pressure also were associated with hypertension in adults explaining up to 2.1% (P=2.0*10−9, using Pdiscovery <0.3) of the variance in hypertension in an adult population (Figure 3a and 3b). Adult based genetic risk scores for systolic and diastolic blood pressure did not significantly explain additional variance in childhood hypertension (see Supplement figure S1a and S1b, please see http://hyper.ahajournals.org).
Figure 3. Increase in explained variance in hypertension by genetic risk scores.
Bars represent the increase in explained variance (%) of hypertension in adults (SBP>140, DBP>90 or the use of anti hypertensive medication), when adding systolic and diastolic genetic risk scores for different p-value thresholds, to the base line model for systolic blood pressure including BMI, sex and age as covariates. The baseline model explained 15.9% of the variance in hypertension in adults. We evaluated the difference in explained variance between two subsequent models by calculating the Akaike Information Criterion (AIC) of each model. The difference in AIC follows a χ2 distribution with one degree of freedom, from which the P-value was derived.
* p < 0.05
** p < 0.001
3a. Systolic blood pressure scores
3b. Diastolic blood pressure scores
To illustrate that these results were not due to chance, we tested whether the blood pressure based risk scores predicted also variation in intracranial volume (in adults) and head circumference (in children). Figures S2a/b and S3a/b show that the genetic risk scores for SBP and DBP did not explain variance of these unrelated traits significantly (please see http://hyper.ahajournals.org). Furthermore, we created random genetic risk scores of sufficient size (~565k SNPs, similar to the SNP set with Pdiscovery < 0.3) and evaluated the additional explained variance. The random genetic risk scores showed a significant increase in explained variance when added to the baseline model (see Table 2). The additional explained variance from the random genetic risk scores was 0.1–0.4% lower as compared to the original genetic risk scores (see Table 2). We also tested whether a set of SNPs from a biological pathway would lead to a higher increase in explained variance than SNP sets with a low P-discovery threshold. We used SNPs in the FGF (fibroblast growth factor) pathway as described by Tomaszewski et al 14. This genetic risk score did not explain additional variance in all phenotypes except for adult hypertension based on the systolic blood pressure score (increase in explained variance 0.3, P=0.014, see Table 3).
Table 2.
Additional explained variance by a random genetic risk score versus original genetic risk score
| Additional explained variance
| |||
|---|---|---|---|
| Phenotype | Random genetic risk score (%) | Original genetic risk score (%) | P-value for difference |
| Adult systolic blood pressure (Rotterdam Study III) | 1.0 † | 1.2 † | 0.07 |
| Adult diastolic blood pressure (Rotterdam Study III) | 1.3 † | 1.7 † | 0.01 |
| Child systolic blood pressure (Generation R) | 0.7 * | 0.8 * | 0.49 |
| Child diastolic blood pressure (Generation R) | 1.4 † | 1.6 † | 0.10 |
| Hypertension – SBP scores (Rotterdam Study III) | 1.1 † | 1.4 † | 0.02 |
| Hypertension – DBP scores (Rotterdam Study III) | 1.7 † | 2.1 † | 0.01 |
P value < 0.05
P value < 0.001
Random Genetic risk score – Genetic risk score calculated on SNP set containing ~565k SNPs which were randomly selected out of the discovery meta-analysis, irrespective of their P-value for association.
Original genetic risk score – Genetic risk score calculated on a SNP set with a Pdiscovery threshold of 0.3. This SNP set contains ~565k SNPs and showed the largest increase in explained variance.
The P-value for difference between the models was obtained by calculating the difference in Akaike Information Criterion between the random model and the original model. This difference follows a χ2 distribution with 1 degree of freedom
Table 3.
Additional explained variance by a genetic risk score based on FGF-signaling pathway 14.
| Phenotype | Additional explained variance by signaling pathway genetic risk score (%) |
|---|---|
| Adult systolic blood pressure (Rotterdam Study III) | 0.1 |
| Adult diastolic blood pressure (Rotterdam Study III) | 0.0 |
| Child systolic blood pressure (Generation R) | 0.1 |
| Child diastolic blood pressure (Generation R) | 0.1 |
| Hypertension – SBP scores (Rotterdam Study III) | 0.3 * |
| Hypertension – DBP scores (Rotterdam Study III) | 0.1 |
P value < 0.05
In a strictly normotensive population the additional explained variance of the genetic risk score including the most significant SNPs (Pdiscovery-threshold 1.0×10 7) increased, while the risk scores with more liberal Pdiscovery-thresholds had a lower additive explained variance compared to the original analysis. Results of sensitivity analyses are shown in the supplement (figure S4a and S4b). We repeated the main analyses after removing SNPs in high linkage disequilibrium. The pattern was similar to the original analysis, but the explained variance was lower. Including more SNPs in the genetic risk score increased the variance explained by that score. The results of these analyses are shown in the supplement (Figure S5a/b, S6a/b and S7a/b, please see http://hyper.ahajournals.org).
DISCUSSION
Our findings indicate that, in addition to the blood pressure variants now identified, large numbers of common genetic variants affecting blood pressure remain to be identified, and that these variants explain a significant part of the variance in blood pressure in adults and children. These non-significant, unidentified SNPs together explain a larger part of the variance than the genome wide significant SNPs only. We also showed that adult based genetic risk scores explained variance in blood pressure in children. This indicates not only that there is a polygenic effect on blood pressure in children, but more importantly, it indicates that there is overlap in variants involved with blood pressure maintenance in adults and children and that these variants act in throughout life.
In this study, we did not remove any SNPs that are in high linkage disequilibrium with each other. Pruned analyses, as presented in the Supplemental Material, do not change our conclusion that adding more non-genome wide significant SNPs to genetic risk scores increases the variance explained by these scores. However, as expected, removing SNPs by LD pruning results in a reduction of the variance explained. Since SNPs in LD are removed randomly, in many cases informative SNPs are taken out of the analyses. Therefore a pruned analysis is expected to underestimate the true effect on the explained variance by the genetic risk scores.
The additive explained variance of genetic risk scores on blood pressure is maximizing around the Pdiscovery-threshold of 0.3 and does not increase with more liberal thresholds. This genetic risk score includes over 550k SNPs. This result suggests that SNPs with a Pdiscovery-value lower than 0.3 in the discovery sample add to the explained variance of blood pressure and that many common variants associated with blood pressure regulation have not been identified yet. SNPs with a Pdiscovery higher than 0.3 are unlikely to be associated with blood pressure. Genetic risk scores of a similar size, consisting of randomly selected SNPs, still resulted in a significant increase in explained variance when added to the baseline model without a genetic risk score. The increase in explained variance based on the random genetic risk scores was lower than the increase based on the original genetic risk score models, although this difference was small and statistically significant in only half of the presented phenotypes. This result suggests that a sufficiently large number of SNPs tags many genes throughout the genome which influence blood pressure regulation. These findings are in line with our hypothesis that blood pressure is polygenic trait and that there are many more genes involved with blood pressure than are found so far in genome wide association studies. Currently, the advantage of using genetic risk scores based on SNPs selected on their P-value in a GWAS discovery sample, as compared to genetic risk scores based on a random set of SNPs, seems to be limited. Larger GWAS discovery samples with identification of new common and rare SNPs might lead to higher explained variance.
Although there have been several mutations described causing dominant, monogenic, forms of hypertension and more of such rare variants may still be undiscovered 6, our results support the hypothesis that hypertension is a polygenic disease, which is in part explained by a large number of genes regulating blood pressure. In our adult population, genetic risk scores including large numbers of SNPs, explain the largest proportion of variance in blood pressure, indicating the involvement of multiple genes in blood pressure regulation.
Risk scores containing highly significant SNPs, identified in large scale genome wide association meta-analyses in adults 7–8, were significantly associated with in blood pressure in adults, but not in children. There are several explanations possible for this finding, including a smaller number of subjects and lower power in the children cohort. However, this finding might also indicate that the genome wide significant SNPs found so far are related to late-onset pathology. It has been long hypothesized that there is a common polygenic regulation of blood pressure in adults and children. This is the first study showing evidence of such a mechanism.
It has been shown in literature that blood pressure tracks from childhood to adulthood 15. This study indicates that genes are explaining a part of the blood pressure tracking over life. We show that the same set of genes, based on an adult discovery sample, explain part of the variation in blood pressure in both adults and children. We also showed that these SNP sets explain variation in hypertension in adults, indicating also a polygenic origin of hypertension. It has been shown that high blood pressure in childhood predisposes to hypertension in adulthood 16. Adult based genetic risk scores do not explain variance in childhood hypertension in children significantly. This fits with the common view that causes of juvenile hypertension are different from adult hypertension 11.
The percentage of explained variance by genetic risk scores is still low, although the heritability has been shown to be substantial 4, yet compared to the variance explained by the genome wide significant SNPs on blood pressure found so far, there is a 4–5 fold increase in explained variance of blood pressure in our target samples.
In the coming years the variance explained by polygenic models may be improved further, using technology, such as whole genome sequencing, which can be used to identify low frequency variants. We used common variants only (MAF >0.01) to create genetic risk scores. Low frequency variants may add to the variance explained by these genetic risk scores. Also, we assumed an additive model, similar to the discovery analysis. We have to recognize that the biology of the genes involved in blood pressure regulation and possible interactions between these genes are unknown. Another possibility would be to construct genetic risk scores based on SNPs included in candidate biological pathways. A genetic risk score including SNPs from the FGF signaling pathway 14 seemed to explain a larger proportion explained variance in hypertension as compared to a genetic risk score including a similar number of SNPs, based on the previous top SNPs from the GWAS. This indicates that prior knowledge on biological models and underlying mechanisms might improve the explained variance by genetic risk scores. Alternative methods of constructing genetic risk scores may be better when further research reveals more of the underlying genetic biology of blood pressure regulation.
Specific common variants that are associated with blood pressure still need to be identified. Much research is still needed to identify more and specific genes associated with blood pressure regulation in adults. If these common variants overlap with blood pressure regulation in children, they could provide clues for early etiology of hypertension.
Supplementary Material
PERSPECTIVES.
At this stage, individual prediction is not yet feasible. Without a doubt, the prediction of blood pressure will improve and might contribute to predicting high blood pressure in the future. Genetic profiling might be a way of identifying subgroups at genetically high risk for increased blood pressure at a population level 17, but whether it will be enough for personalised medicine and early treatment of people at risk for high blood pressure (and possibly also other risk factors for cardiovascular disease), remains to be determined. Our study shows that this may require the identification of many more common variants with small effects on blood pressure.
Acknowledgments
ROTTERDAM STUDY III
Erasmus Medical Center; The Netherlands Organization for Health Research and Development; Dutch Kidney Foundation (C08.2251). The Netherlands Organization for Scientific Research; the Research Institute for Diseases in the Elderly (RIDE); The Netherlands Heart Foundation; The Ministry of Education, Culture and Science; The Ministry of Health Welfare and Sports; The European Commission; and The Municipality of Rotterdam.
Generation R Study
The Generation R study is conducted by the Erasmus Medical Center in close collaboration with the School of Law and Faculty of Social Sciences of Erasmus University Rotterdam; the Municipal Health Service Rotterdam area, Rotterdam; the Rotterdam Homecare Foundation, Rotterdam; and the Stichting Trombosedienst & Artsenlaboratorium Rijnmond, Rotterdam. We gratefully acknowledge the contributions of general practitioners, hospital, midwives, and pharmacies in Rotterdam.
Funding Sources:
ROTTERDAM STUDY III
Support for genotyping was provided by the Netherlands Organization for Scientific Research (NWO Groot, 175.010.2005.011, 911.03.012) and Research Institute for Diseases in the Elderly (014.93.015; RIDE2). This study was supported by the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) project nr. 050-060-810. This study was further financially supported by the Netherlands Heart Foundation grant 2009B102 (BFJV, MAI).
Generation R Study
The first phase of the Generation R Study is made possible by financial support from the Erasmus Medical Center, Rotterdam; Erasmus University Rotterdam; and The Netherlands Organization for Health Research and Development (ZonMw). Additional support was provided by a grant from the Dutch Kidney Foundation (C08.2251) (RT).
The funding sources of other cohorts in the CHARGE consortium are described in the online supplement (please see http://hyper.ahajournals.org)
Footnotes
Financial Disclosure: All the authors declared no competing interests.
Conflict of Interest Disclosures
None
References
- 1.Fields LE, Burt VL, Cutler JA, Hughes J, Roccella EJ, Sorlie P. The burden of adult hypertension in the United States 1999 to 2000: a rising tide. Hypertension. 2004;44:398–404. doi: 10.1161/01.HYP.0000142248.54761.56. [DOI] [PubMed] [Google Scholar]
- 2.Lawes CM, Vander Hoorn S, Rodgers A. Global burden of blood-pressure-related disease, 2001. Lancet. 2008;371:1513–1518. doi: 10.1016/S0140-6736(08)60655-8. [DOI] [PubMed] [Google Scholar]
- 3.Prevention of stroke by antihypertensive drug treatment in older persons with isolated systolic hypertension. Final results of the Systolic Hypertension in the Elderly Program (SHEP). SHEP Cooperative Research Group. JAMA. 1991;265:3255–3264. [PubMed] [Google Scholar]
- 4.Levy D, DeStefano AL, Larson MG, O’Donnell CJ, Lifton RP, Gavras H, Cupples LA, Myers RH. Evidence for a gene influencing blood pressure on chromosome 17. Genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the framingham heart study. Hypertension. 2000;36:477–483. doi: 10.1161/01.hyp.36.4.477. [DOI] [PubMed] [Google Scholar]
- 5.Ehret GB. Genome-wide association studies: contribution of genomics to understanding blood pressure and essential hypertension. Curr Hypertens Rep. 2010;12:17–25. doi: 10.1007/s11906-009-0086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vehaskari VM. Heritable forms of hypertension. Pediatr Nephrol. 2009;24:1929–1937. doi: 10.1007/s00467-007-0537-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Kottgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace-Raso FU, Smith AV, Taylor K, Scharpf RB, Hwang SJ, Sijbrands EJ, Bis J, Harris TB, Ganesh SK, O’Donnell CJ, Hofman A, Rotter JI, Coresh J, Benjamin EJ, Uitterlinden AG, Heiss G, Fox CS, Witteman JC, Boerwinkle E, Wang TJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, van Duijn CM. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, Voight BF, Scott LJ, Zhang F, Farrall M, Tanaka T, Wallace C, Chambers JC, Khaw KT, Nilsson P, van der Harst P, Polidoro S, Grobbee DE, Onland-Moret NC, Bots ML, Wain LV, Elliott KS, Teumer A, Luan J, Lucas G, Kuusisto J, Burton PR, Hadley D, McArdle WL, Brown M, Dominiczak A, Newhouse SJ, Samani NJ, Webster J, Zeggini E, Beckmann JS, Bergmann S, Lim N, Song K, Vollenweider P, Waeber G, Waterworth DM, Yuan X, Groop L, Orho-Melander M, Allione A, Di Gregorio A, Guarrera S, Panico S, Ricceri F, Romanazzi V, Sacerdote C, Vineis P, Barroso I, Sandhu MS, Luben RN, Crawford GJ, Jousilahti P, Perola M, Boehnke M, Bonnycastle LL, Collins FS, Jackson AU, Mohlke KL, Stringham HM, Valle TT, Willer CJ, Bergman RN, Morken MA, Doring A, Gieger C, Illig T, Meitinger T, Org E, Pfeufer A, Wichmann HE, Kathiresan S, Marrugat J, O’Donnell CJ, Schwartz SM, Siscovick DS, Subirana I, Freimer NB, Hartikainen AL, McCarthy MI, O’Reilly PF, Peltonen L, Pouta A, de Jong PE, Snieder H, van Gilst WH, Clarke R, Goel A, Hamsten A, Peden JF, Seedorf U, Syvanen AC, Tognoni G, Lakatta EG, Sanna S, Scheet P, Schlessinger D, Scuteri A, Dorr M, Ernst F, Felix SB, Homuth G, Lorbeer R, Reffelmann T, Rettig R, Volker U, Galan P, Gut IG, Hercberg S, Lathrop GM, Zelenika D, Deloukas P, Soranzo N, Williams FM, Zhai G, Salomaa V, Laakso M, Elosua R, Forouhi NG, Volzke H, Uiterwaal CS, van der Schouw YT, Numans ME, Matullo G, Navis G, Berglund G, Bingham SA, Kooner JS, Connell JM, Bandinelli S, Ferrucci L, Watkins H, Spector TD, Tuomilehto J, Altshuler D, Strachan DP, Laan M, Meneton P, Wareham NJ, Uda M, Jarvelin MR, Mooser V, Melander O, Loos RJ, Elliott P, Abecasis GR, Caulfield M, Munroe PB. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans DM, Visscher PM, Wray NR. Harnessing the information contained within genome-wide association studies to improve individual prediction of complex disease risk. Hum Mol Genet. 2009;18:3525–3531. doi: 10.1093/hmg/ddp295. [DOI] [PubMed] [Google Scholar]
- 11.National High Blood Pressure Education Program Working Group on High Blood Pressure in C Adolescents. The fourth report on the diagnosis, evaluation, and treatment of high blood pressure in children and adolescents. Pediatrics. 2004;114:555–576. [PubMed] [Google Scholar]
- 12.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;19:716–723. [Google Scholar]
- 14.Tomaszewski M, Charchar FJ, Nelson CP, Barnes T, Denniff M, Kaiser M, Debiec R, Christofidou P, Rafelt S, van der Harst P, Wang WY, Maric C, Zukowska-Szczechowska E, Samani NJ. Pathway analysis shows association between FGFBP1 and hypertension. J Am Soc Nephrol. 2011;22:947–955. doi: 10.1681/ASN.2010080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, Wang Y. Tracking of blood pressure from childhood to adulthood: a systematic review and meta-regression analysis. Circulation. 2008;117:3171–3180. doi: 10.1161/CIRCULATIONAHA.107.730366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun SS, Grave GD, Siervogel RM, Pickoff AA, Arslanian SS, Daniels SR. Systolic blood pressure in childhood predicts hypertension and metabolic syndrome later in life. Pediatrics. 2007;119:237–246. doi: 10.1542/peds.2006-2543. [DOI] [PubMed] [Google Scholar]
- 17.Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk of complex disease. Curr Opin Genet Dev. 2008;18:257–263. doi: 10.1016/j.gde.2008.07.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.