Abstract
Dengue virus (DENV), a mosquito-borne member of the family Flaviviridae, is a significant global pathogen affecting primarily tropical and subtropical regions of the world and placing tremendous burden on the limited medical infrastructure that exists in many of the developing countries located within these regions. Recent outbreaks in developed countries, including Australia (Hanna et al., 2009), France (Laruche et al., 2010), Taiwan (Kuan et al., 2010), and the USA (CDC, 2010), lead many researchers to believe that continued emergence into more temperate latitudes is likely. A primary concern is that there are no approved vaccines or antiviral therapies to treat DENV infections. Since the viral NS2B-NS3 protease (DENV NS2B-NS3pro) is required for virus replication, it provides a strategic target for the development of antiviral drugs. In this study, proof-of-concept high-throughput screenings (HTSs) were performed to unambiguously identify dengue 2 virus (DEN2V) NS2B-NS3pro inhibitors from a library of 2000 compounds. Validation screens were performed in parallel to concurrently eliminate insoluble, auto-fluorescing, and/or nonspecific inhibitors. Kinetic analyses of the hits revealed that parallel substrate fluorophore (AMC) interference controls and trypsin inhibition controls were able to reduce false positive rates due to solubility and fluorophore interference while the trypsin inhibition control additionally eliminated non-specific inhibitors. We identified five DEN2V NS2B-NS3pro inhibitors that also inhibited the related West Nile virus (WNV) protease (NS2B-NS3pro), but did not inhibit the trypsin protease. Biochemical analyses revealed various mechanisms of inhibition including competitive and mixed noncompetitive inhibition, with the lowest Ki values being 12 ± 1.5 μM for DEN2V NS2B-NS3pro and 2 ± 0.2 μM for WNV NS2B-NS3pro.
Keywords: dengue virus, West Nile virus, NS2B-NS3, protease, inhibitors, high-throughput screen
1. Introduction
Dengue virus (DENV) is a mosquito-borne virus from the family Flaviviridae. It causes significant disease worldwide, and is considered the most important mosquito-borne viral disease in the world (WHO, 2010). Endemic in more than 100 countries, DENV is estimated to cause over 50 million infections each year, which can result in serious disease including dengue fever (DF), dengue hemorrhagic fever (DHF), dengue shock syndrome (DSS), and death. Complicating matters further, DENV exists as four separate serotypes (DEN1V, DEN2V, DEN3V, and DEN4V) with infection by one serotype not providing protection from infections by the other serotypes. Furthermore, some evidence suggests that subsequent infections by different serotypes may increase the probability of developing more serious forms of the disease like DHF and DSS (Alvarez et al., 2006; Halstead, 2003). Unfortunately, there are no vaccines approved to prevent DENV infection, and no antiviral drugs to treat DENV infection.
DENV is an enveloped, positive-strand RNA virus whose ~11 kb genome is transcribed as a single polyprotein containing three structural (capsid, pre-M, and envelope) proteins at its 5′ end and seven nonstructural proteins at its 3′ end (Fields et al., 1996). The N-terminal 180 residues of the NS3 protein encode the viral protease, and ~40 residues from the central hydrophilic domain of the NS2B protein encode a required protease cofactor (Chambers et al., 1993; Yusof et al., 2000). Along with cellular proteases, the NS2B-NS3 protease complex (NS2B-NS3pro) is responsible for cleavage of the viral polyprotein (Cahour et al., 1992) and has been shown to be required for viral replication (Falgout et al., 1991). As such, NS2B-NS3pro provides a strategic target for inhibition in the development of flavivirus antivirals (Tomlinson et al., 2009a). Several groups have utilized in vitro protease assays to test potential inhibitors (Chanprapaph et al., 2005; Leung et al., 2001; Tomlinson et al., 2009b; Tomlinson and Watowich, 2011; Yin et al., 2006), and recently, a DENV NS2B-NS3pro inhibitor identified by a high-throughput screen (HTS) was reported (Yang et al., 2011).
For this study, active dengue 2 (DEN2V) NS2B-NS3pro and the small fluorogenic peptide substrate BOC-GRR-AMC was used to screen the MicroSource Spectrum library. Previously, we reported protease inhibitors identified from virtual screens (Tomlinson et al., 2009b; Tomlinson and Watowich, 2011) and observed that insolubility and fluorophore interference could give rise to false positive results. A considerable amount of time and resources were expended in validation assays to eliminate false positive results from further consideration. We therefore considered the possibility that it might be beneficial to include replicates and controls at the beginning of a high-throughput screen to reduce the false positive hit rate and limit the amount of time and resources expended pursuing false positive hits in subsequent validation steps. This approach would be beneficial for labs that do not have the ability to rapidly retest every hit from readily available in-house stocks of the library compounds and that therefore must individually purchase a subset of identified hits (i.e. those with the greatest apparent inhibition) for validation. Since solubility problems are often reflected by lack of reproducibility, our HTS proof-of-concept studies were performed in triplicate and only compounds exhibiting similar knock-down in all three replicates were chosen for further testing. Additionally, a fourth compound replicate plate was incubated with the fluorophore AMC to identify and remove false positives due to fluorophore interference that mimic inhibition of AMC-linked substrate cleavage. A preliminary screen of the 2000 compounds that comprise the MicroSource Spectrum collection produced several hits that were reproducible in all three replicates and did not interact with AMC, and subsequent validation revealed that the false positive rate was reduced by almost 80%. In a subsequent screen of the same library, the AMC control plate was replaced with a trypsin inhibition control plate; this internal control reduced the false positive rate by 90% relative to a one-replicate screen. Only soluble DEN2V NS2B-NS3pro inhibitors that did not inhibit trypsin or interfere with the AMC fluorophore were moved forward for additional testing. Detailed biochemical characterization of the validated leads identified five DEN2V NS2B-NS3pro inhibitors that also inhibited WNV NS2B-NS3pro.
2. Materials and Methods
2.1. Chemical compounds for HTS
The 2000 compound MicroSource Spectrum Collection (MicroSource Discovery Systems Inc., Gaylordsville, CT) was obtained from the University of Texas Health Science Center and Gulf Coast Consortium High-throughput Screening Core Facility (Houston, TX). Compounds selected for additional testing were ordered from MicroSource Discovery Systems Inc. (Gaylordsville, CT).
2.2. DEN2V NS2B-NS3pro expression and purification
The expression and purification of the DEN2V NS2B cofactor linked to the protease domain of NS3 (NS2B-NS3pro; plasmid obtained from Dr. S. Pheng, Novartis Institute for Tropical Diseases, Singapore) was performed using a modified version of the Li protocol (Li et al., 2005) as described previously (Tomlinson and Watowich, 2011).
2.3. Construction of high-throughput screen (HTS) was as follows
2.3.a. Preparation of HTS assay plates
A peristaltic pump (Multidrop Combi, Thermo Electron Corporation, West Palm Beach, FL) was used to add 30 μl DEN2Vpro cleavage buffer (200 mM Tris, pH 9.5) to each well of 384 well plates. Each compound was assayed in triplicate, and thus three plates were prepared for each run. Each run required one 96-well enzyme plate and four 96-well compound dilution plates that were ultimately transferred to triplicate 384-well assay plates as well as an additional AMC or trypsin control plate.
2.3.b. Preparation of HTS AMC control plates
AMC (Sigma-Aldrich, St Louis, MO) was diluted in cleavage buffer to a final assay concentration of 1.2 μM, equivalent to the concentration of free AMC after the protease cleavage reaction had proceeded for 45 minutes.
2.3.c. Preparation of trypsin control plates
A stock solution of bovine pancreatic trypsin (Sigma-Aldrich, St Louis, MO) was prepared in trypsin cleavage buffer (67 mM sodium phosphate, pH 7.6) and 40 μl was pipetted into the interior wells of 384-well control plates to a final concentration of 12.5 nM. Control wells contained either (i) cleavage buffer and trypsin, (ii) cleavage buffer, trypsin, and 100 μM ARDP0009 (a previously reported DEN2V NS2B-NS3pro inhibitor [Tomlinson et al., 2009b), (iii) cleavage buffer, trypsin, and 38 μM ARDP0009 (a concentration equivalent to the inhibitor’s Ki1 value), and (iv) cleavage buffer, trypsin, and substrate BOC-GRR-AMC (final concentration 30 μM, which was similar to the substrate’s measured Kd value). A peristaltic pump was used to pipette the cleavage buffer and trypsin mixture into the 384 well plates.
2.3.d. Preparation of HTS compound dilution plates
Compound libraries were stored in DMSO (10 mM) at −80°C in 96-well plate format. A Biomek Nxp robot (Beckman-Coulter, Brea, CA) was used to perform 10-fold dilutions of compounds into aqueous cleavage buffer in 96-well plates. Plate columns 1 and 12 were reserved for controls. These compound dilution plates contained 10x the final assay concentrations of 100 μM inhibitor. Control wells on these plates contained either (i) 10% DMSO in cleavage buffer, which upon addition of either substrate, or enzyme and substrate, provided “substrate alone” and “no inhibitor” controls, respectively, (ii) 1 mM ARDP0009, or (iii) 380 μM ARDP0009. Plates were centrifuged at low speed to remove air bubbles and ensure that all droplets were in the well.
2.3.e. Preparation of HTS enzyme plates
Separate “enzyme” plates were prepared for each run. Frozen aliquots of DEN2V NS2B-NS3pro were thawed on ice and diluted 10-fold with chilled cleavage buffer into 96-well plates. Outside wells were reserved for controls. Cleavage buffer (150 μl) was manually pipetted into each well of a 96-well plate. Plates were centrifuged at low speed to remove air bubbles and ensure that all droplets were in the well. Three enzyme plates were prepared at one time and were kept on ice. Final enzyme concentration was 50 nM.
2.4. HTS DEN2V NS2B-NS3pro inhibition assay
Four compound dilution plates (each with 80 compounds and 16 control wells), one enzyme plate, one control plate (either AMC or trypsin), and three assay plates were transferred to the Biomek FX (Beckman-Coulter, Brea, CA). Automatic solution transfer was accomplished by robotic control to prepare the final triplicate assay plates and control plate, each containing 320 compound wells and 64 internal control wells (located in columns 1, 2, 23, 24). After solution transfers were completed, the plates were covered, shaken (Titer Plate Shaker, Lab-Line Industries) to ensure uniform reagent mixing, and centrifuged at low speed to remove droplets from the sides and lid. Plates were incubated for 30 minutes at room temperature, after which BOC-GRR-AMC substrate was added (final concentration of 100 μM for the assay plates, 30 μM for the trypsin control plates). Plates were incubated at room temperature for an additional 45 minutes. Guanidine hydrochloride (1 M) was added to the assay plates to stop the cleavage reactions as well as to the control plates. Plates were monitored for fluorescence at 380 nm excitation and 465 nm emission using an Infinite M200 spectrofluorometer (Tecan, San Jose, CA).
2.5. Solubility assays
Hits identified in the HTS screen were individually purchased from the vendor and tested for solubility in DMSO and aqueous buffer. Briefly, compounds were dissolved in DMSO at 10 mM and 1 mM. Compounds that appeared soluble by visual inspection were centrifuged at 14000 × g for 30 minutes and inspected for pellet formation. Compounds that were soluble in DMSO were further diluted from the DMSO stock 100-fold into aqueous assay buffer (200 mM Tris [pH 9.0], 20% glycerol) and vortexed. Compounds that appeared soluble by visual inspection were centrifuged as described above and inspected for pellet formation. Compounds that were not fully soluble at either 1 mM (in DMSO) or 10 μM (in aqueous assay buffer, 1% DMSO) were removed from further analysis.
2.6. Single-point DEN2V NS2B-NS3pro inhibition assay to validate hits
Compounds soluble in aqueous assay conditions were tested in a single-point inhibition assay to validate that they inhibited protease-mediated cleavage of fluorophore-linked substrates. Assay conditions were similar to those performed in the original screen and have been previously described (Tomlinson and Watowich, 2011).
2.7. Single-point trypsin inhibition assay to validate selectivity
After the initial screen, each hit was tested for trypsin inhibition to validate specificity. Compounds were diluted to a final concentration of 100 μM in trypsin cleavage buffer (67 mM sodium phosphate, pH 7.6) and vortexed. Bovine pancreatic trypsin and BOC-GRR-AMC substrate were added (as described above in the HTS protocol), and the mixture incubated at room temperature for 30 minutes. Reactions were monitored on a Fluorolog FL3-22 spectrofluorometer (Horiba Jobin Yvon) to quantify fluorescence emitted at 465 nm after excitation at 380 nm. All assays were performed in duplicate. In addition, assays were performed at pH 9.5 to mimic the pH of the cleavage buffer used in the DEN2V assays.
2.8. Steady-state kinetics of HTS-identified inhibitors of DEN2V NS2B-NS3pro
Detailed kinetic analyses were performed as described previously (Tomlinson and Watowich, 2011) to determine the inhibition mechanisms and constants for compounds validated as DEN2V NS2B-NS3pro inhibitors. Briefly, increasing concentrations of each inhibitor were tested for protease inhibition with substrate concentrations ranging from 0 to 1.2 mM. For each substrate concentration, initial reaction velocities were determined and plotted using GraphPad Prism (GraphPad Software, San Diego, CA). Errors associated with initial velocity measurements were calculated and were consistently < 2%. The program Dynafit (Biokin, Watertown, MA) (Kuzmic, 1996) was used to perform kinetic analyses for the velocity vs. substrate concentration data. Kinetic (e.g., kcat) and equilibrium (e.g., Kd) parameters were directly calculated by numerical solution of coupled differential equations and optimized to provide the best global fit of each model to the experimental data. Final model selection was based on fitting accuracy, parameter errors, and model discrimination analysis within the Dynafit program. For each inhibitor, the discrimination analysis selected a substrate inhibition model with kinetic parameters that were reproducible and consistent for all tested inhibitors.
2.9. West Nile virus NS2B-NS3pro expression, purification, and kinetics
Plasmid constructs for WNV NS2B-NS3pro were obtained from Dr. Padmanabhan (Georgetown University) and have been previously described (Mueller et al., 2007). Expression and purification of WNV NS2B-NS3pro were performed as described previously (Tomlinson and Watowich, 2008). To determine the mechanism of inhibition and inhibition constants for lead compounds against WNV NS2B-NS3pro, detailed kinetic analyses were performed using purified WNV NS2B-NS3pro and the DEN2V steady-state kinetic analysis protocol.
2.10. AMC corrections and determination of fluorometer linear response
To correct for systematic variations in instrument response, AMC dilution series were measured in conjunction with each protease reaction. These measurements defined the linear range and response of the spectrofluorometer and also allowed for correction at each inhibitor concentration for variations introduced by compound absorption. Briefly, each concentration of inhibitor, as well as a “no inhibitor” control, was incubated with ~5 two-fold serial dilutions of AMC. Slopes from linear regression (GraphPad Software San Diego, CA) of this data were entered into Microsoft Excel (Redmond, WA) to convert fluorescence intensities to AMC concentrations.
3. Results
3.1. HTS with integrated AMC control
To identify inhibitors of DEN2V NS2B-NS3pro, HTS was performed using the Micro-Source Spectrum Collection, purified DEN2V NS2B-NS3pro, and the fluorophore-linked peptide substrate BOC-GRR-AMC. For each molecule in the screening library, protease assays were performed in triplicate and fluorescence intensity from an AMC internal control experiment was examined to reduce false positives resulting from interference of substrate product fluorescence. This assay configuration repeatedly produced Z′ scores that were > 0.7 and allowed the Microsource library to be screened in a single day. Each library compound was tested at 100 μM final concentration. Though lower concentrations (e.g., 10 μM) might identify less soluble compounds, 100 μM was chosen to increase the number of positive hits identified in the preliminary 2000-member compound library and thus identify protease inhibitors with diverse chemical scaffolds that might be subsequently modified and optimized, as opposed to identifying only a few strong inhibitors. A previous study demonstrated that the Ki1 of a lead dengue protease inhibitor could be improved by ~60-fold after a single round of SAR optimization (Tomlinson and Watowich, 2011).
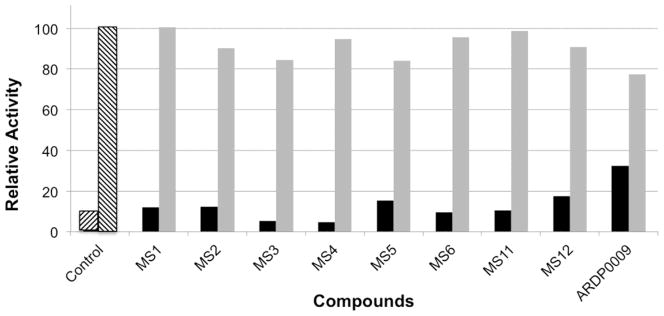
Eight compounds demonstrated > 80% DEN2V NS2B-NS3pro inhibition with little reduction of the AMC control signal (Fig. 1). In many cases, the observed fluorescence signal was comparable to the background signal. In all cases the observed fluorescence signal was lower than that measured with ARDP0009, a previously identified DEN2V NS2B-NS3pro inhibitor (Tomlinson et al., 2009b). Although the replicate and AMC control plate strategy yielded only soluble hits that inhibited DEN2V NS2B-NS3pro and that did not interfere with the AMC fluorophore, six of the hits strongly inhibited trypsin, and were removed from further study. Compounds MS2 and MS5 inhibited DEN2V protease and did not inhibit trypsin and were thus moved forward for further analysis.
Figure 1.
Relative activity of DEN2V NS2B-NS3pro. The black bars display the averaged relative fluorescence of the protease reaction in the presence of inhibitors. The fluorescence intensity was normalized to the averaged signal produced by the protease reaction without inhibitor (hatched bar, second from the left). The grey bars display the averaged relative fluorescence of AMC in the presence of inhibitors, normalized to the signal produced by AMC alone. The first hatched bar on the left-hand side is a control containing substrate alone.
3.2. HTS with integrated trypsin control
To reduce false-positive rates arising from non-selective protease inhibitors, we repeated the HTS replacing the AMC control with an integrated trypsin control. Like the AMC control, this control was designed to identify false-positives arising from compound insolubility, fluorophore interference (same fluorophore-linked substrate as that used in the DEN2V inhibition assay), and protein denaturation while also eliminating nonselective protease inhibitors (i.e. trypsin inhibitors). Interestingly, many assay wells with dramatic reductions in fluorescence signal had corresponding signal reductions in the trypsin control wells, perhaps indicating fluorophore interference, non-selective binding, or protein denaturation. Hits identified in the initial screen with the integrated AMC control were also identified in this screen, but eliminated due to significant signal reduction on the trypsin control plate. The two selective DEN2V protease hits identified in the initial screen with an internal AMC control were validated as “non-inhibitors” on the trypsin control plate.
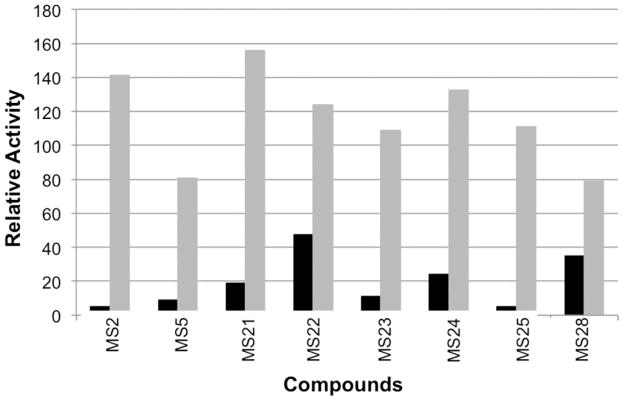
In the current screen, the criteria for moving a compound forward was ≥ 50% reduction of DEN2V NS2B-NS3pro activity and ≤ 20% reduction on the trypsin control plate. Since the dengue protease was found to have a Kd ~ 921 ± 68 μM and a kcat ~ 0.05 ± 0.002 s−1, the “cutoff” criteria would be expected to identify strictly competitive DEN2V NS2B-NS3pro inhibitors with Ki1 ≤ ~90 μM and strictly uncompetitive inhibitors with Ki2 ≤ ~10. In addition, the reagent concentrations used in the screen were chosen to increase the likelihood of identifying compounds with a competitive inhibition component as opposed to compounds with a strong uncompetitive component. Since signal reduction on the trypsin control plate could result from fluorophore interference and/or trypsin inhibition, the 20% selection criterion on the trypsin control removed compounds that displayed modest fluorophore interference (21–100% signal reduction) and/or even weak trypsin inhibition (Ki1 < ~200 μM or Ki2 < ~200 μM). Eight compounds (MS21, MS22, MS23, MS24, MS25, MS28 from the current screen and MS2, MS5 from the original screen) were validated in single-point DEN2V NS2B-NS3pro (Fig. 2) and trypsin inhibition assays. These compounds inhibited DEN2V protease and did not inhibit trypsin and were subsequently tested in detailed kinetic assays. It should be noted that a few of the compounds demonstrated increased signal above that of the “no inhibitor” control, perhaps indicating auto-fluorescence of the compound. Controls in the kinetic analyses (see Methods) corrected for compound interactions with the fluorescent AMC protease reaction product.
Figure 2.
Relative activity of DEN2V NS2B-NS3pro. The black bars display the averaged relative fluorescence of the protease reaction in the presence of inhibitors. The fluorescence intensity was normalized to the averaged signal produced by the protease reaction without inhibitor. The grey bars display the averaged relative fluorescence of AMC in the presence of inhibitors, normalized to the signal produced by AMC alone.
3.3. Kinetic analyses of DEN2V NS2B-NS3pro inhibitors
Kinetic analyses of DEN2V NS2B-NS3pro inhibitors were routinely performed with substrate BOC-GRR-AMC concentrations ranging from 0 to 1.2 mM. As previously reported (Tomlinson and Watowich, 2011), DEN2V NS2B-NS3pro clearly demonstrated substrate inhibition at high substrate concentrations (Fig. 3). Numerous kinetic analysis using a substrate inhibition model and maximal substrate concentrations ranging from 1.2 to 5 mM all produced very similar kinetic parameters. In addition, analyses of several inhibitors showed that similar kinetic and inhibition constants were predicted by Dynafit kinetic modeling irrespective of the maximal substrate concentration being either 1.2 mM or 5 mM. The agreement between inhibition constants determined from using a maximal substrate concentration of either 1.2 mM or 5 mM was obtained without constraining the kinetic parameters, although such constraints generally improved error estimates of the inhibition constants. This comprehensive analysis of the Dynafit program guided the design of our NS2B-NS3pro inhibitor experiments and allowed us to accurately predict inhibition constants from kinetic inhibitor experiments performed using a maximal substrate concentration of 1.2 mM (Fig. 4).
Figure 3.
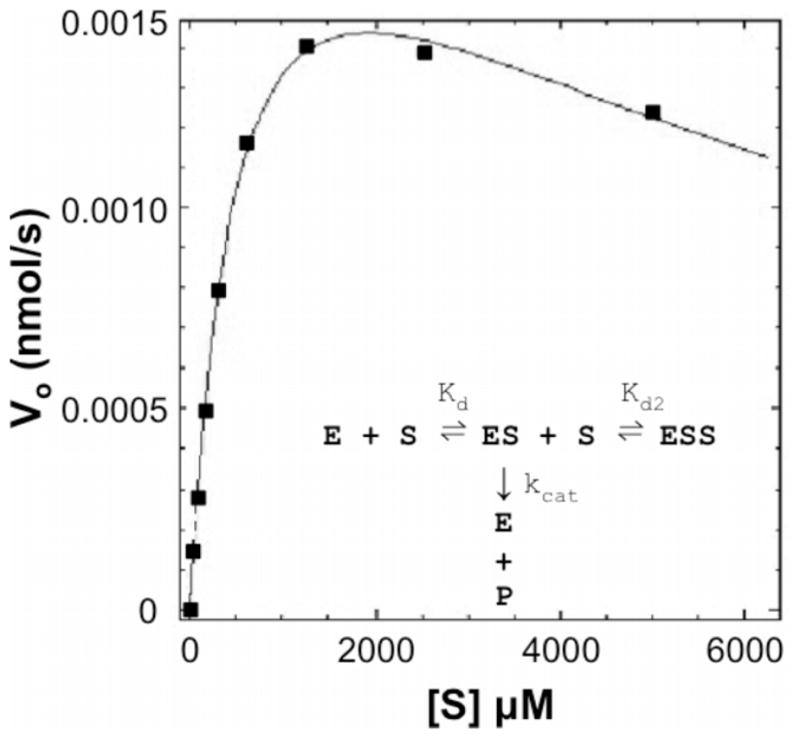
DEN2V protease reaction with the small peptide substrate Boc-Gly-Arg-Arg-AMC demonstrated Michaelis-Menten kinetics with substrate inhibition. The model curve was produced with the program Dynafit, which optimized the kinetic parameters in a substrate inhibition model (insert) to best fit the data points.
Figure 4.
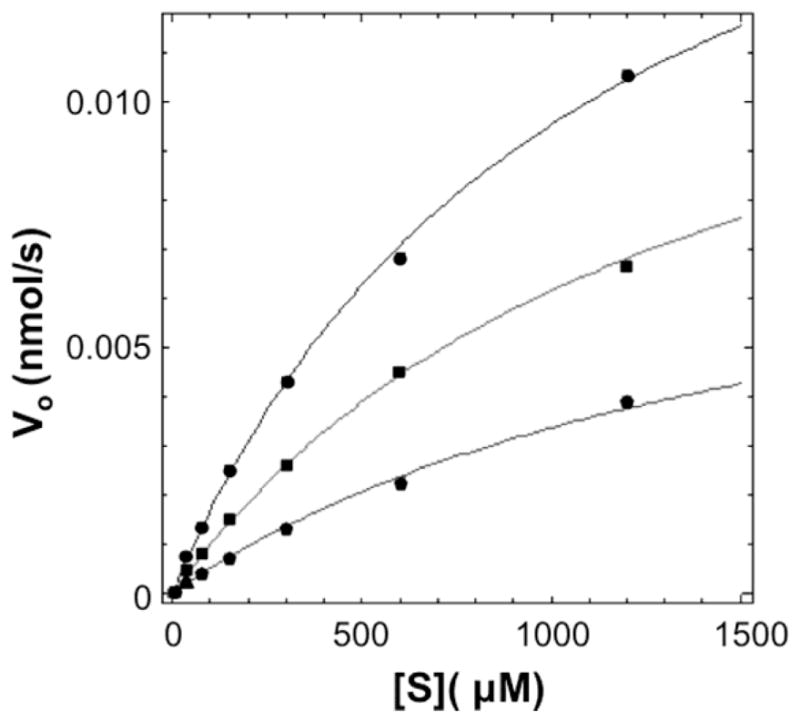
Representative curve for inhibitor MS28 (alexidine hydrochloride), which was modeled as having a mixed inhibition mechanism against DEN2V NS2B-NS3pro. Concentrations of MS28 tested were 0 (circles, top curve), 30 (squares, middle curve), and 100 (pentagons, lower curve) μM. Data were analyzed with the program Dynafit, with curves calculated from global fitting to the data points. Error bars were within the dimensions of the data points.
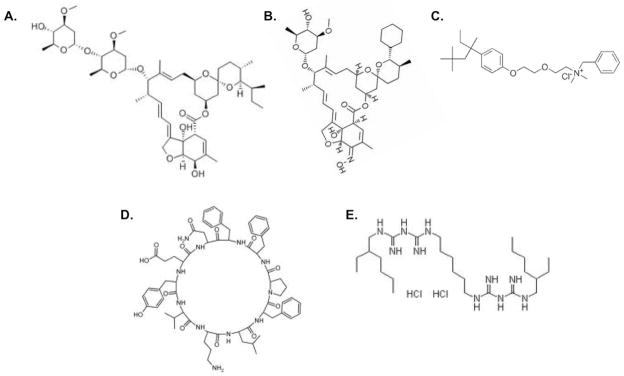
Detailed DEN2V NS2B-NS3pro kinetic analyses were completed for each compound and clearly demonstrated inhibition of DEN2V NS2B-NS3pro. Compounds MS2, MS5, and MS25 were removed from the study due to lack of reproducibility in kinetic experiments, which may have been caused by instability of compounds (i.e. oxidation over time). Kinetic inhibition models demonstrated excellent fits to the experimental data as indicated by the representative curve for MS28 (Fig. 4). Mechanisms of inhibition and inhibition constants for the compounds are provided in Table 1. Competitive inhibition was observed for MS23, and mixed noncompetitive inhibition was observed for the other inhibitors. Inhibition constants were in the low to mid micromolar range. With the exception of MS21 (ivermectin) and MS24 (selamectin), the molecules identified as DEN2V protease inhibitors had chemically distinct structures (Fig. 5).
Table 1.
DEN2V NS2B-NS3pro inhibition constants and mechanisms.
| ID | Common name | Mechanism | Ki1 (μM) | Ki2 (μM) |
|---|---|---|---|---|
| MS21 | Ivermectin | mixed | 79 ± 21 | 35 ± 4 |
| MS22 | Methylbenzethonium chloride | mixed | 322 ± 81 | 160 ± 39 |
| MS23 | Tyrothricin | competitive | 12 ± 1.5 | - |
| MS24 | Selamectin Alexidine | mixed | 63 ± 18 | 45 ± 8 |
| MS28 | hydrochloride | mixed | 41 ± 3 | 84 ± 16 |
Figure 5.
Chemical structures for lead DEN2V protease inhibitors: (A) ivermectin (MS21), (B) selamectin (MS24), (C) methylbenzethonium chloride (MS22), (D) tyrothricin (MS23), and (E) alexidine hydrochloride (MS28).
3.4. Inhibition of WNV NS2B-NS3pro
Since DEN2V and WNV are phylogenetically related (sequence identity ~50% [Erbel 2006]) and have similar NS2B-NS3pro structures, DEN2V NS2B-NS3pro inhibitors were tested for activity against WNV NS2B-NS3pro. All five DEN2V protease inhibitors listed in Table 1 demonstrated activity against WNV NS2B-NS3pro. Moreover, similar inhibition mechanisms and constants were observed for both proteases (Tables 1, 2).
Table 2.
WNV NS2B-NS3pro inhibition constants and mechanisms.
| ID | Common name | Mechanism | Ki1 (μM) | Ki2 (μM) |
|---|---|---|---|---|
| MS21 | Ivermectin | mixed | 55 ± 18 | 16 ± 4 |
| MS22 | Methylbenzethonium chloride | mixed | 141 ± 20 | 307 ± 80 |
| MS23 | Tyrothricin | competitive | 2 ± 0.2 | - |
| MS24 | Selamectin | mixed | 15 ± 9 | 28 ± 6 |
| MS28 | Alexidine hydrochloride | mixed | 12 ± 1.2 | 28 ± 5 |
4. Discussion
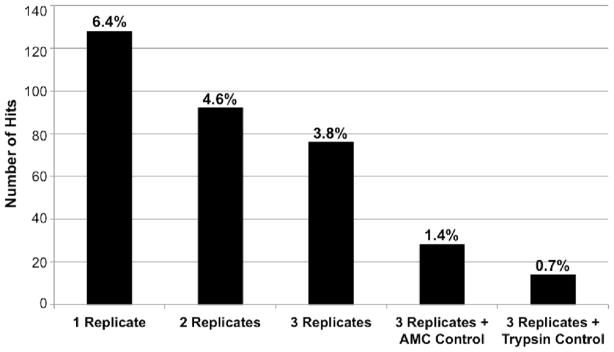
As a test and proof-of-concept for planned high-throughput screens with large numbers (10,000s to 100,000s) of compounds, the MicroSource Spectrum Collection was used to screen for inhibitors of the DEN2V NS2B-NS3pro. This library included 1040 compounds from the US Drug Collection that had reached clinical trials in the US, 800 pure natural products, and 160 natural and synthetic compounds with previously demonstrated toxicity. The screen was performed in triplicate and repeated twice using two different parallel internal controls. One screen used as a control the substrate fluorophore (AMC) alone to identify false positives due to fluorophore interference and insolubility. Previous studies in our lab had shown that solubility artifacts often produced non-reproducible changes in fluorescent signals and in many cases produced false positives that were obvious even in the absence of enzyme. Hits that were reproducible in all three replicates and did not demonstrate significantly reduced signal on the AMC control plate provided greater confidence of identifying true positives. While this strategy was successful at removing compounds that were either insoluble or that interacted with the fluorophore, the majority of the hits from this screen demonstrated strong inhibition of the related cellular protease trypsin in subsequent kinetic assays. A second screen replaced the AMC control with an internal control that simultaneously assayed for trypsin inhibition. Since both DEN2V NS2B-NS3pro and trypsin utilized the same AMC-linked substrate, this control still eliminated false positives from fluorophore interference and compound insolubility. In addition, it also eliminated compounds that strongly inhibited trypsin and potentially compounds that simply denatured the proteases. While using replicates in HTS experiments is not a novel idea, studies reported in the literature have often used only one replicate citing expense as the primary reason. However, by performing assays in triplicate, the hit rate was reduced almost two-fold from that observed for assays performed with a single replicate (Fig. 6). Moreover, inclusion of parallel internal controls in the primary HTS reduced hit rates approximately 10-fold relative to HTS that used a single replicate and no parallel controls (Fig. 6), and eliminated compounds that interfered with the AMC signal or strongly inhibited trypsin.
Figure 6.
Hit rate as a function of replicate number and inclusion of AMC and trypsin internal controls.
This HTS protocol effectively resulted in quadruplicate testing, although overall screening efficiency was improved as less time and money were expended examining false positives. It should be noted that some labs have in-house libraries and resources for performing counter-screens for every hit identified in a HTS. In these situations it might not be beneficial to include all of the replicates and parallel controls described above. However, for environments that do not have the resources to counter-screen every hit identified in an HTS, but must select “best” hits to be individually purchased for validation studies, the approach described above provided an efficient method for successfully screening moderately-sized libraries as less time and resources were spent pursuing false positives.
Subsequent validation and detailed kinetic testing identified five compounds that inhibited DEN2V and WNV NS2B-NS3pro (Tables 1 and 2). Though we prefer to see inhibitors with nanomolar inhibition constants, the inhibitors identified in this proof-of-concept HTS provide a foundation for future optimization studies and are valuable since they have already undergone clinical studies (discussed below). In other studies in our laboratory, we have identified compounds that selectively inhibited DEN2V NS2B-NS3pro, WNV NS2B-NS3pro, or inhibited both proteases. Since WNV NS2B-NS3pro is more phylogenetically distant from DEN2V than the other dengue serotypes (DEN1V, DEN3V, DEN4V), it is likely that inhibitors of both DEN2V and WNV NS2B-NS3pro will inhibit all DENV serotypes. Moreover, inhibitors of both DEN2V and WNV NS2B-NS3pro likely interacted with conserved residues (Tomlinson and Watowich, 2011), though the size and flexibility of these inhibitors precluded docking studies that might have proposed specific interactions. Such broad-spectrum flavivirus inhibitors may delay the emergence of drug-resistant dengue strains. The inhibitors identified in this screen (Fig. 5) have completed clinical trials and have been marketed for other medicinal purposes.
Ivermectin (MS21; Fig. 5A) is a macrocyclic lactone derived from Streptomyces avermitilis. In humans, ivermectin is most often used to treat roundworm infections such as strongyloidiasis and onchocerciasis (river blindness). In canines, ivermectin is used to kill larval heart-worms and sterilize adult heartworms, thus preventing serious infection. The determined mechanism of action associated with roundworm infection is activation of glutamate-gated chloride channels, thus interfering with muscle and nervous system function of the helminthes (Yates and Wolstenholme, 2004). Interestingly, ivermectin has been reported to also block nuclear import of HIV integrase (Wagstaff et al., 2011). Ivermectin can be administered by injection, but is most often administered orally as a tablet. Ivermectin toxicity is documented (Molinari et al., 2009; Xie et al., 2008) for the therapeutic concentration required to treat onchocerciasis, and would need to be evaluated at the concentrations required for DEN2V and WNV NS2B-NS3pro inhibition. Therapeutic use of ivermectin for DEN2V infections may be worth pursuing as ivermectin distribution networks currently exist to provide free onchocerciasis treatment in countries that are endemic with dengue virus (Alleman et al., 2006).
Selamectin (MS24; Fig. 5B), a macrolide lactone related to ivermectin, is also derived from Streptomyces avermitilis. It is marketed as a topical broad-spectrum parasiticide used in dogs and cats to control fleas, ear mites, heartworms, hookworms, and roundworms. It is not, however, approved for human use. Like ivermectin, selamectin is administered orally, but can also be absorbed through the skin, and enter the blood, intestines, and sebaceous glands. Selamectin is reported to have a high safety profile in cats and dogs (including breeding and young animals). Its oral and topical dosages (Pipano, 2002) exceed that of the related ivermectin, and additional studies will indicate if it may be worth pursuing as a DEN2V or WNV protease inhibitor. Related macrolides (e.g., the milbemycins) may also be promising candidates as DEN2V and WNV NS2B-NS3pro inhibitors.
Methylbenzethonium chloride (MS22; Fig. 5C) is a bactericide and antiseptic used to treat diaper rash and has also been evaluated as a treatment for cutaneous leishmaniasis (Kim et al., 2009). As it is a topical treatment, it has not undergone toxicity studies for systemic administration.
Tyrothricin (MS23; Fig. 5D), synthesized by the bacteria Bacillus brevis, is a polypeptide antibiotic mixture of tyrocidins and gramicidins. It was reported to be a very effective antibiotic against gram-positive and gram-negative bacteria (Mogi and Kita, 2009), pathogenic fungi, and nematodes (Otoguro et al., 1988). Tyrothricin had a competitive mechanism of inhibition against both DEN2V and WNV proteases, which may result from the lysine moiety of cyclic decapeptide mimicking the basic residues of the protease substrate and interacting with the active site P1 pocket of the DEN2V and WNV proteases. As tyrothricin is a mixture of cyclic and linear peptides, further work will be necessary to determine which peptide or peptides were responsible for the observed protease inhibitory activity.
Alexidine hydrochloride (MS28; Fig. 5E) is a potent antibiotic that was used in mouthwashes (McDonnell and Russell, 1999), and most recently reported as a potential cancer therapeutic (Yip et al., 2006). Toxicity studies in cells, however, demonstrated low micromolar CC50 (cell viability reduced by 50%) values (Yip et al., 2006). It is possible, however, that the protease inhibitory activity of MS28 requires only half of the symmetrical molecule, and such an inhibitor may have an improved toxicity profile.
5. Conclusion
High-throughput screening of chemical libraries has become an important tool for the identification of lead compounds for therapeutic development. In this study, dual screens of the same library, each with integrated parallel control screens, demonstrated that compounds that inhibited a particular target could be identified with few false positives. Moreover, including multiple replicates and appropriate controls significantly reduced the number of false-positive hits that required subsequent validation effort. By testing compounds that demonstrated inhibition of DEN2V NS2B-NS3pro but not trypsin, we were able to identify five DEN2V NS2B-NS3pro inhibitors that also inhibited the related WNV NS2B-NS3pro, but did not inhibit trypsin. Kinetic analyses revealed Ki1 values as low as 12 ± 1.5 μM for DEN2V NS2B-NS3pro and 2 ± 0.2 μM for WNV NS2B-NS3pro. Since the identified compounds have already been through clinical trials and approved for use in humans and/or animals, and since distribution networks are already available in dengue-endemic countries (ivermectin), further efficacy studies for these compounds as dengue antivirals may be warranted.
Highlights.
presents efficiency improvements to conventional biochemical HTS
parallel HTS enabled discovery of promising DENV and WNV protease inhibitors
kinetic validation showed competitive and mixed noncompetitive inhibition mechanism
inhibitors were specific for the flavivirus serine proteases
of interest to pharmaceutical & academic researchers in antiviral drug discovery
Acknowledgments
We thank Dr. S. Pheng (Novartis Institute for Tropical Diseases) for the plasmid encoding DEN2V NS2B-NS3pro. In addition, we thank Drs. A. Barrett, S. Gilbertson, J. Halpert, and J. Lee for helpful discussions, and Dr. J. Stevens for help with robotic screening at the University of Texas Health Science Center (Houston, TX).
Footnotes
This research was supported in part by NIH/NIAID AI066160 (SJW), the Welch Foundation H-1642 (SJW), and the Dunn Foundation (SJW) and training fellowships to SMT from the Computational and Structural Biology in Biodefense Training Program (NIAID Grant No. 1 T32 AI065396-01) and the National Library of Medicine Training Program (NLM Grant No. T15 LM007093) through the Keck Center for Interdisciplinary Bioscience Training of the Gulf Coast Consortia.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alleman M, Twim-Danso N, Thylefors B. The Mectizan donation program -highlights from 2005. Filaria J. 2006;5:1–11. doi: 10.1186/1475-2883-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez M, Rodriguez-Roche R, Bernardo L, Vazquez S, Morier L, Gonzalez D, Castro O, Kouri G, Halstead SB, Guzman MG. Dengue hemorrhagic fever caused by sequential dengue 1–3 virus infections over a long time interval: Havana epidemic, 2001–2002. Am J Trop Med Hyg. 2006;75:1113–1117. [PubMed] [Google Scholar]
- Cahour A, Falgout B, Lai CJ. Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions are mediated by viral protease NS2B-NS3, whereas NS4A/NS4B may be processed by a cellular protease. J Virol. 1992;66:1535–1542. doi: 10.1128/jvi.66.3.1535-1542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . Locally acquired Dengue--Key West, Florida, 2009–2010. MMWR Morb Mortal Wkly Rep. 2010;59:577–581. [PubMed] [Google Scholar]
- Chambers TJ, Nestorowicz A, Amberg SM, Rice CM. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B-NS3 complex formation, and viral replication. J Virol. 1993;67:6797–6807. doi: 10.1128/jvi.67.11.6797-6807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanprapaph S, Saparpakorn P, Sangma C, Niyomrattanakit P, Hannongbua S, Angsuthana-sombat C, Katzenmeier G. Competitive inhibition of the dengue virus NS3 serine protease by synthetic peptides representing polyprotein cleavage sites. Biochem Biophys Res Commun. 2005;330:1237–1246. doi: 10.1016/j.bbrc.2005.03.107. [DOI] [PubMed] [Google Scholar]
- Erbel P, Schiering N, D’Arcy A, Renatus M, Kroemer M, Lim SP, Yin Z, Keller TH, Vasudevan SG, Hommel U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol. 2006;13:372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- Falgout B, Pethel M, Zhang YM, Lai CJ. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991;65:2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B, Knipe D, Howley P, Chanock R, Melnick J, Monath T, Roizman B, Strauss S. Field’s Virology. 3. Lippincott Williams & Wilkins; Philadelphia: 1996. [Google Scholar]
- Halstead SB. Neutralization and antibody-dependent enhancement of dengue viruses. Adv Virus Res. 2003;60:421–467. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- Hanna JN, Ritchie SA, Richards AR, Humphreys JL, Montgomery BL, Ehlers GJ, Pyke AT, Taylor CT. Dengue in north Queensland, 2005–2008. Commun Dis Intell. 2009;33:198–203. doi: 10.33321/cdi.2009.33.19. [DOI] [PubMed] [Google Scholar]
- Kim DH, Chung HJ, Bleys J, Ghohestani RF. Is paromomycin an effective and safe treatment against cutaneous leishmaniasis? A meta-analysis of 14 randomized controlled trials. PLoS Negl Trop Dis. 2009;3:e381. doi: 10.1371/journal.pntd.0000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan MM, Lin T, Chuang JH, Wu HS. Epidemiological trends and the effect of airport fever screening on prevention of domestic dengue fever outbreaks in Taiwan, 1998–2007. Int J Infect Dis. 2010;14:e693–697. doi: 10.1016/j.ijid.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- La Ruche G, Souares Y, Armengaud A, Peloux-Petiot F, Delaunay P, Despres P, Lenglet A, Jourdain F, Leparc-Goffart I, Charlet F, Ollier L, Mantey K, Mollet T, Fournier JP, Torrents R, Leitmeyer K, Hilairet P, Zeller H, Van Bortel W, Dejour-Salamanca D, Grandadam M, Gastellu-Etchegorry M. First two autochthonous dengue virus infections in metropolitan France, September 2010. Euro Surveill. 2010;15:19676. [PubMed] [Google Scholar]
- Leung D, Schroder K, White H, Fang NX, Stoermer MJ, Abbenante G, Martin JL, Young PR, Fairlie DP. Activity of recombinant dengue 2 virus NS3 protease in the presence of a truncated NS2B co-factor, small peptide substrates, and inhibitors. J Biol Chem. 2001;276:45762–45771. doi: 10.1074/jbc.M107360200. [DOI] [PubMed] [Google Scholar]
- Li J, Lim SP, Beer D, Patel V, Wen D, Tumanut C, Tully DC, Williams JA, Jiricek J, Priestle JP, Harris JL, Vasudevan SG. Functional profiling of recombinant NS3 proteases from all four serotypes of dengue virus using tetrapeptide and octapeptide substrate libraries. J Biol Chem. 2005;280:28766–28774. doi: 10.1074/jbc.M500588200. [DOI] [PubMed] [Google Scholar]
- McDonnell G, Russell AD. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev. 1999;12:147–179. doi: 10.1128/cmr.12.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi T, Kita K. Gramicidin S and polymyxins: the revival of cationic cyclic peptide antibiotics. Cell Mol Life Sci. 2009;66:3821–3826. doi: 10.1007/s00018-009-0129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari G, Soloneski S, Reigosa MA, Larramendy ML. In vitro genotoxic and cytotoxic effects of ivermectin and its formulation on Chinese hamster ovary (CHOK1) cells. J Hazard Mater. 2009;165:1074–1082. doi: 10.1016/j.jhazmat.2008.10.083. [DOI] [PubMed] [Google Scholar]
- Mueller NH, Yon C, Ganesh VK, Padmanabhan R. Characterization of the West Nile virus protease substrate specificity and inhibitors. Int J Biochem Cell Biol. 2007;39:606–614. doi: 10.1016/j.biocel.2006.10.025. [DOI] [PubMed] [Google Scholar]
- Ogoussan KT, Hopkins A. Mectizan procurement and delivery for onchocerciasis mass drug administration programmes. Acta Trop. 2010;120(Suppl 1):S173–6. doi: 10.1016/j.actatropica.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Otoguro K, Liu ZX, Fukuda K, Li Y, Iwai Y, Tanaka H, Omura S. Screening for new nematocidal substances of microbial origin by a new method using the pine wood nematode. J Antibiot (Tokyo) 1988;41:573–575. doi: 10.7164/antibiotics.41.573. [DOI] [PubMed] [Google Scholar]
- Pipano E. Recent developments in the control of ectoparasites and endoparasites of dogs and cats. Israel Journal of Veterinary Medicine. 2002;58:2–3. [Google Scholar]
- Tomlinson SM, Malmstrom RD, Watowich SJ. New approaches to structure-based discovery of dengue protease inhibitors. Infect Disord Drug Targets. 2009a;9:327–343. doi: 10.2174/1871526510909030327. [DOI] [PubMed] [Google Scholar]
- Tomlinson SM, Malmstrom RD, Russo A, Mueller N, Pang YP, Watowich SJ. Structure-based discovery of dengue virus protease inhibitors. Antiviral Res. 2009b;82:110–114. doi: 10.1016/j.antiviral.2009.02.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson SM, Watowich SJ. Substrate inhibition kinetic model for West Nile virus NS2B-NS3 protease. Biochemistry. 2008;47:11763–11770. doi: 10.1021/bi801034f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson SM, Watowich SJ. Anthracene-based inhibitors of dengue virus NS2B-NS3 protease. Antiviral Res. 2011;89:127–135. doi: 10.1016/j.antiviral.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff KM, Rawlinson SM, Hearps AC, Jans DA. An AlphaScreen(R)-based assay for high-throughput screening for specific inhibitors of nuclear import. J Biomol Screen. 2011;16:192–200. doi: 10.1177/1087057110390360. [DOI] [PubMed] [Google Scholar]
- WHO. 2010 http://www.who.int/mediacentre/factsheets/fs117/en/
- Xie J, Nair A, Hermiston TW. A comparative study examining the cytotoxicity of inducible gene expression system ligands in different cell types. Toxicol In Vitro. 2008;22:261–266. doi: 10.1016/j.tiv.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Yang CC, Hsieh YC, Lee SJ, Wu SH, Liao CL, Tsao CH, Chao YS, Chern JH, Wu CP, Yueh A. Novel dengue virus-specific NS2B/NS3 protease inhibitor, BP2109, discovered by a high-throughput screening assay. Antimicrob Agents Chemother. 2011;55:229–238. doi: 10.1128/AAC.00855-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates DM, Wolstenholme AJ. An ivermectin-sensitive glutamate-gated chloride channel subunit from Dirofilaria immitis. Int J Parasitol. 2004;34:1075–1081. doi: 10.1016/j.ijpara.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Yin Z, Patel SJ, Wang WL, Chan WL, Ranga Rao KR, Wang G, Ngew X, Patel V, Beer D, Knox JE, Ma NL, Ehrhardt C, Lim SP, Vasudevan SG, Keller TH. Peptide inhibitors of dengue virus NS3 protease. Part 2: SAR study of tetrapeptide aldehyde inhibitors. Bio-org Med Chem Lett. 2006;16:40–43. doi: 10.1016/j.bmcl.2005.09.049. [DOI] [PubMed] [Google Scholar]
- Yip KW, Ito E, Mao X, Au PY, Hedley DW, Mocanu JD, Bastianutto C, Schimmer A, Liu FF. Potential use of alexidine dihydrochloride as an apoptosis-promoting anticancer agent. Mol Cancer Ther. 2006;5:2234–2240. doi: 10.1158/1535-7163.MCT-06-0134. [DOI] [PubMed] [Google Scholar]
- Yusof R, Clum S, Wetzel M, Murthy HM, Padmanabhan R. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J Biol Chem. 2000;275:9963–9969. doi: 10.1074/jbc.275.14.9963. [DOI] [PubMed] [Google Scholar]