Abstract
The oomycete genus Phytophthora includes many of the world's most destructive plant pathogens, which are generally disseminated by asexual sporangia. To identify factors relevant to the biology of these propagules, genes induced in sporangia of the potato late blight pathogen Phytophthora infestans were isolated using cDNA macroarrays. Of ∼1,900 genes known to be expressed in sporangia, 61 were up-regulated >5-fold in sporangia versus hyphae based on the arrays, including 17 that were induced >100-fold. A subset were also activated by starvation and in a nonsporulating mutant. mRNAs of some genes declined in abundance after germination, while others persisted through the germinated zoospore cyst stage. Functions were predicted for about three-quarters of the genes, including potential regulators (protein kinases and phosphatases, transcription factors, and G-protein subunits), transporters, and metabolic enzymes. Predominant among the last were several dehydrogenases, especially a highly expressed sorbitol dehydrogenase that accounted for 3% of the mRNA. Sorbitol dehydrogenase activity also rose during sporulation and several stress treatments, paralleling the expression of the gene. Another interesting metabolic enzyme resembled creatine kinases, which previously were reported only in animals and trypanosomes. These results provide insight into the transcriptional and cellular processes occurring in sporangia and identify potential targets for crop protection strategies.
Sporulation is central to the life cycles of most plant-pathogenic lower eukaryotes, including true fungi and oomycetes. The latter group encompasses important pathogens, such as Phytophthora, Pythium, and the downy mildews, which infect many economically significant hosts (14). The molecular biology of sporulation has been characterized in detail for several true fungi (1, 11, 28, 43). However, little information exists on oomycetes, which despite their “fungus-like” appearance are more closely related to organisms such as diatoms and brown algae (3).
Asexual sporangia play the major role in spreading Phytophthora infestans, which causes late blight of potato and tomato (17). The roles of nutrients, pH, aeration, light, and humidity in sporulation are well described (33, 41). In older portions of plant lesions and in laboratory cultures, sporangia form upon aerial sporangiophores branched from hyphae. In some members of the genus, such as P. infestans, this occurs spontaneously as cultures age, but in others significant sporulation requires washing nutrients from media (41). In both cases, nuclear divisions rapidly occur within sporangiophores, and then nuclei and cytoplasm quickly flow into terminal swellings, which develop into sporangia (7, 32). These mature by forming a basal plug, which delimits the multinucleate sporangium from the coenocytic hypha, and an apical papilla, which is a future site of germination.
Two modes of germination are possible in most oomycetes, including P. infestans. Indirect germination by way of zoospores predominates at cool temperatures, typically below 12°C, while direct germination is favored at higher temperatures (41). Indirect germination is considered most critical for plant infection. This involves the cleavage of sporangial cytoplasm into six or more zoospores, which swim, encyst, form germ tubes, and develop appressoria on plants or hydrophobic surfaces (20). These are rapid steps, with cytoplasmic cleavage typically visible 30 min after cold induction, zoospore liberation 15 min later, and zoospore encystment occurring seconds after chemical or mechanical stimulation. The factors required for germination and encystment are thought to be preformed, since these stages are not blocked by actinomycin D or cycloheximide (9, 36).
Some themes in sporulation appear to be conserved between oomycetes and true fungi, including the role of starvation as an inducer, the necessity of an aerial environment, a period required for sporulation competence, and the reliance of germination upon preformed RNA and DNA (11, 31, 42). However, most morphological, genetic, and biochemical features are very different. In true fungi, spores (conidia) usually form upon differentiated stalk cells separated from hyphae (sterigmata), while oomycete sporangiophores are contiguous with vegetative hyphae and similar in structure (21). In addition, unlike the desiccated conidia of true fungi, oomycete sporangia remain metabolically and transcriptionally active and are consequently short-lived (35). Acyclic polyols are generally abundant in fungal conidia but not in oomycete sporangia (27, 38). Oomycete sporangia are also distinct in their two modes of germination, although a few true fungi produce zoospores (31, 44).
Studies of sporulation in oomycetes have lagged behind those of true fungi, in part due to the uniqueness and challenges of applying genetic and molecular tools to oomycetes. A few loci that are up-regulated in sporangia have been identified (26, 34), but no comprehensive search for such genes has been reported. Nevertheless, transcripts present in sporangia but not vegetative hyphae of an oomycete such as P. infestans would be predicted to fall into several categories. One class would participate in both early and late stages of sporangium formation, since cytoplasm migrates rapidly into sporangia from sporangiophores and no intervening cell types analogous to sterigmata exist. A second class would help maintain the viability of sporangia and detect signals for germination. Other classes of transcripts would function in forming zoospores and enabling their behaviors, such as chemotaxis and encystment. To identify genes that transact these functions, we performed a cDNA macroarray screen for mRNAs present in sporangia but not hyphae. This resulted in the identification of 61 up-regulated genes and the characterization of their expression during different stages of growth and in a sporulation-defective mutant. One of the most common types of sporulation-induced genes, a putative sorbitol dehydrogenase, was examined in detail.
MATERIALS AND METHODS
Growth of P. infestans.
Developmental stages of isolate 88069 were prepared as described previously (24). Briefly, this involved nonsporulating hyphae from 5-day-old liquid rye broth cultures; sporulating hyphae or purified sporangia from 8- to 18-day-old rye agar cultures; zoospores released from sporangia after incubation in 10°C water for 90 min; germinated zoospore cysts, prepared by vortexing zoospores for 1 min followed by 6 h of incubation in rye broth at 18°C; and hyphae from liquid cultures of defined media (52) either lacking glucose but retaining fumaric acid (carbon starvation) or with 1/10 the normal amount of ammonium sulfate (nitrogen starvation).
Also used was the P. infestans transformant AF193, which shows a 99% reduction in sporangiophore and sporangium formation. It was generated by transformation of a homology-dependent silencing construct of the PiCdc14 gene and will be described separately (2).
Enzyme assays.
A Polytron homogenizer (Brinkmann, Westbury, N.Y.) was used at speed 7 for 30 s to make extracts, which were clarified at 10 × g for 1 min and assayed at 25°C for alcohol dehydrogenase activity as described previously (50). This involved measuring changes in absorbance at 340 nm due to the reduction of NAD in the presence of 1 M ethanol, xylitol, sorbitol, or glycerol. One unit is defined as the enzyme forming 1 μmol of NADH per min. A Coomassie dye-binding method was used to determine protein concentrations.
cDNA library construction.
RNA extracted as described previously (24) from sporangia from rye agar cultures was used to construct a directional cDNA library in the NotI and SalI sites of pSPORT1 (Invitrogen, Carlsbad, Calif.). Plasmids were electrotransformed into Escherichia coli DH5α, and the transformants were stored in 384-well plates.
RNA, DNA, and array analyses.
cDNA inserts were amplified by two-step PCR using SPORT-L (5′-ACGTCGCATGCACGCGTACGTAAGC) and SPORT-R (5′-ACGACTCACTATAGGGAAAGCTGGTACG), a denaturation temperature of 94°C (30 s), an annealing-extension temperature of 72°C (60 s), and a 35-cycle reaction. A fraction of the reaction products were checked on a gel for purity and concentration. The remainder were spotted on nylon membranes using a slotted 96-pin tool delivering 0.5 μl per pin (V and P Scientific, San Diego, Calif.). Also spotted were cDNAs from actin (actA) and elongation factor 1α (EF1) plus controls for nonspecific hybridization. Membranes were treated with 0.4 M NaOH for 10 min and with 3× SSPE (540 mM NaCl, 30 mM NaHPO4, 3 mM EDTA, pH 7.4) for 10 min, UV cross-linked to 1,200 μJ/cm2, boiled for 1 min in 0.1% sodium dodecyl sulfate, and then hybridized with 32P-labeled cDNA as described previously (15). Primary screens for genes up-regulated in sporangia were performed using replicate filters, with each clone represented once. After removal of the probe by heating the membrane to 95°C, reciprocal hybridizations were performed to confirm the expression patterns. Secondary filters which included two replicates of each clone were then prepared and hybridized in the same manner. To control for the quality of each filter and the amount of DNA per spot, the filters were probed with 32P-labeled SPORT-L, using a hybridization temperature of 45°C and washing conditions of 6× SSPE-0.1% sodium dodecyl sulfate at 40°C.
Blot analysis of RNA and DNA electrophoresed on agarose gels was performed as described previously (24). For RNA blots, EF1 and 18S ribosomal DNA probes were used as loading controls.
Hybridization signals were captured by phosphorimager analysis and analyzed using Quantity One version 4.1 software for Macintosh (Bio-Rad, Richmond, Calif.). For arrays, several exposure times were used to ensure that all signals were captured within the linear range of the phosphorimager, and then circular zones were matched to each spot for quantitation. After correction for background, the data were filtered based on the signal-to-noise ratio (a minimum of 3.0 was deemed acceptable to assume expression), normalized to EF1 or 18S rRNA, and analyzed for significant differences by Student's t test. Hierarchical clustering based on average distance (unweighted pair group method with averaging) was performed using EPCLUST (European Bioinformatics Institute [http://ep.ebi.ac.uk/EP/EPCLUST]).
Sequence analysis.
Data from automated DNA sequencing were trimmed of vectors and assembled using Seqman software for Macintosh (DNAStar, Madison, Wis.). GenBank searches were performed using a local implementation of BLASTX. Alignments of protein sequences were performed using a version of ClustalW, as described below.
Nucleotide sequence accession numbers.
The DNA sequences have been deposited in GenBank under accession numbers CF106689 to CF106749.
RESULTS
Identification of genes up-regulated in asexual sporangia.
Sixty-one unique genes expressed at higher levels in sporangia than in nonsporulating hyphae were identified by hybridizing differential probes to cDNA macroarrays. The arrays were made from 5,184 clones from a sporangial cDNA library, some of which had also been used in an expressed sequence tag (EST) project (S. T. Lam, abstract from the Phytophthora genomics consortium, Phytopathology 91:S158, 2001). Sequence assemblies suggested that these clones represented 1,927 genes.
In a primary screen, 329 of the 5,184 clones appeared to display significant up-regulation in sporangia versus hyphae. These candidates were then subjected to a secondary screen using new filters containing replicate spots; representative portions of the arrays are shown in Fig. 1A. To identify and eliminate culture-to-culture variation, the primary and secondary screens employed cDNA probes from independent cultures of P. infestans. After redundant clones revealed by cross-hybridization and comparisons of 5′ and 3′ sequence data were removed, 61 unique up-regulated genes were identified that fit the following criteria: ≥5-fold induction in sporangia versus hyphae as calculated on the final filters, consistent up-regulation in each round of screening (±30%), signal-to-noise ratios of >5.0 in sporangia, and induction that was significant (P = 0.05) based on t test analysis. The genes were named pisp genes (for P. infestans sporangia) (Table 1 and Fig. 2).
FIG.1.
Identifying differentially expressed genes of P. infestans using cDNA macroarrays. (A) Representative portions of macroarrays from the secondary screen. Filters containing two spots of each cDNA on the diagonal were prepared and hybridized with 32P-cDNA from sporangia (spore; filter 1) or nonsporulating hyphae (filter 2). The filters were then stripped and rehybridized with radiolabeled SPORT-L (oligo). Not all spots in this illustration represent up-regulated genes, including the lower right-hand spot pair (EF1) and the lower left-hand pair (a control for nonspecific hybridization) in each portion. (B) RNA blot analysis of representative pisp genes. RNAs from nonsporulating hyphae (NSH) and purified sporangia (SP) were hybridized with probes for the indicated genes. (C) Relative expression of EF1 and 18S RNA in hyphae and sporangia of P. infestans. RNAs from nonsporulating hyphae (5-day-old cultures [NSH]) and from sporangia from cultures 8, 13, and 18 days after inoculation (SP8, SP13, and SP18, respectively) were electrophoresed in the presence of ethidium bromide (EtBr), blotted, and hybridized with probes for EF1 and 18S RNA. Shown at the base of the blot are the ratios of EF1 to 18S, relative to nonsporulating hyphae, as calculated by phosphorimager analysis.
TABLE 1.
P. infestans genes upregulated >5-fold in asexual sporangia
| Gene | Sporangia vs hypha induction ratioa | Relative mRNA abundanceb | Best BLASTX matchc | Cellular functiond | BLAST E |
|---|---|---|---|---|---|
| pisp1 | >100 | 4.40 | No hit | Unknown | |
| pisp2 | >100 | 3.25 | Hypothetical protein (C. elegans CAA98279) | Unknown | 2e-10 |
| pisp3 | >100 | 2.80 | Sorbitol dehydrogenase (N. crassa EAA34038) | Metabolism | 1e-99 |
| pisp4 | >100 | 1.82 | Protein phosphatase 2C (C. elegans NP505702) | Signal transduction | 7e-42 |
| pisp5 | >100 | 0.79 | No hit | Unknown | |
| pisp6 | >100 | 0.49 | No hit | Unknown | |
| pisp7 | >100 | 0.46 | Hypothetical protein (C. elegans T23512) | Unknown | 2e-12 |
| pisp8 | >100 | 0.32 | Sorbitol dehydrogenase (N. crassa EAA34038) | Metabolism | 5e-65 |
| pisp9 | >100 | 0.29 | Sorbitol dehydrogenase (Neurospora crassa EAA34038) | Metabolism | 1e-77 |
| pisp10 | >100 | 0.22 | Hypothetical protein (A. thaliana AAL59988) | Unknown | 6e-08 |
| pisp11 | >100 | 0.15 | Glucose-inhibited protein A (Fusobacterium nucleatum EAA23857) | Unknown | 5e-18 |
| pisp12 | >100 | 0.14 | Cdc14 cell-cycle phosphatase (Homo sapiens NP201569) | Cell cycle | 2e-84 |
| pisp13 | >100 | 0.05 | Hypothetical protein (A. thaliana AF436836) | Unknown | 4e-28 |
| pisp14 | >100 | 0.03 | Nuclear LIM domain interactor (Dictyostelium discoideum AF111941) | Transcription | 2e-39 |
| pisp15 | >100 | 0.02 | Mucin-like protein (Heterodera glycines AF092449) | Cell organization | 4e-19 |
| pisp16 | >100 | 0.02 | Endo-1,4-beta-glucanase (Pyrococcus horikoshii X62582) | Metabolism | 2e-15 |
| pisp17 | >100 | 2.61 | NADH-ubiquinone oxidoreductase (Bos taurus Q02366) | Metabolism | 7e-12 |
| pisp18 | 87 ± 21 | 0.03 | Serine/threonine protein kinase (H. sapiens Q9HBY8) | Signal transduction | 1e-56 |
| pisp19 | 83 ± 11 | 2.35 | Hypothetical protein (H. sapiens AAN37911) | Unknown | 7e-61 |
| pisp20 | 78 ± 13 | 0.05 | Leucine aminopeptidase (Vibrio cholerae B82414) | Protein fate | 2e-17 |
| pisp21 | 74 ± 1 | 0.36 | No hit | Unknown | |
| pisp22 | 68 ± 8 | 0.60 | Aldehyde dehydrogenase (Zea mays AF467541) | Metabolism | 1e-123 |
| pisp23 | 61 ± 11 | 0.07 | Suppressor of actin mutations (H. sapiens AAH16559) | Cell organization | 5e-46 |
| pisp24 | 59 ± 19 | 0.05 | No hit | Unknown | |
| pisp25 | 58 ± 6 | 0.24 | Hypothetical protein (Mus musculus XP236938) | Unknown | 1e-06 |
| pisp26 | 37 ± 4 | 0.13 | Protein phosphatase 2B (Paramecium tetraurelia AAB80918) | Signal transduction | 3e-09 |
| pisp27 | 36 ± 3 | 0.02 | Hypothetical protein (A. thaliana NM101737) | Unknown | 2e-09 |
| pisp28 | 36 ± 2 | 0.03 | Diphosphonucleotide phosphatase (Oryzae japonica BAB86413) | Metabolism | 2e-27 |
| pisp29 | 35 ± 2 | 0.14 | Cathepsin D (M. musculus AF197479) | Protein fate | 1e-8 |
| pisp30 | 34 ± 9 | 0.01 | Homocysteine S-methyltransferase (P. putida NP744920) | Metabolism | 2e-31 |
| pisp31 | 32 ± 10 | 0.02 | No hit | Unknown | |
| pisp32 | 30 ± 8 | 0.03 | Complement receptor (Papio hamadryas AAA62170) | Unknown | 2e-07 |
| pisp33 | 28 ± 4 | 0.06 | Suppressor of bimD6 (H. sapiens NP0038220 | Cell cycle | 3e-34 |
| pisp34 | 26 ± 3 | 0.18 | PEP carboxykinase (Leptospira interrogans AAN47450) | Metabolism | e-138 |
| pisp35 | 25 ± 4 | 0.50 | Elongation factor 3A (S. cerevisiae S65245) | Protein synthesis | 3-178 |
| pisp36 | 22 ± 2 | 0.01 | Hypothetical protein (D. discoideum AAM34324) | Unknown | 5e-15 |
| pisp37 | 19 ± 2 | 0.02 | Origin recognition complex subunit (Schizosaccharomyces pombe U92539) | Cell cycle | 7e-08 |
| pisp38 | 17 ± 1 | 0.01 | Glycoprotein-N-acetylgalactosamine galactosyltransferase (C. elegans AAG36940) | Protein fate | 4e-27 |
| pisp39 | 14 ± 1 | 0.01 | No hit | Unknown | |
| pisp40 | 13 ± 3 | 0.01 | Sexually induced protein 3 (Thalassiosira weissflogii AF154501) | Unknown | 2e-36 |
| pisp41 | 12 ± 2 | 0.01 | Creatine kinase (Ictalurus punctatus AAO25756) | Energy | 2e-94 |
| pisp42 | 11 ± 1 | 0.65 | Glutathione S-transferase (Xenopus laevis AAM82563) | Defense | 4e-28 |
| pisp43 | 11 ± 0 | 0.04 | G protein beta subunit (P. infestans AY050538) | Signal transduction | 2e-77 |
| pisp44 | 10 ± 2 | 0.02 | Growth arrest-specific 11 (H. sapiens AAC69519) | Unknown | 2e-31 |
| pisp45 | 10 ± 0 | 0.02 | Calcium/calmodulin-dependent protein kinase (X. laevis BAC19848) | Signal transduction | 2e-72 |
| pisp46 | 10 ± 2 | 0.04 | Calcium-transporting ATPase (A. thaliana P92939) | Transport | 0 |
| pisp47 | 10 ± 1 | 0.01 | Methionine aminopeptidase (H. sapiens BAA07679) | Protein fate | 7e-26 |
| pisp48 | 9 ± 1 | 0.08 | Mitochodrial succinate-fumarate carrier (A. thaliana NP195754) | Energy | 2e-68 |
| pisp49 | 9 ± 2 | 0.01 | ABC transporter (N. crassa EAA36439) | Transport | 3e-33 |
| pisp50 | 9 ± 0 | 0.04 | Nodulin MtN3 family protein (A. thaliana BAA97235) | Unknown | 8e-21 |
| pisp51 | 9 ± 2 | 0.02 | Vacuolar proton-translocating ATPase (D. discoideum AAO51473) | Transport | 7e-84 |
| pisp52 | 8 ± 2 | 0.02 | Na+ K+ ATPase alpha subunit (Callinectes sapidus AF327439) | Transport | 1e-33 |
| pisp53 | 8 ± 2 | 0.03 | Hypothetical protein (A. thaliana AAM62828) | Unknown | 2e-12 |
| pisp54 | 8 ± 1 | 0.01 | G protein alpha subunit (P. infestans AY050536) | Signal transduction | 2e-91 |
| pisp55 | 7 ± 0 | 0.02 | Phosphoribosylglycinamide synthetase (S. cerevisiae X04337) | Metabolism | 2e-42 |
| pisp56 | 7 ± 1 | 0.02 | Mitochondrial ribosomal protein S13 (P. infestans U17009) | Protein synthesis | 2e-66 |
| pisp57 | 6 ± 1 | 0.14 | Adenosine kinase (Anopheles gambiae EAA02798) | Metabolism | 6e-44 |
| pisp58 | 6 ± 0 | 0.01 | Hypothetical protein (Oryza sativa BAA83585) | Unknown | 3e-40 |
| pisp59 | 6 ± 1 | 0.01 | No hit | Unknown | |
| pisp60 | 5 ± 1 | 0.01 | Ribosomal protein S9 (A. thaliana NP198801) | Protein synthesis | 4e-69 |
| pisp61 | 5 ± 1 | 0.01 | Actin-depolymerizing factor (A. thaliana NP194289) | Cell organization | 5e-19 |
Hybridization signal with sporangia-cDNA probe divided by hyphal signal, normalized by a two-step procedure to 18S rRNA. This involved normalization to internal EF1 controls, followed by correction for differences in 18S and EF1 signals as determined from Northern blot analysis of RNAs from three independent preparations of hyphae and sporangia. Accumulated error (Ea) in the induction ratio (R) was calculated from error (e) in measurements from hyphal (h) and sporangial (s) cDNA hybridizations as follows: Ea = R × ✓([eh/h]2 + [es/s]2).
Compared to EF1 mRNA.
Only matches with an E of <10−5 are shown.
Using functional categories from Munich Information Center for Protein Studies (http://mips.gsf.de/proj/yeast/catalogues/funcat/).
FIG.2.
Abundance of pisp mRNA based on macroarray data. Arrays were hybridized with radiolabeled cDNAs from nonsporulating hyphae, sporangia (SPOR), germinated cysts (GCY), a nonsporulating mutant (MUT), and nitrogen and carbon-limited cultures of hyphae (N-ST and C-ST). Each column shows the ratio of mRNA abundance in the indicated tissue relative to hyphal grown on rich rye medium, normalized to 18S rRNA and scaled as shown at the base of the figure. Black represents no change, and the maximum ratio shown is 20-fold induction. The 61 pisp genes (right) are ranked in decreasing order of their expression in sporangia versus germinated cysts. At the top of the figure is a tree based on hierarchical clustering that portrays the similarity of the results from each probe set.
Of the 61 pisp genes, 47 showed >10-fold induction in sporangia based on the arrays, and 17 were induced >100-fold. The induction ratios of the latter are recorded in Table 1 as >100-fold, since accurate ratios usually could not be determined because the hyphal probe yielded a signal near background. RNA blot analysis was performed for nine representative genes, which yielded data in good agreement with the array results (Fig. 1B). These blots employed preparations of RNA separate from those used with the arrays, thus serving as a third biological replicate of the data.
Several strategies for normalizing the array data were tested, resulting in the choice of standardization to 18S rRNA. Previous RNA blot analyses from our laboratory indicated that traditional standards, such as actin and EF1, varied relative to rRNA in sporangia versus hyphae and thus would be inappropriate for normalization (10). Levels of EF1 were lower in sporangia than in hyphae, for example (Fig. 1C). There was also concern about normalizing to the average of all spots, even after excluding outliers, since the array was enriched for genes induced in sporangia. Consequently, a two-step approach that normalized the level of each mRNA to EF1 was employed, which was then corrected for the 18S rRNA/EF1 ratio in each RNA sample used for probe generation. For example, the EF1 level in RNA from sporangia of the 13-day-old culture used as a probe in the secondary screen was 0.53 times that of EF1 in hyphal RNA (Fig. 1C). Other preparations of hyphal and sporangial RNAs yielded similar ratios (0.48 to 0.55).
The abundance of pisp mRNAs ranged from 1 to 440% of EF1, with a median of 4% (Table 1). This was determined by comparing the hybridization signals from the sporangial cDNA probe with that of 32P-labeled SPORT-L, which was a primer used to amplify each cDNA insert. This also confirmed that the replicated spots usually contained equal amounts of DNA (±5%); in the few exceptions, the SPORT-L signal was used to make the appropriate correction. Based on the frequency of EF1 in a public database of hyphal ESTs (25), and correcting for the reduction of EF1 mRNA in sporangia versus hyphae, the 61 pisp genes accounted for 14% of the mRNA in sporangia. This represents a transcript population with complexity similar to that of conidia of the downy mildew pathogen Bremia lactucae, another oomycete. In B. lactucae, the 39 most highly expressed genes comprised 20% of the mRNA (23) versus 13% for the top 39 from P. infestans.
Gene functions predicted by sequence comparisons.
BLASTX analysis revealed matches against GenBank for 53 of the 61 genes, using an E value threshold of 10−5 (Table 1). This utilized an average of 937 nucleotides (nt) of sequence data per clone, which was obtained by 5′ and 3′ sequencing and primer walking. This relates to a typical full-length P. infestans mRNA of ∼1,200 nt, as predicted from the public EST database for Phytophthora (25).
Classification of the matches revealed hits against proteins involved in a range of functions, including cellular regulation (protein kinases and phosphatases, and transcriptional activators), transport (ion pumps and ABC transporters), and metabolism. The most common group of genes included those for several distinct NADH-dependent ketose reductases (pisp3, -8, -9, and -17), which in BLASTX analysis typically matched proteins called xylitol or sorbitol dehydrogenases. One of the latter (pisp3) was expressed at particularly high levels, accounting for >300 positives on the primary arrays, and is discussed in detail below.
Three genes from Phytophthora had previously been identified. These were α and β G-protein subunits (25) and a cdc14 cell cycle phosphatase (2). The mucin-like gene (pisp15) was distinct from the mucin-like cDNA previously identified in germinated cysts of P. infestans (19).
Expression of genes under other conditions.
To better address the roles of the pisp genes, the arrays were used to measure mRNA levels in germinated zoospore cysts, hyphae undergoing nutrient-limited growth in defined media, and a nonsporulating mutant generated by introducing a homology-dependent silencing construct containing the cdc14 gene of P. infestans (2). In yeasts and metazoans, Cdc14 regulates cytokinesis and the exit from mitosis (51), and we have proposed that the P. infestans orthologue regulates nuclear behavior at an early stage of sporulation (2).
One interesting point of differentiation among pisp genes was their relative expression in ungerminated sporangia versus germinated zoospore cysts (germlings). This criterion was used to order the genes in Fig. 2, where those at the top have the highest ratio of mRNA in sporangia versus germinated cysts, and those at the base have the lowest. Most genes were expressed at higher levels in sporangia than in germlings (i.e., pisp6 and -30), but others were at similar levels (i.e., pisp1 and -2) or higher in germlings (i.e., pisp55 and -56).
Many pisp genes were induced in nonsporulating hyphae grown on media containing limiting amounts of nitrogen or carbon (Fig. 2), although the abundances of their mRNAs were usually lower in starved hyphae than in sporangia. Induction of some pisp genes was expected, since starvation is considered a trigger of sporulation in P. infestans and since some degree of starvation may be experienced by the sporangia once they become delimited from hyphae. Some genes were induced in both nitrogen- and carbon-limited cultures (i.e., pic2 and -3), while others were induced by only one of the treatments (i.e., pic14 and -44). Differences in genetic response to nitrogen and carbon starvation have also been observed in other species (18). However, while it was interesting to check for overlaps between sporulation- and starvation-induced genes, excessive conclusions about the specific responses of P. infestans to the carbon- and nitrogen-limited media should not be drawn from this study. A proper analysis of starvation responses should include additional nutrient manipulations and a less biased set of genes.
The nonsporulating mutant (Fig. 2) provided further information about the staging of gene expression during spore development. This strain was grown under conditions that matched those of the wild-type cultures used to harvest sporangia, except that the mutant produced few if any sporangia or sporangiophores. Despite this sporulation defect, 16 of the pisp genes still were induced >5-fold.
Sorbitol dehydrogenase-like proteins are the most highly expressed.
Of several putative dehydrogenases induced during sporulation, the most abundant was pisp3; its RNA rose in abundance >100-fold upon the formation of sporangia and was 2.8 times more plentiful than that of EF1. In BLASTX analyses, pisp3 best matched a known xylitol dehydrogenase from Candida albicans and a putative sorbitol dehydrogenase from Neurospora crassa (both E values equaled 10−99). These are members of a family of dimeric, zinc-containing NAD-dependent ketose reductases (EC 1.1.1.14) that are typically ∼360 amino acids in size. Such enzymes generally have broad substrate specificity, especially against sugar alcohols. Hence, a protein called a xylitol dehydrogenase is also a sorbitol, mannitol, or glycerol dehydrogenase (29). For simplicity, they will henceforth be described as sorbitol dehydrogenases.
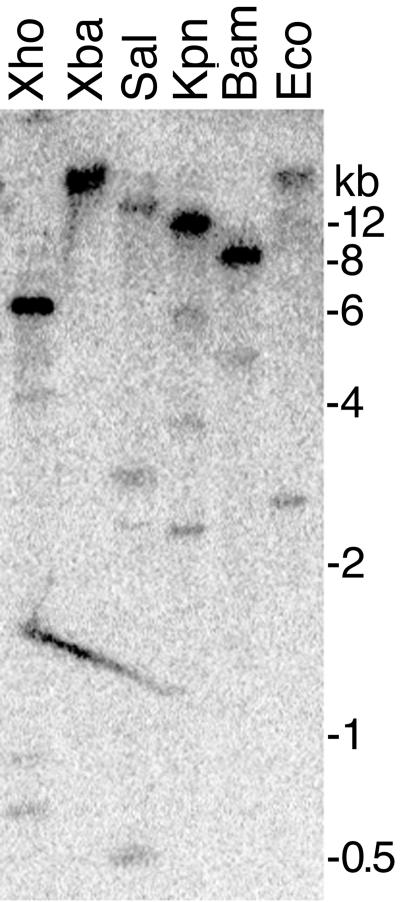
In consideration of their high frequency of detection, the sorbitol dehydrogenases from P. infestans, and in particular pisp3, were examined in more detail. Low-stringency hybridizations of a full-length cDNA clone of pisp3 against genomic DNA digested with BamHI, EcoRI, KpnI, SalI, XbaI, and XhoI (Fig. 3) revealed that it was a member of a small family, since three or four bands were detected in each digest; pisp3 does not contain sites for any of these enzymes. Sequence comparisons of pisp3 and the other sorbitol dehydrogenases (pisp8, -9, and -17) indicated that the former lacked significant similarity to the others (<50% DNA identity), and consequently, the others did not account for the weak bands seen in Fig. 3. pisp8 and pisp9 appeared to be members of the same family, however.
FIG. 3.
DNA blot analysis of pisp3. Genomic DNA of P. infestans was digested with XhoI, XbaI, SalI, KpnI, BamHI, or EcoRI and hybridized with a full-length pisp3 cDNA. Washes were performed at low stringency (melting temperature, −20°C). Size markers are shown on the right.
The weak bands related to pisp3 appeared to represent pseudogenes. Sequencing of >300 pisp3-related cDNA clones from the sporangium library suggested that each cDNA was derived from a single highly expressed locus, since only three sequence differences were detected within the 1,167-nt full-length cDNA. All polymorphisms were C-to-T changes, which probably represent differences between alleles of a single-copy gene. Nevertheless, it is possible that the weak bands represent loci transcribed at an untested developmental stage.
Structure and expression of pisp3.
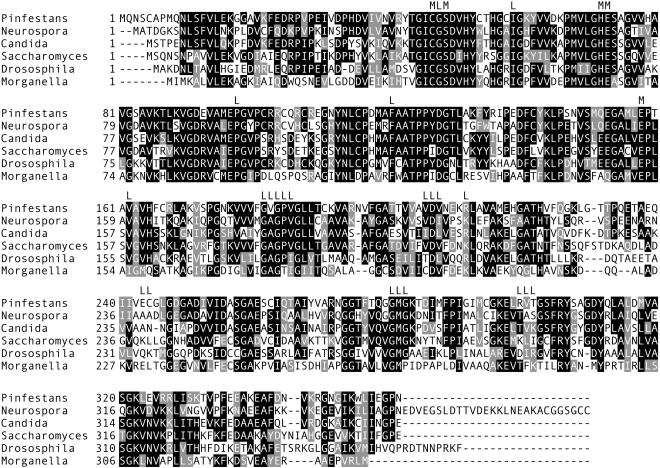
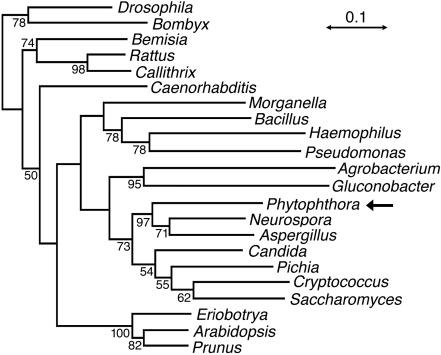
A comparison of the pisp3 cDNA to genomic clones showed that the 1,178 transcription units contained 45- and 46-nt 5′ and 3′ untranslated regions, a 1,167-nt open reading frame, and no introns. The predicted 389-amino-acid gene product aligned closely with other members of the sorbitol dehydrogenase family (Fig. 4). These included residues shown by X-ray crystallography to contact the NAD and zinc cofactors (L and M, respectively [Fig. 4]) (12). A phylogenetic analysis of the P. infestans protein and orthologues from animals, plants, true fungi, and bacteria revealed the highest affinity to proteins from true fungi (Fig. 5). This was surprising, since previous analyses of Phytophthora genes usually revealed better affinity to plant genes, although there were exceptions (49).
FIG. 4.
Alignment of sorbitol dehydrogenase proteins. Shown are the P. infestans protein (from pisp3) and proteins from N. crassa (GenBank accession no. NCU00891), C. albicans (AAC24597), S. cerevisiae (NP0100035), Drosophila melanogaster (AAD00902), and the bacterium Morganella morganii (AAA25324.1). Indicated at the top of the alignment are regions in contact with metal ions (M) or the NAD ligand (L). The solid squares represent blocks of identity, and the shaded squares represent blocks of similar amino acids.
FIG. 5.
Neighbor-joining tree of NAD-dependent sorbitol dehydrogenases. An alignment was developed using a version of ClustalW as implemented in the VectorNTI Suite for Macintosh (Informax, Bethesda, Md.), which was exported to Phylip version 3.57c for Macintosh (16). Bootstrap replicates were generated using SEQBOOT, distances were determined using the PAM option of PROTDIST, neighbor-joining trees were developed using NEIGHBOR, and a consensus tree was made with CONSENSE. The numbers at the nodes indicate the percentages of occurrence in 500 replicates, and the scale represents 0.1 PAM units. In addition to the proteins shown in Fig. 3, others were from Bombyx mori (BAA02634), Bemisia argentifolii (AAD02817), Rattus norvegicus (S38363), Callithrix sp. (AAB69288), Caenorhabditis elegans (CAA94841), Bacillus subtilis (AAA22508), Haemophilus influenzae (Q57517), Pseudomonas putida (AAB58982), Agrobacterium tumefaciens (NP357437), Gluconobacter suboxydans (AAE66066), Aspergillus fumigatis (The Institute for Genomic Research contig 4937), Pichia stipitis (P22144), Cryptococcus neoformans (from the Stanford Genome Technology Center C. neoformans Genome Project, contig cneo011005.C1135, and The Institute for Genomic Research C. neoformans EST k4g08j2.r1), and the plants Eriobotrya japonica (BAA95897), Arabidopsis thaliana (BAB11045), and Prunus persica (BAA94084).
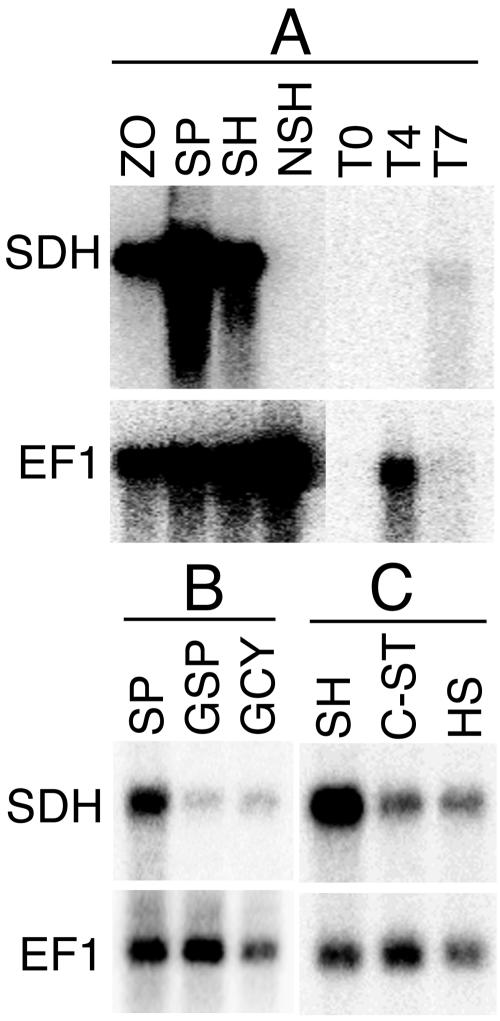
RNA blot analysis confirmed that pisp3 mRNA was absent from nonsporulating hyphae (Fig. 6A) but was expressed in sporulating hyphae, purified sporangia, and zoospores. Greater-than-10-fold reductions in mRNA abundance occurred once hyphae began to reform, after 16 h of direct germination of sporangia (Fig. 6B) or 6 h of germination of zoospore cysts.
FIG. 6.
RNA blot analysis of pisp3. Total RNA was electrophoresed and hybridized with a probe for pisp3 or EF1. (A) RNAs from zoospores (ZO), sporangia (SP), sporulating 10-day-old hyphae (SH), nonsporulating hyphae from a 5-day broth culture (NSH), uninfected tomato leaflets (T0), and tomato 4 or 7 days after inoculation with P. infestans zoospores (T4 and T7). (B) RNAs from sporangia (SP), directly germinated sporangia (GSP), and germinated cysts from zoospores (GCY). (C) RNAs from sporulating hyphae (SH), carbon-starved hyphae (C-ST), and heat-stressed hyphae (HS).
As predicted by the arrays, the gene was also induced in nonsporulating cultures by starvation (Fig. 6C). Growing P. infestans at 26°C, which appears to cause heat stress, since growth is reduced 30% relative to that at 21°C, also induced the gene (Fig. 6B). While the starvation conditions eventually led to sporulation 3 days after tissue was harvested for RNA extraction, the heat-stressed cultures did not sporulate.
The pattern of expression in planta also indicated that pisp3 expression was associated with sporulation (Fig. 6A). No pisp3 RNA was detected in tomato leaflets 4 days after inoculation with zoospores, at which time all P. infestans organisms were within the plant and sporulation had not yet occurred. EF1 expression was easily observed in the 4-day sample, indicating that substantial pathogen RNA was present. At 7 days, when the leaflets showed profuse sporulation, pisp3 mRNA was clearly detected (despite the underloading of RNA in the sample).
Sorbitol dehydrogenase activity parallels pisp3 expression.
The enzymatic activities of sorbitol dehydrogenases during the growth and development of P. infestans paralleled the abundance of pisp3 mRNA and that of its relatives. In nonsporulating hyphae, very little activity was detected (Fig. 7). Activity rose greatly in sporulating hyphae and in purified sporangia, peaking at levels between 3 and 5 U per pg of protein. Enzyme levels also rose in response to nutrient stress. This included growth in defined minimal medium, in which growth is reduced one-third compared to rye medium, and the defined media in which nitrogen and carbon sources were reduced to levels that retarded growth by an extra one-third relative to that in rye medium. Minimal medium appeared sufficient to induce the gene to near-maximal levels.
FIG. 7.
Sorbitol dehydrogenase activity in P. infestans. Assays were performed using 1 M sorbitol and tissue extracts from nonsporulating (nonspor.) hyphae, sporulating (spor.) hyphae, purified sporangia, defined media, and defined media with reduced levels of nitrogen (nitrogen starved) and carbon (carbon starved). Activity is expressed as the number of micromoles of sorbitol oxidized per minute per picogram of protein. The data are averages (plus standard deviations) of assays performed on at least two replicate tissue samples made on different days.
The enzyme assays also showed that the predominant activity in extracts of sporangia exhibited the same biochemical features described for other members of the sorbitol dehydrogenase family (29). This included optimal activity at alkaline pH (pH 9 to 10), a preference for NAD as opposed to NADP as a cofactor, and broad substrate specificity. The induced enzyme displayed similar activities when sorbitol, xylitol, sorbitol, or glycerol was used as a substrate.
DISCUSSION
This study presents the first large-scale screen for genes induced in the asexual sporangia of oomycetes, which is a critical phase in their life and disease cycles. Of the estimated 1,927 genes examined, 3% displayed >5-fold increases in mRNA abundance in sporangia, accounting for 14% of the mRNA, and many appeared to be specific for sporangia. This can be compared to studies of sporogenesis in true fungi, even though the processes are not biologically equivalent. In Saccharomyces cerevisiae, 7.7% of all genes were up-regulated during sporulation, with an average of 1.5% induced >5-fold at any given time (8). In N. crassa, 3% of genes were induced by blue light, which stimulates conidiation (28). The proportion of P. infestans genes that change in expression is certainly higher than the 3% value reported in this study, however. Our focus was on genes showing substantial increases, not decreases, in mRNA abundance in sporangia compared to that in hyphae, even though subtle increases or reductions can also be biologically relevant. Genes declining in mRNA abundance were uncommon in the data; however, these would have been underrepresented in the arrays due to the use of cDNAs from sporangia.
The nature of sporangial development in P. infestans facilitated some aspects of our experiments but may have limited the genes identified. P. infestans, unlike many members of the genus, produces abundant sporangia in culture without requiring starvation treatments. Its sporangia are also decidious, i.e., detachable from sporangiophores. This aided our comparisons of hyphae and sporangia, but genes transiently induced at the onset of sporulation may not have been detected if their mRNAs failed to persist in sporangia. P. infestans does not represent a good system for synchronously inducing sporulation, at least in laboratory culture (32). Better synchrony is observed in species like Phytophthora cinnamomi, in which starvation induces sporangia, although they are not deciduous (34). There may be value in studying the expression of the pisp genes in such species.
Previous observations of the biology of oomycete sporangia can be connected to the predicted functions of many pisp genes. Several genes participate in calcium signaling, which regulates zoosporogenesis and encystment (20). These include pisp45, which encodes a putative calcium-regulated protein kinase, and pisp46, which encodes a calcium pump. As purines were shown also to regulate germination (9), it was interesting to find that pisp55 encodes phosphoribosylglycinamide synthetase, an enzyme in the purine biosynthetic pathway, and that pic57 encodes adenosine kinase, part of the purine salvage pathway.
Many pisp genes were also induced by nutrient limitation. This may reflect the stimulatory effect of starvation on sporulation (41) or a shift to the utilization of nutrient reserves once sporangia form. However, only a subset of genes induced in the nutrient-limited cultures were expressed at significant levels in the nonsporulating mutant, and vice versa. Sporulation is clearly not determined by a simple linear dependent pathway activated by starvation.
After sporulation, the uptake of nutrients does not appear to be possible until after zoospores encyst and germinate (36). Until germinated spores contact laboratory media or establish a feeding relationship with a plant, stored lipids and a β-1,3-glucan called mycolaminarin are used for energy (21). pisp34 and pisp48 may help mobilize carbon stored in the lipids through the citric acid cycle. pisp48 encodes an enzyme involved in transporting citric acid cycle intermediates across the mitochondrial membrane, while pisp34 encodes phosphenolpyruvate carboxykinase, which converts oxaloacetate from the citric acid cycle to phosphoenolpyruvate. Interestingly, phosphoenolpyruvate metabolism was recently suggested to play a role in sporulation in P. cinnamomi (34).
Three other sporulation-induced genes with potential roles in catabolism are pisp20, -29, and -47, which encode proteases. Only pisp20 was induced by starvation, however. Such proteases may cycle proteins during transitions between developmental stages. This would provide a source of amino acids during germination, as suggested for some true fungi (48). Alternatively, these proteases may be formed in anticipation of a role in plant penetration.
One intriguing group of genes induced by both sporulation and starvation, such as pisp3, encoded sorbitol hydrogenases. These are typically broad-spectrum reversible oxidoreductases, in which conversions, such as mannitol to mannose or sorbitol to fructose, are coupled to NAD reduction (29). In true fungi, polyols, such as mannitol, arabitol, and glycerol, accumulate and exhibit multiple cellular functions. These include maintaining osmotic balance (40), generating turgor in appressoria (47), and quenching reactive oxygen generated within stressed fungal tissue or by plants during defense reactions (6, 22). Sugar alcohols may form in plant-pathogenic fungi when their β-fructosidases transform plant sucrose to fructose, which is converted to sorbitol by sorbitol dehydrogenase; biosynthesis rather than direct uptake is more likely for colonizers of the majority of plants, including potato and tomato, which produce little of these compounds (45).
Assessing the role of the sorbitol dehydrogenases in Phytophthora is challenging, since oomycetes were reported in several studies not to accumulate any sugar alcohols. This is a classic criterion used to demonstrate that oomycetes are unrelated to true fungi (4, 30, 38, 39). However, a recent paper reported that P. infestans spores contain arabitol (46). This was at only 7% of the level of glucose, which seems too low to explain why genes like pisp3 account for 5% of the mRNA in sporangia. Consequently, alternative roles for the enzyme must be considered, such as whether it acts on an atypical substrate. Alternatively, the enzyme may shuttle carbon from glucose to fructose, which through the action of aldehyde oxidoreductase (pisp22?), ketohexokinase, and fructose-bisphosphate aldolase would produce dihydroxyacetone phosphate for entry midway through the glycolytic pathway or for lipid biosynthesis. The difficulty in detecting sugar alcohols might be explained if flux through this pathway is rapid.
A final pisp gene worthy of special comment also appears to play an intriguing role in P. infestans metabolism. pisp41, which was also induced during starvation, exhibited high similarity in database searches to genes for phosphagen kinases, with E values in the 10−90 range. Such enzymes transfer phosphate to guanidino group-containing compounds, such as creatine and arginine (13). Creatine kinase in muscles, for example, catalyzes the transfer of a phosphoryl group from phosphocreatine to ADP to form creatine and ATP. This buffers ATP levels in cell types with high energy demands, a class into which zoospores would certainly fall. However, with one exception, this type of pathway has been found only in animals, not in plants or true fungi (5). The exception is interesting: Trypanosoma cruzi, a unicellular parasite that contains flagella, like Phytophthora (37). Addressing the function of this putative phosphogen kinase, as well as the sorbitol dehydrogenases, will require integrating genetics and biochemistry into future studies. Until then, these enzymes will be considered orphans with uncertain substrates and roles in sporulation.
Acknowledgments
This work was supported by awards from the U.S. Department of Agriculture, Syngenta, and the University of California BioSTAR program.
We thank Sam Roberts for technical assistance and Cristina Cvitanich for making helpful comments on the manuscript.
REFERENCES
- 1.Adams, T. H., J. K. Wieser, and J.-H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ah Fong, A., and H. S. Judelson. 2003. Cell cycle regulator Cdc14 is expressed during sporulation but not hyphal growth in the fungus-like oomycete Phytophthora infestans. Mol. Microbiol. 50:487-494. [DOI] [PubMed]
- 3.Baldauf, S. L., A. J. Roger, I. Wenk-Siefert, and W. F. Doolittle. 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290:972-977. [DOI] [PubMed] [Google Scholar]
- 4.Bokhary, H. A., and R. C. Cooke. 1974. Translocation of carbon-14 glucose by Phytophthora cactorum. Trans. Br. Mycol. Soc. 63:535-540. [Google Scholar]
- 5.Canonaco, F., U. Schlattner, P. S. Pruett, T. Wallimann, and U. Sauer. 2002. Functional expression of phosphagen kinase systems confers resistance to transient stresses in Saccharomyces cerevisiae by buffering the ATP pool. J. Biol. Chem. 277:31303-31309. [DOI] [PubMed] [Google Scholar]
- 6.Chaturvedi, V., A. Bartiss, and B. Wong. 1997. Expression of bacterial mtlD in Saccharomyces cerevisiae results in mannitol synthesis and protects a glycerol-defective mutant from high-salt and oxidative stress. J. Bacteriol. 179:157-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christen, J., and H. R. Hohl. 1972. Growth and ultrastructural differentiation of sporangia in Phytophthora palmivora. Can. J. Microbiol. 18:1959-1964. [DOI] [PubMed] [Google Scholar]
- 8.Chu, S., J. Derisi, M. Eisen, J. Mulholland, D. Botstein, P. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 9.Clark, M. C., D. L. Melanson, and O. T. Page. 1978. Purine metabolism and differential inhibition of spore germination in Phytophthora infestans. Can. J. Microbiol. 24:1032-1038. [DOI] [PubMed] [Google Scholar]
- 10.Cvitanich, C., and H. S. Judelson. 2003. A gene expressed during sexual and asexual sporulation in Phytophthora infestans is a member of the Puf family of translational regulators. Eukaryot. Cell 2:465-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebbole, D. J. 1996. Morphogenesis and vegetative differentiation in filamentous fungi. J. Genet. 75:361-374. [Google Scholar]
- 12.Eklund, H., E. Horjales, H. Jornvall, C. I. Branden, and J. Jeffery. 1985. Molecular aspects of functional differences between alcohol and sorbitol dehydrogenases. Biochemistry 24:8005-8012. [DOI] [PubMed] [Google Scholar]
- 13.Ellington, W. R. 2001. Evolution and physiological roles of phosphagen systems. Annu. Rev. Physiol. 63:289-325. [DOI] [PubMed] [Google Scholar]
- 14.Erwin, D. C., and O. K. Ribeiro. 1996. Phytophthora diseases worldwide. APS Press, St. Paul, Minn.
- 15.Fabritius, A.-L., C. Cvitanich, and H. S. Judelson. 2002. Stage-specific gene expression during sexual development in Phytophthora infestans. Mol. Microbiol. 45:1057-1066. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 17.Fry, W. E., and S. B. Goodwin. 1997. Re-emergence of potato and tomato late blight in the United States. Plant Dis. 81:1349-1357. [DOI] [PubMed] [Google Scholar]
- 18.Gasch, A., P. P. Spellman, T. C. Kao, M. O. Carmel-Harel, M. Eisen, B. G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goernhardt, B., I. Rouhara, and E. Schmelzer. 2000. Cyst germination proteins of the potato pathogen Phytophthora infestans share homology with human mucins. Mol. Plant-Microbe Interact. 13:32-42. [DOI] [PubMed] [Google Scholar]
- 20.Hardham, A. R. 2001. The cell biology behind Phytophthora pathogenicity. Aust. Plant Pathol. 30:91-98. [Google Scholar]
- 21.Hemmes, D. E. 1983. Cytology of Phytophthora, p. 9-40. In D. C. Erwin, S. Bartnicki-Garcia, and P. H. Tsao (ed.), Phytophthora, its biology, taxonomy, ecology, and pathology. APS Press, St. Paul, Minn.
- 22.Jennings, D. B., M. Ehrenshaft, D. M. Pharr, and J. D. Williamson. 1998. Roles for mannitol and mannitol dehydrogenase in active oxygen-mediated plant defense. Proc. Natl. Acad. Sci. USA 95:15129-15133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Judelson, H. S., and R. W. Michelmore. 1990. Highly abundant and stage-specific messenger RNA in the obligate pathogen Bremia lactucae. Mol. Plant-Microbe Interact. 3:225-232. [DOI] [PubMed] [Google Scholar]
- 24.Judelson, H. S., and S. Roberts. 2002. Novel protein kinase induced during sporangial cleavage in the oomycete Phytophthora infestans. Eukaryot. Cell 1:687-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamoun, S., P. Hraber, B. Sobral, D. Nuss, and F. Govers. 1999. Initial assessment of gene diversity for the oomycete pathogen Phytophthora infestans based on expressed sequences. Fungal Genet. Biol. 28:94-106. [DOI] [PubMed] [Google Scholar]
- 26.Laxalt, A. M., M. Latijnhouwers, M. van Hulten, and F. Govers. 2002. Differential expression of G protein alpha and beta subunit genes during development of Phytophthora infestans. Fungal Genet. Biol. 36:137-146. [DOI] [PubMed] [Google Scholar]
- 27.Lewis, D. H., and D. C. Smith. 1967. Sugar alcohols in fungi and green plants. I. Distribution, physiology, and metabolism. New Phytol. 66:143-184. [Google Scholar]
- 28.Lewis, Z. A., A. Correa, C. Schwerdtfeger, K. L. Link, X. Xie, R. H. Gomer, T. Thomas, D. J. Ebbole, and D. Bell-Pedersen. 2002. Overexpression of White Collar-1 (WC-1) activates circadian clock-associated genes, but is not sufficient to induce most light-regulated gene expression in Neurospora crassa. Mol. Microbiol. 45:917-931. [DOI] [PubMed] [Google Scholar]
- 29.Lindstad, R. I., P. Koell, and J. S. McKinley-Mckee. 1998. Substrate specificity of sheep liver sorbitol dehydrogenase. Biochem. J. 330:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long, D. E., and R. C. Cooke. 1974. Carbohydrate composition and metabolism of Senecio squalidus leaves infected with Albugo tragopogonis. New Phytol. 73:889-899. [Google Scholar]
- 31.Lovett, J. S. 1975. Growth and differentiation of the water mold Blastocladiella emersonii: cytodifferentiation and the role of ribonucleic acid and protein synthesis. Bacteriol. Rev. 39:345-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maltese, C. E., G. Conigliaro, and D. S. Shaw. 1995. The development of sporangia of Phytophthora infestans. Mycol. Res. 99:1175-1181. [Google Scholar]
- 33.Marks, G. E. 1965. The cytology of Phytophthora infestans. Chromosoma 16:681-692. [DOI] [PubMed] [Google Scholar]
- 34.Marshall, J. S., A. R. Ashton, F. Govers, and A. R. Hardham. 2001. Isolation and characterization of four genes encoding pyruvate, phosphate dikinase in the oomycete plant pathogen Phytophthora cinnamomi. Curr. Genet. 40:73-81. [DOI] [PubMed] [Google Scholar]
- 35.Mizubuti, E. S. G., D. E. Aylor, and W. E. Fry. 2000. Survival of Phytophthora infestans sporangia exposed to solar radiation. Phytopathology 90:78-84. [DOI] [PubMed] [Google Scholar]
- 36.Penington, C. J., J. R. Iser, B. R. Grant, and K. R. Gayler. 1989. Role of RNA and protein synthesis in stimulated germination of zoospores of the pathogenic fungus Phytophthora palmivora. Exp. Mycol. 13:158-168. [Google Scholar]
- 37.Pereira, C. A., G. D. Alonso, M. C. Paveto, A. Iribarren, M. L. Cabanas, N. H. Torres, and M. M. Flawia. 2000. Trypanosoma cruzi arginine kinase characterization and cloning: a novel energetic pathway in protozoan parasites. J. Biol. Chem. 275:1495-1501. [DOI] [PubMed] [Google Scholar]
- 38.Pfyffer, G. E., B. U. Pfyffer, and D. M. Rast. 1986. The polyol pattern, chemotaxonomy, and phylogeny of the fungi. Sydowia 39:160-201. [Google Scholar]
- 39.Pfyffer, G. E., and D. M. Rast. 1980. The polyol pattern of some fungi not hitherto investigated for sugar alcohols. Exp. Mycol. 4:160-170. [Google Scholar]
- 40.Redkar, R. J., R. D. Locy, and N. K. Singh. 1995. Biosynthetic pathways of glycerol accumulation under salt stress in Aspergillus nidulans. Exp. Mycol. 19:241-246. [DOI] [PubMed] [Google Scholar]
- 41.Ribeiro, O. K. 1983. Physiology of asexual sporulation and spore germination in Phytophthora, p. 55-70. In D. C. Erwin, S. Bartnicki-Garcia, and P. H. Tsao (ed.), Phytophthora, its biology, taxonomy, ecology, and pathology. APS Press, St. Paul, Minn.
- 42.Schmit, J. C., and S. Brody. 1976. Biochemical genetics of Neurospora crassa conidial germination. Bacteriol. Rev. 40:1-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sparrow, F. K. 1960. Aquatic phycomycetes, 2nd ed. University of Michigan Press, Ann Arbor.
- 45.Stoop, J. M. H., J. D. Williamson, and D. M. Pharr. 1996. Mannitol metabolism in plants: a method for coping with stress. Trends Plant Sci. 1:139-144. [Google Scholar]
- 46.Tereshina, V. M., A. S. Memorskaya, E. V. Morozova, V. P. Kozlov, and E. P. Feofilova. 2000. Alterations in the carbohydrate composition of the cytosol of fungal spores caused by temperature changes and the storage process. Mikrobiologiya 69:511-517. [PubMed] [Google Scholar]
- 47.Thines, E., R. W. S. Weber, and N. J. Talbot. 2000. MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 12:1703-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas, S. W., S. W. Rasmussen, M. A. Glaring, J. A. Rouster, S. K. Christiansen, and R. P. Oliver. 2001. Gene identification in the obligate fungal pathogen Blumeria graminis by expressed sequence tag analysis. Fungal Genet. Biol. 33:195-211. [DOI] [PubMed] [Google Scholar]
- 49.Torto, T. A., L. Rauser, and S. Kamoun. 2002. The pipg1 gene of the oomycete Phytophthora infestans encodes a fungal-like endopolygalacturonase. Curr. Genet. 40:385-390. [DOI] [PubMed] [Google Scholar]
- 50.Vallee, B. L., and F. L. Hoch. 1955. Zinc, a component of yeast alcohol dehydrogenase. Proc. Natl. Acad. Sci. USA 41:327-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Visintin, R., E. S. Hwang, and A. Amon. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398:818-823. [DOI] [PubMed] [Google Scholar]
- 52.Xu, R. 1982. A defined media for Phytophthora. Acta Mycol. Sin. 1:40-47. [Google Scholar]