Abstract
The early telencephalon shares molecular features with the early mid-hindbrain region. In particular, these two developing brain areas each have a signaling center that secretes FGFs and an adjacent one that secretes WNTs. WNTs and FGFs each play essential roles in regulating cell fates in both the telencephalon and mid-hindbrain. Despite this similarity, telencephalic and mid-hindbrain precursors express distinct genes and ultimately generate different cell types, tissue morphologies, and neural functions. Here we show that genetically increasing the level of β-catenin, a mediator of canonical WNT signaling, in the anterior neural plate causes a loss of telencephalic characteristics and a gain of mid-hindbrain characteristics. These results, together with previous ones demonstrating that increased WNT signaling in the anterior neural plate increases FGF expression, suggest that the levels of WNT and FGF signaling regulate telencephalic versus mid-hindbrain fates.
Keywords: β-catenin, telencephalon, mid-hindbrain boundary, FGF, WNT, mouse
INTRODUCTION
The neural plate is patterned along its anterior-posterior axis into distinct areas, which then develop into different parts of the central nervous system. In the early steps of neural development, signaling centers induce the neural plate whose cells acquire broad anterior or posterior fates (Wilson and Houart, 2004; Stern, 2006; Levine and Brivanlou, 2007). Subsequently, new signaling centers emerge to further subdivide the anterior neural plate into distinct brain areas. Two such signaling centers secrete similar factors despite their different locations. First, at the mid-hindbrain boundary, an isthmus forms that acts as a signaling center with its anterior domain secreting WNT1 and its posterior domain secreting FGFs (Sato et al., 2004; Zervas et al., 2005). Second, in the forebrain region, the ridge between the neural plate and ectoderm, which will form the midline separating the hemispheres of the telencephalon, emerges as a signaling center with its most anterior-medial domain secreting FGFs and its more posterior-lateral domain secreting WNTs (Wilson and Rubenstein, 2000; Monuki and Walsh, 2001; Zaki et al., 2003; Hébert and Fishell, 2008). Despite the molecular similarities between the forebrain and mid-hindbrain signaling centers, they induce markedly different brain structures.
FGFs play similar roles at the onset of both telencephalon and mid-hindbrain development. For the telencephalon, FGFs induce a telencephalic cell fate, maintain telencephalic precursor cells alive, and pattern the telencephalon (Mason, 2007; Hébert and Fishell, 2008). Similarly, for the mid-hindbrain region, FGFs induce cerebellar and midbrain cell fates, maintain the precursor cells for these brain areas alive, and pattern these structures (Sato et al., 2004; Zervas et al., 2005). How FGFs promote telencephalon formation in one case and mid-hindbrain development in the other is unknown.
WNT signaling also plays a crucial role in the early development of both the telencephalon and mid-hindbrain region. At the mid-hindbrain boundary, Wnt1 maintains Fgf8 expression, and vice versa, and loss of either one leads to elimination of the cerebellum and midbrain due to cell death of the precursors (Sato et al., 2004; Zervas et al., 2005). In the anterior neural plate, prior to telencephalon induction, canonical WNT signaling must be low or absent for FGFs to be expressed and for a telencephalon to form (Houart et al., 2002). Once the telencephalon is induced, however, WNT signaling becomes required to maintain FGF signaling and consequently cell survival in the anterior neural plate (Paek et al., 2009; Paek et al., 2011; Wang et al., 2011). At this stage, genetically increasing the level of canonical WNT signaling by expressing a stabilized form of β-catenin, a mediator of WNT signaling, leads to abnormally widespread FGF expression and signaling (Paek et al., 2011; Wang et al., 2011). However, the effect of increased β-catenin and FGFs on the fate of anterior neural precursors has not been examined. Here we show that increasing β-catenin levels in the anterior neural plate, which increases FGF signaling, promotes mid-hindbrain characteristics at the expense of telencephalic ones.
RESULTS AND DISCUSSION
To determine the effect of increasing β-catenin activity in the anterior neural plate, mice carrying an allele of β-catenin, Ctnnb1Δex3, in which exon 3 is floxed (Harada et al., 1999), were crossed to mice carrying the Foxg1Cre allele, which conveys recombination specifically in telencephalic precursor cells starting at ~E8.5 (Hébert and McConnell, 2000). Recombination of the Ctnnb1Δex3 allele leads to expression of a stabilized β-catenin protein and increased canonical WNT signaling (Harada et al., 1999). The control embryos used in all the experiments described here were littermates that carried either Foxg1Cre or Ctnnb1Δex3 alone. Mutants were recovered at Mendelian ratios until E10.5, after which their numbers decline rapidly (due presumably to heart defects since Foxg1Cre can lead to recombination in this organ; Hébert and McConnell, 2000).
Using a Tcf-lacZ reporter line, we previously showed that Foxg1Cre/+;Ctnnb1Δex3/+ mutants have 5-fold higher levels of canonical WNT signaling in the anterior neural plate by E9 (Paek et al., 2011). We also showed that markers for cell death (TUNEL and p21Cip1) and mitosis (phospho-histone H3) were not significantly changed at E9 (Paek et al., 2011). However, the fate of the mutant cells beyond E9 and whether elevated β-catenin had an effect on cell identity was not previously examined.
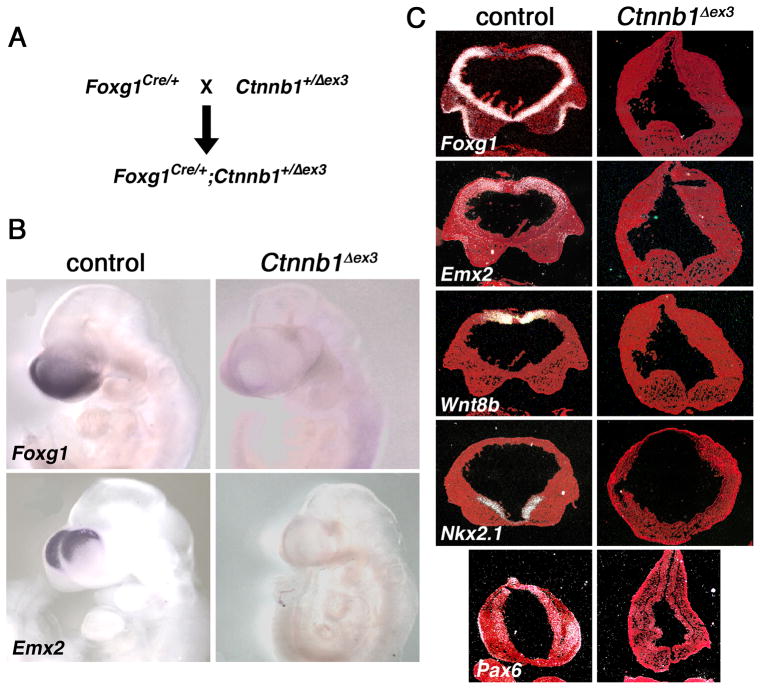
The anterior neural tube of E9.5 and E10.5 Foxg1Cre/+;Ctnnb1Δex3/+ mutants, rather than forming bilateral ventricles characteristic of the telencephalon, form a single abnormally formed ventricle (Fig. 1,2). Single ventricles are typical of more caudal brain areas. The abnormal morphology of the mutants could be due to a fate change of anterior precursor cells from telencephalic to more caudal identities. Consistent with a caudal transformation, anterior cells no longer express telencephalic markers. Whereas control E9.5 and E10.5 embryos express Foxg1 throughout the telencephalon, Emx2 and Pax6 in the dorsal telencephalon, Wnt8b in the dorso-medial area, and Nkx2.1 in the ventro-medial area, examination of whole embryos and serial sections throughout the anterior neural tissue of mutants revealed that expression of these genes is absent (Fig. 1 and data not shown).
Figure 1. Expression of telencephalic markers is absent in Foxg1Cre/+;Ctnnb1+/Δex3 mutants.
(A) Cross used to generate mutant and control embryos (only the mutant genotype is shown). (B) Whole mount DIG-labeled RNA in situ hybridization of E10.5 embryos. Foxg1 and Emx2 expression (purple) are undetectable in the anterior region of the mutant embryos. (C) 35S-labeled RNA in situ hybridization of E10.5 (except E9.75 for Pax6) control and Ctnnb1+/Δex3 embryos on coronal sections through the anterior neural tissue. Expression of telencephalic markers (white) is undetectable in the mutant.
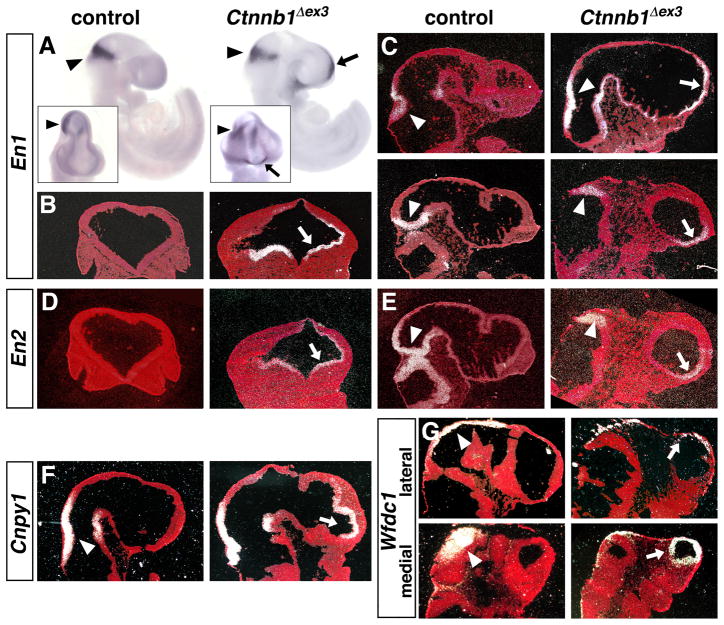
Figure 2. The Foxg1Cre/+;Ctnnb1+/Δex3 mutants ectopically express Engrailed genes.
(A–E) Expression of En1 and En2 by whole mount DIG-labeled RNA in situ hybridization of E9.5 embryos (A) and 35S-labeled RNA in situ hybridization of E10.5 control and Ctnnb1+/Δex3 embryos on coronal (B,D) and sagittal sections (C,E, with anterior to the right). Insets in (A) are frontal views. (F–G) Expression of Cnpy1 and Wfdc1 by 35S-labeled RNA in situ hybridization of E10.5 control and Ctnnb1+/Δex3 sagittal sections. For all probes, arrowheads point to the normal mid-hindbrain region of expression whereas arrows point to ectopic expression in the anterior neural tissue.
If the anterior neural plate of mutants does not acquire a telencephalic identity, then perhaps it adopts a more caudal fate. To test this possibility, the anterior brain region of the activated β-catenin mutant was examined for expression of diencephalic, midbrain, and mid-hindbrain markers. A diencephalic fate was ruled out because expression of the diencephalic markers Tcf4b and Foxd1 was absent from the anterior of both E10.5 control and mutant brains (data not shown). Moreover, expression of Nkx2.1, which in controls is found in both the ventral diencephalon and telencephalon, could not be detected in mutants (Fig. 1C). Hence anterior neural precursor cells that express elevated levels of β-catenin no longer adopt a forebrain fate, either telencephalic or diencephalic.
The possibility that the anterior neural tissue in the mutants adopted a midbrain or hindbrain fate was assessed by examining the expression of En1, En2, Mrp4, Hes3, and Cnpy1, markers for the mid-hindbrain boundary region, Pax2, a marker for the mid-hindbrain region and hindbrain, Gbx2, a marker for the hindbrain, and Wfdc1, a marker expressed in a high anterior to low posterior gradient in the midbrain (Sato et al., 2004; Zervas et al., 2005; Jukkola et al., 2006). Expression of En1, En2, and Cnpy1, which are completely undetectable throughout the control telencephalon, were readily and consistently detected at E10.5 in the anterior neural tissue of all mutants examined (Fig. 2A–F). Coronal and sagittal sections revealed strongest expression in the anterior ventral areas. The ectopic expression of En1, En2, and Cnpy1 suggests that the anterior neural tissue of mutants had adopted characteristics of the mid-hindbrain region.
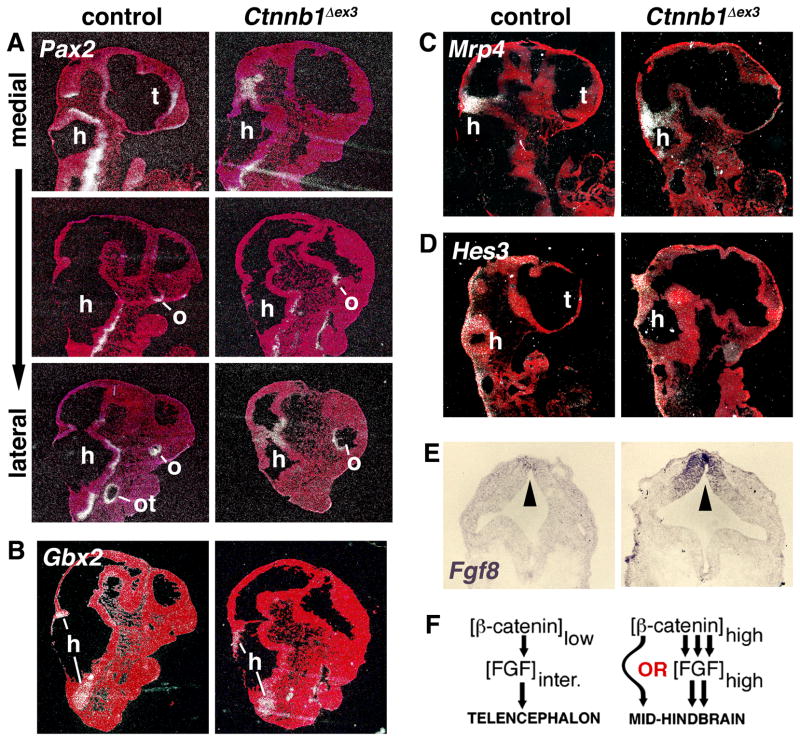
Unlike En1, En2, and Cnpy1 whose expression extends somewhat anteriorly in the midbrain, Mrp4 and Hes3, although also expressed at the mid-hindbrain boundary, only have a narrow band of expression in the posterior midbrain (Jukkola et al., 2006). Interestingly, Mrp4 and Hes3 expression was low or undetectable in the anterior neural tissue of mutants (Fig. 3C,D), suggesting that this tissue might have adopted more midbrain rather than hindbrain characteristics.
Figure 3. The Foxg1Cre/+;Ctnnb1+/Δex3 mutants do not ectopically express Pax2, Gbx2, Mrp4, or Hes3.
(A–D) 35S-labeled RNA in situ hybridization of E10.5 control and Ctnnb1+/Δex3 embryos on sagittal sections. (A) Medial to lateral sections showing the different areas of Pax2 expression. Pax2 is not detected in the anterior neural tissue of mutants. (B–D) Likewise, expression of Gbx2, Mrp4 and Hes3 is undetectable in the anterior of controls or mutants. (E) DIG-labeled RNA in situ hybridization on horizontal sections of E9.5 embryos showing increased Fgf8 expression (dark purple) at the anterior neuroepithelium (arrowhead) of the mutant as previously reported (Paek et al., 2011). (F) Model for how β-catenin, with or without FGF signaling, could promote telencephalic versus mid-hindbrain fates. Abbreviations: h, hindbrain; o, optic stalk or vesicle; ot, otic vesicle; t, telencephalon.
Our findings with Pax2 and Gbx2 are consistent with this possibility. In controls, Pax2 was expressed as expected in the hindbrain region and expression was also consistently detected in the anterior medial telencephalon and in the optic stalk and vesicle (Fig. 3A). In the mutant, although expressed in the hindbrain area, Pax2 was absent throughout the anterior neural tissue, including the anterior medial areas, but was still detected in the optic stalk and enlarged optic vesicle. Similarly, expression of Gbx2 was strong in the hindbrain of controls and mutants as expected, but no expression of this gene could be detected throughout the telencephalon of controls or the anterior tissue of mutants (Fig. 3B). WNT and FGF signaling at the mid-hindbrain isthmus induces primarily midbrain and cerebellar fates (Sato et al., 2004; Zervas et al., 2005). The lack of Gbx2 and Pax2 expression in the anterior tissue of mutants is therefore consistent with the likelihood that WNTs and/or FGFs are inducing features that are characteristic of the mid-hindbrain boundary or midbrain region, rather than general hindbrain features.
Moreover, expression of the midbrain marker Wfdc1 could be readily detected in the anterior neural tissue of mutant embryos with especially high levels of expression in lateral areas (Fig. 2G). Together, the data presented in this study indicate that elevating the level of active β-catenin, and consequently canonical WNT signaling (Paek et al., 2011), in the anterior neural plate leads to the loss of telencephalic features and the acquisition of certain mid-hindbrain boundary or midbrain characteristics such as expression of En1, En2, Cnpy1, and Wfdc1.
The neural plate is initially patterned along its anterior-posterior axis with high levels of FGF, WNT, and retinoic acid signaling leading to caudalization. Shortly thereafter, several signaling centers emerge in the developing brain, including the isthmus at the mid-hindbrain boundary and the ridge at the anterior most edge of the neural plate. These provide new sources of FGFs and WNTs. Although levels of FGF signaling are more difficult to assess, levels of WNT signaling assessed with lacZ reporter mice are higher in the mid-hindbrain region than in the anterior most region of the neural plate (Maretto et al., 2003; Mohamed et al., 2004; Paek et al., 2011).
Therefore, the change of fate that we observe in the Foxg1+/Cre;Ctnnb1+/Δex3 mutants from telencephalic to more mid-hindbrain-like suggest that what is important in distinguishing the outcome of WNT and FGF signaling at the mid-hindbrain boundary region versus the telencephalon is the level of WNT signaling. However, in both locations normal levels of WNT signaling are necessary to maintain FGF signaling and increased WNT signaling is sufficient to promote increased FGF signaling (Fig. 3E; Lee et al., 1997; Paek et al., 2011; Wang et al., 2011). Therefore it is also possible that higher levels of FGF signaling, or the combination of higher FGF and WNT signaling, promotes mid-hindbrain rather than telencephalic fates (Fig. 3F). In support of a role for FGFs, ectopic En1, En2, and Cnpy1 expression is strongest in the anterior ventral-medial area of the mutant closest to the highest levels of FGF expression (Fig. 2A–F, 3E). From the data presented here, it is not possible to distinguish between these possibilities. To do so would require elevating WNT signaling while keeping the level of FGF signaling constant, and vice versa, which may be technically difficult due to the cross-regulatory nature of these signaling pathways. Interestingly, and consistent with our data, loss of Apc, which leads to increased WNT/β-catenin signaling, at slightly later stages of neocortical development also leads to the ectopic expression of more posterior dorsal markers including midbrain ones (Ivaniutsin et al., 2009).
EXPERIMENTAL PROCEDURES
Mice
Mice carrying the conditional Ctnnb1Δex3 allele were kindly provided by Harald von Boehmer (Dana Farber Cancer Institute, Boston, U.S.A.) and Makoto Mark Taketo (Kyoto University, Kyoto, Japan; Harada et al., 1999) and mice carrying the Cre recombinase knocked into the Foxg1 locus were previously described (Hébert and McConnell, 2000). Embryos were obtained from timed pregnancies with noon of the vaginal plug date counted as embryonic day 0.5 (E0.5). All the experiments described herein have been approved by, and meet the strictest standards of the IACUC of the Albert Einstein College of Medicine.
RNA in situ hybridization
35S-labeled RNA in situ hybridizations on sections and digoxygenin (DIG)-labeled RNA hybridizations on whole embryos were carried out as described previously (Paek et al., 2009). DIG-labeled RNA in situ hybridization on sections using DIG-labeled probes was performed on 10μm thick cryosections as described previously (Hirsch et al., 2007). At least three embryos from each genotype were analyzed for each probe. Coronal sections comparing controls and mutants are positionally matched along the antior-posterior axis between the front most point of the anterior neural tissue and the diencephalon.
Acknowledgments
We are grateful to Makoto Mark Taketo and Harald von Boehmer for mice and Outi Kostia and Juha Partanen for mid-hindbrain probes. J.M.H. was supported by the NIH (MH083804) and the Hirschl/Weill-Caulier Foundation.
References
- Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. Embo J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert JM, Fishell G. The genetics of early telencephalon patterning: some assembly required. Nat Rev Neurosci. 2008;9:678–685. doi: 10.1038/nrn2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev Biol. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- Hirsch MR, Glover JC, Dufour HD, Brunet JF, Goridis C. Forced expression of Phox2 homeodomain transcription factors induces a branchio-visceromotor axonal phenotype. Dev Biol. 2007;303:687–702. doi: 10.1016/j.ydbio.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Houart C, Caneparo L, Heisenberg C, Barth K, Take-Uchi M, Wilson S. Establishment of the telencephalon during gastrulation by local antagonism of Wnt signaling. Neuron. 2002;35:255–265. doi: 10.1016/s0896-6273(02)00751-1. [DOI] [PubMed] [Google Scholar]
- Ivaniutsin U, Chen Y, Mason JO, Price DJ, Pratt T. Adenomatous polyposis coli is required for early events in the normal growth and differentiation of the developing cerebral cortex. Neural Dev. 2009;4:3. doi: 10.1186/1749-8104-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukkola T, Lahti L, Naserke T, Wurst W, Partanen J. FGF regulated gene-expression and neuronal differentiation in the developing mid-hindbrain region. Dev Biol. 2006;297:141–157. doi: 10.1016/j.ydbio.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Lee SMK, Danielian PS, Fritzsch B, McMahon AP. Evidence that FGF8 signalling from the midbrain-hindbrain junction regulates growth and polarity in the developing midbrain. Development. 1997;124:959–969. doi: 10.1242/dev.124.5.959. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Brivanlou AH. Proposal of a model of mammalian neural induction. Dev Biol. 2007;308:247–256. doi: 10.1016/j.ydbio.2007.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci U S A. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason I. Initiation to end point: the multiple roles of fibroblast growth factors in neural development. Nat Rev Neurosci. 2007;8:583–596. doi: 10.1038/nrn2189. [DOI] [PubMed] [Google Scholar]
- Mohamed OA, Clarke HJ, Dufort D. Beta-catenin signaling marks the prospective site of primitive streak formation in the mouse embryo. Dev Dyn. 2004;231:416–424. doi: 10.1002/dvdy.20135. [DOI] [PubMed] [Google Scholar]
- Monuki ES, Walsh CA. Mechanisms of cerebral cortical patterning in mice and humans. Nat Neurosci. 2001;4:1199–1206. doi: 10.1038/nn752. [DOI] [PubMed] [Google Scholar]
- Paek H, Gutin G, Hébert JM. FGF signaling is strictly required to maintain early telencephalic precursor cell survival. Development. 2009;136:2457–2465. doi: 10.1242/dev.032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek H, Hwang JY, Zukin RS, Hébert JM. β-catenin-dependent FGF signaling sustains cell survival in the anterior embryonic head by countering Smad4. Dev Cell. 2011;20:689–699. doi: 10.1016/j.devcel.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Joyner AL, Nakamura H. How does Fgf signaling from the isthmic organizer induce midbrain and cerebellum development? Dev Growth Differ. 2004;46:487–494. doi: 10.1111/j.1440-169x.2004.00769.x. [DOI] [PubMed] [Google Scholar]
- Stern CD. Neural induction: 10 years on since the “default model”. Curr Op Cell Biol. 2006;18:692–697. doi: 10.1016/j.ceb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Wang Y, Song L, Zhou CJ. The canonical Wnt/b-catenin signaling pathway regulates Fgf signaling for early facial development. Dev Biol. 2011;349:250–260. doi: 10.1016/j.ydbio.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Wilson SW, Houart C. Early steps in the development of the forebrain. Dev Cell. 2004;6:167–181. doi: 10.1016/s1534-5807(04)00027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SW, Rubenstein JLR. Induction and dorsoventral patterning of the telencephalon. Neuron. 2000;28:641–651. doi: 10.1016/s0896-6273(00)00171-9. [DOI] [PubMed] [Google Scholar]
- Zaki PA, Quinn JC, Price DJ. Mouse models of telencephalic development. Curr Opin Genet Dev. 2003;13:423–437. doi: 10.1016/s0959-437x(03)00084-4. [DOI] [PubMed] [Google Scholar]
- Zervas M, Blaess S, Joyner AL. Classical embryological studies and modern genetic analysis of midbrain and cerebellum development. Curr Top Dev Biol. 2005;69:101–138. doi: 10.1016/S0070-2153(05)69005-9. [DOI] [PubMed] [Google Scholar]