Abstract
Selective isotopic labeling provides an unparalleled window within which to study the structure and dynamics of RNAs by high resolution NMR spectroscopy. Unlike commonly used carbon sources, the asymmetry of 13C-labeled pyruvate provides selective labeling in both the ribose and base moieties of nucleotides using Escherichia coli variants, that until now were not feasible. Here we show that an E. coli mutant strain that lacks succinate and malate dehydrogenases (DL323) and grown on [3-13C]-pyruvate affords ribonucleotides with site specific labeling at C5′ (~95%) and C1′ (~42%) and minimal enrichment elsewhere in the ribose ring. Enrichment is also achieved at purine C2 and C8 (~95%) and pyrimidine C5 (~100%) positions with minimal labeling at pyrimidine C6 and purine C5 positions. These labeling patterns contrast with those obtained with DL323 E. coli grown on [1, 3-13C]-glycerol for which the ribose ring is labeled in all but the C4′ carbon position, leading to multiplet splitting of the C1′, C2′ and C3′ carbon atoms. The usefulness of these labeling patterns is demonstrated with a 27-nt RNA fragment derived from the 30S ribosomal subunit. Removal of the strong magnetic coupling within the ribose and base leads to increased sensitivity, substantial simplification of NMR spectra, and more precise and accurate dynamic parameters derived from NMR relaxation measurements. Thus these new labels offer valuable probes for characterizing the structure and dynamics of RNA that were previously limited by the constraint of uniformly labeled nucleotides.
Electronic supplementary material
The online version of this article (doi:10.1007/s10858-011-9582-5) contains supplementary material, which is available to authorized users.
Keywords: Site selective labeling, DL323, Pyruvate-3, Ribose and nucleobase, RNA, Structure and dynamics
Introduction
RNA is increasingly viewed not only as an intermediary between DNA and protein, but also as a key regulator of gene expression and catalysis (Boisvert et al. 2007; Korostelev et al. 2008; Steitz 2008; Breaker 2009; Wahl et al. 2009; Newman and Nagai 2010). Methods to obtain very high-resolution structures and site specific dynamics of RNA molecules are therefore critical. Nuclear magnetic resonance (NMR) spectroscopy has emerged as an effective method for solving RNA 3D structures and for probing their motions. However, RNA molecules that are bigger than 40 nucleotides are difficult to characterize using NMR spectroscopy because of severe chemical shift overlap and rapid signal decay. Selective labeling techniques, first introduced to extend the utility of NMR spectroscopy for protein analysis, are increasingly being applied to RNA as well (Dayie 2008; Lu et al. 2010).
Of the three primary methods for incorporating selective isotopic labels into nucleic acids, biomass production using E. coli variants (Johnson et al. 2006; Johnson and Hoogstraten 2008; Dayie and Thakur 2010; Thakur et al. 2010a, b) appears to be more general and cost-effective than chemical synthesis (Milecki 2002) or de novo biosynthesis (Schultheisz et al. 2008, 2011). Selective introduction of 13C isotopes into the ribose ring using chemical synthesis entails difficult regio- and stereo-selective synthesis with low yields (Milecki 2002). The de novo biosynthetic method requires a large number of enzymes of which only a few are commercially available (Schultheisz et al. 2008, 2011). Additionally, the de novo method requires prohibitively expensive precursors that are limited in the labeling pattern that can be achieved with the biomass methods.
Unfortunately a biomass method based on E. coli metabolism, until recently, suffered from the disadvantage of isotopic scrambling in both the ribose and nucleobase atoms. This scrambling is caused by the admixture of the metabolic flux from both the oxidative and non-oxidative pentose phosphate pathways (noPPP) and a very efficient TCA cycle (Johnson et al. 2006; Johnson and Hoogstraten 2008). Hoffman and Holland earlier showed that wild-type E. coli grown using selectively labeled acetate enables the incorporation of 13C-labels at either the protonated or non-protonated carbon sites in the RNA ribose and nucleobases. LeMaster and coworkers also showed that a modified bacterial strain, in which the TCA cycle enzymes succinate dehydrogenase and malate dehydrogenase were disabled (DL323), could be grown on selectively labeled glycerol to provide alternative site labeling that removed the 13C–13C coupling in protein NMR spectra (LeMaster and Richards 1982; LeMaster and Kushlan 1996). Hoogstraten and coworkers then showed that the DL323 strain grown on selectively labeled glycerol also enabled site selective labeling within the ribose ring (Johnson et al. 2006; Johnson and Hoogstraten 2008). Earlier, Pardi and colleagues introduced labeling using 13C-formate in a background of 12C-glucose which site specifically labels purine C8 position exclusively; results were not presented for purine C2 position (Latham et al. 2005). Building on these previous ideas, we showed that an isolated two spin system could be achieved for both base and ribose depending on the strain and complementation of the growth media with formate (Dayie and Thakur 2010; Thakur et al. 2010a, b). Even under these newer conditions, the purine C2 positions were only labeled to ~26%, indicating a general limitation of the formate supplementation methodology (Dayie and Thakur 2010; Thakur et al. 2010a, b).
However until now, most biomass production of nucleotides had focused on using symmetric carbon sources such as glycerol, glucose, and acetate. For instance if glucose is the carbon source, then at the onset of the TCA cycle this input carbon source is already diluted to 50% by glycolysis and similarly the purine precursor 3-phosphoglycerate (3PG) is only 50% labeled. Similarly if [1, 3-13C]-glycerol is the carbon source, selectivity in the ribose and base regions are again reduced because of the symmetric nature of glycerol such that the purine C5 and all the ribose C1′ have substantial residual coupling that can render relaxation measurements inaccurate (Johnson et al. 2006; Dayie and Thakur 2010). This residual coupling arises from the C-1 carbon of three carbon precursors such as pyruvate and glycerol, and thus [1-13C]-pyruvate will similarly introduce coupling between C1′–C2′–C3′ and C2′–C3′. We, therefore, reasoned that the asymmetry of 13C-labeled pyruvate would provide selective labeling in both the ribose and base moieties of nucleotides that would be impossible with symmetric carbon sources.
Unlike the previous result with the use of [1, 3-13C]-glycerol (Johnson et al. 2006; Thakur et al. 2010a, b), the use of [3-13C]-pyruvate affords selective 13C enrichment of the ribose C1′ and C5′, of the purine C2 and C8, and of the pyrimidine C5 without any of the residual couplings described above. NMR T1 relaxation experiments and 13C-methylene TROSY or HSQC on a site selectively 13C-labeled 27-nt A-site RNA demonstrate that accurate T1 relaxation parameters and higher resolution spectra can be obtained. Thus, the technology to incorporate site specific labels into both the base and ribose moieties of purine and pyrimidine nucleotides promises to expand the scope of NMR studies to regions hitherto inaccessible.
Materials and methods
Bacterial strains
The mutant strains DL323 (CGSC # 7538, F-, sdh-1, &lambda-, mdh-2, rph-1) (Hansen and Juni 1979; LeMaster and Kushlan 1996) and K10-15-16 (CGSC # 4858 Hfr fhuA22, zwf-2, relA1, T2R, pfk-10; Fraenkel 1968; referred to here as K10zwf) and the wild-type K12 strain (Clowes 1968; Soupene et al. 2003; CGSC # 4401:F+) used in this work were obtained from the Yale Coli Genetic Stock Center (CGSC). Dr. Paliy kindly provided the wild-type K12 NCM3722 (Soupene et al. 2003).
Isotopes
Isotopic labeled compounds were purchased from Cambridge Isotope Laboratory (Andover, MA) and Isotec-Sigma-Aldrich (Miamisburg, OH): [3-13C]-pyruvate (99%), 15N-(NH4)2SO4 (99%).
Media for E. coli growth
Luria–Bertani (LB) media was prepared as described (Sambrook and Russell 2001) and LeMaster-Richards (LMR) minimal media was prepared as described (LeMaster and Richards 1982; Dayie and Thakur 2010; Thakur et al. 2010a, b). The LMR media contains 176 mM KH2PO4, 25 mM NaOH, 10 μl H2SO4, 12.6 mM (NH4)2SO4, 2 mM MgSO4, 10 μM FeSO4 and 0.2% trace metals, supplemented with the [3-13C]-pyruvate and 15N–(NH4)2SO4 enriched nitrogen source.
Method for E. coli growth optimization
The growth evaluation of the E. coli mutant (DL323) was performed as described previously (Thakur et al. 2010a, b). Briefly, a 5 ml starter culture in LeMaster media supplemented with unlabeled carbon sources was inoculated from a single colony of DL323 E. coli grown on LB agar plates at 37°C overnight (~16 h). The overnight culture was pelleted, cells were washed twice in 1× phosphate-buffered saline (PBS), and re-suspended in fresh 5 ml of LMR medium; 1 ml of this re-suspension was added to a 50 ml culture in LMR medium for another overnight growth at 37°C. The overnight culture was pelleted prior to complete saturation of these cells, the cells were washed twice in 1×PBS, resuspended in a 50 ml of LMR medium with no carbon supplements, and then 5 ml from this resuspension was added to a 500 ml LMR medium supplemented with [3-13C]-pyruvate and 15N–(NH4)2SO4, and the solution incubated at 37°C.
P1 nuclease digestion, nucleotide separation and purification
The ribonucleoside monophosphates (rNMPs) were isolated from E. coli (DL323) mutant culture as described earlier (Batey et al. 1992; Thakur et al. 2010a, b). The nucleic acid mixture was digested with P1 nuclease and separated into individual ribo- or deoxy-monophosphates using a cis-diol boronate affinity column chromatography as described (Batey et al. 1992; Thakur et al. 2010a, b). The site specific labeling pattern of rNMPs was verified by NMR prior to re-phosphorylation to rNTPs.
Enzymatic phosphorylation of rNMPs
The detailed procedure for enzymatic phosphorylation of rNMP was described previously (Nyholm et al. 1995; Thakur et al. 2010a, b). Briefly, the individual rAMP, rCMP, rGMP or rUMP and kinases were freshly prepared and pre-incubated at 37°C for 10 min prior to mixing. The enzymatic conversion of the labeled rNMPs to rNTP was complete in 5 h. The progression of the phosphorylation reaction was monitored on a TARGA C18 column using conditions similar to those in the purification step (Supplementary Figure S1). Collected rNTP fractions were pooled and dialyzed overnight and lyophilized.
Site specific enrichment of A-Site RNA and in vitro transcription
A-Site RNA (5′-GGC GUC ACA CCU UCG GGU GAA GUC GCC-3′) was synthesized using in vitro transcription (Milligan et al. 1987; Milligan and Uhlenbeck 1989), with a His-tagged mutant (P266L) T7 RNA polymerase (Guillerez et al. 2005) using a mixture of uniformly labeled guanine and cytosine triphosphates (rGTP and rCTP) (Sigma Aldrich) or site specific labeled rGTP and rCTP. Another set of samples were made using a mixture of uniformly labeled adenosine triphosphates (rATP) (Sigma Aldrich) or site specific labeled rATP. Two terminal 2′-O-methyl modifications in the template strand were introduced to substantially reduce the amount of transcripts with extra nucleotides at the 3′-end (Kao et al. 1999). The optimal transcription conditions were as follows: 10 mM total NTP and 15 mM Mg2+. The reactions were carried out in a transcription buffer A (40 mM Tris–HCl, pH 8.1, 1 mM spermidine, 5 mM dithiothreitol (DTT), 0.01% Triton X-100, 80 mg/ml PEG 8000), 300 nM each DNA strand, and 1.5 μl of 2.4 mg/ml T7 RNAP (optimized amount) per 40 μl of transcription volume. After 3 h of incubation at 310 K, the RNA from the transcription reactions were purified and dialyzed as described (Dayie 2005). After dialysis, the RNA was lyophilized, and resuspended into NMR buffer (100 mM KCl, 10 mM potassium phosphate pH 6.2, 8 or 100% D2O), with or without 3 mM MgCl2, and a trace of sodium azide). The concentration of each NMR sample in Shigemi tubes in 250 μl of buffer was ~0.1 mM.
NMR experiments
All NMR experiments were run on a four channel Bruker Avance III 600 MHz spectrometer equipped with actively shielded z-axis gradient triple resonance probe. The experiments were conducted at temperatures ranging from 15° to 45°C. The NMR data sets were processed and the peak positions and intensities were analyzed with Bruker’s TOPSPIN 2.1 as described previously (Dayie and Thakur 2010; Thakur et al. 2010a, b). One dimensional (1D) 13C spectra and two-dimensional non-constant-time (1H, 13C) heteronuclear single quantum correlation (HSQC) spectra (Bodenhausen and Ruben 1980; Bax et al. 1990) were acquired to analyze the rNMP fractions extracted from each bacterial strain. The fractional 13C enrichment at each carbon site was quantified directly by 1D proton methods or indirectly using 2-bond (2JHN) HSQC as described previously (Dayie and Thakur 2010; Thakur et al. 2010a, b).
To demonstrate the usefulness of site selective labeling 2D 1H–13CH2 TROSY (Meissner et al. 1998; Miclet et al. 2004) spectra were measured using non-constant time evolution in the carbon dimension. Methylene CH2-TROSY experiments (Miclet et al. 2004) were run with the following modifications that dispense with selective decoupling: the WURST-4 decoupling waveform was omitted during the carbon t 1 evolution period, and a non-selective 180° pulse replaced the selective pulse on the C5′ carbon. To compare the usefulness of the modified CH2-TROSY experiments, the normal CH2-methylene optimized HSQC (Schleucher et al. 1994; Sattler et al. 1995) were also run under identical conditions: for each data set, 16 scans and 1,024 complex points were collected in t 2 and 512 complex points were collected in t 1 using the Echo-Anti echo method (Palmer et al. 1991; Kay et al. 1992) for quadrature detection; proton and carbon carrier was placed at 4.7 and 64 ppm respectively as described (Thakur et al. 2010a, b). The time domain data sets were zero filled in t 1 and t 2 before Fourier transformation to give a final real matrix size of 2,048 × 1,024 points.
Longitudinal (R1) relaxation rates were measured for ribose C1′ and cytosine nucleobase C5 carbons using TROSY detected experiments (Hansen and Al-Hashimi 2007). To compare the usefulness of the selective labels relative to uniform labels, the R1 relaxation experiments were run under identical conditions: for each data set, 64 scans and 1,024 complex points were collected in t 2, and 128 complex points were collected in t 1 using the Echo-Anti echo method (Palmer et al. 1991; Kay et al. 1992) for quadrature detection. Proton and carbon carrier was placed at 4.7 and 93 ppm respectively. The time domain data sets were zero filled in t 1 and t 2 before Fourier transformation to give a final real matrix size of 2,048 × 1,024 points. For the 2D longitudinal (R1) experiments, relaxation delays of 21.1 (2x), 61.4, 141.1, 301.2, 381.3, 461.3 (2x), 781.6 ms were used. The longitudinal relaxation decay curves were fitted using the Levenberg–Marquardt algorithm by assuming monoexponential decay.
Results
Incorporation of13C into ribose and base of nucleotides
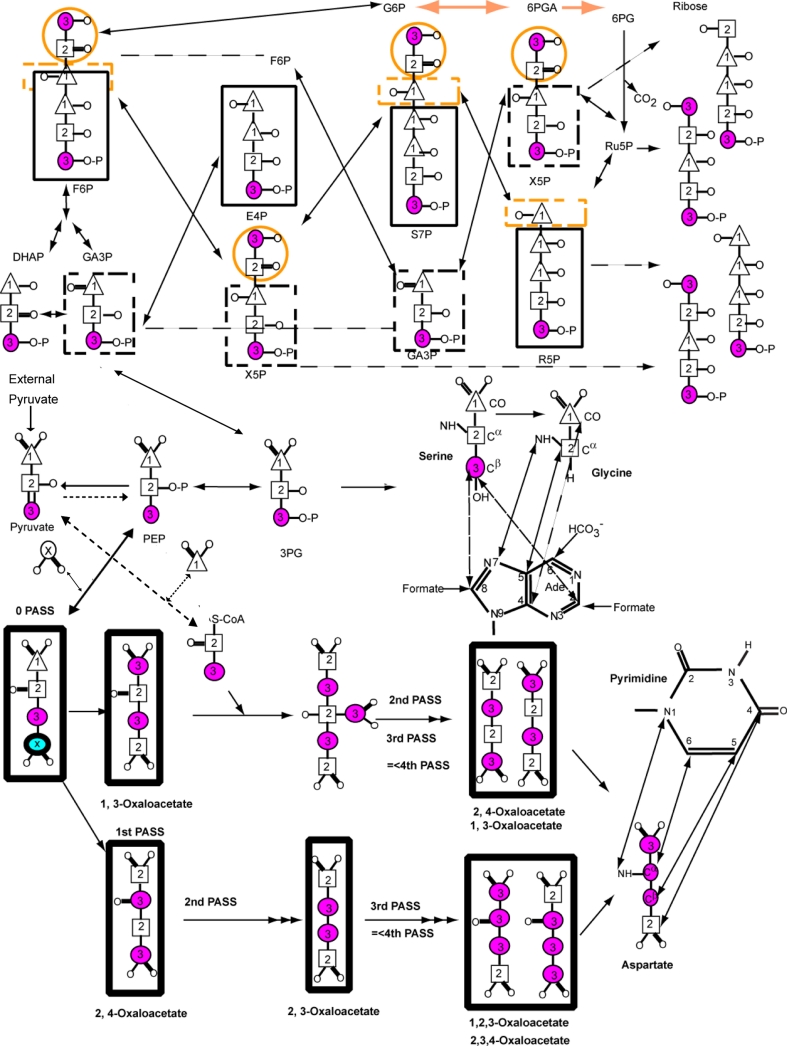
To test the hypothesis that an asymmetric carbon source such as [3-13C]-pyruvate will provide superior site-selective labeling, we compared the growth of wild type and two mutant E. coli strains on [3-13C]-pyruvate (Fig. 1 and Table 1). To place our results in context, we outline nucleotide metabolism in E. coli via glycolysis, gluconeogenesis, and the Kreb cycle using pyruvate as the sole carbon source. If glucose is used as the sole carbon source, the conversion of PEP (phosphoenolpyruvate) to pyruvate by pyruvate kinase is an irreversible reaction under physiological conditions (Fig. 1). However, under minimal growth conditions in E. coli using pyruvate as the sole carbon source, PEP synthetase (PPS) can convert pyruvate directly to PEP (Fig. 1), and PEP carboxylase (PPC) can convert PEP to an oxaloacetate intermediate (Chao et al. 1993; Schilling et al. 2001). In eukaryotes, pyruvate is converted to PEP via a two-step process with an oxaloacetate intermediate catalyzed by pyruvate carboxylase and PEP carboxykinase. However, pyruvate carboxylase is a mitochondrial protein not operative in E. coli, and so we expect all PEP will be derived from the externally supplied pyruvate without dilution by the TCA cycle for the DL323 E. coli strain. Consequently purine and pyrimidine moieties derived from oxaloacetate and serine should have enrichment levels quite near that of the initial pyruvate. Similarly the labeling pattern in the ribose ring derived from PEP, which is not diluted by the TCA cycle, should label only C1′ and C5′ in 1:2 ratio and all other sites should have minimal label.
Fig. 1.
Major metabolic pathways involved in the production of nucleic acid nucleotides from pyruvate, including key steps in glycolysis, gluconeogenesis and several passes through the tricarboxylic (TCA) cycle as derived from Covert and Palsson (2002). With the E. coli strain lacking succinate and malate dehydrogenases (DL323), the oxidative branch of the pentose phosphate pathway remains intact but the TCA cycle is severed in two places such that the oxaloacetate is derived exclusively from carboxylation of phosphoenolpyruvate (PEP) and the resulting label is not diluted by the TCA cycle. Atom labels for the terminal (3) carbon (magenta and thin circle), the central (2) carbon (square), and the terminal (1) carbon (triangle) of pyruvate are highlighted. Positions that are enriched due to the presence of 13CO2 in the growth medium are shown with a circled x. Pyrimidine base derived from the oxaloacetate (OAA) produced by carboxylation of PEP is shown via the aspartate intermediate. This OAA cannot be used as a substrate in the first and subsequent rounds of the TCA cycle because of the two mutations. Consequently OAA derived aspartate amino acid can be produced with 13C labeling at only the Cα (C6) position if [2-13C]-pyruvate is used. If [3-13C]-pyruvate is used only Cβ (C5) position is labeled. In either case carboxylation of PEP leads to labeling of the Cγ (C4) position. Similarly the labeling pattern of purines from glycine derived from 3PG are shown such that if [2-13C]-pyruvate is used only the Cα position of Gly and therefore C5 position of the purine ring is labeled. Otherwise if [3-13C]-pyruvate is used the CO of Gly and therefore C4 of purine ring is labeled, and the labeling of the Cβ position of Ser also leads to labeling of the purine C2 and C8 positions
Table 1.
13C enrichment levels at various carbon positions within ribonucleotides harvested from K12, DL323, and K10zwf E. coli strains grown on [3-13C]-pyruvate
| Carbon position labeled | DL323 E. coli strain | K10zwf E. coli strain | K12 E. coli strain |
|---|---|---|---|
| Purinea | |||
| Ade C2 | 99 ± 1 | 85 ± 4 | 95 ± 2 |
| C8 | 93 ± 1 | 57 ± 1 | 98 ± 2 |
| Pyrimidinea | |||
| C5 | 98 ± 4 | 81 ± 4 | 91 ± 1 |
| C6 | 15 ± 1 | 60 ± 4 | 64 ± 1 |
| Ribose | |||
| C1′a | 42 ± 4 | 82 ± 2 | 83 ± 2 |
| C2′b | 4 | 9 ± 1 | 21 ± 1 |
| C3′b | <2 | <1 | 8 |
| C4′b | 3 | 9 ± 1 | 30 ± 1 |
| C5′c | 95 ± 5 | 95 | 95 ± 1 |
aThe percentage label is calculated as an average of two methods: (i) the ratio of the sum of the intensities of satellite peaks to the sum of the intensities of the satellite and center peaks using the 2-bond 15N HSQC without 13C decoupling during acquisition (Dayie and Thakur 2010) and (ii) the ratio of the sum of the intensities of satellite peaks to the sum of the intensities of the satellite and center peaks using the 1D 1H experiment without 13C decoupling during acquisition as described in the text
b,cThe percentage label (Plabel) is calculated as in (a) but this time with only method (ii)
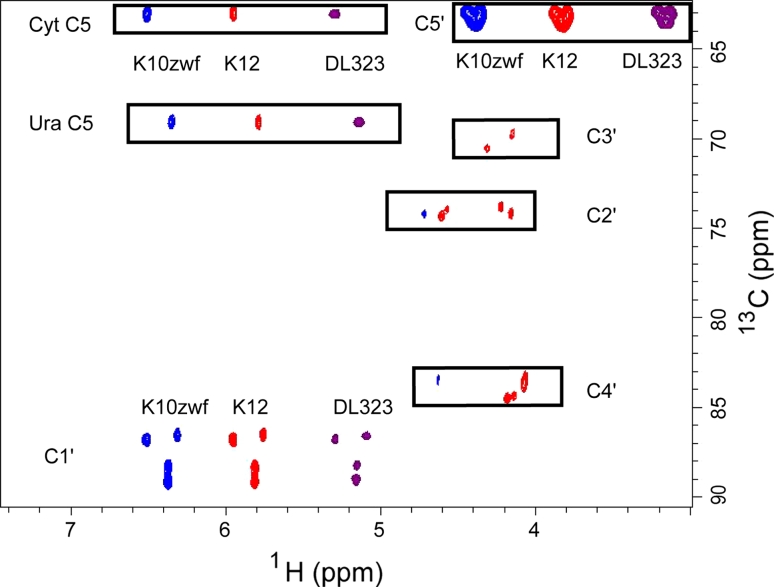
In agreement with this metabolic analysis (Fig. 1) and for DL323 E. coli grown on [3-13C]-pyruvate, the ribose ring is labeled only in the C1′ and C5′ carbon positions at ~42 and 98% respectively (Fig. 2 and Table 1). Thus these two positions remain singlet. This labeling pattern is very different from what was observed for DL323 E. coli grown on [1, 3-13C]-glycerol (Johnson and Hoogstraten 2008; Thakur et al. 2010a, b). In this case of growth on glycerol, the ribose ring is labeled in all but the C4′ carbon position, so that the C2′ and C3′ positions suffer from multiplet splitting by carbon–carbon coupling and the C1′ position has some admixture of residual C1′–C2′ coupling (Thakur et al. 2010a, b).
Fig. 2.
2D non-constant time HSQC spectra of all four labeled nucleotides extracted from K10zwf (blue contours, left shifted) or K12 (red contours) or DL323 (purple contours, right shifted) E. coli strains grown on [3-13C]-pyruvate. Growth on [3-13C]-pyruvate results in label at only C1′ and C5′ for DL323 and K10 strains, whereas growth of K12 leads to some residual label at C2′, C3′, and C4′ in addition to the major labels at the C1′ and C5′ carbons. The cytosine (Cyt) and Uracil (Ura) C5 resonances at 96.67 and 102.69 ppm respectively are folded into the spectrum
Similar to DL323, K10zwf E. coli grown on [3-13C]-pyruvate leads to exclusive labeling of the C1′ and C5′ carbon position without 13C–13C coupling evident (Fig. 2). Importantly, in this case the abundance of 13C label at C1′ is higher than that obtained with DL323 (Table 1). Again similar to DL323 and K10zwf E. coli, wildtype K12 grown on [3-13C]-pyruvate also leads to labeling of the C1′ and C5′ carbon positions, but unlike the other two strains, sufficient residual labeling of the C2′ and C3′ carbon atoms leads to 13C–13C coupling (Figs. 1, 2). This residual labeling presumably arises from PEP derived from the mixture of [1, 2, 3-13C]-oxaloacetate and [2, 3, 4-13C]-oxaloacetate intermediates (Fig. 1).
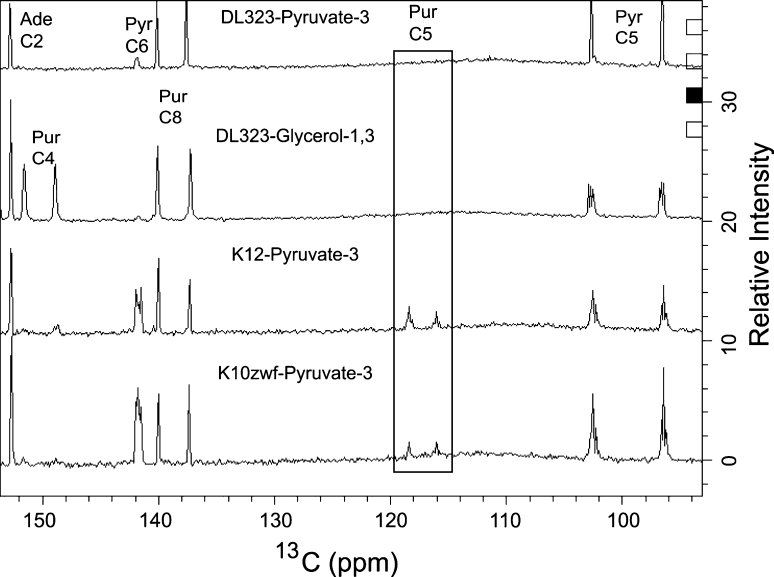
The dramatic improvement of the labeling afforded by using [3-13C]-pyruvate is most visible in the base region. For nucleotides derived from K10zwf and K12 cultures, both the protonated C5 and C6 carbon positions of pyrimidines are substantially labeled (≥60%) because the flux through the TCA cycle is efficient in these two E. coli strains. This efficient flux also ensures that the purine C4 and C5 positions are labeled. A single pass through the TCA cycle leads to an equal probability of carrying the 13C label into either the pyrimidine C5 or C6 position because the oxaloacetate is generated by [1, 3-13C]-malate or [2, 4-13C]-malate (Fig. 1). If the precursor is [1, 3-13C]-oxaloacetate, then the desired label at pyrimidine C5 or C6 is diluted after the first and subsequent rounds of the TCA cycle (Fig. 1). This dilution of labeling is observed for the nucleotides derived from the cultures of E. coli K10zwf and K12 strains (Table 1). Again, if the precursor is [2, 4-13C]-oxaloacetate, then the second pass through the TCA cycle leads to a [2, 3-13C]-oxaloacetate and subsequent cycles leads to a mixture of [1, 2, 3-13C]-oxaloacetate and [2, 3, 4-13C]-oxaloacetate. The net effect of the TCA cycle is to thoroughly scramble the labels arising from oxaloacetate such that, in addition to the desired selective label at either the pyrimidine C5 or C6, undesired pyrimidine C5–C6 or C4–C5–C6 labeled spin pairs would be obtained. Thus the significant coupling observed between pyrimidine C4 and C5 or C5 and C6 (Fig. 3; Supplementary Figure S2) is a consequence of this scrambling by the TCA cycle. These labeling patterns, similar to those observed with [1, 3-13C]-glycerol grown on DL323, have the undesirable consequence of substantive 13C–13C couplings that degrade the resolution and the accuracy of measured relaxation parameters (as shown below). In contrast to K10zwf and K12, E. coli strain DL323 provides a much cleaner labeling pattern that is devoid of unwanted labeling at the pyrimidine C6 and purine C4 and C5 positions (Fig. 3; Supplementary Figure S2). For the DL323 strain, the flux through the TCA cycle is reduced essentially to zero, thereby preventing the dilution of the labels arising from oxaloacetate described above.
Fig. 3.
Labeling pattern of a mixture of four rNMPs isolated from K10zwf, K12, and DL323 E. coli grown on a [3-13C]-Pyruvate background, and DL323 E. coli grown on a [1, 3-13C]-glycerol background. The direct carbon detection 1D spectrum shows all the labeled carbon positions. A long recycle delay of 5 s was used to allow for sufficient magnetization recovery and proton decoupling was limited to the acquisition period only. The residual 13C–13C coupling observed in a [1, 3-13C]-glycerol is absent with the [3-13C]-Pyruvate only in DL323; K10 and K12 grown on [3-13C]-Pyruvate still retain the residual 13C–13C coupling
As expected, all of the C2 and C8 atoms of purine are substantially labeled to >90% for nucleotides derived from DL323 and K12 cultures, but those from K10zwf have these labels reduced to 85 and 57% respectively partly because the scrambling from the TCA cycle apparently feeds more efficiently into the purine biosynthetic pathway for K10zwf than K12 (Figs. 2, 3; Supplementary Figure S2). The origin of these differences is not clear. Nonetheless, some of the scrambled malate or oxaloacetate from the TCA cycle is likely fed back into PEP such that purine C4, purine C5, and pyrimidine C6 positions are labeled in K10zwf and K12 but not DL323 (Fig. 3). As a result, K10zwf and K12 provide 13C–13C spin pairs in the purine and pyrimidine base moieties that can potentially hinder accurate extraction of relaxation parameters (shown below).
Applications of site selective labels for 13C NMR study of nucleic acids
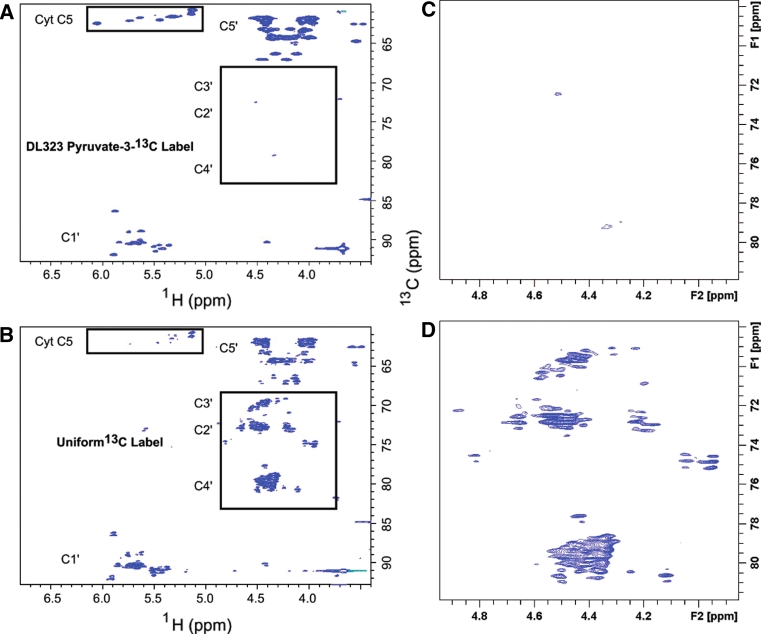
Spectra obtained using our selective labeling methodology is of high quality (Figs. 4 and 5). A 27-nt A-site ribosomal RNA fragment derived from the 30S ribosomal subunit (Purohit and Stern 1994; Fourmy et al. 1996; Kaul et al. 2006; Wirmer and Westhof 2006; Schmeing and Ramakrishnan 2009) was transcribed using either the uniformly 13C/15N-labeled GTP and CTP or the site selectively-labeled GTP and CTP derived from DL323 E. coli cells grown on [3-13C]-pyruvate as described above. For the C,G-uniformly 13C-labeled sample, the peaks are not only broader, but they also overlap extensively (Fig. 4). These unwanted splitting can also be removed using either constant time experiments, adiabatic band selective decoupling schemes, or maximum entropy reconstruction-deconstruction; as discussed previously, these have their own limitations (Dayie and Thakur 2010; Thakur et al. 2010a, b). In particular, the length of the constant time period (T) limits the acquisition times to multiples of the homonuclear coupling constant. To obtain reasonable digital resolution, large values of T are needed and this leads to significant signal attenuation for RNA molecules larger than 30 nucleotides (Dayie 2005). But with the selective labels, one is not forced to compromise resolution for improved sensitivity.
Fig. 4.
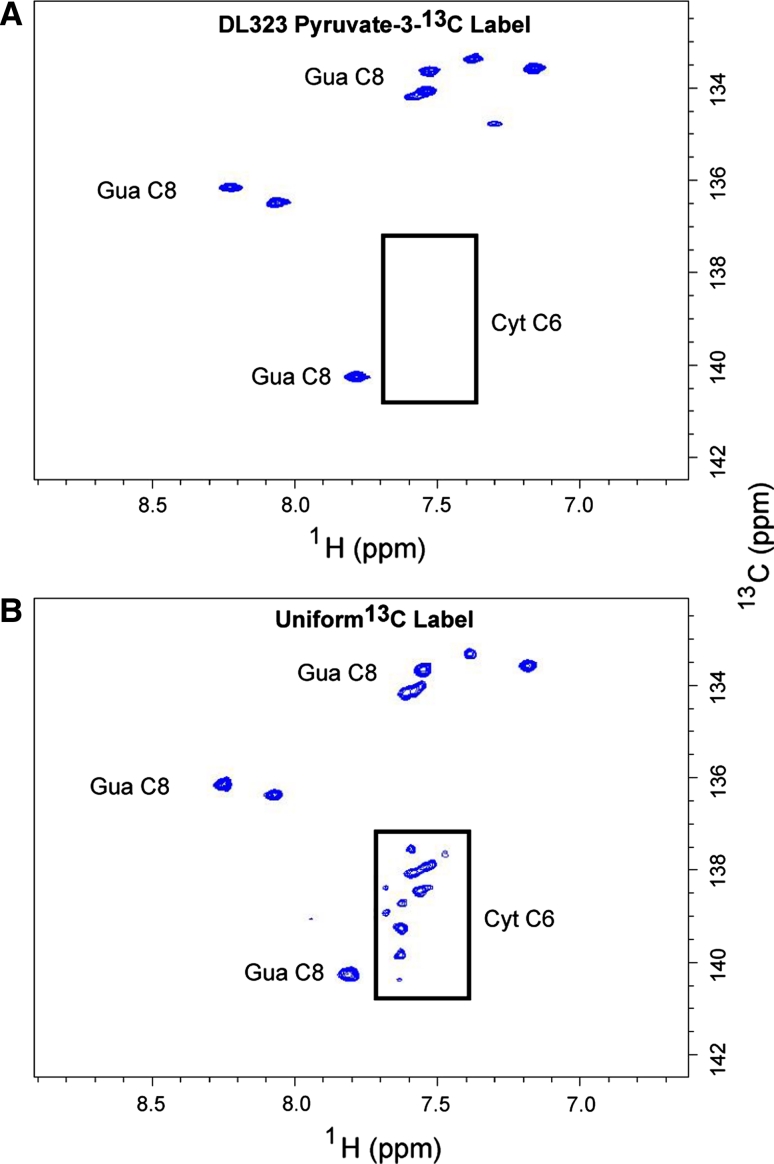
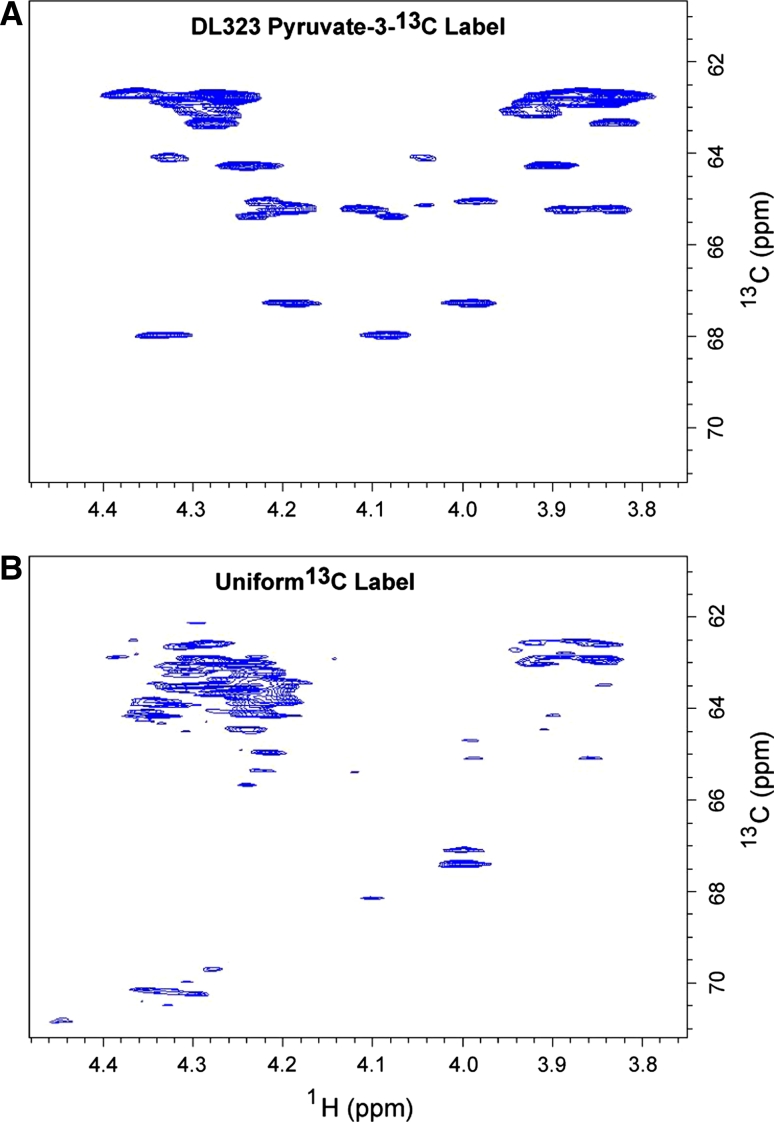
NMR spectra showing enhanced resolution afforded by site selective labeling of A-Site RNA bound to paromomycin. The experiments were performed on the A-Site RNA a site-selectively 13C-GTP and CTP labeled and b uniformly 13C-GTP and CTP labeled. 2D non-constant time HSQC spectra of the ribose region. The cytosine (Cyt) and Uracil (Ura) C5 resonances at 96.67 and 102.69 ppm respectively are folded into the spectrum and are boxed to highlight the reduced signal from the uniformly labeled sample. The C2′, C3′, and C4′ regions are boxed to highlight the absence of labeling in selectively labeled sample. Blown-up views of the C2′, C3′, and C4′ regions boxed in (a) and (b) are depicted in (c) and (d) respectively
Fig. 5.
NMR spectra showing enhanced resolution afforded by site selective labeling of A-Site RNA bound to paromomycin. The experiments were performed on the A-Site RNA a site-selectively 13C-GTP and CTP labeled and b uniformly 13C-GTP and CTP labeled. 2D non-constant time HSQC spectra of the base region. The cytosine (Cyt) and Uracil (Ura) C6 resonances are boxed to highlight the absence of labeling in selectively labeled sample
Use of the site selectively labeled NTPs removes the crowding due to the splitting of signals from J-coupling and eliminates the C2′, C3′, and C4′ resonances while preserving only the C1′ and C5′ signals (Fig. 4). In addition to improving resolution, sensitivity is also greatly enhanced. For instance, cytosine C5 has a large chemical shielding anisotropy (CSA) and so decays rapidly, and as expected most of the C5 resonances are absent in the spectra of the uniformly labeled sample. These signals are preserved in the spectra of the site selectively labeled sample (Fig. 4).
In the C8 purine and C6 pyrimidine regions, the selective labels once again improve the resolution and sensitivity (Fig. 5). By eliminating the C6 pyrimidine signals, the guanosine C8 signals can be readily identified without concerns about potential overlap with cytosine C6, especially for a sample that is labeled with all four nucleotides. These observations underscore the difference selective labeling makes on sensitivity and resolution.
A non-constant time version of the 13CH2 TROSY experiment (Miclet et al. 2004) that enables the rescue of the slowest relaxing multiplet component was next performed to demonstrate the usefulness of these site selective labels. Compared to uniform labeling, the site-selective labels shows two clear advantages of improved resolution and sensitivity (Fig. 6a, b) of a spectrum of a 27-nt A-Site RNA fragment labeled only in G and C. Compared to the normal CH2-methylene optimized experiment (Schleucher et al. 1994; Sattler et al. 1995), 14 out of the expected 18 GC C5′ resonances are visible in the TROSY experiment but most of these are either overlapped, or weak, or absent in the uniformly labeled sample. Thus, it is anticipated that these and other new experiments that incorporate the 13CH2 TROSY module can be designed to probe RNA-ligand interactions at very high resolution using the site specific labels described here.
Fig. 6.
CH2-TROSY spectra depicting the C5′ region of A-Site RNA bound to paromomycin. Experiments were performed on the A-Site RNA a site-selectively 13C-GTP and CTP labeled and b uniformly 13C-GTP and CTP labeled. Spectra are plotted at identical contour levels. Notice the dramatic gain in sensitivity and resolution with the site selectively labeled sample
Site selective labeling affords more accurate relaxation rate measurements using non-constant time non-selective pulse experiments
To place the results of the relaxation in context, it is instructive to examine how spins are dipolar coupled to each magnetization of interest in a relaxation measurement. For instance, the longitudinal relaxation of the 13C5 spin during a relaxation period T for a uniformly labeled RNA sample has the following time dependency (Yamazaki et al. 1994; Dayie and Wagner 1997; Boisbouvier et al. 2003; Hansen and Al-Hashimi 2007):
| 1 |
ΔCi are the deviations of each carbon’s longitudinal magnetization from its equilibrium magnetization, ΔH5 is the deviation of the H5 proton magnetization from its equilibrium magnetization, and the autorelaxation and cross relaxation rates are given by RC5-X(z) and RX(x → C5) respectively. Similar equations can be written for the ribose C1′ spin and all its dipolar coupled neighbors. These rates are given as follows (Eldho and Dayie 2007):
| 2 |
| 3 |
| 4 |
| 5 |
Here X = C4 or C6. The physical constants γH and γC are the gyromagnetic ratios of 1H and 13C respectively (γC = 6.7283 × 107 rad (Ts)−1, γH = 26.75 × 107 rad (Ts)−1), rHC is the length of the internuclear 1H–13C bond vector, rCC is the length of the internuclear 13C–13C bond vector, and
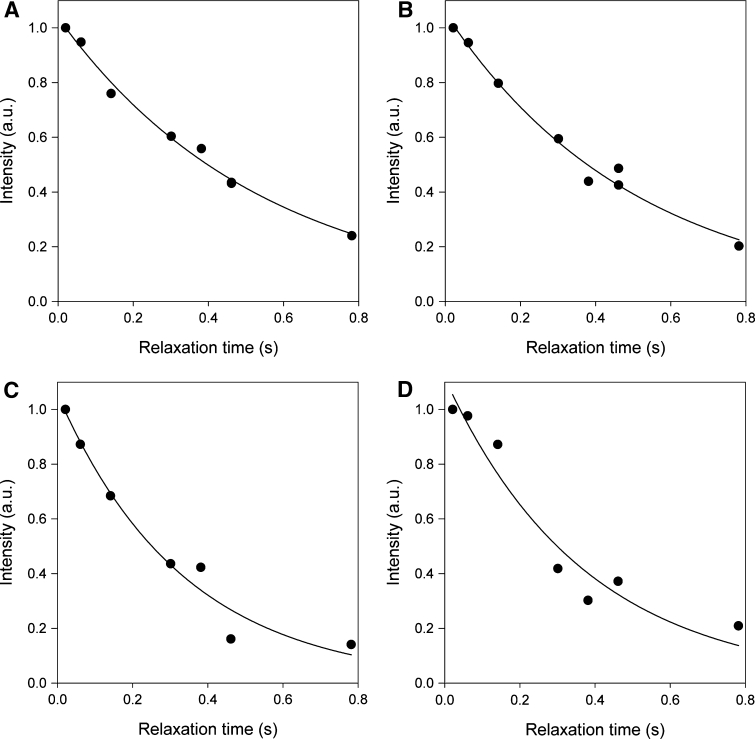
We expected therefore for the case of a uniformly labeled cytosine or uridine, the H5, C4, C6 spins will contribute to the longitudinal relaxation of the 13C5 nucleus, and H1′ and C2′ spins will contribute to the relaxation of the 13C1′ nucleus. For the site selectively labeled nucleotides, only H5 will contribute to the longitudinal relaxation of the 13C5 nucleus, and only H1′ will contribute to the relaxation of the 13C1′ nucleus. Thus, the labeling pattern of rNMPs derived from DL323 grown on [3-13C]-pyruvate is particularly attractive for relaxation studies of the ribose C1′ and the base C5 (as well as C2 and C8 positions that are not discussed further here). Each of these positions is essentially singlet. The isolation of the C1′ ribose and the C5 base positions from labeled adjacent neighbors means that the interference described above that arises from strong 13C–13C magnetic interactions in the base and ribose rings is no longer a hindrance for extracting accurate relaxation parameters. Therefore as a third demonstration of the usefulness of the site selectively labeled versus the uniformly labeled nucleotides for quantifying dynamics in RNA, R1 experiments were carried on the 27–nt A-site ribosomal RNA fragment transcribed using uniformly 13C/15N-labeled GTP and CTP and site selectively-labeled GTP and CTP derived from DL323 E. coli cells grown on [3-13C]-pyruvate. Examples of 13C R1 decay curves for both ribose C1′ and base carbon C5 sites are depicted in Fig. 7 and Supplementary Figure S4. The decay curves fit well to a mono-exponential function for the C1′ position of a site selective ATP- labeled sample (Fig. 7a), but not as well for a uniform ATP-labeled sample (Fig. 7b). The discrepancy between R1 measured for uniform (1.8 ± 0.08 s−1) and site-selective (2.0 ± 0.12 s−1) labeled sample is 0.2 s−1 (Fig. 7a). Similarly, the fit to a monoexponential decay function is only applicable to the site selectively CTP-labeled RNA sample (Fig. 7c). Again fit to a monoexponential decay function is only applicable to the C5 position of the site selectively CTP-labeled RNA sample. This discrepancy is also worse for the C5 carbons with relaxation rates of 3.4 ± 1.0 s−1 for the uniformly labeled sample and 2.9 ± 0.4 s−1 for the selectively labeled sample, is part because of the expected dipolar coupled interactions, but also because of substantial decrease in signal-to noise for the uniformly labeled sample (Supplementary Figure 4). These observations suggest that the contributions from the cross-relaxation terms in (1), (3), and (5) are not negligible for the uniformly labeled sample.
Fig. 7.
Representative longitudinal R1 relaxation decay curves showing deviation from monoexponential decay for uniformly labeled samples. Ribose C1′ R1 relaxation measurements at 25°C for the A-site RNA labeled with a site selectively-labeled ATP and b uniformly 13C/15N-labeled ATP. Ribose C1′ R1 relaxation measurements at 25°C for A-site RNA labeled with c site selectively-labeled GTP and CTP and d uniformly 13C/15N-labeled GTP and CTP
Discussion
Previous labeling technologies using wild type E. coli strains and symmetric carbon sources spurred the development of new solution NMR methods to characterize the structure and dynamics of small to medium sized uniformly labeled RNA molecules (Batey et al. 1995; Hall 1995; Fürtig et al. 2003; Latham et al. 2005; Dayie 2008; Lu et al. 2010). A number of alternative labeling approaches have been proposed since then, and it is important therefore to discuss the advantages and limitations of these different methods. First, uniform labeling introduces direct one-bond and residual dipolar couplings that prevents accurate measurement of 13C relaxation rates (Johnson et al. 2006; Dayie and Thakur 2010; Thakur et al. 2010a, b) and decreases the resolution and sensitivity of NMR experiments. For instance, growth of wildtype E. coli strain K12 on [1-13C]-acetate yields 13C label at the ribose C1′, C2′, and C3′ atomic sites but very little label at C4′ and C5′ (Hoffman and Holland 1995). As a result, C2′ and C3′ have substantial multiplet structure and even C1′ has residual multiplet structure (Supplementary Figure S3). In addition, the protonated aromatic carbons (purine C2 and C8 and pyrimidine C5 and C6) are not labeled, making this particular labeling pattern less useful for routine NMR applications. In contrast, growth of wildtype E. coli strain K12 on [2-13C]-acetate yields 13C label at all five ribose carbon positions of C1′, C2′, C3′, C4′ and C5′ (Hoffman and Holland 1995). Since each site is highly enriched, all five positions have substantial multiplet structure (Supplementary Figure S3). Also for the aromatic carbons, the protonated carbon C5 and C6 of pyrimidines and C2 and C8 of purines are labeled at very high level (Hoffman and Holland 1995; Supplementary Figure S3). Similarly, growth of wildtype E. coli strain K12 on [2-13C]-glucose yields 13C label only at C1′, C2′, and C4′. No label is observed at C3′ and C5′. For the aromatic carbons, the protonated carbon C5 and C6 of pyrimidine and C2 and C8 of purine are also labeled at very high level (Hoffman and Holland 1995). These second set of labeling patterns are therefore very useful for NMR applications that require substantial coupling for transfer of coherences, but problematic for applications that require suppression of these coupling networks (Supplementary Figure S3 E).
To remove these coupling networks, alternate 13C–12C labeling schemes have been shown to be important for tackling these difficulties in RNA molecules (Johnson et al. 2006; Johnson and Hoogstraten 2008; Dayie and Thakur 2010; Thakur et al. 2010a, b). However some of the earlier schemes had their unique problems. For instance, the labeling pattern of rNTPs derived from DL323 grown on [1,3-13C]-glycerol is not attractive for relaxation studies because the ribose C2′ position is doublet, the C1′ retains some residual doublet arising from 13C2′–13C1′ isotopomers, and the C4′ has no label (Thakur et al. 2010a, b). In the base region the pyrimidine C5 site has multiplet structure, again precluding its use for accurate relaxation measurements (Thakur et al. 2010a, b). The use of G6PDH mutant strains rather than DL323 using 2-13C-glycerol was also recommended in previous work (Johnson et al. 2006).
Recently de novo biosynthetic methods have been proposed that potentially resolve these problems (Schultheisz et al. 2008, 2011). However, the new method requires 29 enzymes for making ATP and GTP and 18 enzymes for making CTP and UTP. Of these enzymes only a few are commercially available, which makes this method excellent for adaptation by companies but not by individual laboratories. Additionally, the de novo method requires selectively labeled glucose and serine which are not readily available commercially, or prohibitively expensive, or both. For instance the de novo method of labeling ribose C1′ and C5′ in the context of purine C2, C5, and C8, indicated above by our method, requires 13C-2,6-glucose that is not commercially available and 13C-3-serine that costs ~$4,000/g; the biomass method is able to make these labels readily.
Of greater interest the main advantage of using the E. coli strain DL323 grown on a non-symmetric carbon source such as [3-13C]-pyruvate is that it provides isolated two-spin systems that are ideal for relaxation measurements as well as improving resolution and sensitivity. For example in the ribose region, the C1′ and C5′ labels are completely isolated from C2′ and C4′ spins respectively. Similarly in the base region, the purine C2 and C8 and pyrimidine C5 spins are isolated from the network of carbon spins within the aromatic ring. For instance in a uniformly labeled sample, adenine C2 has substantial 2JC2C5 coupling of 11 Hz to the C5 carbon, and the purine C8 carbons have 2JC4C8 coupling of ~9 Hz to the C5 carbon (Wijmenga and van Buuren 1998). In addition, the carbon atoms C4 (149–154 ppm)/C5 (116–120 ppm)/C6 (156–161 ppm)/C8 (131–142 ppm) resonate within a chemical shift range that precludes effectively decoupling C8 from the C4, C5, or C6 carbon atoms. These complications again prevent accurate relaxation measurements using CPMG experiments. Formate supplementation has been proposed as a potential cost-effective solution (Latham et al. 2005; Dayie and Thakur 2010; Thakur et al. 2010a, b). However, the C8 is labeled to ~90% and C2 to only ~25%, limiting the general usefulness of formate labeling. Thus the confounding effects of scalar coupling from adjacent labeled sites are eliminated using the current method proposed in this work, unlike the case for previous labels.
Conclusions
Any RNA sequence can be site specifically labeled with site alternate 13C-enriched isotopes efficiently prepared from site-specifically labeled rNTPs using in vitro transcription by T7 RNA polymerase. We have presented an efficient method for preparing these ribonucleotides containing site specific distributions of 13C enrichment that has significant advantages in terms of cost, flexibility, resolution and sensitivity over previously reported procedures. These labels will be particularly beneficial to investigators using heteronuclear NMR spectroscopy to study the structure and dynamics of RNA and RNA complexes of increasingly large sizes, currently an important and active area of research.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported in part by the University of Maryland Nano-Biotechnology Award and the National Institutes of Health grant GM077326 to T.K.D. We are grateful to Professors Jon Dinman and Steve Rokita for helpful discussions.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- AMP
Adenosine 5′-monophosphate
- CMP
Cytidine 5′-monophosphate
- UMP
Uridine 5′-monophosphate
- GMP
Guanosine 5′-monophosphate
- R5P
Ribose-5-phosphate
- FBP
Fructose-6-bisphosphate
- F6P
Fructose-6-phosphate
- GA3P
Glyceraldehyde-3-phosphate
- G6PDH
Glucose-6-phosphate dehydrogenase
- K10zwf
Glucose-6-phosphate dehydrogenase mutant
- Gly
Glycine
- Ser
Serine
- noPPP
Non-oxidative pentose phosphate pathway
- OAA
Oxaloacetate
- DHAP
Dihydroxyacetone phosphate
- oPPP
Oxidative pentose phosphate pathway
- rNTPs
Ribonucleoside triphosphates
- TIM
Triosephosphate isomerase
- PEP
Phosphoenolpyruvate
- G6P
Glucose-6-phosphate
- Ru5P
Ribulose-5-phosphate
- X5P
Xylulose-5-phosphate
- S7P
Sedoheptulose-7-phosphate
- E4P
Erythrose-4-phosphate
- 3PG
3-phosphoglycerate
- 6PG
6-phosphogluconate
- 6PGA
6-phosphogluconate-δ-lactone
Footnotes
This article was published twice due to a technical mishap. In a citation following information is correct: Thakur CS, Dayie TK (2011) Asymmetry of 13C labeled 3-pyruvate affords improved site specific labeling of RNA for NMR spectroscopy. J Biomol NMR 51:505-517 DOI: http://dx.doi.org/10.1007/s10858-011-9581-6
References
- Batey RT, Inada M, Kujawinski E, Puglisi JD, Williamson JR. Preparation of isotopically labeled ribonucleotides for multidimensional NMR spectroscopy of RNA. Nucleic Acids Res. 1992;20:4515–4523. doi: 10.1093/nar/20.17.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Battiste JL, Williamson JR. Preparation of isotopically enriched RNAs for heteronuclear NMR. Methods Enzymol. 1995;261:300–322. doi: 10.1016/S0076-6879(95)61015-4. [DOI] [PubMed] [Google Scholar]
- Bax A, Ikura M, Kay LE, Torchia DA, Tschudin R. Comparison of different modes of two-dimensional reverse correlation NMR for the study of proteins. J Magn Reson. 1990;86:304–318. [Google Scholar]
- Bodenhausen G, Ruben DJ (1980) Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem Phys Lett 69:185–189
- Boisbouvier J, Wu Z, Ono A, Kainosho M, Bax A. Rotational diffusion tensor of nucleic acids from 13CNMR relaxation. J Biomol NMR. 2003;27:133–142. doi: 10.1023/A:1024931619957. [DOI] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Breaker RR. Riboswitches: from ancient gene-control systems to modern drug targets. Future Microbiol. 2009;4:771–773. doi: 10.2217/fmb.09.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao YP, Patnaik R, Roof WD, Young RF, Liao JC. Control of gluconeogenic growth by pps and pck in Escherichia coli. J Bacteriol. 1993;175:6939–6944. doi: 10.1128/jb.175.21.6939-6944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowes R (1968) In: Hayes W (ed) Experiments in microbial genetics. Wiley, New York
- Covert MW, Palsson BO. Transcriptional regulation in constraints-based metabolic models of Escherichia coli. J Biol Chem. 2002;277:28058–28064. doi: 10.1074/jbc.M201691200. [DOI] [PubMed] [Google Scholar]
- Dayie KT. Resolution enhanced homonuclear carbon decoupled triple resonance experiments for unambiguous RNA structural characterization. J Biomol NMR. 2005;32:129–139. doi: 10.1007/s10858-005-5093-6. [DOI] [PubMed] [Google Scholar]
- Dayie KT. Key labeling technologies to tackle sizeable problems in RNA structural biology. Int J Mol Sci. 2008;9:1214–1240. doi: 10.3390/ijms9071214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayie TK, Thakur CS. Site-specific labeling of nucleotides for making RNA for high resolution NMR studies using an E. coli strain disabled in the oxidative pentose phosphate pathway. J Biomol NMR. 2010;47:19–31. doi: 10.1007/s10858-010-9405-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayie KT, Wagner G. Carbonyl carbon probe of local mobility in 13C, 15N-enriched proteins using high resolution NMR. J Am Chem Soc. 1997;119:7797–7806. doi: 10.1021/ja9633880. [DOI] [Google Scholar]
- Eldho NV, Dayie KT. Internal bulge and tetraloop of the catalytic domain 5 of a group II Intron ribozyme are flexible: implications for catalysis. J Mol Biol. 2007;365:930–944. doi: 10.1016/j.jmb.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Fourmy D, Recht MI, Blanchard SC, Puglisi JD. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- Fraenkel DG. Selection of Escherichia coli mutants lacking glucose-6-phosphate dehydrogenase or gluconate-6-phosphate dehydrogenase. J Bacteriol. 1968;95:1267–1271. doi: 10.1128/jb.95.4.1267-1271.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürtig B, Richter C, Wöhnert J, Schwalbe H. NMR spectroscopy of RNA. ChemBioChem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- Guillerez J, Lopez PJ, Proux F, Launay H, Dreyfus M. A mutation in T7 RNA polymerase that facilitates promoter clearance. Proc Natl Acad Sci USA. 2005;102:5958–5963. doi: 10.1073/pnas.0407141102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall KB. Uses of 13C- and 15N-labeled RNA in NMR of RNA-protein complexes. Methods Enzymol. 1995;261:542–559. doi: 10.1016/S0076-6879(95)61024-3. [DOI] [PubMed] [Google Scholar]
- Hansen AL, Al-Hashimi HM. Dynamics of large elongated RNA by NMR carbon relaxation. J Am Chem Soc. 2007;129:16072–16082. doi: 10.1021/ja0757982. [DOI] [PubMed] [Google Scholar]
- Hansen EJ, Juni E. Properties of mutants of Escherichia coli lacking malic dehydrogenase and their revertants. J Biol Chem. 1979;254:3570–3575. [PubMed] [Google Scholar]
- Hoffman DW, Holland JA. Preparation of C-13-labeled ribonucleotides using acetate as an isotope source. Nucleic Acids Res. 1995;23:3361–3362. doi: 10.1093/nar/23.16.3361-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Hoogstraten CG. Extensive backbone dynamics in the GCAA RNA tetraloop analyzed using 13C NMR spin relaxation and specific isotope labeling. J Am Chem Soc. 2008;130:16757–16769. doi: 10.1021/ja805759z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Julien KR, Hoogstraten CG. Alternate-site isotopic labeling of ribonucleotides for NMR studies of ribose conformational dynamics in RNA. J Biomol NMR. 2006;35:261–274. doi: 10.1007/s10858-006-9041-x. [DOI] [PubMed] [Google Scholar]
- Kao C, Zheng M, Rüdisser S. A simple and efficient method to reduce non template nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA. 1999;5:1268–1272. doi: 10.1017/S1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul M, Barbieri CM, Pilch DS. Aminoglycoside-induced reduction in nucleotide mobility at the ribosomal RNA A-site as a potentially key determinant of antibacterial activity. J Am Chem Soc. 2006;128:1261–1271. doi: 10.1021/ja056159z. [DOI] [PubMed] [Google Scholar]
- Kay LE, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc. 1992;114:10663–10665. doi: 10.1021/ja00052a088. [DOI] [Google Scholar]
- Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr Opin Chem Biol. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham MR, Brown DJ, McCallum SA, Pardi A. NMR methods for studying the structure and dynamics of RNA. Chembiochem. 2005;6:1492–1505. doi: 10.1002/cbic.200500123. [DOI] [PubMed] [Google Scholar]
- LeMaster DM, Kushlan DM. Dynamical mapping of E. coli thioredoxin via 13C NMR relaxation analysis. J Am Chem Soc. 1996;118:9255–9264. doi: 10.1021/ja960877r. [DOI] [Google Scholar]
- LeMaster DM, Richards FM. Preparative-scale isolation of isotopically labeled amino acids. Anal Biochem. 1982;122:238–247. doi: 10.1016/0003-2697(82)90275-5. [DOI] [PubMed] [Google Scholar]
- Lu K, Miyazaki Y, Summers MF. Isotope labeling strategies for NMR studies of RNA. J Biomol NMR. 2010;46:113–125. doi: 10.1007/s10858-009-9375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Schulte-Herbrueggen T, Briand J, Sorensen OW. Double spin-state-selective coherence transfer. Application for two-dimensional selection of multiplet components with long transverse relaxation times. Mol Phys. 1998;96:1137–1142. [Google Scholar]
- Miclet E, Williams DC, Jr, Clore GM, Bryce DL, Boisbouvier J, Bax A. Relaxation-optimized NMR spectroscopy of methylene groups in proteins and nucleic acids. J Am Chem Soc. 2004;126:10560–10570. doi: 10.1021/ja047904v. [DOI] [PubMed] [Google Scholar]
- Milecki J. Specific labelling of nucleosides and nucleotides with 13C and 15N. J Label Compd Radiopharm. 2002;45:307–337. doi: 10.1002/jlcr.553. [DOI] [Google Scholar]
- Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acid Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AJ, Nagai K. Structural studies of the spliceosome: blind men and an elephant. Curr Opin Struct Biol. 2010;20:82–89. doi: 10.1016/j.sbi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm T, Andang M, Hotchkiss G, Hard T, Baumann H, Larsson S, Ahrlund-Richter L. A method for production of 13C/15N double labelled RNA in E. coli, and subsequent in vitro synthesis of ribonucleotide 5′ triphosphates. J Biochem Biophys Methods. 1995;30:59–68. doi: 10.1016/0165-022X(94)00067-N. [DOI] [PubMed] [Google Scholar]
- Palmer AG, Cavanagh J, Wright PE, Rance M. Sensitivity improvement in proton-detected two-dimensional heteronuclear correlation NMR spectroscopy. J Magn Reson. 1991;93:151–170. [Google Scholar]
- Purohit P, Stern S. Interactions of a small RNA with antibiotic and RNA ligands of the 30S subunit. Nature. 1994;370:659–662. doi: 10.1038/370659a0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular cloning: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- Sattler M, Schwendinger MG, Schleucher J, Griesinger C. Novel strategies for sensitivity enhancement in heteronuclear multi-dimensional NMR experiments employing pulsed field gradients. J Biomol NMR. 1995;5:11–22. doi: 10.1007/BF00417487. [DOI] [PubMed] [Google Scholar]
- Schilling CH, Edwards JS, Letscher D, Palsson BØ. Combining pathway analysis with flux balance analysis for the comprehensive study of metabolic systems. Biotechnol Bioeng. 2001;71:286–306. doi: 10.1002/1097-0290(2000)71:4<286::AID-BIT1018>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Schleucher J, Schwendinger M, Sattler M, Schmidt P, Schedletzky O, Glaser SJ, Sørensen OW, Griesinger C. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J Biomol NMR. 1994;4:301–306. doi: 10.1007/BF00175254. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- Schultheisz HL, Szymczyna BR, Scott LG, Williamson JR. Pathway engineered enzymatic de novo purine nucleotide synthesis. ACS Chem Biol. 2008;3:499–511. doi: 10.1021/cb800066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheisz HL, Szymczyna BR, Scott LG, Williamson JR. Enzymatic De Novo pyrimidine nucleotide synthesis. J Am Chem Soc. 2011;133:297–304. doi: 10.1021/ja1059685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soupene E, van Heeswijk WC, Plumbridge J, Stewart V, Bertenthal D, Lee H, Prasad G, Paliy O, Charernnoppakul P, Kustu S. Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J Bacteriol. 2003;185:5611–5626. doi: 10.1128/JB.185.18.5611-5626.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz TA. A structural understanding of the dynamic ribosome machine. Nat Rev Mol Cell Biol. 2008;9:242–253. doi: 10.1038/nrm2352. [DOI] [PubMed] [Google Scholar]
- Thakur CS, Brown ML, Jackson ME, Sama JN, Dayie TK. Growth of wildtype and mutant E. coli strains in minimal media for optimal production of nucleic acids for preparing labeled nucleotides. Appl Microbiol Biotechnol. 2010;88:771–779. doi: 10.1007/s00253-010-2813-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur CS, Sama JN, Jackson ME, Chen B, Dayie TK. Selective 13C labeling of nucleotides for large RNA NMR spectroscopy using an E. coli strain disabled in the TCA cycle. J Biomol NMR. 2010;48:179–192. doi: 10.1007/s10858-010-9454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Werbelow LG (1994) (Tycko R, ed). Kluwer Academic Publishers, Boston, pp 223–263
- Wijmenga S, van Buuren B. The use of NMR methods for conformational studies of nucleic acids. Prog Nucl Magn Reson Spectrosc. 1998;32:287–387. doi: 10.1016/S0079-6565(97)00023-X. [DOI] [Google Scholar]
- Wirmer J, Westhof E. Molecular contacts between antibiotics and the 30S ribosomal particle. Methods Enzymol. 2006;415:180–202. doi: 10.1016/S0076-6879(06)15012-0. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Muhandiram R, Kay LE. NMR experiments for the measurement of carbon relaxation properties in highly enriched, uniformly 13C, 15N-labeled proteins: application to 13Cα carbons. J Am Chem Soc. 1994;114:8266–8278. doi: 10.1021/ja00097a037. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.