Summary
APOBEC3G (A3G) is a host cytidine deaminase that inhibits retroviruses. HIV and related primate lentiviruses encode Vif, which counteracts A3G by inducing its degradation. This Vif-mediated A3G inhibition is species-specific, suggesting that the A3G-Vif interaction has evolved as primate lentiviruses have adapted to their hosts. We examined the evolutionary dynamics of the A3G-Vif interaction within four African Green Monkey (AGM) subspecies, which are each naturally infected with a distinct simian immunodeficiency virus (SIV). We identified single amino acid changes within A3G in two AGM subspecies that render it resistant to Vif proteins, except for Vif from the viruses that naturally infect these subspecies. Moreover, experimental infection of AGMs shows that Vif can rapidly adapt to these arising Vif-resistant A3G genotypes. These data suggest that despite being generally non-pathogenic in its natural host, SIV infection selects for Vif-resistant forms of A3G in AGM populations, driving Vif counter-evolution and functional divergence.
Introduction
Human Immunodeficiency Virus-type 1 (HIV-1) and HIV-2 originated from multiple cross-species transmissions of SIVs that naturally infect nonhuman primates in Africa (Hahn et al., 2000). Although many natural SIV infections exhibit species-specificity, suggestive of coevolution between virus and host, new viral lineages arise through cross-species transmission and subsequent rounds of virus-host adaptation (Charleston and Robertson, 2002; Holmes, 2008). The adaptation of primate lentiviruses to new hosts is influenced by cell-intrinsic host restriction factors that impose species-specific barriers to viral replication (Malim and Emerman, 2008).
One such restriction factor is APOBEC3G (A3G), a cytidine deaminase that inhibits a broad range of retroviruses and retrotransposons (Sheehy et al., 2002). Of the seven members of the APOBEC3 family in humans (A3A-A3H), A3G exhibits the most robust antiviral activity against HIV-1 and is actively expressed in relevant host cells targeted by the virus (Koning et al., 2009; Miyagi et al., 2010; Vetter et al., 2009). On the other hand, retroviruses have evolved various mechanisms to overcome the antiviral activity of A3G; for example, most lentiviruses encode the accessory protein Vif, which carries out the accelerated degradation of A3G via the proteasome by linking it to a cellular E3 ubiquitin ligase complex (Goila-Gaur and Strebel, 2008). Antagonism of A3G by Vif can occur in a species-specific manner, suggesting that the specificity of Vif reflects adaptation to the host (Bogerd et al., 2004; Mariani et al., 2003; Xu et al., 2004).
The natural SIV infections of African Green Monkeys (AGM) provide a unique opportunity to assess how host antiviral genes impact the evolution of lentiviruses and vice versa. AGM are the most abundant non-human primates in Africa, occupying ranges spanning most of the continent south of the Sahara (Wolfheim, 1983). The AGM species (Chlorocebus aethiops) is subdivided into four subspecies commonly known as sabaeus, vervet, tantalus, and grivet monkeys. Despite sharing a recent common ancestor 1-3 million years ago (Perelman et al., 2011; Wertheim and Worobey, 2007), each subspecies is naturally infected with a distinct subtype of SIV, aptly named after the specific host it infects: SIVagm.Sab, SIVagm.Ver, SIVagm.Tan, SIVagm.Gri, respectively (Peeters and Courgnaud, 2002). SIVagm infection is widely believed to result in an asymptomatic, nonpathogenic chronic infection in its natural host (Sodora et al., 2009). Therefore, the host-virus associations in these primates provide a setting to study how viral subtypes have diverged over time as a result of host-specific adaptation in the absence of severe pathogenesis.
In this study, we examine the co-evolution of A3G-Vif interactions. We identify adaptive SNPs in A3G that allow the antiviral factor to evade destruction by SIVagm Vif, which in turn has triggered Vif counter-evolution in a subspecies-specific manner. These data argue that SIV infection selects for Vif-resistant forms of A3G in AGM. Furthermore, in an in vivo experimental evolution study, we show that SIVagm Vif proteins evolve adapted specificity that reflects the A3G genotype of the host population, implicating the viral gene as a key accessory to the process of host-specific viral adaptation. Our data highlight the conflict-driven dynamics of host-virus arms races and prompts a reexamination of how primate populations are impacted by natural, “non-pathogenic” lentivirus infections.
Results
APOBEC3G is highly polymorphic among AGM subspecies
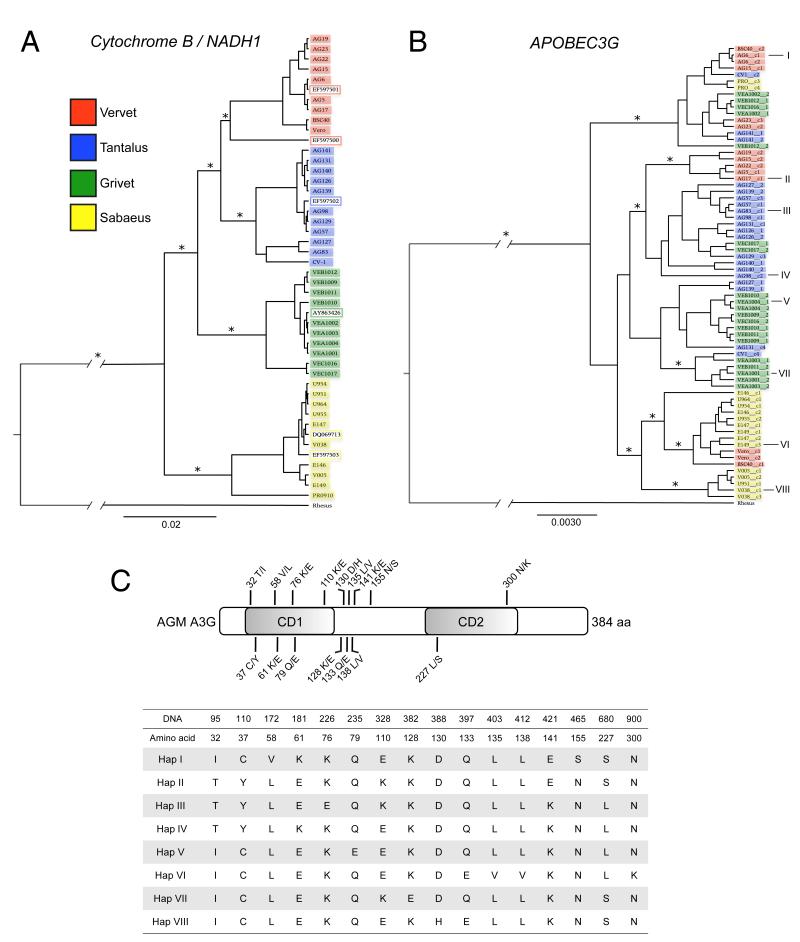
Since Vif sequences are among the most divergent viral genes across all lineages of SIV (Peeters and Courgnaud, 2002), we hypothesized that Vif divergence takes place as an adaptive response to differences in the A3G repertoire of primate hosts. To determine the degree of A3G diversity in different AGM subspecies, we sequenced the gene from 40 unique AGM individuals representing the four major subspecies (Table S1). Mitochondrial genes (mtDNA) were also sequenced in order to confirm the subspecies of origin of each AGM sample. AGM mtDNA sequences exhibited monophyly; sequences derived from the same AGM population clustered together to form clades related by common ancestry (Figure 1a). In contrast, A3G sequences from any given AGM subspecies are paraphyletic (Figure 1b), indicating that the evolutionary history of A3G is incongruent with the species phylogeny.
Figure 1. Phylogenetic analysis of partial mitochondrial genomes and APOBEC3G.
The evolutionary relationship between AGM samples was inferred by Bayesian MCMC phylogenetic reconstruction as described in the Methods. Sequence IDs are color-coded according to legend to indicate AGM subspecies of origin. Rhesus macaque DNA was used as an outgroup. Ancestral outgroup branch length is shortened due to space constraint. Scale bars indicate an arbitrary unit of time, relative to 1. (A) Partial mitochondrial DNA sequences. Asterisks (*) mark nodes with posterior probability scores of ≥0.9. (B) Full-length A3G sequences from AGM. Asterisks (*) mark nodes with posterior probability scores of ≥0.5. Eight distinct A3G haplotypes selected for functional analysis are indicated by I-VIII. (C) Sixteen nsSNPs observed at a frequency ≥5% are indicated according to amino acid position, relative to the N-terminal cytidine deaminase domain (CD1) and the C-terminal cytidine deaminase domain (CD2). The eight AGM A3G haplotypes selected for functional analysis encode proteins that are differentiated by amino acid changes at these positions. See also Table S1.
Virtually every polymorphism in AGM A3G yields an amino acid replacement—we identified 28 non-synonymous Single Nucleotide Polymorphisms (nsSNPs) present in more than one individual, 16 of which were observed with a minor allele frequency of 5% or greater (Figure 1c). In contrast, only one prevalent synonymous SNP was found, and it was tightly linked with a neighboring nsSNP. The heavy bias of non-synonymous changes in AGM A3G is suggestive of adaptive evolution by natural selection. Therefore, to address the consequences of AGM A3G polymorphism, we selected 2 haplotypes from each of the four subspecies for functional analysis (Figure 1b and 1c). A3G haplotypes were co-transfected with proviral lentiviruses in single-round infectivity experiments to measure their inherent antiviral activity. In this assay, the infectivity of each virus produced in the absence of A3G is normalized to 100%. Hereafter, the proteins encoded by A3G haplotypes are referred to as A3G variants.
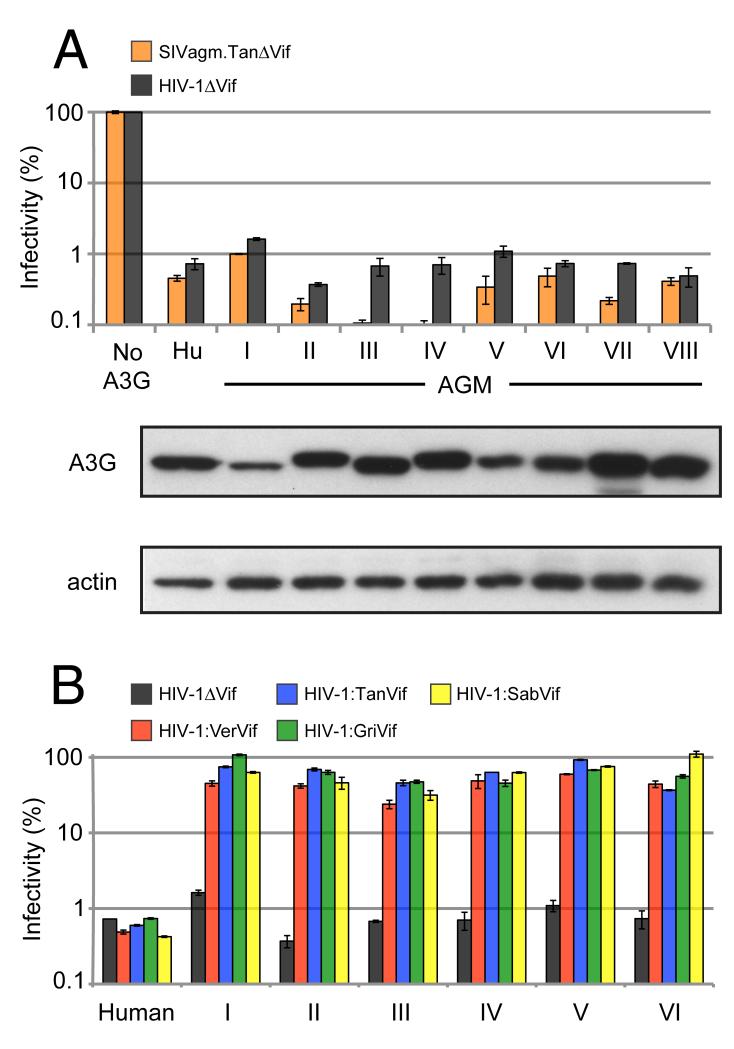
Our results indicate that each A3G variant exhibits strong restriction of SIVagm.TanΔVif and HIV-1ΔVif (Figure 2a), decreasing viral infectivity of HIV-1ΔVif on average by approximately 150-fold. For comparison, HIV-1ΔVif infectivity was inhibited about 140-fold by human A3G. These data suggest that there is extensive polymorphism in AGM A3G, and that this diversity does not affect intrinsic antiviral activity against lentiviruses.
Figure 2. AGM A3G variants maintain potent restriction of lentiviruses.
Single-round viral infectivity assays were performed with virus produced in the presence of absence of A3G proteins. Error bars indicate standard deviation from the mean of three infection replicates. (A) Infectivity of Vif-deficient SIVagm.Tan and HIV-1 (SIVagm.TanΔVif and HIV-1ΔVif). Infectivity in the absence of A3G was normalized to 100% (No A3G), while infectivity in the presence of human A3G served as a positive control for virus restriction. Anti-HA Western blot analysis was used to measure expression of AGM A3G variants in 293T cells. Anti-β-actin served as protein loading controls. (B) Infectivity of HIV-1ΔVif and HIV-1 expressing SIVagm Vif in the presence of AGM A3G variants I-VI. HIV-1:SIVagm.Ver Vif in red bars; HIV-1:SIVagm.Tan Vif in blue bars; HIV-1:SIVagm.Gri Vif in green bars; and HIV-1:SIVagm.Sab Vif in yellow bars. See also Table S2.
Evidence for antagonism-driven evolution of A3G in AGM
In order to address whether the polymorphism in AGM A3G affects susceptibility to the viral antagonist Vif, we constructed four recombinant HIV-1 proviruses expressing Vif from the following SIVagm subtype viruses: SIVsab-1 (Jin et al., 1994), SIVgri.667 (Fomsgaard et al., 1991), SIVver90 (Hirsch et al., 1995), and SIVtan.1 (Soares et al., 1997). A3G haplotypes were co-transfected with HIV-1 provirus expressing SIVagm Vif and virus infectivity was assayed in single-cycle infections. The antiviral activity of A3G variants I-VI is readily counteracted by viruses encoding each SIVagm Vif protein, resulting in a rescue of viral infectivity that approaches levels observed in the absence of A3G (Figure 2b). This finding suggests that the SNPs contained in haplotypes I-VI do not affect sensitivity to Vif.
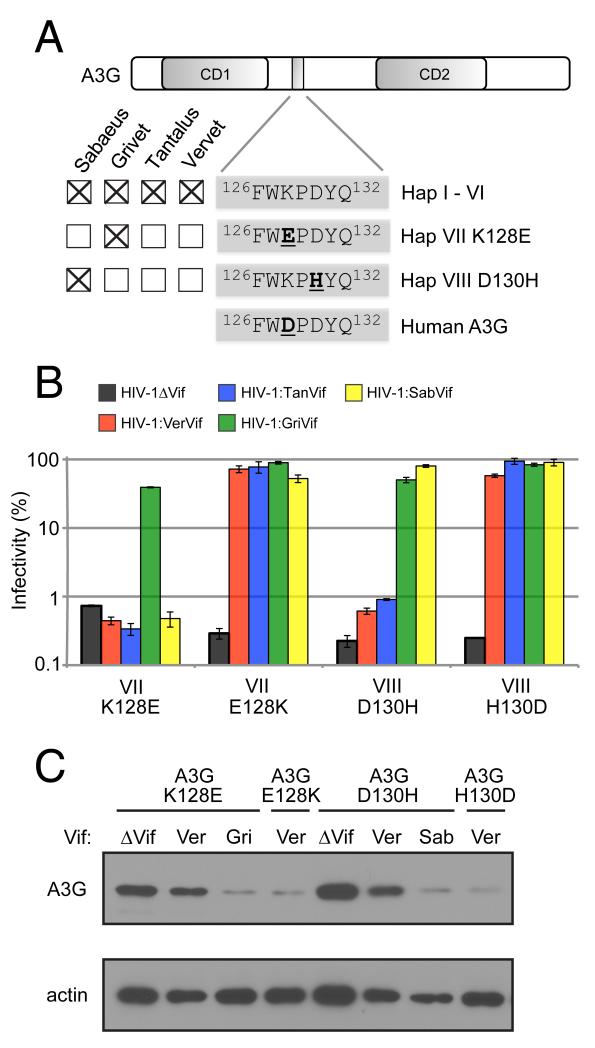
Haplotypes VII and VIII are unique in that they encode single amino acid changes at codons 128 and 130, respectively (Figure 3a), which in the context of the motif 126FWKPDYQ132, have been implicated in the electrostatic interaction between human A3G and HIV-1 Vif (Huthoff and Malim, 2007; Xu et al., 2004). We found that variant VII is resistant to Vif from SIVagm.Ver, SIVagm.Tan, and SIVagm.Sab (sensitive only to SIVagm.Gri Vif) while variant VIII is also resistant to Vif from SIVagm.Ver and SIVagm.Tan (Figure 3b). Interestingly, haplotypes VII and VIII are confined to single subspecies of AGM (the grivet and sabaeus monkeys, respectively) at an allele frequency of 20% (4/20 chromosomes per subspecies) (Figure 3a).
Figure 3. Vif-resistance phenotype of AGM A3G maps to residues 128 and 130 of the putative Vif binding site.
(A) The presence of A3G haplotypes in AGM subspecies is indicated by checked boxes. AGM A3G haplotypes VII and VIII, exclusive to grivet and sabaeus monkeys, respectively, are distinguished by a negatively-charged glutamic acid at residue 128, and a positively-charged histidine at residue 130, respectively. Human A3G encodes a negatively-charged aspartic acid at residue 128. (B) Single-round viral infectivity assays were performed with HIV-1ΔVif and HIV-1 expressing SIVagm Vif in the presence of AGM A3G variants VII K128E and VIII D130H. Error bars indicate standard deviation from the mean of three infection replicates. Mutagenesis of sites 128 and 130 restored Vif sensitivity to Vif-resistant AGM A3G variants VII K128E and VIII D130H, respectively. (C) Anti- HA Western blot analysis of A3G from cell lysates co-transfected with HIV-1 encoding different Vif proteins. Anti-β-actin served as protein loading controls.
To demonstrate that the Vif-resistance exhibited by these two variants of A3G (henceforth referred to as variants VII K128E and VIII D130H) is conferred by single residue changes at sites 128 and 130, respectively, we performed site-directed mutagenesis to revert the amino acid at these sites to the more common residues present in variants I-VI. An E128K change rendered variant VII K128E fully susceptible to all four SIVagm Vif proteins, confirming the importance of this site for Vif recognition (Figure 3b). Similarly, a H130D mutation rendered variant VIII D130H fully sensitive to SIVagm Vif (Figure 3b). Western blot analysis shows that antagonism of A3G variants by Vif results in depletion of intracellular A3G protein. A3G variants VII K128E and VIII D130H are effectively degraded by SIVagm.Gri Vif and SIVagm.Sab Vif, respectively, but not by SIVagm.Ver Vif (Figure 3c). Furthermore, the revertant mutations E128K and H130D confer sensitivity to degradation by SIVagm.Ver Vif, mirroring the results of the infectivity rescue experiments (Figure 3c). Thus, standing variation in the A3G locus of AGM is adaptive, as it allows the antiviral factor to evade destruction by the viral antagonist, Vif. Furthermore, the different specificities exhibited by SIVagm Vif proteins demonstrate that SIVagm lineages have co-adapted to distinct versions of A3G in AGM subspecies.
Experimental adaptation of SIVagm to Vif-resistant forms of AGM A3G in vivo
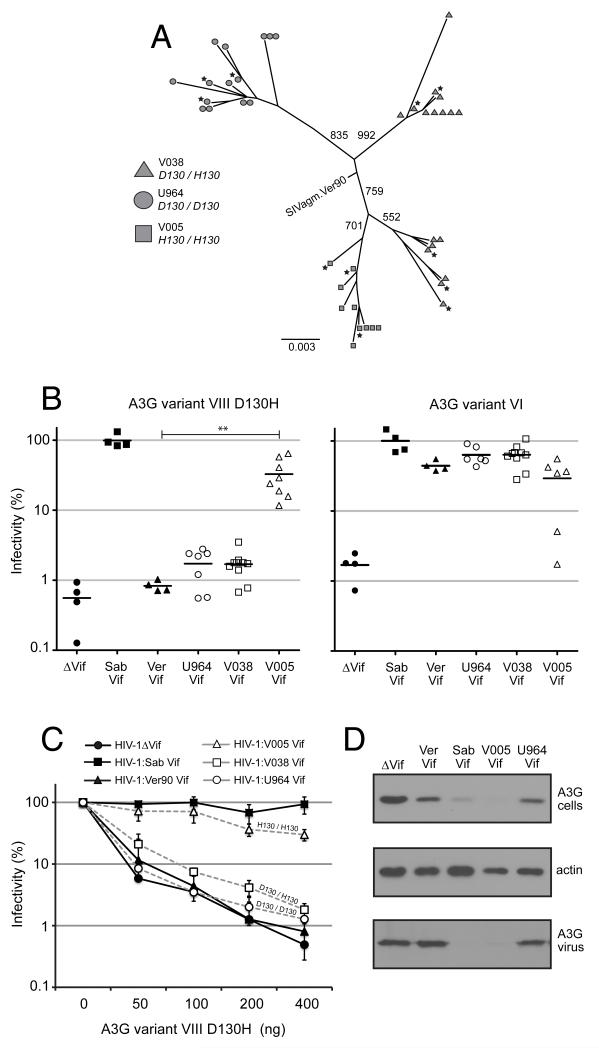
Retrospective studies of virus evolution allow one to infer adaptations to a host, but do not address the timeframe in which this occurs. On the other hand, prospective studies in which the time of infection is known can be powerful tools in understanding the dynamics of viral adaptation. We took advantage of an experimental evolution study in which the impact of A3G variation on SIVagm evolution could be evaluated within the timeframe of single infected animals. In a previous study, sabaeus monkeys were experimentally infected with SIVagm.Ver90 and plasma viremia was monitored in infected sabaeus monkeys for nearly two years (Goldstein et al., 2006). We genotyped the A3G locus in these animals and found that monkey V005 is homozygous for A3G haplotype VIII D130H, which is resistant to SIVagm.Ver90 Vif (Figure 3b), while monkey V038 is heterozygous for D130H. Moreover, monkey U964 is homozygous for A3G D130, which is found in A3G variants that are sensitive to SIVagm.Ver90 Vif (Figure 2b). As this Vif is derived from the same virus used to inoculate the monkeys experimentally, we anticipated a scenario in which restriction by a Vif-resistant form of A3G would select for a functional form of Vif in vivo. Thus, we had the opportunity to track Vif evolution in monkeys expressing different A3G variants.
We amplified Vif from plasma viral RNA isolated 6-12 months post-infection. Phylogenetic analysis of >10 viral sequences per monkey demonstrates that Vif isolates sort by A3G genotype, with Vif from monkeys V005 (H130/H130) and U964 (D130/D130) exhibiting monophyly and Vif from V038 (D130/H130) forming two distinct clades (Figure 4a). An alternate topology in which V038 Vif share a single ancestor was rejected by the SH-test (p = 0.006) (Shimodaira and Hasegawa, 1999). The SIVagm.Ver90 Vif sequence lies near the hypothetical ancestor generated by the analysis. Three to five unique Vif sequences that were representative of the diversity seen within each monkey were cloned into an HIV-1 backbone and their ability to antagonize variant VIII D130H in single-round infectivity assays was measured (Figure 4a, black stars). All three Vif isolates derived from an infected monkey expressing two copies of A3G variant VIII D130H (V005) exhibited robust antagonism of the this variant, conferring a 38-fold increase in viral infectivity on average relative to the unadapted control SIVagm.Ver90 Vif (p < 0.001)(Figure 4b, left). By comparison, Vif isolated from monkeys expressing one or zero copies of the haplotype VIII D130H (monkeys V038 and U964, respectively) did not demonstrate elevated antagonism of this A3G variant relative to SIVagm.Ver90 Vif (p > 0.05). We found that all of the in vivo Vif isolates antagonized the AGM A3G haplotype VI encoding the ancestral D130 allele, with the exception of some Vif genes derived from monkey V005 (Figure 4b, right). Together, these results demonstrate that the Vif sequences tested encode functional proteins, and moreover, that observed differences in antagonism are due to changes that alter specificity for the A3G substrate.
Figure 4. Experimental adaptation of SIVagm to Vif-resistant forms of AGM A3G in vivo.
(A) Vif sequences amplified by nested RT-PCR were aligned and subjected to phylogenetic analysis. An unrooted, neighbor-joining plot was generated after 1000 iterations, with bootstrap values indicated above branches. The scale bar indicates the number of nucleotide substitutions per site. Vif sequences were isolated from two time points post-infection: 6 months (circles) and 12-months (triangles). The sequence for SIVagm.Ver90 Vif is included to show proximity to the hypothetical ancestor. Blacks stars indicate sequences cloned into the HIV-1ΔVif background and tested for activity against A3G. Three to five unique Vif sequences amplified from each of three sabaeus monkeys were cloned into the HIV-1ΔVif background. HIV-1ΔVif is denoted by black circles. See also Figure S1 for Vif alignments. (B) Left: Single-round viral infectivity assays were performed with virus produced by co-expressing these HIV-1:SIV Vif proviral plasmids with AGM A3G variant VIII D130H. The plotted points per column correspond to infectivity experiment replicates performed with viruses encoding Vif isolated from monkey plasma (each virus replicated two to four times) except in the case of No Vif, SIVsagm.Sab Vif, and SIVagm.Ver Vif, in which plotted points represent three independent experiments using the same virus. Vif from SIVagm.Sab is denoted by black squares; SIVagm.Ver, black triangles; V005 (two copies of D130H), white triangles; V038 (one copy of D130H), white squares; U964 (zero copies of D130H), white circles. Horizontal bars indicate the mean relative infectivity. p-values were calculated using a one-way ANOVA test, relative to SIVagm.Ver90 Vif. Right: Single-round viral infectivity assays were performed with virus produced by co-expressing the same HIV-1:SIV Vif proviral plasmids with Vif-sensitive AGM A3G variant VI, encoding D130. (C) Single-round viral infectivity assays were performed with virus produced by co-expressing HIV-1ΔVif or HIV-1:SIV Vif proviral plasmids in the presence of decreasing concentrations of AGM A3G variant VIII D130H. Each point corresponds to the mean infectivity of 3-5 viruses encoding different Vif proteins, each replicated 2-4 times each, except in the case of No Vif, SIVagm.Sab Vif and SIVagm.Ver Vif. (D) Western blot analysis was used to measure A3G protein expression in cells (upper panel) and encapsidated in virions (lower panel) after co-transfection with HIV-1 encoding Vif derived from animals V005 and U964. Anti-β-actin served as cellular protein loading controls.
We also expressed Vif isolates in the presence of different amounts of A3G variant VIII D130H. Again, we observed that Vif from a monkey homozygous for this haplotype (V005) exhibits robust antagonism at all concentrations of A3G (Figure 4c, white triangles). However, the adaptation is partial, as infectivity is not rescued to the same degree as SIVagm.Sab Vif, the fully adapted control that has undergone repeated transmission cycles within wild sabaeus populations (Figure 4b and 4c, black squares). Conversely, Vif from the heterozygous monkey (V038) and the monkey homozygous for D130 (U964) do not antagonize variant VIII D130H at any concentrations relative to SIVagm.Ver90 Vif (Figure 4c, white squares and white circles versus black triangles). Consistent with the infectivity data, V005 Vif effectively degrades A3G variant VIII D130H and excludes it from virions, while Vif from U964 does not (Figure 4d). Therefore, there is evidence of adaptation in the Vif gene upon infection of a single animal, the extent of which is influenced by the host A3G genotype.
Amino acid alignment of all Vif sequences isolated from these monkeys reveal a number of recurrent mutations that arose during the time course of infection, and in some cases, the same mutation appeared independently in different animals (Figure S1a). However, only one mutation (Y84C) was observed in 100% (10/10) of Vif isolates from monkey V005 but not in a single Vif obtained from the other two monkeys. Interestingly, the equivalent change is not observed in SIVagm.Sab Vif, demonstrating that Vif may adapt via a number of distinct evolutionary trajectories (Figure S1b).
Discussion
We present evidence that A3G is adaptively diversifying within AGM as a result of antagonism-driven evolution, suggesting a history of ongoing host-virus conflict. Specifically, we found two sites located in the Vif binding site of A3G that are functionally polymorphic in AGM: high frequency, charge-altering SNPs at residues 128 and 130 alter the sensitivity of A3G to Vif, allowing the antiviral factor to evade antagonism. In response to changes in host A3G, we demonstrate using both natural isolates and experimental evolution in vivo that the viral-encoded Vif protein must itself adapt to re-target the restriction factor and carry out its destruction. We predict that the genetic conflict between these two proteins drives continuous evolution in Vif, which contributes to the species-specificity of natural SIV infections.
Selection costs incurred by apparently “non-pathogenic” viruses
Evolutionary studies indicate that A3G is rapidly evolving in different primate species through a process of positive selection (Ortiz et al., 2006; Sawyer et al., 2004). The Vif-resistant SNPs K128E and D130H were observed at 20% frequency in A3G of the grivet and sabaeus subspecies, respectively, suggesting that they have emerged recently relative to other polymorphisms that were identified in multiple AGM subspecies. Despite the evidence that contemporary infections result in a non-pathogenic outcome, the appearance of adaptive genetic changes in the host implies that SIV infection has exerted selective pressure in at least some AGM populations. It is possible that selection imparted by SIVagm may manifest overtly as progressive disease and immunodeficiency in some monkeys. In support of this, immunodeficiency has been observed in captivity among models of “natural” SIV infection (Ling et al., 2004; Pandrea et al., 2009; Traina-Dorge et al., 1992). The virus may also affect fitness more subtly by reducing female fertility, influencing mate choice, or increasing infant mortality (Keele et al., 2009). Alternatively, SIVagm may have been pathogenic to AGM hosts in the past. Even so, our characterization of the ongoing genetic conflict between SIVagm and AGM suggests that other “non-pathogenic” primate lentiviruses may select for adaptive changes in their respective host species as well.
Paraphyletic phylogeny of A3G in African Green Monkey subspecies and Vif selection
The failure of A3G sequences to fully segregate by AGM subspecies may result from demographic and/or stochastic processes. For example, male-biased migration of AGM between neighboring reproductive groups can lead to rampant gene flow in nuclear genes but not in mtDNA, which is maternally-inherited from the more sedentary AGM females (Isbell et al., 1993). In addition, incomplete lineage sorting could result in alternate retention of ancestral A3G alleles, leading to an apparent paraphyly. Alternatively, the incongruent A3G phylogeny may be explained by selection. The fact that residues in A3G that interact with Vif are undergoing positive selection in primates (Ortiz et al., 2006) is further evidence that Vif-mediated antagonism is capable of selecting for genetic change in A3G. In addition, long-term balancing selection can maintain ancestral gene polymorphism across diverged populations due to a survival/reproductive advantage conferred to heterozygous individuals.
The adaptive inferiority of Vif isolated from monkey V038, despite expressing one copy of A3G variant VIII D130H (Figure 4), suggests that A3G heterozygosity may apply an adaptive constraint on the virus. The relative lack of viral adaptation in this animal demonstrates that the broadening of Vif specificity in order to accommodate multiple A3G variants may come at a considerable cost, which is likely to decelerate viral adaptation. This is supported by the branching order of the Vif phylogenetic tree, in which V038 Vif sequences are split into two clades (Figure 4a, triangles). We postulate that a lack of directional selection in these viral sequences results from the requirement that Vif must counteract two distinct A3G variants in a heterozygous animal.
Host-virus adaptation
A recent report demonstrated that the outcome of experimental cross-species transmission of SIV in rhesus macaques is largely dependent on the TRIM5α genotype of the recipient monkey (Kirmaier et al., 2010). Furthermore, TRIM5α-resistant virus emerged in some of the infected monkeys, demonstrating that adaptation in the capsid (CA) region of Gag facilitates escape from TRIM5α-mediated restriction in vivo. We employed a similar approach to demonstrate that Vif counter-evolves in vivo to degrade a formerly Vif-resistant variant of A3G. While experimental infections are unlikely to reproduce the natural dynamics of host-virus coevolution, we show that A3G-driven evolution of Vif is a naturally occurring phenomenon because the specificity of Vif proteins from SIVagm subtypes reflects adaptation to subspecies-specific A3G variants in AGM populations exposed to SIV in the wild rather than in the laboratory.
There is not a direct correlation between A3G genotype and viral load in this small cohort (Goldstein et al., 2006). Thus, A3G is not the sole determinant of clinical outcome in this virus-host setting. Nonetheless, by tracking viral evolution in SIV-infected monkeys, we have shown that adaptation in Vif occurs upon infection of monkeys expressing naturally occurring variants of A3G, demonstrating that A3G has the potential to set the course of viral evolution in vivo. By analyzing intraspecies A3G variation in a natural host of SIV, we have now demonstrated a case of contemporary selection and adaptation between primate lentiviruses and their natural primate hosts.
Experimental Procedures
PCR amplification of primate genes
Sources of AGM samples are described in Table S1. Whole RNA and genomic DNA (gDNA) were extracted from AGM peripheral blood mononuclear cells or AGM-derived continuous cell lines using the RNeasy Mini Kit (Qiagen) and the Whole Blood DNA Isolation Kit (Qiagen), respectively. Partial mitochondrial genes Cytochrome B and NADH1 were PCR amplified (see Table S2 for oligo sequences) from gDNA and bulk PCR products were sequenced. A3G was amplified via two-step RT-PCR, in which cDNA was synthesized from whole RNA using the SuperScript II Reverse-Transcriptase Kit (Invitrogen) and oligo-dT primers (Invitrogen). cDNA was used as template in PCR using OWM A3G-specific primers. Bulk PCR product was subcloned and six to ten clones were sequenced. If two distinct A3G sequences were detected from a single RNA sample, the individual was considered to be heterozygous. When samples were limited to gDNA only, A3G coding exons (1-8) were amplified and sequenced individually using primers targeted to non-coding regions of the locus.
PCR amplification of SIVagm Vif genes
Vif genes were PCR-amplified from full-length molecular clones SIVsab-1 and SIVtan.1 and from plasma derived from monkeys infected with SIVgri.667 and SIVver90. In the latter cases, viral RNA was isolated using the QIAamp Viral RNA Mini Kit (Qiagen) and used as template in Two-Step RT-PCR with Vif-specific primers containing Mlu1 and Xba1 restriction sites (Supplemental Table 2). SIVver90 Vif derived from experimentally infected sabaeus monkeys was amplified using a nested RT-PCR approach. One to two mL of plasma was concentrated by centrifugation for 30 min at 16,000 rpm. Viral RNA was isolated using the QIAamp Viral RNA Mini Kit (Qiagen). Using specific primers that were targeted to SIVagmVer90 integrase and vpr, Vif was amplified using a one-step RT-PCR. Products were amplified in a second round of PCR using nested primers containing Mlu1 and Xba1 restriction sites.
Phylogenetic analysis
A3G and mtDNA sequences were subjected to phylogenetic analysis using a Bayesian Monte Carlo Markov Chain (MCMC) approach implemented in BEAST v1.6.1. Sequence alignments were executed using ClustalX. Partial Cytochrome B (273 bp) and NADH1 (298 bp) sequences were concatenated and subjected to 50,000,000 MCMC generations using the HKY85 substitution model, Gamma site heterogeneity model, and estimated base frequencies. The following AGM mtDNA GenBank submissions were included to guide classification efforts: EF597500, EF597501, EF597502, EF597503, AY863426, DQ069713. We used the constant population size coalescent as the tree prior. The resulting phylogenies were annotated using TreeAnnotator, with a “burnin” of 40 and a posterior probability limit of 0.5. A3G sequences were treated similarly, except 100,000,000 MCMC generations were performed. Additionally, phylogenetic analysis by Maximum Likelihood (ML) was performed using the Phylogeny.fr web service. ML trees were imported into PAUP* and trees were scored for log likelihood using the Shimodaira-Hasegawa (SH) test. Test parameters were estimated using 1000 RELL replicates.
A3G expression plasmids
Full-length reconstruction of haplotypes originating from grivet monkeys (hap V and VII) was performed by mutagenesis of other AGM A3G haplotypes using the Quikchange Site-Directed Mutagenesis Kit (Stratagene). Haplotypes I-VIII were appended with a 5′ hemagluttinin (HA) tag by PCR (Supplemental Table 2) and cloned into the mammalian expression vector pcDNA3.1. Site-directed mutagenesis was used to generate the E128K and H130D A3G mutants.
Recombinant HIV-1 proviral plasmids
SIVagm Vif sequences were appended with a 5′ Kozac sequence and 5′ Mlu1 and 3′ Xba1 restriction sites by PCR (Supplemental Table 2) and cloned into the HIV-1ΔVif molecular clone (pLai3ΔEnvLuc2ΔVif, generated after NdeI-StuI deletion in pLai3ΔEnvLuc2). The resulting proviral plasmids lack Env, have a firefly luciferase gene inserted into Nef, and encode SIVagm Vif in the context of the HIV-1 backbone.
Single-round Viral Infectivity Assays
293T cells were plated in 1 mL in 12-well plates at 2.5 × 105 cells/mL. The following day, cells were cotransfected with 0.4 μg of A3G expression plasmid or an empty expression plasmid, 0.1 μg of L-VSV-G (vesicular stomatitis virus glycoprotein, for pseudotyping), and 0.6 μg of proviral plasmid in a 100 μL transfection volume with TransIT-LT1 lipid transfection reagent (Mirus Bio). Virus supernatants were harvested at 48 h and clarified by centrifugation for 5 min at 1,800 rpm. The total amount of virus was quantified by p24 Gag enzyme-linked immunosorbent assay. Four ng of virus was used to infect supT1 cells plated at 3.8 × 105 cells/mL in the presence of 20 μg/mL DEAE-Dextran. Infections were performed in triplicate for 48 h. Luciferase activity was measured in 100 μL of Bright-Glo Luciferase Assay Reagent (Promega).
Supplementary Material
Highlights.
SIV selects for variants of a restriction factor, Apobec3G, in its primate host.
SIV adapts to polymorphism in Apobec3G through evolution of the viral gene, Vif
Lentiviruses thought to be non-pathogenic still cause selection in their natural host
Acknowledgements
We are grateful to the following investigators and their staff for providing invaluable samples: M. Muller-Trutwin (AGM PBMC), C. Apetrei (AGM PBMC), and N. Freimer and A. Jasinska (AGM DNA). We thank S. Whitted (NIH) and W. Switzer (CDC) for preparing AGM samples for inspection and shipment. We thank the NIH Nonhuman Primate Research Resource for providing the V038 and AG23 AGM cells and SIVagm.Gri+ plasma. We thank E. Matsen, V. Minin, and C. Carlson for helpful discussions on phylogenetics and population genetics. We thank H. Malik, M. Levine, M. Metzger, and members of the Emerman lab for critical reading of the manuscript.
AC and ME designed the experiments. AC performed the experiments. VH provided AGM PBMC, SIV+ AGM plasma samples and guidance on experimental design. AC and ME wrote the paper. This work was supported by NIH R01 AI30937 (to ME), an NSF Graduate Research Fellowship (to AC) and an NIH Training Grant in Viral Pathogenesis T32AI083203 (to AC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bogerd HP, Doehle BP, Wiegand HL, Cullen BR. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3770–3774. doi: 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charleston MA, Robertson DL. Preferential host switching by primate lentiviruses can account for phylogenetic similarity with the primate phylogeny. Systematic biology. 2002;51:528–535. doi: 10.1080/10635150290069940. [DOI] [PubMed] [Google Scholar]
- Fomsgaard A, Hirsch VM, Allan JS, Johnson PR. A highly divergent proviral DNA clone of SIV from a distinct species of African green monkey. Virology. 1991;182:397–402. doi: 10.1016/0042-6822(91)90689-9. [DOI] [PubMed] [Google Scholar]
- Goila-Gaur R, Strebel K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology. 2008;5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein S, Brown CR, Ourmanov I, Pandrea I, Buckler-White A, Erb C, Nandi JS, Foster GJ, Autissier P, Schmitz JE, et al. Comparison of simian immunodeficiency virus SIVagmVer replication and CD4+ T-cell dynamics in vervet and sabaeus African green monkeys. Journal of virology. 2006;80:4868–4877. doi: 10.1128/JVI.80.10.4868-4877.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: scientific and public health implications. Science (New York, NY) 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- Hirsch VM, Dapolito G, Johnson PR, Elkins WR, London WT, Montali RJ, Goldstein S, Brown C. Induction of AIDS by simian immunodeficiency virus from an African green monkey: species-specific variation in pathogenicity correlates with the extent of in vivo replication. Journal of virology. 1995;69:955–967. doi: 10.1128/jvi.69.2.955-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC. Evolutionary history and phylogeography of human viruses. Annual review of microbiology. 2008;62:307–328. doi: 10.1146/annurev.micro.62.081307.162912. [DOI] [PubMed] [Google Scholar]
- Huthoff H, Malim MH. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. Journal of virology. 2007;81:3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isbell LA, Cheney DL, Seyfarth RM. ARE IMMIGRANT VERVET MONKEYS, CERCOPITHECUS-AETHIOPS, AT GREATER RISK OF MORTALITY THAN RESIDENTS. Animal Behaviour. 1993;45:729–734. [Google Scholar]
- Jin MJ, Hui H, Robertson DL, Müller MC, Barré-Sinoussi F, Hirsch VM, Allan JS, Shaw GM, Sharp PM, Hahn BH. Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. The EMBO journal. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Jones JH, Terio KA, Estes JD, Rudicell RS, Wilson ML, Li Y, Learn GH, Beasley TM, Schumacher-Stankey J, et al. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–519. doi: 10.1038/nature08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmaier A, Wu F, Newman RM, Hall LR, Morgan JS, O’Connor S, Marx PA, Meythaler M, Goldstein S, Buckler-White A, et al. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS biology. 2010;8 doi: 10.1371/journal.pbio.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. Journal of virology. 2009;83:9474–9485. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling B, Apetrei C, Pandrea I, Veazey RS, Lackner AA, Gormus B, Marx PA. Classic AIDS in a sooty mangabey after an 18-year natural infection. Journal of virology. 2004;78:8902–8908. doi: 10.1128/JVI.78.16.8902-8908.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell host & microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Mariani R, Chen D, Schröfelbauer B, Navarro F, König R, Bollman B, Münk C, Nymark-McMahon H, Landau NR. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- Miyagi E, Brown CR, Opi S, Khan M, Goila-Gaur R, Kao S, Walker RC, Hirsch V, Strebel K. Stably expressed APOBEC3F has negligible antiviral activity. Journal of virology. 2010;84:11067–11075. doi: 10.1128/JVI.01249-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz M, Bleiber G, Martinez R, Kaessmann H, Telenti A. Patterns of evolution of host proteins involved in retroviral pathogenesis. Retrovirology. 2006;3:11. doi: 10.1186/1742-4690-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandrea I, Silvestri G, Apetrei C. AIDS in african nonhuman primate hosts of SIVs: a new paradigm of SIV infection. Current HIV research. 2009;7:57–72. doi: 10.2174/157016209787048456. [DOI] [PubMed] [Google Scholar]
- Peeters M, Courgnaud V. Overview of primate lentiviruses and their evolution in non-human primates in Africa. HIV Sequence Database. 2002 Available: http://www.hiv.lanl.gov/content/hiv-db/REVIEWS/PEETERS2002/Peeters2002.html.
- Perelman P, Johnson W, Roos C, Seuánez H, Horvath J, Moreira M, Kessing B, Pontius J, Roelke M, Rumpler Y, et al. A Molecular Phylogeny of Living Primates. PLoS genetics. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS biology. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molecular biology and evolution. 1999;16:1114–1116. [Google Scholar]
- Soares MA, Robertson DL, Hui H, Allan JS, Shaw GM, Hahn BH. A full-length and replication-competent proviral clone of SIVAGM from tantalus monkeys. Virology. 1997;228:394–399. doi: 10.1006/viro.1996.8387. [DOI] [PubMed] [Google Scholar]
- Sodora DL, Allan JS, Apetrei C, Brenchley J, Douek DC, Else JG, Estes JD, Hahn BH, Hirsch V, Kaur A, et al. Toward an AIDS vaccine: lessons from natural simian immunodeficiency virus infections of African nonhuman primate hosts. Nature Medicine. 2009;15:861–865. doi: 10.1038/nm.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traina-Dorge V, Blanchard J, Martin L, Murphey-Corb M. Immunodeficiency and lymphoproliferative disease in an African green monkey dually infected with SIV and STLV-I. AIDS research and human retroviruses. 1992;8:97–100. doi: 10.1089/aid.1992.8.97. [DOI] [PubMed] [Google Scholar]
- Vetter M, Johnson M, Antons A, Unutmaz D, D’Aquila R. Differences in APOBEC3G expression in CD4+ T helper lymphocyte subtypes modulate HIV-1 infectivity. PLoS Pathogens. 2009;5:e1000292. doi: 10.1371/journal.ppat.1000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Worobey M. A challenge to the ancient origin of SIVagm based on African green monkey mitochondrial genomes. PLoS Pathogens. 2007;3:e95. doi: 10.1371/journal.ppat.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfheim J. Primates of the World: Distribution, Abundance, and Conservation. University of Washington Press; Seattle, WA: 1983. [Google Scholar]
- Xu H, Svarovskaia ES, Barr R, Zhang Y, Khan MA, Strebel K, Pathak VK. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:5652–5657. doi: 10.1073/pnas.0400830101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.