Abstract
A biofilm is an organized, resilient group of microbes where individual cells acquire properties, such as drug resistance, that are distinct from those observed in suspension cultures. Here we describe and analyze the transcriptional network controlling biofilm formation in the pathogenic yeast Candida albicans, whose biofilms are a major source of medical device-associated infections. We have combined genetic screens, genome-wide approaches, and two in vivo animal models to describe a master circuit controlling biofilm formation, composed of six transcription regulators that form a tightly woven network with ~1000 target genes. Evolutionary analysis indicates that the biofilm network has rapidly evolved: genes in the biofilm circuit are significantly weighted towards genes that arose relatively recently with ancient genes being underrepresented. This circuit provides a framework for understanding many aspects of biofilm formation by C. albicans in a mammalian host. It also provides insights into how complex cell behaviors can arise from the evolution of transcription circuits.
Keywords: biofilm, transcriptional regulation, biofilm network, biofilm circuitry, Candida albicans, evolution of transcription circuits, evolution of biofilm formation, evolution of multicellularity
Introduction
Biofilms are organized communities of surface-associated microorganisms embedded in a matrix of extracellular polymers. In this paper, we analyze how C. albicans, the predominant fungal pathogen of humans, forms biofilms. Biofilms are a predominant microbial growth form in natural environments (Kolter and Greenberg, 2006), and a leading cause of persistent human infection (Costerton et al., 1999). These infections are typically seeded from biofilms that form on implanted medical devices, such as intravascular catheters, and become resistant to drug and mechanical treatments (Donlan and Costerton, 2002). The mechanisms behind biofilm development are thus important to our understanding of microbial ecology (since mixed species biofilms are common) as well as infectious disease.
C. albicans biofilm formation can be partitioned into four basic stages, based on studies carried out in vitro (Baillie and Douglas, 1999; Chandra et al., 2001; Douglas, 2003; Hawser and Douglas, 1994); (Nobile et al., 2009; Uppuluri et al., 2010; Uppuluri et al.). These are (i) attachment and colonization of yeast-form (nearly spherical) cells to a surface, (ii) growth and proliferation of yeast-form cells to allow formation of a basal layer of anchoring microcolonies, (iii) growth of pseudohyphae (ellipsoid cells joined end to end) and extensive hyphae (chains of cylindrical cells) concomitant with the production of extracellular matrix material, and (iv) dispersal of yeast-form cells from the biofilm to seed new sites. At least some of these features of biofilm formation have also been observed in vivo. For example, C. albicans biofilms from denture stomatitis patients confirm the presence of yeast, hyphae and extracellular matrix (Ramage et al., 2004). Furthermore, biofilm architectures in two animal catheter models and a denture model include numerous yeast cells in the basal region, as well as hyphae and extracellular matrix extending throughout the biofilm (Andes et al., 2004; Nett et al., 2010; Schinabeck et al., 2004).
Here, we combine “classical” genetics, genome-wide approaches, RNA deep sequencing technology, and two in vivo animal models to comprehensively map the transcriptional circuitry controlling biofilm formation in C. albicans. The circuit has led to many new predictions about genes involved in biofilm formation, and we have validated a set of these predictions by confirming the roles of several of these genes in biofilm development. The circuit also provides insight into how biofilm formation may have evolved in the C. albicans lineage.
Results
Identification and phenotypic characterization of biofilm-defective transcription regulator mutants in vitro
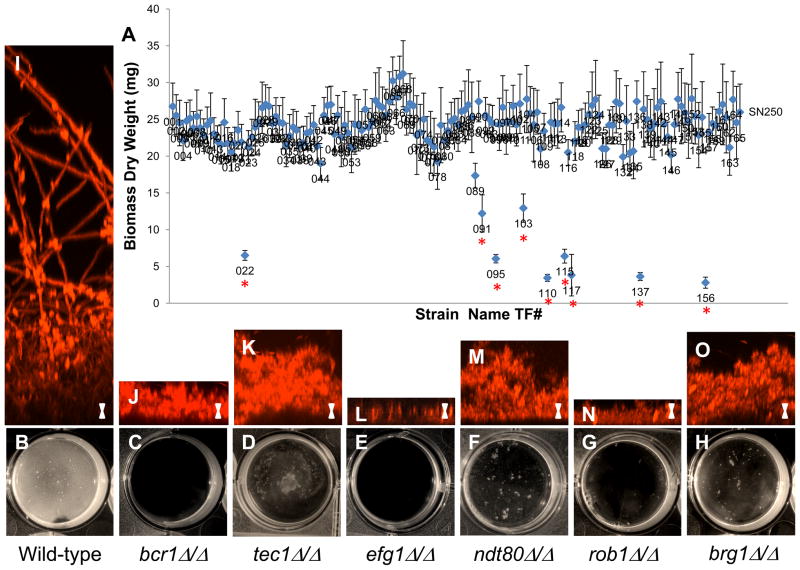
Transcription regulators (defined here as sequence-specific DNA-binding proteins that regulate transcription) play important roles in the control of many developmental pathways; often, they define a group of co-regulated target genes that function together to carry out a specific function in the cell. Hence, transcription regulators represent a powerful entry point to understanding a biological process. Using information on transcription regulators taken from a wide variety of species, we constructed a C. albicans library of 165 fully vetted transcription regulator (TR) deletion mutants consisting of two independently constructed mutants for each strain (Homann et al., 2009). This library was screened for biofilm formation on the surface of serum-treated polystyrene plates under a standard set of biofilm-inducing conditions (Nobile et al., 2006a; Nobile and Mitchell, 2005; Nobile et al., 2006b). The screening was based on biofilm dry weight biomass, visual, and microscopic (confocal) inspection (Figure 1). The screen revealed nine mutants with deficiencies in forming biofilms (Figure 1A; Dataset S1; Figure S2A). Three of these mutants were not analyzed further because they exhibited either general growth defects in suspension cultures or a wide variety of other phenotypes in suspension cultures (Supplemental Experimental Procedures). The remaining six transcription regulator deletion mutants (bcr1Δ/Δ, tec1Δ/Δ, efg1Δ/Δ, ndt80Δ/Δ, rob1Δ/Δ, and brg1Δ/Δ) have the following characteristics: 1) they were significantly compromised in biofilm formation (P<0.0005) (Figure 1B–H), 2) they did not exhibit general growth defects, and 3) they did not show extensive phenotypes aside from defects in biofilm formation. Of these six transcription regulators, three are newly identified as biofilm regulators (Ndt80/Orf19.2119, Rob1/Orf19.4998; named for Regulator Of Biofilms, and Brg1/Orf19.4056; named for Biofilm ReGulator), and three had been previously implicated in biofilm formation (Bcr1 (Nobile and Mitchell, 2005), Tec1 (Nobile and Mitchell, 2005), and Efg1 (Ramage et al., 2002)). The screen was carried out blindly, and our identification of all previously identified regulators serves as an internal control for both the library construction and the screen.
Figure 1.
Screening and characterization of in vitro biofilm-defective transcription regulator mutants. Biofilm biomass (dry weight) determinations of the entire transcription regulator (TR) mutant library (165 strains) are shown in panel A. The average total biomass +/− standard deviation for each TR mutant strain grown under standard biofilm conditions (Experimental Procedures) was calculated from five independent samples of each strain. Statistical significance (P values) was calculated with a Student’s one-tailed paired t test, and is represented by the red asterisk under the nine regulator strains (TF022, TF091, TF095, TF103, TF110, TF115, TF117, TF137, and TF156) with biomasses significantly deviating (P<0.0005) from the reference strain (SN250). Phenotypic characterization of the mutants is shown in panels B–O. Panels B–H show the visual appearance after 48 h of growth on polystyrene plates. Panels I–O are CSLM side view images of the wild-type and six biofilm-defective mutant strains. Scale bars represent 20 μm. See also Dataset S1, Figure S1 and Figure S2.
We further characterized the morphology of the six biofilm-defective mutant strains by confocal scanning laser microscopy (CSLM), using silicone squares as the substrate (Figure 1I–O). By CSLM, the wild-type reference strain formed a biofilm with typical architecture and thickness (Chandra et al., 2001; Douglas, 2003; Nobile and Mitchell, 2005) of ~250 μm in depth, containing both round budding yeast-form cells adjacent to the substrate, and hyphal cells extending throughout the biofilm (Figure 1I) (see also Figure S1 for CSLM visualization of each regulator mutant over a time course of biofilm development). In all six mutants only rudimentary biofilms of approximately 20–80 μm in depth were formed, although the detailed phenotypes of the mutants differ (Figure 1J–O; Figure S1). Reintroduction of an ectopic copy of the wild-type allele back into each mutant reversed the biofilm-formation defect of each mutant (Figure S2B). Thus, BCR1, TEC1, EFG1, NDT80, ROB1, and BRG1 are required for wild-type biofilm formation in vitro.
Because hyphal development is an important step in normal biofilm development, we assessed the ability of our six biofilm-defective transcription regulator mutants to form normal hyphae when they were not in the context of a biofilm. We found that, with the exception of the efg1Δ/Δ strain, true hyphae could be detected in the medium surrounding the biofilm (Figure S3A) as well as in suspension cultures using the same medium as that used for biofilm formation (Figure S3B). We also observed hyphal development for all strains except the efg1Δ/Δ strain in a variety of suspension culture media, although the fraction of hyphal cells was often reduced relative to the parental strain (Figure S3B). Thus, for all of these mutants (with the possible exception of efg1Δ/Δ), the defect in biofilm formation was not due to an intrinsic inability to form hyphae.
Characterization of biofilm-defective transcription regulator mutants in two in vivo animal models
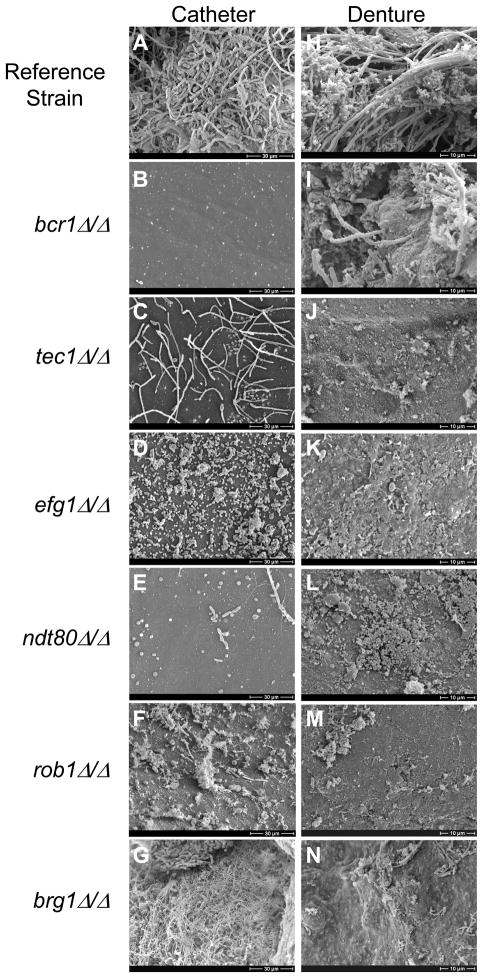
Biofilm formation in vivo is the cause of the majority of new infections in humans, and it is widely appreciated that the conditions for biofilm formation in vivo differ considerably from those in standard in vitro assays (Nett and Andes, 2006). For example, many additional elements are present in vivo, such as liquid flow, host factors, and components of the host immune response. Because biofilm-based catheter infections are a major clinical problem (Kojic and Darouiche, 2004), we used a well-established rat venous catheter model of infection (Andes et al., 2004) to test the six mutants for biofilm formation in vivo. We inoculated the catheters with C. albicans cells intraluminally, allowed biofilm formation to proceed for 24 h, removed the catheters, and visualized the catheter luminal surfaces by scanning electron microscopy (SEM) (Figure 2A–G; Figure S4A). The wild-type reference strain formed a thick, mature biofilm on the rat catheter, consisting of yeast and hyphal cells and extracellular matrix material (Figure 2A). Of the six transcription regulator mutants, five (bcr1Δ/Δ, tec1Δ/Δ, efg1Δ/Δ, ndt80Δ/Δ, and rob1Δ/Δ) were unable to form biofilms (Figure 2B–F); bcr1Δ/Δ had been previously shown to be defective in this model (Nobile et al., 2008). The sixth mutant (brg1Δ/Δ) formed a thick biofilm consisting of many adherent cells and a large amount of extracellular matrix material (Figure 2G), but appeared morphologically distinct from the reference strain in that considerably fewer hyphae were observed within the biofilm (compare Figures 2A and 2G).
Figure 2.
Biofilm formation in two in vivo rat models – a catheter model and a denture model. For the catheter model, the wild-type reference strain SN250 (panel A), and the six transcription regulator mutant strains (panels B–G) were inoculated into rat intravenous catheters; resulting biofilms were visualized after 24 h of growth by scanning electron microscopy (SEM). SEM catheter Images show the catheter luminal surfaces at magnifications of 1000X (see also Figure S4A). For the denture model, the wild-type reference strain SN425 (panel H), and the six transcription regulator mutant strains (panels I–N) were inoculated into rat dentures, and the resulting biofilms were visualized after 24 h of growth by SEM. SEM denture images show the denture surfaces at magnifications of 2000X. See also Figure S4B for SEM images of denture surfaces containing extensive bacterial biofilms, which formed in the presence of the mutant C. albicans strains.
The most common form of oral candidiasis is denture stomatitis, prevalent largely in the elderly population, and affecting up to 70% of denture wearers (Webb et al., 1998; Wilson, 1998). Denture stomatitis occurs by biofilm colonization and growth over the surface of a denture, leading to inflammation of the palatal mucosa (Ramage et al., 2004). Because biofilm growth on dentures represents a completely different host environment from that of an intravenous catheter, we also screened our six biofilm-defective regulator mutants in a recently established in vivo rat denture model, which was developed to mimic and assess C. albicans biofilm formation in denture stomatitis (Nett et al., 2010). In particular, this oral model includes host salivary components, host commensal bacteria, salivary flow dynamics, and direct contact between the denture biofilm and the host mucosal surface (Nett and Andes, 2006). We inoculated the rat dentures with C. albicans cells, permitted biofilm formation to proceed for 24 h, removed the dentures, and visualized the denture surfaces by SEM (Figure 2H–N). The wild-type reference strain formed a thick, mature biofilm on the surface of the rat denture, consisting predominantly of hyphal C. albicans cells interspersed with C. albicans yeast-form cells, various host commensal oral bacteria, and extracellular matrix material (Figure 2H). In contrast, the genetically matched mutant strains all showed significant defects in biofilm formation. In particular, tec1Δ/Δ, efg1Δ/Δ, ndt80Δ/Δ, rob1Δ/Δ, and brg1Δ/Δ were severely defective (Figure 2J–N), while the bcr1Δ/Δ mutant, which has previously been shown to be defective in this model (Nett et al., 2010), had less pronounced defects than the other five mutants (Figure 2I). We note that, extensive bacterial biofilms consisting of both cocci and rods were seen on the dentures of the six C. albicans biofilm-defective mutants (Figure S4B), suggesting a competition between biofilm formation by C. albicans and biofilm formation by the native bacteria present in the mouth.
In summary, BCR1, TEC1, EFG1, NDT80, ROB1, and BRG1 are each required for normal biofilm formation in vivo in both the rat denture and catheter models. The effects of certain deletion mutants (brg1Δ/Δ and bcr1Δ/Δ) differed to varying degrees between the two models (Figure 2G, N), likely reflecting the influence of the host environment in biofilm formation. The results, taken as a whole, indicate that performing genetic screens and analyzing biofilm formation in vitro is a valid approach to understanding clinically relevant C. albicans biofilm formation.
Developing transcriptional relationships among biofilm regulators
To identify genes directly regulated by Bcr1, Tec1, Efg1, Ndt80, Rob1, and Brg1, we performed full genome chromatin immunoprecipitation microarray (ChIP-chip) to map the position across the genome to which each of the six transcription regulators are bound during biofilm formation. Based on this analysis (see Supplemental Experimental Procedures for details; Dataset S2 for a complete list of all significantly bound locations for each regulator, and Dataset S3 for MochiView image plots of every called significant peak for each regulator), we calculate the following number of intergenic regions bound by each regulator: 211 for Bcr1, 76 for Tec1, 328 for Efg1, 558 for Ndt80, 95 for Rob1, and 283 for Brg1 (Dataset S2). 831 intergenic regions are bound by one or more regulators, 350 intergenic regions are bound by two or more, 186 intergenic regions are bound by three or more, 111 intergenic regions are bound by four or more, 55 intergenic regions are bound by five or more, and 18 intergenic regions are bound by all six of the biofilm regulators (Dataset S2). We noticed two unusual characteristics for the intergenic regions bound by the biofilm regulators. First, the average length of intergenic regions bound by the biofilm regulators is over twice that of the remainder of the genome (1540 bp compared with 693 bp); this trend is true for all six biofilm regulators (see Table “Length of intergenic regions bound for the biofilm regulators” in Supplemental Experimental Procedures). Second, binding peaks are distributed throughout the intergenic regions of the regulator-bound target genes rather than being clustered a fixed distance upstream of the transcription start site (Dataset S5), as is common for many yeast target genes (Lin et al., 2010).
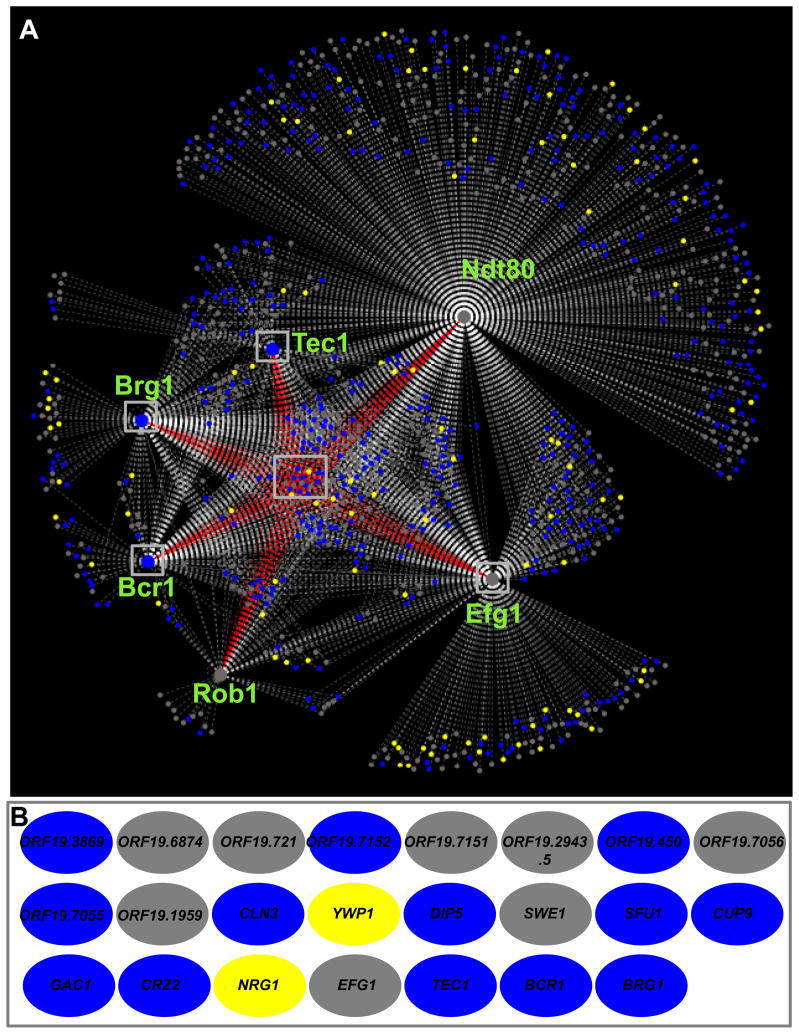
If we convert bound intergenic regions to genes likely to be controlled (for example a single bound intergenic region between divergently transcribed genes is counted as two genes), our analysis suggests the network is composed of 1,061 target genes that are bound in their promoter regions by at least one of the six biofilm regulators (Figure 3; Dataset S4). This regulatory network is shown in Figure 3. Based on the ChIP-chip data, the high degree of overlap between target genes among biofilm regulators suggests that the biofilm regulatory network is considerably interwoven; that is, many of the target genes are controlled by more than one regulator.
Figure 3.
The biofilm regulatory network. The six master biofilm regulators are represented by the six large circular hubs. Smaller circles represent target genes, which are connected to their respective regulators by dashed lines, indicating a direct interaction as determined by genome-wide ChIP-chip. Genes that are differentially regulated as determined by expression data (using a 2-fold cutoff) in biofilm compared to planktonic cells are shown in blue for those genes upregulated in biofilms, in yellow for those downregulated, and in grey for those with no change. Grey boxes are drawn around the 23 target genes bound by all six regulators, and are connected to their respective regulators by red dashed lines (panel A). The identity of these 23 genes are indicated as the colored ovals in panel B (blue ovals are genes that are upregulated, and yellow ovals are genes that are downregulated in biofilm compared to planktonic cells). Overall, 23 genes are bound by all six, 77 are bound by five or more, 165 are bound by four or more, 265 are bound by three or more, and 458 are bound by two or more of the biofilm regulators. See also Dataset S4.
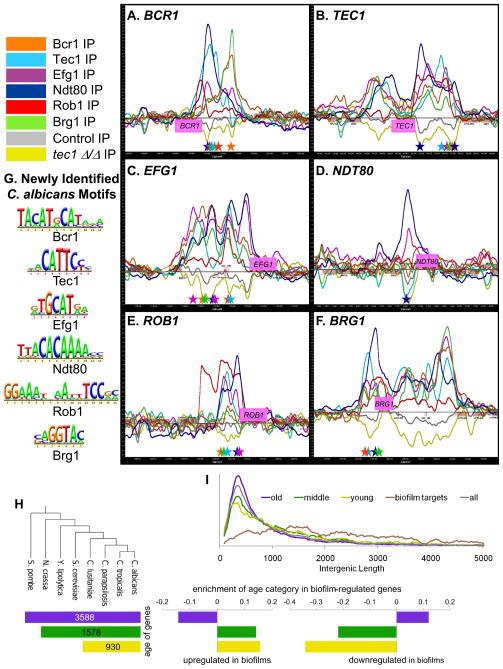
The results also indicate that the six regulators originally identified in the genetic screen control each other’s expression: all six of the regulators bind to the upstream promoter regions of BCR1 (Figure 4A), TEC1 (Figure 4B), EFG1 (Figure 4C), and BRG1 (Figure 4F), four of the regulators (Tec1, Efg1, Ndt80, and Rob1) bind to the upstream promoter region of ROB1 (Figure 4E), and two of the regulators (Efg1 and Ndt80) bind to the upstream promoter region of NDT80 (Figure 4D).
Figure 4.
Chromatin immunoprecipitation (ChIP) mapping and motif identification of the six master biofilm regulators. All six regulators bind to each another’s upstream promoter regions (panels A–F). Immunoprecipitation (IP) binding data for Bcr1-Myc (orange line), Tec1-custom antibody (light blue line), Efg1-Myc (magenta line), Ndt80-Myc (dark blue line), Rob1-Myc (red line), Brg1-Myc (green line), untagged wild-type/control IP (grey line), and tec1Δ/Δ (yellow line) strains are shown. The ChIP-chip microarray binding data was mapped and plotted onto the chromosomes containing BCR1 (panel A), TEC1 (panel B), EFG1 (panel C), NDT80 (panel D), ROB1 (panel E), and BRG1 (panel F) using MochiView. The promoters of these genes show significant peak enrichments for the binding of the indicated biofilm regulators. The X-axis represents ORF chromosomal locations. The Y-axis gives the Agilent normalized enrichment value (log2) post-smoothing for the binding of each regulator. Genes (pink boxes) plotted above the bold line read in the sense direction; genes plotted below the bold line read in the antisense direction. Using de novo motif-finding based on our ChIP-chip data, we identified significantly enriched core binding motifs for all six of our biofilm regulators (panel G). Motifs were identified using MochiView, independently verified using MEME, and motif graphics were generated with MochiView. See Datasets S5 and S2 for details on motif analysis and bound motif locations, respectively, for each regulator. Colored stars corresponding to the colors of the regulators indicate the location of strong instances of the indicated biofilm regulator motifs under the enrichment peaks in panels A–F. Panel H shows the evolutionary age of target genes in the biofilm network. Genes were divided into three categories based on when they arose during evolution, with the numbers in each bar giving the number of C. albicans genes that fall into that age category. The enrichment of each age category in biofilm-regulated genes is log10 of the observed divided by the expected (for all age categories, P<1.23E-9). Panel I shows a histogram of the length of the intergenic regions between tandem and divergent gene pairs targeted by the biofilm regulators. Each category was normalized to the total number of intergenic regions in that category. See also Dataset S7.
De Novo Motif Finding for the Six Master Biofilm Regulators
A test of the self-consistency of ChIP-chip data is the non-random occurrence of cis-regulatory sequences (motifs). Based on several hundred significant binding events from our ChIP-chip data, we were able to identify statistically significant motifs for all six of the biofilm regulators (Figure 4G; Dataset S5; Dataset S2). This motif generation was based solely on the ChIP-chip data and did not incorporate data from any other experiment or from any other species. We note that the motif generated for Ndt80 (TTACACAAAA) is very similar to the reported binding motif for its homolog, Ndt80, in S. cerevisiae (GMCACAAAA) (Zhu et al., 2009). The motif for Tec1 (RCATTCY) is identical to that determined for its homolog, Tec1, in S. cerevisiae (Harbison et al., 2004; Madhani and Fink, 1997). (This Tec1 motif, generated from 107 bound intergenic regions, does not closely resemble the Tec1 motif recently reported in the white-specific pheromone response element (WPRE) (AAAAAAAAAAGAAAG) in C. albicans, which was generated from a much smaller set of data (Sahni et al., 2010).) Finally, the Efg1 motif derived from our ChIP-chip data (RTGCATRW) closely resembles the “TGCAGNNA” consensus sequence of the S. cerevisiae ortholog, Sok2 (Harbison et al., 2004). Thus, for three of the biofilm regulators, the motifs developed from our C. albicans ChIP-chip data can be independently verified by their similarities to the motifs recognized by their S. cerevisiae orthologs. This analysis provides independent support for both the motif analysis and for the validity of the full genome ChIP data. For the other three regulators, we were able to determine statistically significant motifs, but we were not able to independently verify them by comparison with S. cerevisiae because either the orthology relationships are uncertain (Rob1 and Brg1) or because the orthologous S. cerevisiae regulator has not been characterized (Bcr1).
Exploring the transcriptional patterns of biofilms
Although the ChIP-chip experiments reveal the genomic positions where each regulator binds, they do not indicate whether these binding events are associated with differences in gene transcription. We first consider control of the regulators themselves, as they are all bound by one or more of the other regulators. We deleted each regulator and measured the mRNA levels of the other five (Figure S7A). This analysis revealed that each regulator positively regulates each of the other regulators. We also examined the effect of each regulator on its own synthesis by fusing its upstream region to an mCherry reporter, and measuring levels of the reporter in the absence and presence of the regulator (Figure S7B). In all cases, a given regulator activates its own synthesis. Thus, the connections among the six biofilm regulators are primarily, if not exclusively, positive.
To assess the relationship of regulator binding and transcription across the entire circuit, we performed both RNA-seq and gene expression microarray analyses of cells grown in biofilm and planktonic conditions. From our RNA-seq data, we generated 46 million mappable strand-specific sequence reads, expanding our previous gene annotation (Tuch et al., 2010) by newly identifying 622 “novel transcriptionally active regions” (nTARs), and 161 nTARs that overlap, at least partially, transcribed regions identified in other recent genome-wide experimental annotations (Bruno et al., 2010; Sellam et al., 2010) (Dataset S6). We know from previous work that nTARs identified by RNA-seq include both non-coding RNAs (Mitrovich et al., 2010) as well as transcripts that encode for proteins too short to have been identified in previous genome annotations (Tuch et al., 2010).
We used our RNA-seq data in addition to our gene expression microarray data to obtain a complete set of genes (coding and non-coding) differentially expressed between planktonic and biofilm conditions (Dataset S6). Combining the RNA-seq and microarray data, we find 1,599 genes upregulated and 636 genes downregulated at least twofold in biofilm compared to planktonic cells (Dataset S6). By analyzing the overlap between our ChIP-chip data and our gene expression data (Dataset S7), we find a strong correlation between transcription regulator binding and differential gene expression. For example, if we consider regions bound by at least four transcription regulators, approximately 60% of these regions are associated with differentially expressed transcripts. This is significantly greater than that expected by chance (P<0.0001), and suggests – at least broadly – that binding of the regulators is associated with differential transcription in biofilm versus planktonic cultures. For the correlation between the binding of given, single transcription regulator and differential gene expression, we find a range of 38–56%, comparable to, or greater than, the associations documented for other C. albicans transcription regulators (Askew et al., 2011; Lavoie et al., 2010; Nobile et al., 2009; Sellam et al., 2009; Tuch et al., 2010).
We examined the evolutionary history of genes that are differentially regulated under biofilm conditions. To do this, we categorized each C. albicans gene into an age group based on orthology mappings across the Ascomycota, a large group of yeasts that include both C. albicans and S. cerevisiae ((Wapinski et al., 2007); Supplemental Experimental Procedures). Gene ages were defined using orthology assignments from The Fungal Orthogroups Repository (http://www.broad.mit.edu/regev/orthogroups/). The oldest genes are present in distantly related yeast clades, whereas the youngest are found only in C. albicans. Young genes can arise in several ways, including relatively rapid mutation that obscures the relation to an ancient gene, horizontal gene transfer, and de novo gene formation (Long et al., 2003). We found that genes upregulated in biofilms are enriched for young and middle-aged genes, and depleted in old genes. The opposite trend was observed for genes that are downregulated in biofilms (Figure 4H). Genes that were not differentially expressed were not strongly enriched for any age category (see Table “Age of biofilm target genes correlated with expression data” in Supplemental Experimental Procedures). Young genes typically show longer intergenic regions than old genes (Sugino and Innan, 2011), and this trend may help to explain the unusually long intergenic regions of biofilm circuit genes. However, biofilm genes exhibited significantly longer intergenic regions even when compared to other young genes (P<2.2E-16) (Figure 4I).
Identifying Functionally Relevant Target Genes of the Master Biofilm Network
To understand the connections between the six regulators and biofilm development, we performed gene expression microarray experiments of all six regulator mutants compared to a reference strain under biofilm-forming conditions. In interpreting this data, it is important to keep in mind that the mutant strains do not form mature biofilms under these conditions, so that many of the transcriptional effects may be indirect consequences of defective biofilms. Consistent with this idea, the transcriptional responses to deletion of each of the biofilm transcription regulators tended to encompass a relatively large set of genes (Dataset S4). For example, we found 234 genes that were downregulated and 173 genes that were upregulated in the bcr1Δ/Δ mutant relative to the isogenic parent (threshold of (log2 > 0.58, and log2 < −0.58)) (Dataset S4). Of these genes, Bcr1 binds directly to the promoters of 46 (11%) of them, a number significantly higher than that predicted by chance (P=0.0002). Nonetheless, the results indicate that most of the effects of deleting Bcr1 are indirect. Of the genes directly bound by Bcr1, half were downregulated and half were upregulated in the bcr1Δ/Δ mutant, indicating that Bcr1 can act as both an activator and repressor of its direct target genes. Similar analysis (Dataset S4; Supplemental Experimental Procedures) indicates that Efg1, Ndt80, Rob1, and Brg1 are all both activators and repressors of their biofilm-relevant direct target genes, and that Tec1 is primarily an activator of its biofilm-relevant direct target genes.
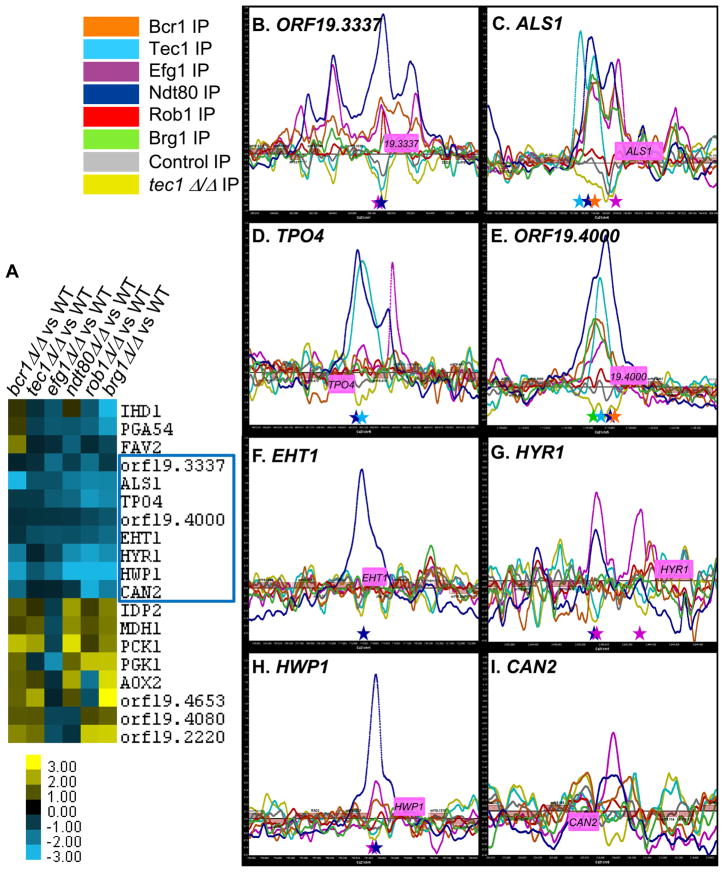
From these large data sets, we attempted to identify a set of target genes that might be expected to have important roles in biofilm formation. Using hierarchical cluster analysis to characterize genes with similar patterns of expression in each of the six biofilm regulator mutants compared to a reference strain under biofilm conditions, we found nineteen target genes that were differentially regulated in all six data sets (threshold of (log2 > 0.58, and log2 < −0.58) (Figure 5A; Dataset S4). Eight of these target genes (ORF19.3337, ALS1, TPO4, ORF19.4000, EHT1, HYR1, HWP1, and CAN2) were expressed at lower levels in all six of the biofilm regulator mutants compared to the reference strain (Figure 5A); seven of these genes were also expressed at higher levels in biofilm compared to planktonic wild-type cells (Dataset S4). Additionally, all of these eight target genes were bound in their upstream promoter regions by at least one of the six biofilm regulators; most were bound by multiple regulators (Figure 5B–I).
Figure 5.
Core candidate biofilm target genes. Using hierarchical cluster analysis of our gene expression microarray data, we identified a set of nineteen candidate target genes (IHD1, PGA54, FAV2, ORF19.3337, ALS1, TPO4, ORF19.4000, EHT1, HYR1, HWP1, CAN2/ORF19.111, IDP2, MDH1, PCK1, PGK1, AOX2, ORF19.4653, ORF19.4080, and ORF19.2220) that were differentially regulated (log2 > 0.58, and log2 < −0.58) in all gene expression array experiments that compared each biofilm regulator mutant to a reference strain under biofilm conditions (panel A). Eight of these targets were differentially regulated in the same direction (all down in the mutants), and were chosen for further functional analyses (panel A, as indicated by the blue square). ChIP-chip enrichment data for the binding of the six biofilm regulators in the promoters of these eight candidate target genes (panels B–I). IP binding data for Bcr1-Myc (orange line), Tec1-custom antibody (light blue line), Efg1-Myc (magenta line), Ndt80-Myc (dark blue line), Rob1-Myc (red line), Brg1-Myc (green line), untagged wild-type/control IP (grey line), and tec1Δ/Δ (yellow line) strains are shown. The ChIP-chip microarray binding data was mapped and plotted onto the chromosomes containing ORF19.3337 (panel B), ALS1 (panel C), TPO4 (panel D), ORF19.4000 (panel E), EHT1 (panel F), HYR1 (panel G), HWP1 (panel H), and CAN2 (panel I) using MochiView. The promoters of these genes show significant peak enrichments for the binding of the indicated biofilm regulators: ORF19.3337 by Bcr1, Efg1, Ndt80, and Rob1 (panel B); ALS1 by Bcr1, Tec1, Efg1, Ndt80, and Brg1 (panel C); TPO4 by Tec1, and Ndt80 (panel D); ORF19.4000 by Bcr1, Tec1, Efg1, Ndt80, and Brg1 (panel E); EHT1 by Ndt80 (panel F); HYR1 by Efg1 (panel G); HWP1 by Ndt80 (panel H); and CAN2 by Efg1 (panel I). The X-axis represents ORF chromosomal locations. The Y-axis is the Agilent normalized enrichment value (log2) post-smoothing for the binding of each regulator. Genes (pink boxes) plotted above the bold line read in the sense direction; genes plotted below the bold line read in the antisense direction. Colored stars corresponding to the colors of the regulators indicate the location of strong instances of the indicated biofilm regulator motifs under the enrichment peaks.
Further analysis of the regulation of these eight target genes helps to reconcile their expression patterns with the chromatin IP results. As indicated in Figure S5, the transcriptional effects of deleting each one of the six regulators can be accounted for by 1) direct binding and transcriptional activation by that regulator on the target gene, and 2) direct binding and activation of a different regulator, which, in turn, binds directly to and activates the target gene (Figure S5). This “hierarchical cascade” between the biofilm regulators and target genes, applied more broadly, can explain much of the expression data (Figure S5, Dataset S4; Supplemental Experimental Procedures).
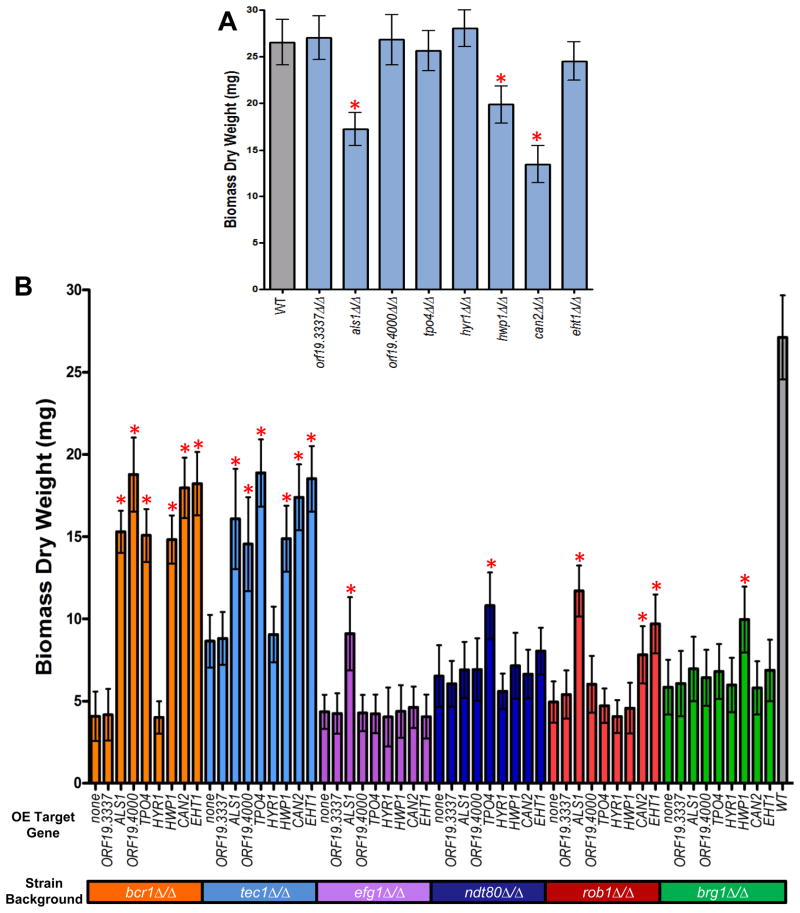
To determine whether the eight target genes identified by this analysis affected biofilm formation, we constructed homozygous deletion strains for each of the eight target genes. We observed significant biofilm defects for als1Δ/Δ (P=0.01), hwp1Δ/Δ (P=0.01), and can2Δ/Δ (P=0.003) mutant strains compared to the reference strain, with the can2Δ/Δ strain the most defective (Figure 6A). Although all three of these mutants were capable of forming partial biofilms, these biofilms were less stable than those of the wild-type and often detached from the substrate; partial biofilm defects have been previously reported for als1Δ/Δ and hwp1Δ/Δ mutant strains (Nobile et al., 2006a; Nobile et al., 2006b; Nobile et al., 2008), while can2Δ/Δ is new to this study. The other five knockout strains did not show any obvious biofilm defects under the conditions tested, and we hypothesized that their roles may be masked by genetic redundancy. To explore this idea, we created ectopic expression strains where each of the eight target genes was ectopically expressed in strains where each transcription regulator was deleted. In other words, in a grid of 6×8 = 48 constructed strains, we determined whether ectopic expression of the target genes could suppress the defect of the original transcription regulator deletion. Overexpression of several of the candidate target genes was able to significantly rescue biofilm formation to varying degrees depending on the target gene-mutant background combination (P<0.0005) (Figure 6B; see Figure S6 for CSLM images of the rescued biofilms). For example, overexpression of ORF19.4000, CAN2 or EHT1 in the bcr1Δ/Δ mutant strain background was able to rescue biofilm formation to near wild-type levels of biomass (although the biofilms are fragile) (Figure 6B; Figure S6), implicating these genes in biofilm formation. Taken as a whole, our data suggest that six of the original set of eight candidate target genes have direct roles in biofilm formation. Of course, there are more than 1000 additional target genes, and their analysis is a future challenge.
Figure 6.
Functionally relevant biofilm target genes. Biofilm biomass (dry weight) determinations were measured for the eight core candidate biofilm target gene deletion mutants (panel A), and the strains in which each of the eight target genes was ectopically expressed in the background of each regulator mutant (panel B). The average total biomass +/− standard deviation for each strain grown under standard biofilm conditions was calculated from five independent samples of each strain. Statistical significance (P values) was calculated with a Student’s one-tailed paired t test, and is represented by the red asterisks above the strains with biomasses significantly deviating (P<0.0005) from either the reference strain (WT) for panel A or the corresponding mutant strain for panel B.
Discussion
A Master Transcription Network Controlling Biofilm Formation in C. albicans
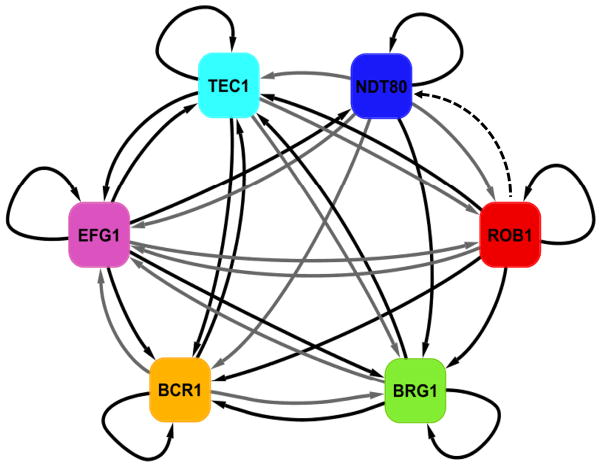
We have described a master circuit of six transcription regulators that controls biofilm formation by C. albicans in vitro and in two different animal models. C. albicans biofilms are an organized structure of three types of cells (yeast, pseudohyphae and hyphae) enclosed in an extracellular matrix. The transcription regulators form an elaborate, interconnected transcriptional network: each regulator controls the other five and most target genes are controlled by more than one master regulator (Figure 3). The circuit appears to be based largely, if not exclusively, on positive regulation (Figure 7; Figure S7A and S7B). Taking into consideration all of the target genes of the six regulators, the biofilm network comprises about 15% of the genes in the genome.
Figure 7.
Regulatory network model for biofilm formation. Panel A shows the biofilm network model based on our ChIP-chip and expression data. Solid arrows indicate direct binding interactions determined by our ChIP-chip analysis. Solid black arrows indicate experimentally validated regulatory interactions (as determined by expression profiling data and validated by qPCR) in addition to direct binding interactions (as determined by ChIP-chip data), and solid grey arrows indicate direct binding interactions only. The dashed black arrow indicates an indirect regulatory interaction only.
Circuit Complexity
Although the circuit is large and complex (~1,000 genes and twice that many connections), this level of complexity is not without precedent. For example, circuits that control osmotic stress and pseudohyphal growth pathways of S. cerevisiae (Borneman et al., 2006; Ni et al., 2009), competence and spore formation in Bacillus subtilis (de Hoon et al., 2010; Hamoen et al., 2003; Losick and Stragier, 1992; Suel et al., 2006), the hematopoietic and embryonic stem cell differentiation pathways of mammals (Wilson et al., 2010; Young, 2011), and the regulation of circadian clock rhythms in Arabidopsis thaliana (Alabadi et al., 2001; Locke et al., 2005) show certain similarities: they all consist of a core group of master transcription regulators that control each other and – working together – control a large set of additional target genes.
Several possibilities might account for the complexity of the biofilm network. The regulators we have described can orchestrate biofilm formation in two very different niches of the human host, the bloodstream and the oral cavity; it seems likely that the same circuit also controls biofilm formation in other host niches (for example, in the vagina and gastrointestinal tract). Thus, the biofilm circuit responds to many environmental conditions, such as temperature, nutrient availability, flow rate, surface-type, other microbial species, and components of the host immune system. One possibility is that the complex circuit we have described can integrate a wide range of environmental cues to produce a stereotyped morphological and functional output under many different conditions. Consistent with this idea is the finding that one regulator (Bcr1) plays an important role in biofilm formation in the catheter model but has a less pronounced role in the denture model, while another regulator (Brg1) shows the opposite behavior. It is also possible that the complex structure of the network (consisting of many direct and indirect feedback loops, many feed-forward loops, and highly overlapping regulons) is responsible for a form of cell memory that acts over generations to ensure coordinated cooperation among cells in maintaining the biofilm state. A third possibility, as has been suggested for the ribosomal protein gene regulation (Muller and Stelling, 2009), is that the more complex the regulatory architecture of a network, the more precisely the dynamics of gene expression can be regulated.
A consideration of the evolution of the biofilm network might also help to explain why it differs structurally from simple regulatory schemes. Incorporation of genes one at a time into a network requires a gain of a binding site upstream of each gene; however bringing a regulatory protein gene into a network instantly incorporates all of that regulator’s targets into the network. Thus, the interconnectedness of the biofilm network may reflect the ease by which many genes can be simultaneously incorporated into an existing circuit. Finally, it is formally possible that the complexity per se of a transcriptional network is not, in itself, adaptive; rather some aspects of the network complexity could simply be the result of neutral (non-adaptive) evolution (Fernandez and Lynch, 2011).
Evolutionary Conservation of the Biofilm Network
Only a few of the many (probably over a million) fungal species can proliferate and cause disease in humans. These pathogenic species are widely distributed over the fungal lineage indicating that survival in a human host probably evolved independently multiple times. Although many fungal species can form aggregates (flocs, mats, biofilms, etc.), it seems likely that C. albicans is one of very few fungal species that can efficiently form biofilms in a healthy mammalian host. How, then, did the biofilm circuit evolve in the C. albicans lineage?
Several lines of evidence suggest that the biofilm network in C. albicans has undergone extensive evolutionary change relatively recently. First, as described in the Results section, “young” genes are enriched in the biofilm circuit and “old” genes are underrepresented (Figure 4H). For example, approximately 120 C. albicans genes appear to have arisen (or at least have changed extensively) after the common ancestor of C. albicans and Candida tropicalis (a closely related species), and one third of these are part of the biofilm circuit. Second, if we map (when possible) the C. albicans biofilm circuit target genes to other species, we find the motifs of two of the master regulatory proteins (Ndt80 and Efg1) only sporadically enriched in these genes (Figure S7C). Thus, the regulator-target gene connections are not strongly conserved outside of C. albicans itself. (This analysis could not be meaningfully performed for the other regulators due to a lack of predictive power of their motifs (See Supplemental Experimental Procedures)). Third, the intergenic regions targeted by biofilm regulators are much longer than average (Figure 4I), possibly providing a larger mutational target for the gain of binding sites. In combination with short motifs, this may help to explain how new genes have quickly become incorporated into the network. Finally, as we discuss in more detail below, the functions of the master transcription regulators in C. albicans have diverged significantly from their “assignments” in S. cerevisiae. Our data and analyses suggest that the biofilm networks of other CTG clade species (species that translate the CUG codon into serine instead of the conventional leucine, e.g. C. tropicalis, Candida parapsilosis, Lodderomyces elongisporus, Debaryomyces hansenii, Candida guiermondii, and Candida lusitaniae) will likely be comprised of different transcription regulators and/or different target genes, or both.
Evolutionary Reassignment of Transcription Regulators
A direct comparison between C. albicans and its non-pathogenic relative S. cerevisiae provides additional insight into how the biofilm network evolved. We can ask, for example, whether the six master transcription regulators of biofilm formation in C. albicans have clear orthologs in S. cerevisiae and – if so – what processes they regulate in S. cerevisiae. To explore orthology relationships for the master biofilm regulators, we used SYNERGY and INPARANOID mappings, in addition to hand-annotation using constructed gene trees. Details are given in Supplemental Experimental Procedures.
Overall, this analysis indicates that the biofilm circuit consists of two regulators (Tec1 and Efg1) whose broad function – regulation of cell morphology – is deeply conserved in the fungal lineage. However, the set of target genes controlled by these regulators differ significantly between S. cerevisiae and C. albicans (Supplemental Experimental Procedures). A third regulator (Ndt80) is deeply conserved in the fungal lineage but its function appears completely different between S. cerevisiae and C. albicans. In the former, it regulates meiosis (Hepworth et al., 1998) and in the latter, biofilm formation. Two regulators (Rob1 and Brg1) are detectable only in species closely related to C. albicans, and the sixth biofilm regulator (Bcr1) has orthologs in S. cerevisiae, but they have not been characterized. Given that the DNA binding specificity of Tec1, Efg1, and Ndt80 are strongly conserved, extensive gains and losses of cis-regulatory sequence must be responsible – at least in part – for the evolution of the biofilm circuit in the C. albicans lineage. The Rob1 and Brg1 proteins appear to have undergone extensive changes in the C. albicans lineage such that their direct connection to the ancestor of C. albicans and S. cerevisiae (if any) has been obscured. Thus, it seems likely that extensive changes in both regulators and cis-regulatory sequences were necessary for the evolution of the modern C. albicans biofilm circuit. These considerations, in combination with our analysis of “young” versus “old” genes, indicate that the C. albicans biofilm circuit evolved relatively recently, and we suggest that this development had an important role in the ability of C. albicans to adapt to its human host.
Experimental Procedures
Strain Construction
Primer sequences and C. albicans strains are listed and described in Supplemental Experimental Procedures; strains were constructed in isogenic backgrounds.
In vitro Biofilm Growth, Confocal Microscopy, and Biomass Determination
In vitro biofilm growth assays were carried out in Spider medium as described in detail in Supplemental Experimental Procedures. The average total biomass for each strain was calculated from five independent samples. Statistical significance (P values) was calculated with a Student’s one-tailed paired t test.
In vivo Rat Catheter Biofilm Model
A rat central-venous catheter infection model (Andes et al., 2004) was used for in vivo biofilm modeling to mimic human catheter infections, as described in detail in Supplemental Experimental Procedures. Catheters were removed after 24 h of C. albicans infection to assay biofilm development on the intraluminal surface by scanning electron microscopy (SEM).
In vivo Rat Denture Biofilm Model
A rat denture stomatitis infection model (Nett et al., 2010) was used for in vivo biofilm modeling to mimic human denture infections, as described in Nett et al., with certain modifications described in Supplemental Experimental Procedures. Dentures were removed after 24 h post C. albicans infection to assay biofilm development on the denture surface by SEM.
RNA Sample Preparation and Extraction
Biofilms for gene expression microarray and RNA-seq analysis were grown in Spider medium at 37°C directly on the bottom of 6-well polystyrene plates. Planktonic cells for gene expression microarrays were grown in Spider medium at 37°C to an OD600 of 1.0, and planktonic cells for RNA-seq were grown in SC+Uri medium at 30°C to an OD600 of 1.0. Further details on growth, cell harvesting, RNA extraction, and treatment of the biofilm and planktonic cells used for gene expression microarray and RNA-seq analysis are described in Supplemental Experimental Procedures.
Gene Expression Microarray Design and Analysis
We used custom-designed oligonucleotide microarrays, containing at least two independent probes for each ORF from the C. albicans Assembly 21 genome (http://www.candidagenome.org/); printed by Agilent Technologies (AMADID #020166). Expression microarray data are reported in Dataset S4 as the median of three independent experiments. We used a cutoff of twofold in both directions (log2 > 1.0, and log2 < 1.0) for the differential expression of biofilm versus planktonic cells, and 1.5-fold in both directions (log2 > 0.58, and log2 < −0.58) for the differential expression of mutant over wild-type. Raw gene expression array data are available at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo, accession # GSE30474).
Full Genome Chromatin Immunoprecipitation Tiling Microarray (ChIP-chip)
Each transcription regulator was tagged with a Myc tag at the C- or N-terminal end of the protein in a wild-type reference strain background. (In the case of Tec1, tagging the protein at either the C- or N- terminal end interfered with the protein’s activity, and we used a custom-designed polyclonal antibody against an epitope near the C terminus of the Tec1 protein.) The tagged strains were grown under standard biofilm conditions (because the tags do not compromise function, the strains form normal biofilms), and harvested the biofilm cells for chromatin immunoprecipitation. After precipitation using the commercially available Myc antibody or the custom Tec1 antibody, the immunoprecipitated DNA and whole-cell extract were amplified and competitively hybridized to custom whole-genome oligonucleotide tiling microarrays. The ChIP-chip microarrays were designed by tiling 181,900 probes of 60 bp length across 14.3 Mb included in the C. albicans Assembly 21 genome (http://www.candidagenome.org/), as previously described (Tuch et al., 2008); printed by Agilent Technologies (AMADID #016350). The ChIP-chip experiments were performed as previously described (Nobile et al., 2009) with two independent biological replicates for each strain. Normalized enrichment values were determined for every probe on the microarray by LOWESS normalization using Agilent Chip Analytics. Display, analysis and identification of the binding events were determined using MochiView (Homann and Johnson, 2010). Raw ChIP-chip data are available at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo, accession # GSE29785).
Motif Analysis
Motif analysis was performed using MochiView. MEME was also used to independently verify the motifs found by MochiView. See Datasets S5, S2, and Supplemental Experimental Procedures for details.
RNA Sequencing (RNA-seq) of Biofilm and Planktonic Cells
Strand-specific, massively-parallel SOLiD System sequencing of RNA from wild-type C. albicans biofilm and planktonic cells and mapping of resulting reads were performed as previously described (Tuch et al., 2010). Library amplification and sequencing resulted in 18 million planktonic and 28 million biofilm ~50 nt strand-specific sequence reads mappable to the C. albicans genome. RNA sequence data are available at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo, accession # GSE21291).
Identification of Novel Transcriptionally Active Regions (nTARs) in Biofilms
nTARs were identified using MochiView. A previously published transcript annotation (Tuch et al., 2010) was used as a starting scaffold, and additional transcribed regions were identified, as described in Supplemental Experimental Procedures. This approach identified 783 biofilm nTARs distinct from those in the previous annotation (Dataset S6).
Differential Expression Analysis of RNA-seq Data
For every transcribed region in our expanded biofilm genome annotation, mean per-nucleotide sequence coverage was extracted from both biofilm and planktonic datasets, transformed into pseudo-RPKM values, and transcripts differentially expressed between the two datasets were determined as described in Supplemental Experimental Procedures. The union of the RNA-seq and microarray datasets was used to determine the final set of differentially expressed genes (Dataset S6). Statistical significance (P values) for the association of binding and differential transcription was calculated using a two-tailed Fisher’s exact test.
Association of Transcription Regulator Binding Sites with Adjacent Transcripts
To determine the association between transcription regulator binding and differential gene expression, the binding peaks identified by ChIP-chip were mapped to immediately adjacent, divergently transcribed genes. A transcription regulator binding site was considered to be associated with differential expression if at least one divergent flanking transcript was differentially expressed in either the microarray or the RNA-seq comparison.
Exploring Orthology Relationships and Defining Gene Age Categories
Orthologs of the C. albicans and S. cerevisiae biofilm regulators and their direct targets were identified using freely available orthology mapping programs and by hand annotation using gene trees (See Supplemental Experimental Procedures). C. albicans gene age categories were defined as follows: “old” are members of gene families found in all Ascomycetes, “middle-aged” are members of gene families that arose after the divergence of S. pombe and S. japonicus but before the divergence of the CTG clade, “young” are found only in CTG clade species. Overlap of age categories with biofilm-induced genes is described by the hypergeometric distribution (See Supplemental Experimental Procedures).
Supplementary Material
Acknowledgments
We thank Oliver Homann for developing MochiView, Christopher Baker and Isabel Nocedal for help with evolutionary analysis, Francisco De La Vega for making possible the RNAseq analysis, Chiraj Dalal for computational advice, Lauren Booth for comments on the manuscript, and Sudarsi Desta, Jeanselle Dea, and Jorge Mendoza for technical assistance. We are grateful for the advice of Kurt Thorn in the acquisition of the CSLM images at the Nikon Imaging Center at UCSF. This study was supported by NIH grants R01AI073289 (D.R.A.) and R01AI083311 (A.D.J.). C.J.N. was supported by NIH fellowships T32AI060537 and F32AI088822. The content is the responsibility of the authors and does not necessarily represent the views of the NIH.
Footnotes
Accession Numbers. All data have been deposited into the NCBI Gene Expression Omnibus (GEO) portal under the accession numbers GSE21291 (RNA-seq), GSE29785 (ChIP-chip), and GSE30474 (GE Array).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun. 2004;72:6023–6031. doi: 10.1128/IAI.72.10.6023-6031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askew C, Sellam A, Epp E, Mallick J, Hogues H, Mullick A, Nantel A, Whiteway M. The zinc cluster transcription factor Ahr1p directs Mcm1p regulation of Candida albicans adhesion. Mol Microbiol. 2011;79:940–953. doi: 10.1111/j.1365-2958.2010.07504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie GS, Douglas LJ. Role of dimorphism in the development of Candida albicans biofilms. J Med Microbiol. 1999;48:671–679. doi: 10.1099/00222615-48-7-671. [DOI] [PubMed] [Google Scholar]
- Borneman AR, Leigh-Bell JA, Yu H, Bertone P, Gerstein M, Snyder M. Target hub proteins serve as master regulators of development in yeast. Genes Dev. 2006;20:435–448. doi: 10.1101/gad.1389306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno VM, Wang Z, Marjani SL, Euskirchen GM, Martin J, Sherlock G, Snyder M. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res. 2010;20:1451–1458. doi: 10.1101/gr.109553.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 2001;183:5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- de Hoon MJ, Eichenberger P, Vitkup D. Hierarchical evolution of the bacterial sporulation network. Curr Biol. 2010;20:R735–745. doi: 10.1016/j.cub.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas LJ. Candida biofilms and their role in infection. Trends Microbiol. 2003;11:30–36. doi: 10.1016/s0966-842x(02)00002-1. [DOI] [PubMed] [Google Scholar]
- Fernandez A, Lynch M. Non-adaptive origins of interactome complexity. Nature. 2011;474:502–505. doi: 10.1038/nature09992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoen LW, Venema G, Kuipers OP. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology. 2003;149:9–17. doi: 10.1099/mic.0.26003-0. [DOI] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawser SP, Douglas LJ. Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect Immun. 1994;62:915–921. doi: 10.1128/iai.62.3.915-921.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Friesen H, Segall J. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:5750–5761. doi: 10.1128/mcb.18.10.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann OR, Dea J, Noble SM, Johnson AD. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009;5:e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann OR, Johnson AD. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol. 2010;8:49. doi: 10.1186/1741-7007-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojic EM, Darouiche RO. Candida infections of medical devices. Clin Microbiol Rev. 2004;17:255–267. doi: 10.1128/CMR.17.2.255-267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R, Greenberg EP. Microbial sciences: the superficial life of microbes. Nature. 2006;441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- Lavoie H, Hogues H, Mallick J, Sellam A, Nantel A, Whiteway M. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 2010;8:e1000329. doi: 10.1371/journal.pbio.1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Wu WS, Liang H, Woo Y, Li WH. The spatial distribution of cis regulatory elements in yeast promoters and its implications for transcriptional regulation. BMC Genomics. 2010;11:581. doi: 10.1186/1471-2164-11-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JC, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, Turner MS, Millar AJ. Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol. 2005;1:0013. doi: 10.1038/msb4100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betran E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- Losick R, Stragier P. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature. 1992;355:601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- Mitrovich QM, Tuch BB, De La Vega FM, Guthrie C, Johnson AD. Evolution of yeast noncoding RNAs reveals an alternative mechanism for widespread intron loss. Science. 2010;330:838–841. doi: 10.1126/science.1194554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D, Stelling J. Precise regulation of gene expression dynamics favors complex promoter architectures. PLoS Comput Biol. 2009;5:e1000279. doi: 10.1371/journal.pcbi.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett J, Andes D. Candida albicans biofilm development, modeling a host-pathogen interaction. Curr Opin Microbiol. 2006;9:340–345. doi: 10.1016/j.mib.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Nett JE, Marchillo K, Spiegel CA, Andes DR. Development and validation of an in vivo Candida albicans biofilm denture model. Infect Immun. 2010;78:3650–3659. doi: 10.1128/IAI.00480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Bruce C, Hart C, Leigh-Bell J, Gelperin D, Umansky L, Gerstein MB, Snyder M. Dynamic and complex transcription factor binding during an inducible response in yeast. Genes Dev. 2009;23:1351–1363. doi: 10.1101/gad.1781909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, Edwards JE, Filler SG, Mitchell AP. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006a;2:e63. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol. 2005;15:1150–1155. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- Nobile CJ, Nett JE, Andes DR, Mitchell AP. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot Cell. 2006b;5:1604–1610. doi: 10.1128/EC.00194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault JS, Nantel A, Andes DR, Johnson AD, Mitchell AP. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 2009;7:e1000133. doi: 10.1371/journal.pbio.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, Mitchell AP. Complementary adhesin function in C. albicans biofilm formation. Curr Biol. 2008;18:1017–1024. doi: 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G, Tomsett K, Wickes BL, Lopez-Ribot JL, Redding SW. Denture stomatitis: a role for Candida biofilms. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2004;98:53–59. doi: 10.1016/j.tripleo.2003.04.002. [DOI] [PubMed] [Google Scholar]
- Ramage G, VandeWalle K, Lopez-Ribot JL, Wickes BL. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol Lett. 2002;214:95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- Sahni N, Yi S, Daniels KJ, Huang G, Srikantha T, Soll DR. Tec1 mediates the pheromone response of the white phenotype of Candida albicans: insights into the evolution of new signal transduction pathways. PLoS Biol. 2010;8:e1000363. doi: 10.1371/journal.pbio.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinabeck MK, Long LA, Hossain MA, Chandra J, Mukherjee PK, Mohamed S, Ghannoum MA. Rabbit model of Candida albicans biofilm infection: liposomal amphotericin B antifungal lock therapy. Antimicrob Agents Chemother. 2004;48:1727–1732. doi: 10.1128/AAC.48.5.1727-1732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellam A, Hogues H, Askew C, Tebbji F, van Het Hoog M, Lavoie H, Kumamoto CA, Whiteway M, Nantel A. Experimental annotation of the human pathogen Candida albicans coding and noncoding transcribed regions using high-resolution tiling arrays. Genome Biol. 2010;11:R71. doi: 10.1186/gb-2010-11-7-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellam A, Tebbji F, Nantel A. Role of Ndt80p in sterol metabolism regulation and azole resistance in Candida albicans. Eukaryot Cell. 2009;8:1174–1183. doi: 10.1128/EC.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- Sugino RP, Innan H. Natural selection on gene order in the genome reorganization process after whole genome duplication of yeast. Mol Biol Evol. 2011 doi: 10.1093/molbev/msr118. [DOI] [PubMed] [Google Scholar]
- Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 2008;6:e38. doi: 10.1371/journal.pbio.0060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuch BB, Mitrovich QM, Homann OR, Hernday AD, Monighetti CK, De La Vega FM, Johnson AD. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet. 2010;6:e1001070. doi: 10.1371/journal.pgen.1001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppuluri P, Chaturvedi AK, Srinivasan A, Banerjee M, Ramasubramaniam AK, Kohler JR, Kadosh D, Lopez-Ribot JL. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 2010;6:e1000828. doi: 10.1371/journal.ppat.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppuluri P, Pierce CG, Thomas DP, Bubeck SS, Saville SP, Lopez-Ribot JL. The transcriptional regulator Nrg1p controls Candida albicans biofilm formation and dispersion. Eukaryot Cell. 2009;9:1531–1537. doi: 10.1128/EC.00111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- Webb BC, Thomas CJ, Willcox MD, Harty DW, Knox KW. Candida-associated denture stomatitis. Aetiology and management: a review. Part 2. Oral diseases caused by Candida species. Aust Dent J. 1998;43:160–166. doi: 10.1111/j.1834-7819.1998.tb00157.x. [DOI] [PubMed] [Google Scholar]
- Wilson J. The aetiology, diagnosis and management of denture stomatitis. Br Dent J. 1998;185:380–384. doi: 10.1038/sj.bdj.4809821. [DOI] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.