SUMMARY
Following synthesis, integral membrane proteins dwell in the endoplasmic reticulum (ER) for variable periods that are typically rate limiting for plasma membrane delivery. In neurons, the ER extends for hundreds of microns as an anastomosing network throughout highly branched dendrites. However, little is known about the mobility, spatial scales, or dynamic regulation of cargo in the dendritic ER. Here we show that membrane proteins, including AMPA-type glutamate receptors, rapidly diffuse within the continuous network of dendritic ER but are confined by increased ER complexity at dendritic branch points and near dendritic spines. The spatial range of receptor mobility is rapidly restricted by type I mGluR signaling through a mechanism involving protein kinase C (PKC) and the ER protein CLIMP63. Moreover, local zones of ER complexity compartmentalize ER export and correspond to sites of new dendritic branches. Thus, local control of ER complexity spatially scales secretory trafficking within elaborate dendritic arbors.
INTRODUCTION
Most neurons possess highly branched dendrites with thousands of synaptic contacts, a geometry that allows for spatial segregation and integration of diverse signals (Spruston, 2008). The functional properties of dendrites are highly compartmentalized, with individual dendritic segments possessing distinct electrophysiological properties, synaptic inputs, and molecular composition (Losonczy et al., 2008; Matsuzaki et al., 2004). The partitioning of dendrites into functionally distinct segments relies on local regulation of receptor and ion channel distribution in the dendritic membrane (Nicholson et al., 2006). Although regulated transport of membrane components is a requisite for neuronal morphogenesis, synapse modification, and dendritic plasticity (Kennedy and Ehlers, 2006), the organelles and cellular machinery for spatially restricted dendritic trafficking are poorly understood.
Local dendritic synthesis and secretory processing of membrane cargo, and in particular α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate (AMPA)-type glutamate receptors provide attractive mechanisms for spatially delimited modification of specified dendritic segments (Sutton et al., 2006). However, unlike cytoplasmic proteins, which require only mRNA and ribosomes, local production of membrane proteins requires the entire complement of secretory organelles (Hanus and Ehlers, 2008). Yet, little is known about the local dynamics and regulation of AMPA receptors and other dendritic membrane cargo in early secretory compartments.
Recent studies indicate that the entire cast of secretory organelles is present in some dendrites (Gardiol et al., 1999; Horton and Ehlers, 2003). In particular, the endoplasmic reticulum (ER) exists as a continuous anastomosing network in dendrites (Cooney et al., 2002; Spacek and Harris, 1997; Terasaki et al., 1994). For numerous synaptic receptors and notably AMPA receptors (Penn et al., 2008), ER export is rate-limiting for plasma membrane delivery. For slowly exported cargo, diffusion within the ER determines the spatial range of cargo transport to the plasma membrane (Herpers and Rabouille, 2004). Interestingly, AMPA receptors dwell in the ER for variable periods up to several hours (Greger et al., 2002), raising the possibility that dwell time and ER mobility determine the spatial domain over which nascent receptors are delivered to the dendritic plasma membrane. Receptor diffusion at the cell surface emerged as a critical feature of synaptic physiology (Newpher and Ehlers, 2008). However, the internal diffusion of receptors within the dendritic ER has not been examined, despite a potentially large influence on the spatial scale over which dendritic membrane composition is modified.
Here we show that newly synthesized membrane proteins, including AMPA receptors, rapidly diffuse within the continuous network of the dendritic ER. Quantitative photobleaching, super-resolution imaging, and ultrastructural analysis reveal that nascent cargo diffusion is inhomogeneous, with zones of diffusional confinement corresponding to local domains of increased ER complexity near dendritic spines, at dendritic branch points (BPs), and near Golgi outposts (GOs). The spatial range of receptor mobility in the ER becomes progressively limited over neuronal development and is rapidly restricted by group I mGluR signaling through a mechanism involving protein kinase C (PKC) and the microtubule-binding ER protein CLIMP63. Site-specific phosphorylation of CLIMP63 increases ER complexity, thereby trapping nascent membrane proteins within smaller domains of diffusional confinement. Such local zones of ER complexity correspond to domains of ER export and sites of new dendritic branch formation. Correspondingly, manipulations that increase or decrease ER complexity alternately promote or reduce dendritic branching. Finally, increased ER complexity promotes AMPA receptor surface expression and increases synaptic strength. Taken together, these results define a novel mechanism for compartmentalized trafficking in geometrically complex cells.
RESULTS
Rapid but Spatially Restricted Diffusion of Membrane Proteins in the Dendritic ER
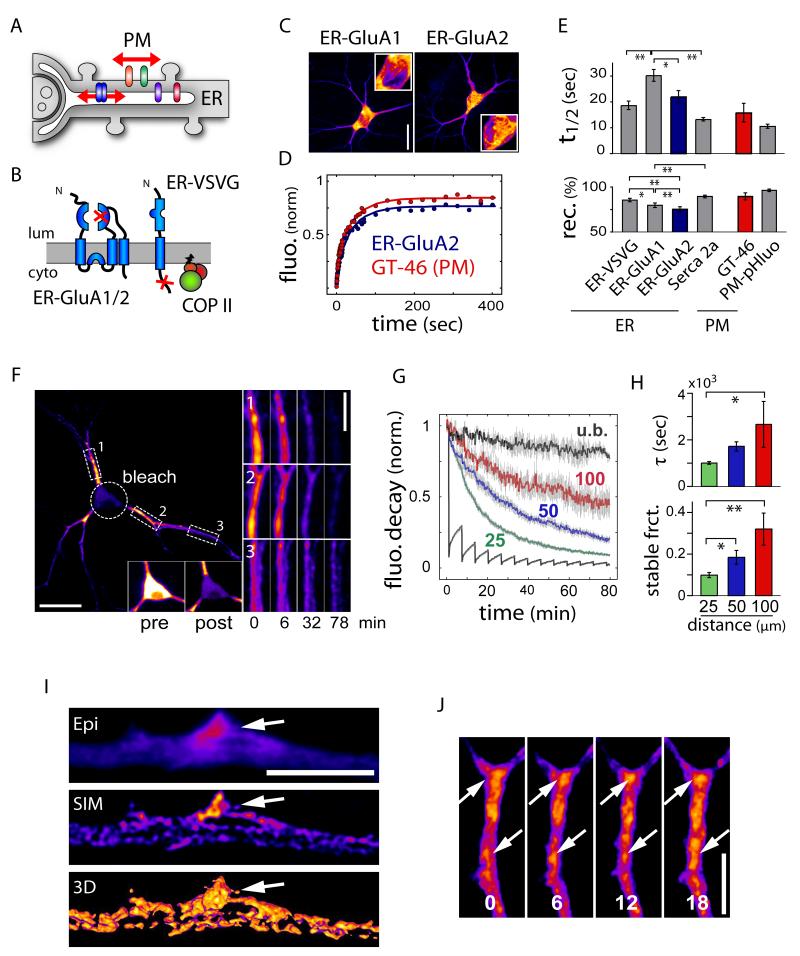
Prior to reaching the plasma membrane, newly synthesized membrane proteins are assembled in the somatodendritic ER where their mobility is unknown. To compare the diffusion of nascent AMPA receptors to that of freely diffusing ER and cell-surface membrane proteins, we engineered various integral membrane GFP fusions and measured fluorescence recovery after photobleaching (FRAP) in hippocampal neurons (Figure 1A, Movie S1). GFP-fusions of ER-retained GluA1 and GluA2 subunits mimicking nascent receptors (ER-GluA1 and ER-GluA2) were generated by introducing point mutations in their glutamate binding sites (Figures 1B, 1C and S1A-D). The dynamics of ER-GluA1/2 in dendrites were compared to that of ER-VSVG (Figures 1D, 1E, S1E-S1H and S2), an ER-retained version of the vesicular stomatitis virus glycoprotein (VSVG) that freely diffuses in the ER membrane, to the ER calcium-pump SERCA2a whose dynamics have been characterized (Fukatsu et al., 2004), and to two freely moving plasma membrane proteins, PM-phluo and GT-46 (Figures S1I and S1J).
Figure 1.
Membrane Protein Dynamics in the Dendritic ER.
(A) Model illustrating the mobility of membrane proteins in the ER and at the cell surface of a neuronal dendrite. PM, plasma membrane.
(B) ER-retained GluA1 and GluA2 subunits (ER-GluA1/2) mimicking nascent receptors were generated by point mutation in the glutamate binding site (see also Figure S1). A constitutively ER-retained version of VSVG (ER-VSVG) was generated by mutagenesis of VSVG COPII binding motifs. Lum, lumen; cyto, cytosol.
(C) Distribution patterns of ER-GluA1 and ER-GluA2 in DIV15 hippocampal neurons. Insets show higher magnification of cell bodies. Scale bar, 30 μm.
(D) Examples of raw (filled circles) and fitted (lines) FRAP plots obtained for GluA2 in the ER (ER-GluA2) and GT46 at the plasma membrane (PM).
(E) Average recovery halftime (t1/2) and fractional recovery measured for integral membrane proteins in the ER or at the cell surface (PM) in DIV17-22 neurons. Mean±SEM, n = 64, 38, 28, 44, 14, and 22 cells, for ER-VSVG, ER-GluA1, ER-GluA2, SERCA2a, GT46 and PM-pHluo, respectively, 3-4 experiments each; *p < 0.05, **p < 0.01; ANOVA.
(F) Loss of dendritic ER-VSVG fluorescence upon continuous bleaching of the soma. Time lapse sequences are shown for proximal (1, 2) and distal (3) dendritic segments. Insets show ER-VSVG fluorescence before (pre) and after (post) photobleaching the soma. Scale bars, 30 μm and 5 μm (inset).
(G) Fluorescence decay in the soma (grey) and in dendritic segments at 25 μm (green), 50 μm (blue) and 100 μm (red) from the soma in bleached neurons and at 100 μm from the soma in unbleached neurons (u.b., black). n = 7-11 dendrites in 9 cells, 3 experiments.
(H) Decay constants (τ) and stable fractions in dendrites as a function of distance from the soma. Mean±SEM; *p < 0.05, **p < 0.01; ANOVA.
(I) Super-resolution image of ER-VSVG obtained by structured illumination microscopy (SIM) showing spatial heterogeneities within the dendritic ER image. Epi, epifluorescence image; SIM, SIM image; 3D, 3D reconstruction. Scale bar, 5 μm.
(J) Time-lapse sequence of ER-VSVG. Shown are z-stack projections illustrating the relative stability of ER spatial heterogeneities (arrows). Times shown in minutes. Scale bar, 5 μm.
As expected, ER-GluA1/2 and ER-VSVG were diffusely distributed throughout the dendritic volume (Figures 1C, S1B, S1F and S1M), consistent with an ER distribution (Cooney et al., 2002; Gardiol et al., 1999; Spacek and Harris, 1997). FRAP showed that GluA2, VSVG and SERCA2a had similar mobility in the dendritic ER, while GluA1 had slightly slower recovery kinetics (Figure 1E, top panel). Determination of apparent diffusion coefficients using a three-dimensional diffusion model (Figure S1K) confirmed the rapid motion of nascent AMPA receptors (Dapp: ER-GluA1, 0.038 ± 0.004 μm2/s; ER-GluA2, 0.069 ± 0.013 μm2/s; ER-VSVG, 0.081 ± 0.004 μm2/s; SERCA2a, 0.092 ± 0.009 μm2/s). Although slightly smaller than that of VSVG, the exchangeable fractions of GluA1 and GluA2 in the ER were quite high (ER-VSVG, 85 ± 2%; ER-GluA1, 79 ± 2%; ER-GluA2, 75 ± 3%), indicating that most AMPA receptors are exchangeable and mobile in the ER (Figure 1E, bottom panel). The measured recovery kinetics, mobile fractions, and apparent diffusion rates were comparable to those of receptors in the extrasynaptic plasma membrane (Ehlers et al., 2007) and mirrored the dynamics of the freely diffusing membrane proteins GT-46 and PM-pHluo at the dendritic surface (Figures 1D and 1E, Table S1). Thus, mobility of nascent receptors in the ER is nearly as rapid as that of mobile receptors at the PM.
To directly examine the rate and spatial extent of ER membrane protein mobility throughout entire living neurons, we performed fluorescence loss in photobleaching (FLIP) experiments in neurons expressing ER-VSVG (Figure 1F). Continuous bleaching of the soma led to a progressive loss of dendritic fluorescence (Figures 1F, 1G and S1L), demonstrating functional continuity of the somatodendritic ER. Whereas fluorescence loss occurred within minutes in proximal dendrites, distal dendrites were more slowly depleted and retained a significant fraction of their initial fluorescence (Figures 1F-H). Thus, although highly mobile over micron scales, nascent receptors in the dendritic ER exhibit spatially restricted exchange over longer distances.
Membrane Cargo Mobility in the Dendritic ER Diminishes Over Development
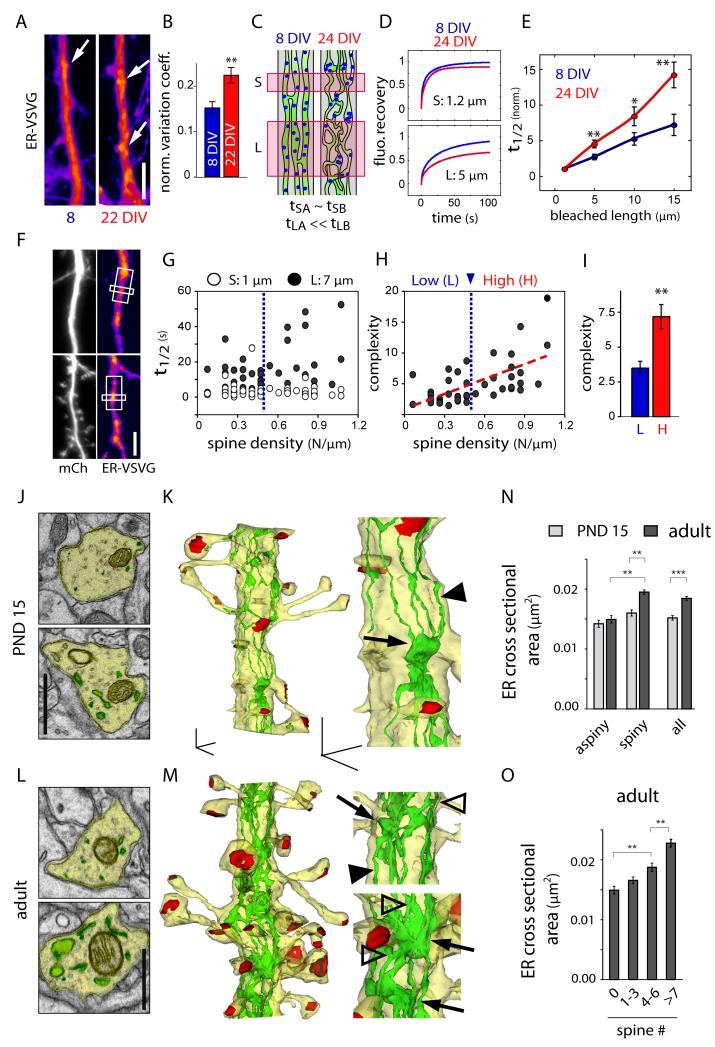
Although ER-VSVG appears diffuse in low-resolution FRAP experiments (Figures S1F and S2G), super resolution structured illumination microscopy (SR-SIM) (Schermelleh et al., 2010) revealed subcellular heterogeneities in the dendritic ER (Figure 1I). Most neurons displayed clusters of increased fluorescence distributed along dendrites (Figure 1I), which were stable for many minutes (Figure 1J), and became more prominent over neuronal development (Figures 2A and 2B). Membrane protein mobility in the ER is likely restricted by structural features such as anastomoses and cisterns. Since these structures cannot be resolved by conventional fluorescence microscopy (Spacek and Harris, 1997), we developed a FRAP-based method to quantitatively assess diffusional restriction and ER complexity within local dendritic segments. We took advantage of the fact that the size of the bleached area affects fluorescence recovery rates (Sprague and McNally, 2005). Specifically, since the volume of any object increases with size faster than its surface area, increasing the size of the bleached area decreases the surface area to volume ratio and hence decreases the interface available for exchange of bleached and non-bleached molecules. Thus, for any exchanging molecule, measured recovery rates become progressively slower as a function of the effective complexity of the fluorophore-accessible volume as the size of the bleached area increases (Figures 2C and S2). Hence, by comparing the ratio of fluorescence recovery after bleaching a large area to that after bleaching a small area, one obtains a functional measure of the effective complexity of the fluorophore-accessible volume (Figure S2).
Figure 2.
Increased ER Complexity and Reduced ER Mobility in Older Spiny Dendrites.
(A) Distribution pattern and (B) variance of ER-VSVG fluorescence along dendrites of young (DIV8-11, left) and older (DIV22, right) neurons. Arrows show spots of increased ER-VSVG intensity. Mean±SEM, n = 13; **p < 0.01; t-test. Scale bar, 5 μm.
(C-D) FRAP-based measurement of ER complexity (see also Figure S2). (C) Schematic of experimental approach. FRAP measurements in small dendritic segments (tS) provide information on intrinsic molecular properties independent of ER geometry (e.g., freely diffusing fraction, binding kinetics, etc.). FRAP measurements in larger dendrite segments (tL) depend on ER geometry and the fluorophore-accessible volume. (D) Fitted plots of ER-VSVG fluorescence recovery after bleaching small (S, 1.2 μm) or large (L, 5 μm) segments in young (DIV8, blue) and old (DIV24, red) neurons.
(E) Recovery halftimes (normalized t1/2) as a function of the length of bleached dendritic segments in young (blue) and older (red) neurons. Mean±SEM, n = 20, 21 cells in 4 experiments, respectively; *p < 0.05, **p < 0.01; t-test.
(F) Images of hippocampal neurons (DIV15) expressing an mCherry cell fill (mCh, left) and ER-VSVG (right). White boxes illustrate the small (1 μm) and large (7 μm) bleached areas. Scale bar, 5 μm.
(G) Fluorescence recovery halftimes (t1/2) as a function of dendritic spine density after sequential bleaching of small 1 μm (S, open circles) and large 7 μm segments (L, filled circles) in older neurons (DIV17-19). n = 42 cells, 3 experiments.
(H) ER complexity index (t1/2, L / t1/2, S) as a function of dendritic spine density. The dashed red line indicates the linear fit. A cutoff for high (H) and low (L) spine density was set at 0.5 spines/μm (dashed blue line).
(I) Mean complexity index of dendritic segments with low spine densities under (L, blue) or high spine densities above (H, red) 0.5 spines/μm. Mean±SEM, n = 17, 25 cells in 3 experiments, respectively; *p < 0.05, **p < 0.01; t-test.
(J-M) Electron micrographs (J, L) and three-dimensional reconstructions (K,M) at low and higher magnification, showing the cytoplasm (J,L) or the plasma membrane (K,M; yellow), the ER (green) and excitatory postsynaptic differentiations (red) in postnatal day 15 (PND 15; J, K) and adult (L, M) CA1 pyramidal neuron apical dendrites. Note the elongated longitudinal ER tubules (arrow heads), ER branching (empty arrow heads) and local increase of ER volume at the base of dendritic spines (arrows). In adult neurons, note the increased ER branching (empty arrowheads) and cisternae (arrows). Scale bar, 1 μm (J,L), 500 nm (K,M).
(N-O) Average ER cross-sectional area in aspiny, spiny and spiny plus aspiny segments (all, N) of PND 15 or adult dendrites, and (O) correlation with spine number in adult dendrites. Mean±SEM, n=528-539 sections in PND 15 aspiny and spiny segments, and n = 433-544 sections in adult aspiny and spiny segments, n = 10-17 dendrites from 2-4 animals; **p < 0.01, ***p < 0.001; ANOVA or t-test.
Using this assay, we found that membrane protein mobility in the dendritic ER declines significantly over neuronal development (Figures 2C-E). As the length of bleached dendritic segment was increased, the slowing of recovery times was more pronounced in older neurons (Figure 2D and 2E), indicating increased effective complexity (Figures 2C and S2). This restricted mobility in older neurons was not due to a heterogeneous distribution of ER-VSVG binding sites as recovered fractions and diffusion anomaly were identical in young and mature neurons (Figure S2J).
Zones of ER Confinement Localize Near Dendritic Spines and Dendritic Branch Points
To determine whether ER membrane protein mobility is reduced near excitatory synapses (Spacek and Harris, 1997), we measured FRAP recovery rates derived from small 1 μm (t1/2,S) and larger 7 μm (t1/2,L) dendritic segments (Figure 2F), and calculated an ER complexity index as the ratio between t1/2,L and t1/2,S obtained from DIV18 hippocampal neurons with varying spine densities. In contrast to t1/2,S which remained constant, t1/2,L increased with spine density (Figure 2G), resulting in a positive correlation between spine density and ER complexity (Figures 2H and 2I). To examine the ultrastructural organization of ER over dendritic development and its spatial relationship with spines in vivo, we performed serial section 3D reconstruction in CA1 pyramidal neurons. We compared ER morphology in dendrites displaying varying spine densities in postnatal day 15 (PND15) or adult neurons. In aspiny segments of PND15 dendrites, ER consisted primarily of multiple elongated longitudinal tubules connecting regions of increased ER volume found near dendritic spines (Figures 2J and 2K). In older dendrites ER volume was overall more heterogenous with more branched tubules and larger distended cisternae in zones of high spine density (Figures 2L and 2M). As an indirect metric for ER complexity, we assessed ER volume variation along dendrites by comparing ER cross-sectional area in aspiny and spiny dendritic segments with varying spine number (Figure S2K). Although ER cross-secional area was higher in spiny dendrites both in PND15 and adult neurons, this difference was more pronounced in adult neurons due to higher spine density (Figure 2N), resulting in a globally increased ER complexity in adults (Figure 2N). In agreement with previous studies (Spacek and Harris, 1997), variation of ER volume along the dendritic shaft of adult neurons was tightly correlated with local spine density (Figure 2O), providing a structural basis for reduced membrane cargo mobility. Consistent with EM reconstructions where spines rarely contained ER (Figures 2K and 2M), fluorescence microscopy showed that ER complexity is not due to ER entry into spines (Figures S2L-N).
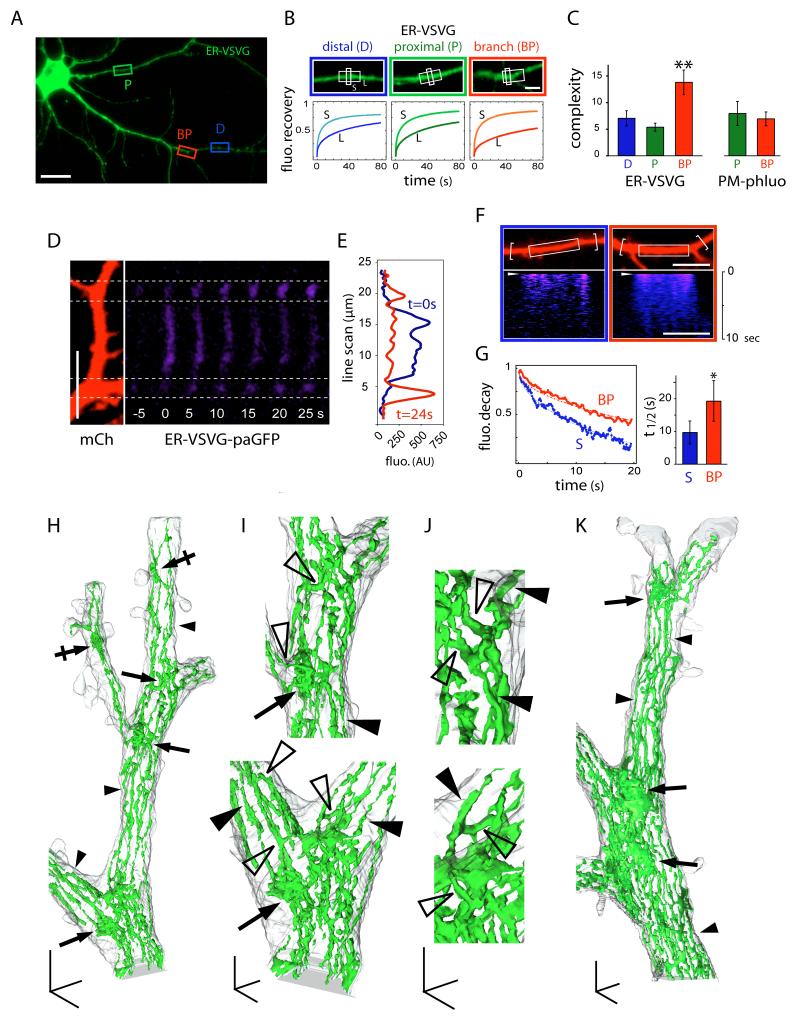
We next performed FRAP measurements at various locations along dendrites (Figure 3A). We found no quantitative difference in submicron diffusion rates or ER complexity (t1/2,L / t1/2,S) between proximal and distal dendritic segments (Figures 3B, 3C and Table S1). In contrast, ER complexity was significantly higher at dendritic BPs (Figures 3B and 3C). Importantly, the increased complexity at BPs was specific for membrane proteins in the ER and was not observed at the cell-surface (Figure 3C).
Figure 3.
Diffusional Trapping of ER Membrane Cargo at Dendritic Branch Points.
(A) ER-VSVG expression in a DIV15 hippocampal neuron. Shown are examples of proximal (P), distal (D), and branch point (BP) regions subjected to photobleaching. Scale bar, 30 μm.
(B) Averaged FRAP plots obtained after sequential bleaching of small 1 μm (S, light colors) and large 7 μm dendritic segments (L, dark colors) in distal dendrites (D, blue), proximal dendrites (P, green), and dendritic BPs (red). Scale bar, 5 μm.
(C) Average complexity (t1/2, L / t1/2, S) measured for ER-VSVG in the ER and PM-phluo at the plasma membrane (PM). D, distal; P, proximal; BP, branch point. Mean±SEM, n = 12-18 cells in 2-3 experiments each; **p < 0.01; ANOVA.
(D) Photoactivation of ER-VSVG-paGFP in a region of dendrite between two BPs. Photoactivated ER-VSVG-paGFP (blue, right) diffused away from the photoactivated region but was trapped at adjacent BPs. mCh was co-expressed as a cytosolic cell fill (left). Scale bar, 10 μm.
(E) Line-scan of ER-VSVG-paGFP fluorescence at two time points following photoactivation.
(F) Photoactivation of ER-VSVG-paGFP in a straight dendritic segment (left, white box) or in a dendritic segment flanked by BPs (right, white box) (see also Figure S3). Bottom panels show kymographs of fluorescence intensity along the dendrite. Scale bar, 10 μm.
(G) Fluorescence decay plots (left) and average decay half times (right) following ER-VSVG-paGFP photoactivation measured in straight (S) dendritic segments or dendritic segments of equal length flanked by BPs. Fluorescence measurements were made from larger regions that encompassed BPs as indicated by the brackets in (F). Mean±SEM, n = 10 cells, 3 experiments; *p < 0.05; t-test.
(H-K) Three-dimensional reconstructions of electron micrographs showing the plasma membrane (grey) and the ER (green) at different magnifications. Note the presence of ER cisternal sheets at dendritic BPs (arrows) and the extensive branching of the ER membrane (bridging tubules, empty arrowheads), contrasting with ER tubules (filled arrowheads) in straight dendritic segments. The morphological complexity of the ER was also increased in areas with high spine density (crossed arrows in H). Scale bars, 1 μm (H and K), 500 nm (I) and 250 nm (J).
The restricted mobility of ER membrane cargo at BPs was further supported by photoactivation experiments (Figures 3D-G). Specifically, ER-VSVG-paGFP photoactivated in dendritic segments accumulated and dwelled for extended periods at flanking BPs (Figures 3D and 3E). Conversely, decay rates of photoactivated VSVG-paGFP measured in 15-30 μm dendritic segments flanked by and encompassing BPs were significantly slower than in straight dendritic segments (Figures 3F and 3G). Taken together, these data show that BPs act as diffusion traps restricting ER membrane protein mobility.
Compartmentalized Diffusion of Nascent AMPA Receptors in the Dendritic ER
To determine how lateral diffusion impacts the size of the dendritic domain explored by newly synthesized membrane cargo in the ER, we simulated the spread of GluA1 in dendrites (Figures S3A and S3B) using measured and extrapolated diffusion coefficients (Table S2) derived from straight segments and dendritic BPs. Exploration maps obtained over a total duration of 12 h assuming a constant rate of synthesis at a given location (e.g. BP), indicate that nascent AMPA receptors concentrate within dendritic subcompartments (Figures S3A, S3B and Movie S2). This accumulation occurred over hours and, by 12 h, produced a nearly 10-fold increase in receptor density centered at the synthesis site. Compartmentalization was particularly marked in complex dendrites (Figures S3C and S3D), due largely to the reduced mobility at BPs. If the reduced diffusion at BPs was excluded, the size of explored dendritic domains was significantly increased (Figure S3D). The additive contribution of individual dendritic branches to nascent receptor confinement was further confirmed by simulations in dendritic templates with defined geometries (Figure S3E), where each bounding branch point induced a ~20% increase in local receptor concentration (Figure S3F). These data show that cargo mobility within the ER tunes the local reservoir of available nascent receptors and is strongly influenced by local dendritic geometry.
Increased ER Complexity and Preferential ER Export at Dendritic Branch Points
The increased diffusional confinement coupled with the unchanged short-range fluorescence recovery kinetics at BPs (Figure 3B) suggested local increase in the geometric complexity of ER structure. To address this, we performed serial section 3D reconstruction in CA1 pyramidal neurons. In straight segments of dendrite, the ER consisted of elongated longitudinal tubules connected by occasional transverse bridging tubules (Figures 3H and 3K). At BPs, ER was much more complex, with multiple anastomosing tubules and flattened cisternae (Figures 3H-3K), providing a structural basis for reduced membrane cargo mobility.
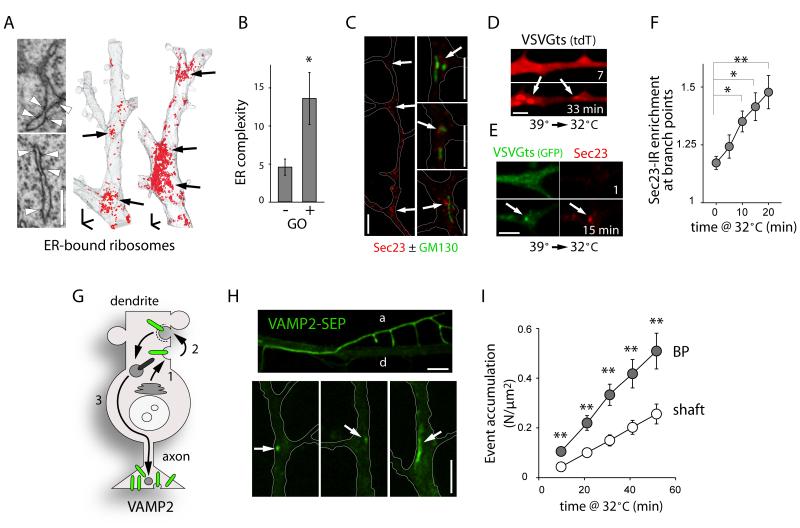
To test whether ER structural complexity corresponds to sites of secretory protein biosynthesis, we examined the distribution of ER-bound ribosomes and found that they are concentrated at dendritic BPs (Figure 4A). This suggests that BPs constitute the entry site of locally translated proteins into the secretory pathway. Indeed, FRAP experiments indicated a significant reduction in secretory cargo mobility in ER domains near Golgi outposts (GOs, Figure 4B), which are themselves concentrated at BPs (Horton et al., 2005; Ye et al., 2007). Moreover, ER-exit sites (ERES) marked by Sec23-immunolabeling were enriched at BPs and locally clustered around GOs (Figure 4C). This suggests that local ER complexity facilitates local export and processing of secretory cargo. To investigate this, we imaged VSVGts045, a model cargo that can be released from the ER in a thermosensitive manner, to monitor synchronized cargo progression in the secretory pathway (Horton and Ehlers, 2003). Following release from the ER, VSVGts rapidly accumulated at BPs (Figures 4D, 4E, S4A and S4B), leading in turn to a rapid and local increase of Sec23 (Figures 4E and 4F), indicative of local ER export (Figure S4C) (Farhan et al., 2008). These data demonstrate a spatial correlation between increased ER complexity, decreased cargo mobility, and preferential ER export at BPs.
Figure 4.
Preferential ER Entry and ER Exit at Dendritic Branch Points.
(A) Three-dimensional EM reconstructions showing ER-bound ribosome distribution in segments of CA1 dendrites. Note the concentration of ER-bound ribosomes at BPs (arrows). Scale bar, 1 μm.
(B) ER complexity index in dendritic segments lacking (−) or containing (+) Golgi outposts (GOs). Mean±SEM, n = 10 cells in 2 experiments; *p < 0.05; t-test.
(C) Sec23 immunoreactivity showing the distribution of ER exit sites in dendrites of DIV7 neurons. Scale bars, 5 μm.
(D) Accumulation of VSVGts at BPs (arrows) 7 and 33 min after temperature-induced release of ER exit blockade (39°C to 32°C shift) (see also Figure S4). Scale bar 5 μm.
(E-F) Images (E) and quantification (F) of the preferential increase of Sec23 at BPs after VSVGts release from the ER by shift to 32°C. Scale bar, 5 μm. Mean±SEM, n = 8-24 cells, 2-4 experiments for each; *p < 0.05, **p < 0.01; ANOVA.
(G-I) Preferential exocytosis at BPs. (G) VAMP2 is first secreted in the somatodendritic membrane (1), internalized (2) and subsequently targeted to the axon (3). (H) VAMP2-SEP fluorescence in a dendrite and an axon (d and a, upper) and discrete exocytosis events at BPs (arrows, lower). Scale bar, 5 μm. (I) Cumulation of exocytosis events at BPs and in the dendritic shaft. Mean±SEM, n= 30 dendrites in 15 neurons; *p < 0.05, **p < 0.01; ANOVA.
We then sought to determine whether zones of increased ER complexity correspond to sites of secretory exocytosis. We took advantage of the transcytosic itinerary of the presynaptic protein VAMP2, which is first exocytosed in dendrites, internalized, and then targeted to and enriched in axon terminals (Sampo et al., 2003), allowing clear spatial segregation of exocytosis from secretory and endosomal compartments. In parallel with VSVGts release from the ER, secretory exocytosis of superecliptic-GFP tagged VAMP2 (VAMP2-SEP) occurred preferentially at BPs (Figures 4G-I and Movie S3).
Acute Regulation of Dendritic ER Complexity by an mGluR/PKC/CLIMP63 Pathway
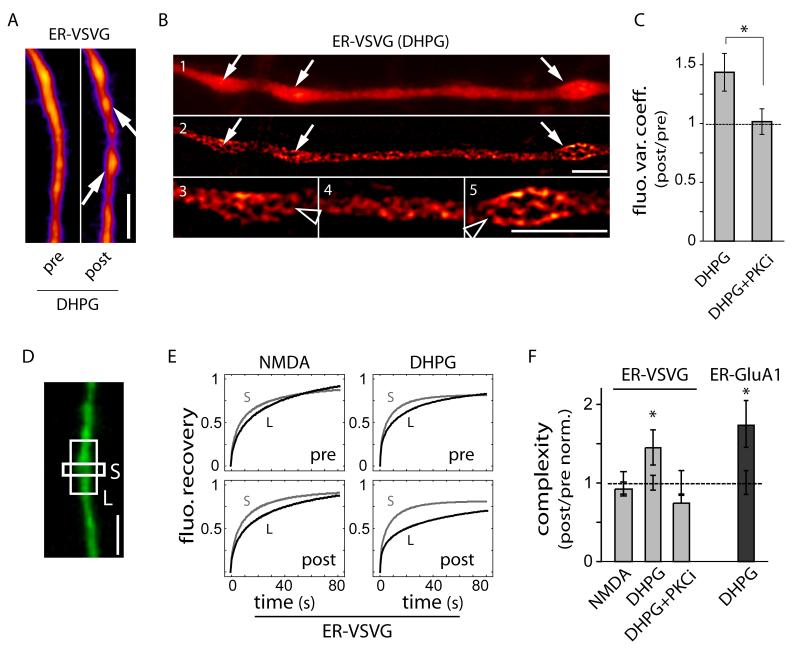
The developmental increase in dendritic ER complexity and its correlation with dendritic spine density (Figure 2) suggested regulation by emergent synaptic activity. This prompted us to assess whether activation of postsynaptic glutamatergic receptors controls ER morphology and effective complexity. In dendrites where ER fine structure could be well-resolved, ER-localized membrane cargo was noticeably less uniform and concentrated in ER nodules following activation of group I metabotropic glutamate receptors (mGluRs) using the selective agonist (S)-3,5-dihydroxyphenylglycine (Figures 5A-5C). As confirmed by super-resolution imaging, these structures correspond to regions of convoluted ER (Figure 5B). Although these nodules were clearly visible only in ~60% of neurons at diffraction-limited resolution, DHPG increased fluorescence variation along dendrites regardless of nodule appeareance (Figure 5C). Correspondingly, DHPG caused a significant increase in dendritic ER complexity measured by FRAP (Figures 5D-F), whereas NMDA receptor activation had no effect on this parameter. The mGluR-induced increase in ER complexity occurred without measurable ER stress (Figure S5) and mGluR activation did not change submicron diffusion rates (Figures 5D, 5E and Table S1), indicating that the effect was not due to a compositional change in the ER. The group I mGluRs (mGluR1, mGluR5) couple to Gq and thereby trigger protein kinase C (PKC) and inositol 1,4,5-trisphosphate (IP3)-dependent signaling pathways. Correspondingly, the DHPG-induced increase in ER morphological and effective complexity was abolished by inhibiting PKC (Figures 5C and 5F). The effect of mGluR activation was not cargo selective, as the apparent diffusional complexity index for both ER-VSVG and ER-GluA1 was increased to the same extent (Figure 5F) despite different absolute mobilities (Figure 1E). Thus, postsynaptic mGluR signaling via PKC rapidly confines nascent receptor diffusion in the dendritic ER.
Figure 5.
Group I mGluR Signaling Modifies ER Morphology and Reduces ER Cargo Mobility.
(A) Increased heterogeneity of ER-VSVG distribution after a 30 min exposure to DHPG (arrows) (See also Figure S5). Scale bar, 5 μm.
(B) Structured illumination microscopy images of a dendrite showing ER-VSVG fluorescence before (1) and after SIM imaging (2-5) at low (1-2) and higher (3-5) magnifications. Note the extensive branching (arrowheads) of ER membranes in DHPG-induced nodules (arrows). Scale bar, 5 μm.
(C) Increased variation in ER-VSVG fluorescence along dendrites in cells as shown in (A) in the absence (DHPG) or presence of a PKC inhibitor (PKCi, 50 nM bisindoylmalemide). Data are normalized to values measured prior to drug application, Mean±SEM. DHPG, n = 7; DHPG + PKC inhibitor, n = 8 cells; 1-2 experiments each; *p < 0.05; t-test.
(D-E) Representative FRAP plots obtained from sequential bleachings of small 1 μm (S, grey) and large 7 μm (L, black) dendritic segments (DIV14) before (pre) and after (post) exposure to NMDA or DHPG.
(F) Effective complexity (t1/2, L / t1/2, S) in individual dendrites exposed to NMDA, DHPG and DHPG + PKCi. ER complexity values measured by FRAP following stimulation with indicated reagents are normalized to values measured prior to drug application (PKCi, 100 nM bisindoylmalemide), Mean±SEM. n = 34, 42, 16 cells for ER-VSVG in NMDA, DHPG, and DHPG + PKCi treatment, respectively; n = 19 cells for ER-GluA1 in DHPG treatment; 2-4 experiments; *p < 0.05 (relative to 1.0); t-test.
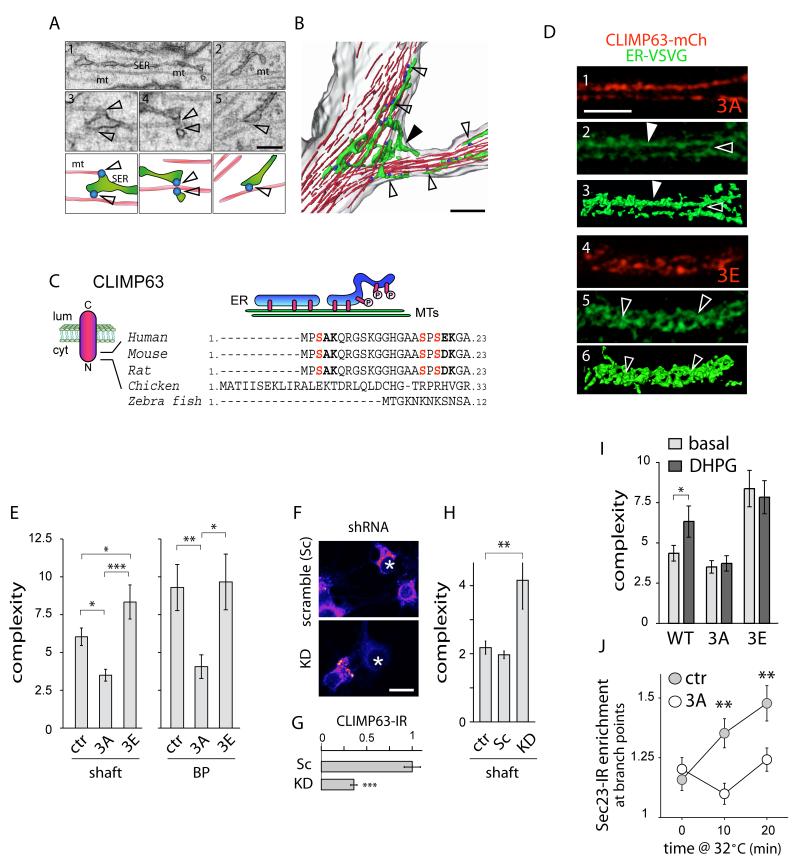
In fibroblasts, ER interaction with microtubules is an important determinant of ER morphology (Vedrenne and Hauri, 2006). We thus examined ER ultrastructure in vivo and found that whereas elongated ER tubules frequently contacted microtubules (Figure 6A), convoluted ER membranes seen at BPs were much more loosely associated with microtubules (Figures 6A and 6B). Among the potential targets of PKC at the ER, CLIMP63 is an ER integral membrane protein whose binding to microtubules is regulated by PKC-dependent phosphorylation (Klopfenstein et al., 1998; Schweizer et al., 1993; Vedrenne et al., 2005). When bound to microtubules in fibroblasts, CLIMP63 promotes elongation of ER tubules to the cell periphery along microtubule tracts (Klopfenstein et al., 1998; Vedrenne et al., 2005). Phosphorylation of one or more N-terminal serine residues (S3, S17, and S19) abrogates microtubule binding (Figure 6C), eventually resulting in ER collapse and membrane stacking around the nucleus (Vedrenne et al., 2005). CLIMP63 is highly expressed in brain (Farah et al., 2005), and given the regulation of dendritic ER complexity by mGluR/PKC (Figure 5), we wondered whether the increased ER complexity seen in neurons after mGluR activation involved CLIMP63 phosphorylation. To test this, we generated phosphodeficient (CLIMP63-S3A, S17A, S19A; referred to as 3A) and phosphomimetic (S3E, S17E, S19E; referred to as 3E) mutant forms of CLIMP63 that either strongly bind (3A) or fail to bind (3E) microtubules (Vedrenne et al., 2005). In neurons, CLIMP63-3A was uniformly distributed in straight tubular structures throughout dendrites (Figures 6D, S6B, and Movie S4), consistent with tight microtubule binding. In contrast, phosphomimetic CLIMP63-3E was present in nonuniform clusters (Figures 6D, S6B, and Movie S4), consistent with a decreased binding to microtubules. The ER in neurons expressing CLIMP63-3E was still present throughout dendrites (Figure S6B), but became much more convoluted (Figure 6D and Movie S4), similar to changes in the ER observed upon mGluR activation (Figure 5B).
Figure 6.
Phosphorylation-Dependent Binding of CLIMP63 to Microtubules Controls ER Morphology and Cargo Mobility.
(A-B) Three-dimensional reconstructions of electron micrographs (A) showing the plasma membrane (grey), ER (green), microtubules (MT, red) and ER-MT contacts (blue) at a dendritic branch point (B). Scale bars, 500 nm. In (B), note the looser association of bunched ER membranes (filled arrowhead) at the branch point compared to elongated tubules aligned with MTs in the dendritic shaft (empty arrowheads).
(C) Topology of CLIMP63 in the ER membrane (left) and regulation of ER microtubule binding by phosphorylation (right). Three serine residues in the N-terminus whose mutation prevents PKC-dependent phosphorylation are shown (red). Numbers indicate amino acids. Lum, lumen; cyt, cytosol.
(D) Super-resolution imaging of ER-VSVG (green) and CLIMP63-S3A, S17A, S19A (3A) or CLIMP63-S3E, S17E, S19E (3E) (red) in hippocampal neuron dendrites (DIV15) (see also Figure S6). Elongated tubules (arrowheads) or branched ER membranes (empty arrowheads) are indicated. Three-dimensional surface renderings (3, 6) were derived from ER-VSVG images (2, 5). Scale bar, 5 μm.
(E) Apparent ER complexity measured by FRAP in the dendritic shaft or at BPs in neurons (DIV13-14) co-expressing ER-VSVG with either mCherry (ctr), mCh-CLIMP63-3A (3A), or mCh-CLIMP63-3E (3E). Mean±SEM, n = 64, 39 and 43 cells for dendritic shaft, n = 18, 14, 9 cells for BPs, respectively, 4-5 experiments for each; *p < 0.05, ***p < 0.01; ANOVA.
(F-G) CLIMP63 knockdown by RNA interference. (F) CLIMP63 immunoreactivity (IR, pseudocolored fluorescence) in untransfected and transfected (asterisks) neurons (somatas) expressing scramble (Sc) or target (KD) short hairpin RNAs (shRNA). (G) Normalized CLIMP63 levels in Sc and KD dendrites, Mean±SEM. n = 19 cells, 2 experiments; ***p < 0.001; t-test.
(H) CLIMP63 knockdown increases ER complexity. FRAP as in (E) in dendrites of neurons expressing scrambled (Sc) or CLIMP63 (KD) shRNAs or reporter only (tdTomato, ctr) with ER-VSVG. Mean±SEM, n = 25, 16 and 17 cells, respectively, 3-5 experiments for each; **p < 0.01; ANOVA.
(I) CLIMP63-3A blocks and CLIMP63-3E occludes the DHPG-induced increase in ER complexity. Mean±SEM, n = 44, 39 and 43 cells, respectively, 4-5 experiments for each; *p < 0.05; t-test.
(J) CLIMP63-3A abrogates the preferential increase of Sec23 at BPs after VSVGts release from the ER. Scale bar, 5 μm. Mean±SEM, n = 11-18 cells, 2 experiments for each; **p < 0.01; t-test. Ctr, control.
Along with changes in ER morphology (Figure 6D), expression of CLIMP63-3E significantly increased ER diffusional complexity in the dendritic shaft (Figure 6E). Conversely, expression of CLIMP63-3A decreased ER complexity in the shaft (Figure 6E). Notably, the increased ER complexity at BPs was abrogated by CLIMP63-3A but was not further increased by CLIMP63-3E (Figure 6E). These effects on ER effective complexity occurred with minor modifications in the local, absolute mobility of ER-VSVG (Table S1). Consistently, knockdown of endogenous CLIMP63 by RNA interference (RNAi) phenocopied the effect of CLIMP63-3E (Figures 6F-6H and S6C). Moreover, whereas CLIMP63-3A blocked the DHPG-induced increase in ER effective complexity, CLIMP63-3E quantitatively occluded the effect (Figure 6I). In addition to normalizing ER complexity between BPs and adjacent dendritic shaft, expression of CLIMP63-3A abolished the preferential ER export of cargo at BPs induced by acute ER release of VSVGts (Figure 6J; compare to Figure 4F). These data provide strong evidence that mGluR signaling increases ER complexity and reduces nascent receptor mobility via PKC-dependent phosphorylation of CLIMP63.
Endoplasmic Reticulum Complexity Regulates Dendritic Morphogenesis and Synaptic Strength
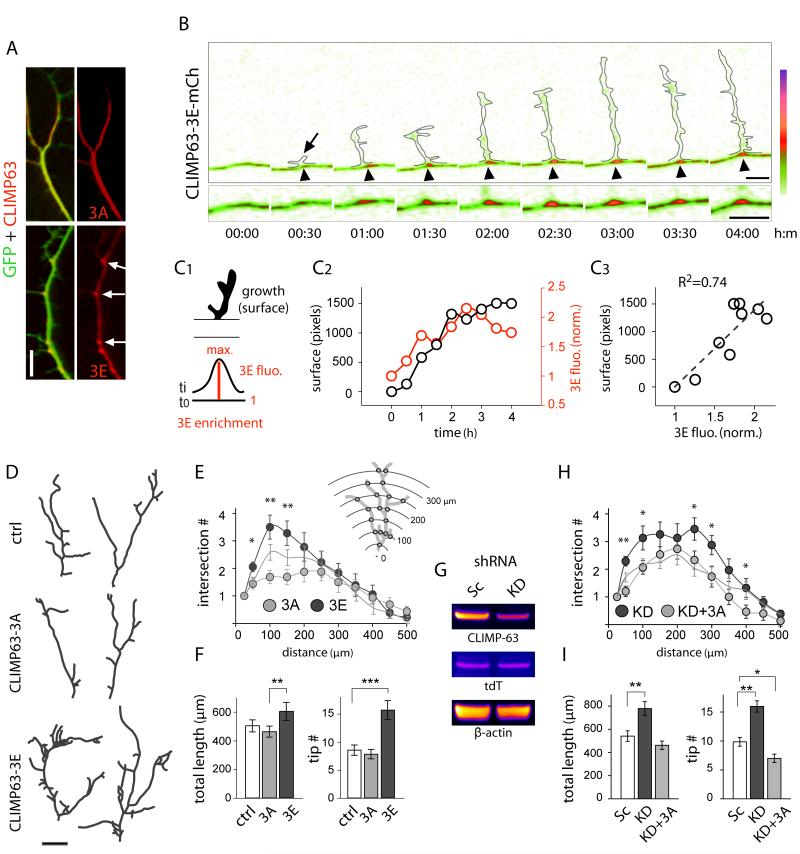
Previous studies identified dendritic GOs as hubs for forward secretory trafficking, locally providing membrane components to growing dendrites (Horton et al., 2005; Ye et al., 2007). Whether and how nascent secretory cargo is confined within the dendritic ER has been unclear. To address this, we first used CLIMP63-3E as a marker for zones of complex ER (Figure 6). In DIV 7 neurons, a period of intense dendritic growth and remodeling (Horton et al., 2005), CLIMP63-3E was concentrated at BPs (Figure 7A). This local concentration was correlated with the emergence and growth of new dendritic branches (Figures 7B, 7C, and S7A). 72% ± 9% of CLIMP63-3E clusters in primary dendrites (n = 40 in 6 neurons) corresponded to sites of new branch formation, suggesting that local zones of increased ER complexity promote dendritic branching. To address this further, we measured the effect of CLIMP63 mutants on the morphology of pyramidal neuron primary dendrites, whose growth and morphology is tightly coupled to secretory trafficking (Horton et al., 2005). Expression of CLIMP63-3A resulted in fewer proximal branches (Figures 7D-7F). In contrast, expression of CLIMP63-3E triggered a large increase in dendritic branching (Figures 7D-7F). Moreover, RNAi knockdown of CLIMP63 (Figure 7G) phenocopied the effect of CLIMP63-3E (Figures 7H-I). The increase in dendritic branching induced by CLIMP63 knock down was reversed by expression of CLIMP63-3A (Figures 7H and 7I).
Figure 7.
Phosphorylation of CLIMP63 Regulates Dendritic Branching.
(A) Distribution of mCherry-tagged CLIMP63-3A and CLIMP63-3E (red) in DIV10 neurons expressing GFP (green). Note CLIMP63-3E enrichment at BPs (arrows). Scale bar, 5 μm.
(B) Zones of ER complexity labeled by CLIMP63-3E concentrate at sites of new dendritic branches (arrow) (see also Figure S7). Outlines represent GFP expressing emerging dendritic branch. Time in hour:min. Scale bar, 5 μm.
(C) CLIMP63-3E clustering precedes and marks sites of dendritic branching. Schematic of method to quantify dendritic branch growth and CLIMP63-3E enrichment index (C1), their variation with time (C2) and correlation (C3) after emergence of the branch shown in (B).
(D-E) Camera lucida drawings (D) and Sholl analysis (E) of primary dendrites from DIV10 hippocampal pyramidal neurons expressing mCherry (ctrl), CLIMP63-3A (3A), or CLIMP63-3E (3E). Scale bar, 5 μm. Mean±SEM, n = 16-18 cells, 2 experiments for each; *p < 0.05, **p < 0.01; t-test (3A vs. 3E).
(F) Primary dendrite length (μm) and branch tip numbers for neurons expressing mCherry (ctrl), CLIMP63-3A (3A), or CLIMP63-3E (3E). Mean±SEM, **p < 0.01, ***p < 1×10−4; ANOVA.
(G) CLIMP63 knockdown after viral transduction. Shown are immunoblots for endogenous CLIMP63, tdTomato (tdT) and β-actin from hippocampal culture lysates after transduction with scramble (Sc) or CLIMP63 (KD) shRNA.
(H-I) Sholl analysis (H) and length and tip numbers (I) of primary dendrites from DIV10 hippocampal neurons. Neurons were transduced with scrambled (Sc) or CLIMP63 (KD) shRNA with or without an RNAi resistant mCherry-tagged CLIMP63-3A (3A). Mean±SEM, n = 15-31 cells, 2-4 experiments for each; *p < 0.05, **p < 0.01; t-test (H) or ANOVA (I).
In addition, we sought to determine whether ER complexity controls the abundance of AMPA receptors at the dendritic plasma membrane. Compared to CLIMP63-3A, CLIMP63-3E expression increased both surface and synaptic GluA1 (Figures S7B and S7C). Moreover, whole cell patch-clamp recordings revealed increased amplitude and frequency of miniature excitatory postsynaptic currents (mEPSCs) in neurons expressing CLIMP63-3E (Figures S7D and S7E). These data show that increases in ER complexity produce corresponding increases in surface and synaptic levels of AMPA receptors.
DISCUSSION
The complex cellular geometry determines the computational properties of neurons (Spruston, 2008) and requires highly compartmentalized delivery of membrane cargo (Kennedy and Ehlers, 2006). In Drosophila oocytes, the range of ER diffusion dictates the location of cargo secretion and is crucial for polarized signaling (Herpers and Rabouille, 2004). In mammalian neurons, dendritic spines act as diffusion sinks restricting cytoplasmic protein diffusion (Santamaria et al., 2006). In both cases, geometric complexity produces functional compartmentalization without direct immobilization. Our results suggest that nascent secretory proteins in dendrites are confined by local ER complexity near spines and BPs, directly coupling organelle structure to dendritic geometry for local control of membrane production and composition.
Local synthesis of membrane proteins has been proposed to mediate diverse forms of neuronal plasticity. However, the spatial range with which locally synthesized receptors modify synapses is not clear. Newly synthesized GluA1 and GluA2 reach the Golgi apparatus with time constants of ~3 h and ~12 h, respectively (Greger et al., 2002). Such variable and prolonged dwell time in the ER is characteristic of multisubunit receptors and neuronal ion channels, and suggests the potential for tight regulation of ER export kinetics which, together with lateral ER diffusion, will determine the spatial range over which ER export occurs. Here we have shown that the overall geometry and complexity of ER membranes can restrict the spatial mobility of membrane cargo near spines and BPs, providing a general mechanism to tune the spatial dimensions of secretory trafficking independent of cargo identity. Our results show that increased ER complexity promotes AMPA receptor surface expression and accumulation at synapses, providing a potential basis for dendritic segment-specific variation in AMPA receptor abundance (Nicholson et al., 2006).
Group I mGluR activation promotes the dendritic accumulation of AMPA receptor subunit mRNAs (Grooms et al., 2006), increases dendritic ER-export (Aridor et al., 2004), and triggers the dendritic translation of AMPA receptors (Mameli et al., 2007). Here we have shown that mGluR activation reduces the lateral mobility of nascent receptors by increasing ER complexity. Such a mechanism provides a novel pathway that couples local secretory processing of nascent receptors with signaling events controlling protein translation.
Extending for hundreds of microns, it has been unclear how the ER is elaborated in dendrites, what controls its structure or morphology, and how ER morphology impacts neuronal development and function. Here we have shown that bidirectional regulation of ER complexity through CLIMP63 phosphorylation provides a direct mechanism to tune the spatial dimensions of ER cargo mobility in dendrites. Notably, the concentration of complex ER domains at dendritic BPs is accompanied by enrichment of ribosomes, ER exit sites, and GOs. This shows that the entire ensemble of secretory machinery becomes compartmentalized at BPs, and indicates that zones of increased ER complexity control dendritic branching by facilitating nascent cargo confinement and ER-export at sites of new branch formation.
Ultimately, spatially directed secretion together with retention of receptors and other secretory cargo in dendritic subdomains could both acutely augment and persistently confine new molecular components for locally modifying dendrite function. More broadly, local regulation of ER complexity may provide a general paradigm for stimulus-dependent regulation of local membrane composition in geometrically complex cells.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection and shRNA Knockdown of CLIMP63
Rat hippocampal cultures were derived from E18 rat embryos and transfected as previously described (Horton and Ehlers, 2003). FRAP and photoactivation experiments were performed 12-24 h post-transfection in neurons displaying low to moderate levels of exogenous protein expression. For shRNA knockdown of CLIMP63, the sequence UCAACCGUAUUAGUGAAGUUCUACA was cloned in pLentilox3.7 and expressed for 5 to 10 d.
Live Cell Imaging and Drug Treatments
Hippocampal neurons were monitored at 37°C in E4 medium (150 mM NaCl, 3 mM KCl, 15 mM glucose, 10 mM HEPES, pH 7.4) supplemented with 2 mM CaCl2, 2 mM MgCl2 (standard E4) or 4 mM CaCl2, 0 mM MgCl2 (low Mg E4), for a maximal duration of 90 min. DHPG, TTX, NMDA, and AP5 (Tocris) were used at final concentrations of 100 μM, 2 μM, 50 μM and 100 μM, respectively. ER complexity was measured by FRAP analysis in low Mg E4 before and 25-35 min after addition of DHPG, or in low Mg E4 supplemented with TTX before and 15-30 min after a 3 min exposure to NMDA (Ehlers, 2000). The PKC inhibitor bisindolylmaleimide I (Calbiochem) was used at 100 nM. The 39° to 32°C and 39°-20°-32°C temperature-shift experiments with VSVGts045 were done as previously described (Horton and Ehlers, 2003).
Computational Analysis of ER Diffusion
Fluorescence recovery in dendrite segments was analyzed using a custom three-dimensional anomalous diffusion model. The diffusion of photoactivated molecules was quantified using the same model. For fluorescence loss in photobleaching (FLIP) experiments, fluorescence decay was quantified using monoexponential curve fitting after correction for photobleaching during imaging acquisition, which was determined on nearby unbleached dendrites.
Serial Section Electron Microscopy, 3D Reconstructions, and Analysis
Serial electron micrographs (EM) were obtained from the stratum radiatum in the CA1 area of male rat hippocampus as previously described (Spacek and Harris, 1997). The RECONSTRUCT™ software (Fiala and Harris, 2001) was used to align, reconstruct, quantify and visualize SER within the three-dimensional dendrite reconstructions using previously described identification criteria (Spacek and Harris, 1997).
ER cross sectional area in aspiny or spiny dendrites was calculated as the average of total cross-sectional ER area per section through spans of serial sections beginning with the first section where the base of a spine started to emerge and ended with the section where the base of the last spine was no longer detectable (Figure S2K). A dendritic segment was considered aspiny if it spanned two or more serial sections without a spine emerging.
Super-Resolution Imaging
Structured illumination fluorescence imaging was performed using a 63× 1.4 NA objective on a Zeiss Super resolution-SIM inverted microscope (Carl Zeiss, Inc.). 3D reconstructions were generated in Amira (Visage Imaging, Inc.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ben Arenkiel, Ian Davison, Juliet Hernandez, Matt Kennedy, Tom Newpher, and Rui Peixoto for critical reading of the manuscript. We thank Larry Lindsey and Cam Robinson for help with EM data processing. We thank Irina Lebedeva and Marguerita Klein for technical assistance. We thank Zeiss USA for use of their structured illumination microcopy system. Work in the laboratory of M. D. E. was supported by NIH grants NS039402, MH064748, and MH086339, and the Howard Hughes Medical Institute. Work in the laboratory of K. M. H. is supported by NIH grants NS021184 and EB002170.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION Supplemental information includes Extended Experimental Procedures, seven figures and four movies.
REFERENCES
- Aridor M, Guzik AK, Bielli A, Fish KN. Endoplasmic reticulum export site formation and function in dendrites. J Neurosci. 2004;24:3770–3776. doi: 10.1523/JNEUROSCI.4775-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney JR, Hurlburt JL, Selig DK, Harris KM, Fiala JC. Endosomal compartments serve multiple hippocampal dendritic spines from a widespread rather than a local store of recycling membrane. J Neurosci. 2002;22:2215–2224. doi: 10.1523/JNEUROSCI.22-06-02215.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron. 2000;28:511–525. doi: 10.1016/s0896-6273(00)00129-x. [DOI] [PubMed] [Google Scholar]
- Ehlers MD, Heine M, Groc L, Lee MC, Choquet D. Diffusional trapping of GluR1 AMPA receptors by input-specific synaptic activity. Neuron. 2007;54:447–460. doi: 10.1016/j.neuron.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah CA, Liazoghli D, Perreault S, Desjardins M, Guimont A, Anton A, Lauzon M, Kreibich G, Paiement J, Leclerc N. Interaction of microtubule-associated protein-2 and p63: a new link between microtubules and rough endoplasmic reticulum membranes in neurons. J Biol Chem. 2005;280:9439–9449. doi: 10.1074/jbc.M412304200. [DOI] [PubMed] [Google Scholar]
- Farhan H, Weiss M, Tani K, Kaufman RJ, Hauri HP. Adaptation of endoplasmic reticulum exit sites to acute and chronic increases in cargo load. EMBO J. 2008;27:2043–2054. doi: 10.1038/emboj.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala JC, Harris KM. Extending unbiased stereology of brain ultrastructure to three-dimensional volumes. J Am Med Inform Assoc. 2001;8:1–16. doi: 10.1136/jamia.2001.0080001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu K, Bannai H, Zhang S, Nakamura H, Inoue T, Mikoshiba K. Lateral diffusion of inositol 1,4,5-trisphosphate receptor type 1 is regulated by actin filaments and 4.1N in neuronal dendrites. J Biol Chem. 2004;279:48976–48982. doi: 10.1074/jbc.M408364200. [DOI] [PubMed] [Google Scholar]
- Gardiol A, Racca C, Triller A. Dendritic and postsynaptic protein synthetic machinery. J Neurosci. 1999;19:168–179. doi: 10.1523/JNEUROSCI.19-01-00168.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger IH, Khatri L, Ziff EB. RNA editing at arg607 controls AMPA receptor exit from the endoplasmic reticulum. Neuron. 2002;34:759–772. doi: 10.1016/s0896-6273(02)00693-1. [DOI] [PubMed] [Google Scholar]
- Grooms SY, Noh KM, Regis R, Bassell GJ, Bryan MK, Carroll RC, Zukin RS. Activity bidirectionally regulates AMPA receptor mRNA abundance in dendrites of hippocampal neurons. J Neurosci. 2006;26:8339–8351. doi: 10.1523/JNEUROSCI.0472-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanus C, Ehlers MD. Secretory outposts for the local processing of membrane cargo in neuronal dendrites. Traffic. 2008;9:1437–1445. doi: 10.1111/j.1600-0854.2008.00775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herpers B, Rabouille C. mRNA localization and ER-based protein sorting mechanisms dictate the use of transitional endoplasmic reticulum-golgi units involved in gurken transport in Drosophila oocytes. Mol Biol Cell. 2004;15:5306–5317. doi: 10.1091/mbc.E04-05-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton AC, Ehlers MD. Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. J Neurosci. 2003;23:6188–6199. doi: 10.1523/JNEUROSCI.23-15-06188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton AC, Racz B, Monson EE, Lin AL, Weinberg RJ, Ehlers MD. Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron. 2005;48:757–771. doi: 10.1016/j.neuron.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Kennedy MJ, Ehlers MD. Organelles and trafficking machinery for postsynaptic plasticity. Annu Rev Neurosci. 2006;29:325–362. doi: 10.1146/annurev.neuro.29.051605.112808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein DR, Kappeler F, Hauri HP. A novel direct interaction of endoplasmic reticulum with microtubules. EMBO J. 1998;17:6168–6177. doi: 10.1093/emboj/17.21.6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losonczy A, Makara JK, Magee JC. Compartmentalized dendritic plasticity and input feature storage in neurons. Nature. 2008;452:436–441. doi: 10.1038/nature06725. [DOI] [PubMed] [Google Scholar]
- Mameli M, Balland B, Lujan R, Luscher C. Rapid synthesis and synaptic insertion of GluR2 for mGluR-LTD in the ventral tegmental area. Science. 2007;317:530–533. doi: 10.1126/science.1142365. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher TM, Ehlers MD. Glutamate receptor dynamics in dendritic microdomains. Neuron. 2008;58:472–497. doi: 10.1016/j.neuron.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson DA, Trana R, Katz Y, Kath WL, Spruston N, Geinisman Y. Distance-dependent differences in synapse number and AMPA receptor expression in hippocampal CA1 pyramidal neurons. Neuron. 2006;50:431–442. doi: 10.1016/j.neuron.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Penn AC, Williams SR, Greger IH. Gating motions underlie AMPA receptor secretion from the endoplasmic reticulum. EMBO J. 2008;27:3056–3068. doi: 10.1038/emboj.2008.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampo B, Kaech S, Kunz S, Banker G. Two distinct mechanisms target membrane proteins to the axonal surface. Neuron. 2003;37:611–624. doi: 10.1016/s0896-6273(03)00058-8. [DOI] [PubMed] [Google Scholar]
- Santamaria F, Wils S, De Schutter E, Augustine GJ. Anomalous diffusion in Purkinje cell dendrites caused by spines. Neuron. 2006;52:635–648. doi: 10.1016/j.neuron.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Heintzmann R, Leonhardt H. A guide to super-resolution fluorescence microscopy. J Cell Biol. 2010;190:165–175. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer A, Ericsson M, Bachi T, Griffiths G, Hauri HP. Characterization of a novel 63 kDa membrane protein. Implications for the organization of the ER-to-Golgi pathway. J Cell Sci. 1993;104(Pt 3):671–683. doi: 10.1242/jcs.104.3.671. [DOI] [PubMed] [Google Scholar]
- Spacek J, Harris KM. Three-dimensional organization of smooth endoplasmic reticulum in hippocampal CA1 dendrites and dendritic spines of the immature and mature rat. J Neurosci. 1997;17:190–203. doi: 10.1523/JNEUROSCI.17-01-00190.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague BL, McNally JG. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Spruston N. Pyramidal neurons: dendritic structure and synaptic integration. Nat Rev Neurosci. 2008;9:206–221. doi: 10.1038/nrn2286. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Ito HT, Cressy P, Kempf C, Woo JC, Schuman EM. Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell. 2006;125:785–799. doi: 10.1016/j.cell.2006.03.040. [DOI] [PubMed] [Google Scholar]
- Terasaki M, Slater NT, Fein A, Schmidek A, Reese TS. Continuous network of endoplasmic reticulum in cerebellar Purkinje neurons. Proc Natl Acad Sci U S A. 1994;91:7510–7514. doi: 10.1073/pnas.91.16.7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedrenne C, Hauri HP. Morphogenesis of the endoplasmic reticulum: beyond active membrane expansion. Traffic. 2006;7:639–646. doi: 10.1111/j.1600-0854.2006.00419.x. [DOI] [PubMed] [Google Scholar]
- Vedrenne C, Klopfenstein DR, Hauri HP. Phosphorylation controls CLIMP-63-mediated anchoring of the endoplasmic reticulum to microtubules. Mol Biol Cell. 2005;16:1928–1937. doi: 10.1091/mbc.E04-07-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Zhang Y, Song W, Younger SH, Jan LY, Jan YN. Growing dendrites and axons differ in their reliance on the secretory pathway. Cell. 2007;130:717–729. doi: 10.1016/j.cell.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.